Abstract
Tektins are important components of flagella. Alterations in the expression of or mutations in mouse tektins are correlated with defective sperm motility, a cause of male infertility. Our proteomic studies of flagellar accessory structures previously identified a novel tektin, TEKT5, whose function is unknown. To understand the role of TEKT5 in mouse sperm, we characterized the expression of the mouse Tekt5 gene and the presence of TEKT5 in spermatogenic cells and spermatozoa. A complete cDNA encoding the Tekt5 transcript was assembled following RT-PCR and 3'-RACE and predicted that TEKT5 is a 62,730 Dalton protein with an unusual, long C-terminus. Tekt5 mRNA was highly expressed during late stages of spermiogenesis. Among examined tissues, Tekt5 mRNA was only present in testis and brain and quantitative RT-PCR showed that the expression level of mRNA in testis was 6.8-fold higher than that of brain. At the protein level, TEKT5 was present in sperm and was enriched in the accessory structures of flagella. Immunofluorescence confirmed that TEKT5 was localized throughout the sperm tail in flagellar accessory structures. The expression pattern suggests that TEKT5 plays an important role in flagella formation during spermiogenesis as well as being implicated in sperm motility.
Keywords: Sperm, sperm motility, flagellum, tektin, flagellar accessory structures
Introduction
A properly formed flagellum is required for sperm motility and is essential for bringing the sperm to the egg for fertilization. At the most basic level, flagellar and ciliary motility is driven by a core structure, the axoneme, which contains a characteristic “9 + 2” microtubular doublet configuration and is remarkably conserved during evolution. In mammalian sperm, however, structural studies have demonstrated that the flagellum is a complex and compartmentalized organelle that is composed of three different segments, midpiece, principal piece, and end piece. The axoneme extends throughout the complete flagellum. Surrounding the axoneme in the various segments are three types of accessory structures (Supplemental Figure 1). In the midpiece, outer dense fibers 1–9 emanate from each outer doublet of the axoneme. The mitochondrial sheath surrounds the outer dense fibers and the axoneme and defines the extent of the midpiece. In the principal piece, outer dense fibers 3 and 8 are replaced by the longitudinal columns of the fibrous sheath, which are held together by transverse ribs of this structure. The end piece lacks accessory structures and is remarkable only for the presence of the axoneme. All regions of the flagellum are encased by the plasma membrane. Previously, it was thought that the accessory structures mainly existed to provide mechanical support for the axoneme. However, in the past decade and a half, evidence has accumulated showing that these structures are involved in a variety of critical physiological processes such as signal transduction, protection against oxidative damage, and energy production (Carrera et al, 1994, Godeas et al, 1997, Miki et al, 2004). More recently, our proteomic study identified additional signaling and metabolic proteins in the accessory structures (Cao et al, 2006a).
Along with known proteins of the mitochondrial sheath, outer dense fibers, and fibrous sheath, several tektins – members of a group of proteins generally considered to be axonemal proteins – were also identified in the flagellar accessory structures. Extensive studies with sea urchin sperm flagella (which lack outer dense fibers and fibrous sheaths in their flagella) and cilia identified three tektins (A, B, C) found in association with tubulin of axonemal outer microtubular doublets. These tektins form heteropolymeric protofilaments (tektins A and B) or homodimers (tektin C) and are considered as structural components of outer doublet microtubules in cilia and flagella (Pirner and Linck, 1994). In contrast to sea urchin sperm axonemes, we recently found two additional tektins in the accessory structures of mouse sperm (Cao et al, 2006a). Furthermore, since 2004, there have been several functional studies demonstrating tektins are critical for sperm motility (Roy et al, 2007, Roy et al, 2004, Tanaka et al, 2004). These studies imply tektins play important roles in sperm function.
Among all tektins, TEKT5 was unknown at the time of our earlier proteomic study. Since the publication of our 2006 report, a recent analysis of the rat orthologue demonstrated that rat TEKT5 is a 62,837 Dalton protein (Murayama et al, 2008). For the mouse, multiple entries in the genomic database paint a confusing picture concerning the cDNA sequence and the resulting deduced protein structure. Moreover, all the sequences available in the databases for mouse TEKT5 are in silico genomic predictions or are based upon high-throughput transcriptomic studies (Carninci et al, 2005, Strausberg et al, 2002) without experimental confirmation. Our proteomic analysis on sperm indicated TEKT5 is present in flagellar accessory structures. The actual localization study needs to be done to refine the TEKT5 localization. Toward a better understanding of the role of TEKT5 in mouse sperm, we carried out a detailed characterization of a Tekt5 cDNA, examined the expression of the RNA and protein during spermatogenesis, and determined that TEKT5 is localized throughout the midpiece and principal piece of the sperm flagellum.
Methods
Sperm and germ cell preparation
All animal procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Mixed germ cells were prepared from decapsulated testes of adult male mice (CD1 retired breeders, Charles River Laboratories, Wilmington, MA) by sequential dissociation with collagenase and trypsin-DNase I (Bellvé et al, 1977). To purify populations of pachytene spermatocytes, round spermatids, and condensing spermatids, the mixed germ cells were separated at unit gravity in a 2–4% bovine serum albumin (BSA) gradient in enriched Krebs-bicarbonate medium (Romrell et al, 1976). Both the pachytene spermatocyte and round spermatid populations were at least 85% pure as determined by microscopic examination and differential counting with a hemocytometer, while the condensing spermatid population was approximately 40%–50% pure with the balance primarily being anucleate residual bodies and round spermatids.
Sperm were collected by mincing the caudae epididymides and allowing the sperm to swim out in PBS (2.68 mM KCl, 136.09 mM NaCl, 1.47mM KH2PO4, 8.07 mM Na2HPO4, pH 7.4). The supernatant was gently aspirated and the sperm in the suspension (>99% pure as assessed by light microscopy) were collected by centrifugation at 800 × g for 5 min at room temperature. The sperm were then homogenized in 1% sodium dodecyl sulfate (SDS), 75 mM NaCl, 24 mM EDTA, pH 6.0 (S-EDTA), layered onto a 1.6 M sucrose cushion in S-EDTA, and centrifuged at 5000 × g for 1 h at room temperature (O'Brien and Bellvé, 1980). The SDS-resistant tail structures (flagellar accessory structures) were collected from the interface. The purity was visually assessed by light microscopy, electron microscopy, and immunoblotting as performed in our previous studies (Cao et al, 2006a, Cao et al, 2006b).
Preparation of RNA, RT-PCR, 3'-RACE, cloning of Tekt5, and cDNA sequence analysis
RNA was prepared from germ cells, testes, and various somatic tissues using TRI Reagent (Sigma-Aldrich Corp., St. Louis, MO). Reverse transcription using 1 μg mRNA was performed using SuperScript II Reverse Transcriptase according to the manufacturer's instructions (Invitrogen Corp., Carlsbad, CA). Products were amplified using the appropriate primers with Ex TaqTM DNA polymerase (Takara Co., Tokyo, Japan) for short products or Easy-ATM High-Fidelity PCR Master Mix (Stratagene, La Jolla, CA) for long products. Amplicons were then cloned into the pCR2.1-TOPO vector (Invitrogen Corp.). Plasmid DNA was prepared and sequenced as directed by the manufacturer's instruction manuals. The initial primer designs were based on the peptides identified in the mass spectrum of the proteomics study as well as the available computer-predicted sequence. The RLM-RACE kit (Ambion, Austin, TX) was used for 3'-RACE and 1 μg mixed germ cell total RNA aliquots were used as templates for reverse transcription with the supplied 3'-RACE adapter (5'-GCG AGC ACA GAA TTA ATA CGA CTC ACT ATA GGT 12VN-3'). The cDNA was then subjected to PCR using the 3'-RACE primers, which were complementary to the anchored adapter (5'-GCG AGC ACA GAA TTA ATA CGA CT-3'), and a primer specific for Tekt5 (5'-GCG TGG AGA AGT TTG ACG GAA C-3'). Gradient PCR was used to optimize annealing temperatures. All PCR products were sequenced in both directions. The cloned sequences were spliced by Sequencher 4.5 (Gene Codes Corp., Ann Arbor, MI). Different tektin isoforms were aligned by Genedoc (http://www.psc.edu/biomed/genedoc). The deduced amino acid sequence was analyzed by the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/).
Quantitative PCR analyses
For quantitative RT-PCR assays, primers were designed using Primer Express 1.5 Taqman Primer Design software (Applied Biosystems, Foster City, CA). The Tekt5 primers, 5'-TGA GGA GAC AGA TGT GAA GAA CAA G-3' (forward) and 5'-TGG CCC TCT CCA GCA ACA-3' (reverse), were each used at a concentration of 22.5 nM and yielded a product of 101 bp. The ribosomal protein S16 [Rps16, (GenBank accession number: BC082286)] primers, 5'-AGA TGA TCG AGC CGC GC-3' (forward) and 5'-GCT ACC AGG GCC TTT GAG ATG GA-3' (reverse), were each used at a concentration of 11.25 nM and yielded a product of 163 bp (Jeong et al, 2005). Products were amplified with the SYBR Green PCR Master Mix (Applied Biosystems Inc, Foster City, CA) and analyzed with the ABI 7900 HT Sequence Detection system. The following PCR protocol was used: 1) denaturation (50°C for 2 min, 95°C for 10 min), 2) amplification and quantification (95°C for 15 sec, 60°C for 1 min) repeated for 40 cycles, 3) a dissociation curve program (95°C for 15 sec, 60°C for 15 sec, 95°C for 15 sec), and 4) cooling at 4°C. Amplicons were analyzed by generating a dissociation curve and determining the threshold cycle (CT) value for each transcript. The relative quantification of gene expression was analyzed by the 2-−ΔΔCT method (Livak and Schmittgen, 2001). The mRNA corresponding to Rps16 was used as a control (Jeong et al, 2005).
Northern blotting
Total RNAs from mixed germ cell and round spermatid (20 μg each) were separated by electrophoresis in a 1% agarose gel and transferred to Hybond N+ membranes (GE Healthcare, Little Chalfont, Bucks, UK). The probe was derived from the initial PCR product (P1–P2, see Table 1, product size 202 bp), which was purified with the QIAquick Gel extraction kit (Qiagen Inc., Valencia, CA). The probe was then quantified by comparison to a Hind III ladder (New England Biolabs, Ipswich, MA) and labeled with [32P]dCTP using the RadPrime DNA Labeling System (Invitrogen Corp.), followed by centrifugation through a NucleoSpin Extraction Spin column (Clontech, Mountain View, CA) to remove the unincorporated nucleotides. The probe was later used for overnight hybridization at 42°C to Hybond N+ membranes. Membranes were washed to a stringency of 0.1x SSC and 0.1% SDS up to 55°C. Relative sample loading was assessed using radiolabeled cDNAs encoding AKAP4 (accession number NM_001042542).
Table 1.
List of Primers Used in Cloning
| Primer name | Sequence 5–3' | Gene related | Direction |
|---|---|---|---|
| P1 | GCGTGGAGAAGTTTGACGGAAC | common | Forward |
| P2 | TCAGAGATGCGAGCGTTGAAGG | common | Reverse |
| P3 | CAGCACTAAGTCACAGAGGC | XM_156349 5' side genomic sequence | Forward |
| P4 | CTGGCCCCTTGGGTAAAGTT | common to XM_156349, XM_908332, BC115969 | Forward |
| P5 | ACTTCCAGAAACCGAGGTC | common | Reverse |
| P6 | AAGACCTCATTCACGAGCC | specific to XM_908332 | Reverse |
| P7 | TGCTTTTCAGTTGCTCGTC | specific to XM_980627 | Reverse |
| P8 | TCACCTGAACTGTGGCACATC | common to XM_908332, XM_980627, BC115969 | Reverse |
| P9 | TAGTCTCCTCTCACCCCAAC | XM_156349 3' side genomic sequence | Reverse |
| p10 | GCGAGCACAGAATTAATACGACT | 3'-RACE outer primer | Reverse |
| p11 | TTTGAGCCTTTTCCAGCG | specific to XM_908332 | Reverse |
| P12 | GCTGATGGGGAAAGATGGAA | common | Forward |
Generation of TEKT5 Antibody and Immunological Procedures
A region of the Tekt5 cDNA, encoding the C-terminal 72 amino acids of TEKT5 (amino acids 486–557, accession number XP_913425), was cloned into a prokaryotic expression vector, pGEX-6p-1 (GE Healthcare). A glutathione S-transferase (GST) fusion protein, GST-TEKT5486-557, was expressed in large quantities in E. coli and isolated using a GSTrapTM FF column (GE Healthcare) according to the manufacturer's instructions, followed by electrophoretic purification in an SDS-polyacrylamide gel. The purified GST-TEKT5486-557 was then sent to Cocalico Biologicals (Reamstown, PA) to generate antibody against TEKT5. The antiserum was used in immunoblot assays at a 1:1000 dilution. To decrease the background in immunohistochemical analyses, we purified the IgG fraction from the antiserum further using HiTrapTM Protein A HP column (GE Healthcare). The purified antibody was only used in immunolocalization studies.
Gel electrophoresis and immunoblot analyses
The sperm and flagellar accessory structures were concentrated by centrifugation, washed in 1 ml of PBS, resuspended in sample buffer (62.5 mM Tris-HCl, pH 6.8, 1.67% SDS, 10% glycerol), and boiled for 5 min. Other tissues were homogenized, sonicated, and boiled in the sample buffer. After centrifugation, the supernatants were recovered and saved. After protein concentrations were determined by the BCA protein assay (Pierce Chemical Co., Rockford, IL), dithiothreitol (DTT) and bromophenol blue were added to final concentrations of 100 mM and 0.002%, respectively. The samples were boiled for 5 min, and protein samples (10 μg per lane) were separated by SDS-PAGE in 10% polyacrylamide gels (Laemmli, 1970). The gels were then transferred to PVDF membranes (Millipore, Bedford, MA) (Towbin et al, 1979). After the membranes were blocked with TBST (125 mM NaCl; 0.1% Tween 20; 25 mM Tris-HCl, pH 8.0) containing 5% BSA, they were incubated with primary antibody (rabbit anti-GST-TEKT5486-557 serum, 1:1000 in 5% BSA-TBST) for 1 h. After washing with TBST, the blots were incubated for 1 h with secondary antibody (donkey anti-rabbit IgG conjugated with horseradish peroxidase [GE Healthcare], 1:5000 in 5% BSA-TBST) and, after washing with TBST, the bound enzyme was developed with the ECL kit (GE Healthcare) according to the manufacturer's directions and exposed to film. As a control, the anti-TEKT5 was neutralized by adding a 10-fold molar excess of the peptide in 200 μl 5% BSA in TBST and incubating the mixture at 4°C overnight. After centrifugation to remove any particulate material, the supernatant was used for immunoblotting.
Two-dimensional Gel Electrophoresis and Mass Spectrometry
Two-dimensional gel electrophoresis was performed with the Ettan IPGphor II and Ettan DALTsix equipment with PlusOne™ reagents from GE Healthcare. Samples (900 μg of protein in 200 μl of sample buffer) were mixed with 250 μl of rehydration buffer (8 M urea, 2% (w/v) CHAPS, 1% IPG buffer [pH 3–11, non-linear], 2 mg/ml DTT) and loaded in the IPGphor strip holder. Immobiline Drystrips (pH 3–11 non-linear, 24 cm) were placed in the holder and overlaid with ~4 ml of DryStrip cover fluid (GE Healthcare). Strips were hydrated under 50 V for 24 h and focused afterward on the IPGphor IEF system for a total of 80 kV-h at 20°C. After electrophoresis, each strip was incubated with 15 ml of equilibration buffer A (6 M urea; 30% (v/v) glycerol; 2% (w/v) SDS; 1% DTT; 100 mM Tris-HCl, pH 8.8) on a rocking platform for 15 min and then with 15 ml of equilibration buffer B (6 M urea; 30% (v/v) glycerol; 2% (w/v) SDS; 2.5% iodoacetamide; 100 mM Tris-HCl, pH 8.8) for an additional 15 min. For the second dimension, the strips then were placed on top of 10% polyacrylamide gels containing SDS and the proteins separated using the Ettan DALTsix apparatus. After electrophoresis, proteins were stained with colloidal Coomassie Blue. Two-dimensional gels were scanned with a Typhoon 9400 scanner (GE Healthcare), and the spots were analyzed with DeCyder software (GE Healthcare). Protein spots were selected and the corresponding gel pieces were cored manually. After the cored gel pieces were digested with trypsin, a portion of the digest was analyzed directly by MALDI-TOF for molecular weight determination of released peptides and MALDI-TOF/TOF for sequence information using a Voyager 4700 proteomics analyzer mass spectrometer (Applied Biosystems). Another portion of the digest was subjected to nanoLC/Qstar-XL analysis (Applied Biosystems). The data were acquired and analyzed with Analyst QS. The protein identification and database search were performed with Mascot.dll script of Analyst QS; the combined MS and MS/MS data were used for the Mascot database search. A protein score of >70 with a confidence identification of >95% was considered acceptable.
Indirect immunofluorescence analyses
Cauda epididymal sperm were collected, processed with the S-EDTA solution to solubilize the sperm, cytoplasm, membranes and the axoneme, resulting in separate head and tail remnants (i.e., flagellar accessory structures). The heads and flagellar accessory structures were washed 3 times in PBS, then attached to slides, and fixed with 4% paraformaldehyde in PBS for 15 min. The slides were washed with PBS and the samples were incubated with 10% horse serum in PBS (blocking solution) for 1 h at room temperature and then with the mixed solution of primary antibody (10 μg/ml rabbit anti-GST-TEKT5486-557 antibody) in blocking solution for 1 h at room temperature. For a control, an equivalent amount of primary antibody pre-absorbed with GST-TEKT5486-557 fusion protein was substituted. After washing with PBS, the samples were incubated for 1 h at room temperature with secondary antibody (1 μg/ml of goat anti-rabbit IgG linked with Alexa Fluo-568 [Molecular Probes, Eugene, OR]) in blocking solution. After washing with PBS, the samples were mounted with coverslips using Fluoromount-G (Southern Biotechnology Associates, Inc., Birmingham, AL), examined using an inverted microscope (Nikon Eclipse TE 2000-U, Nikon Corp., Melville, NY) and photographed with a Scion Corporation (Frederick, MD) CFW-1610C digital FireWire camera using the NIH ImageJ Imaging Software (Rasband, 2007). As a positive control for ODFs, goat anti-ODF2 polyclonal antibody (Invitrogen) was used as a primary antibody (0.2 μg/ml), the secondary antibody is (1 μg/ml of rabbit anti-goat IgG linked with Alexa Fluo-488 [Molecular Probes])
Results
Proteomic Identification of TEKT5 in mouse sperm and in silico analysis
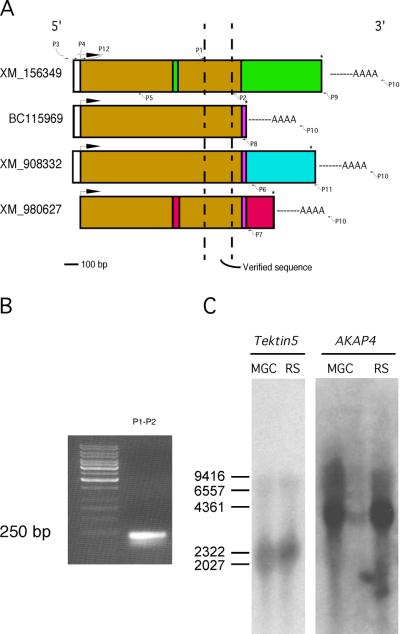
Previously we performed two-dimensional gel electrophoresis of flagellar accessory structures and selected about 200 protein spots for analysis (Cao et al, 2006a). Several of these spots were determined to be tektin family members, TEKT1–4 (Fig.1). In addition, two protein spots (A and B in Fig. 1) were determined to be related variants of a new tektin family member. Each protein spot yielded a peptide identified by mass spectrometry (Table 2) that contained the first 8 amino acids of the tektin nonapeptide signature sequence (RPNVELCRD), confirming that this novel protein belongs to the tektin family. Spots A and B yielded 17 and 27 peptides, respectively. Sixteen peptides recovered from each of the two protein spots were identical and the seventeenth peptide from spot A overlapped with a peptide of spot B. Further in silico analysis demonstrated that all of the peptides from protein spots A and B were included within the deduced translation product encoded by the RIKEN cDNA 3300001K11Rik. By using BLAST to compare the peptides with the NCBI database at that time, we identified several other cDNA sequences encoding some but not all of the experimentally identified peptides. These sequences were predicted to encode variously sized proteins and had also been designated as Tekt5 cDNA clones (Fig. 2A). Thus, it was necessary to determine experimentally the true sequence of the protein expressed in the testis. To address this problem, we isolated and characterized a full-length Tekt5 cDNA.
Figure 1.
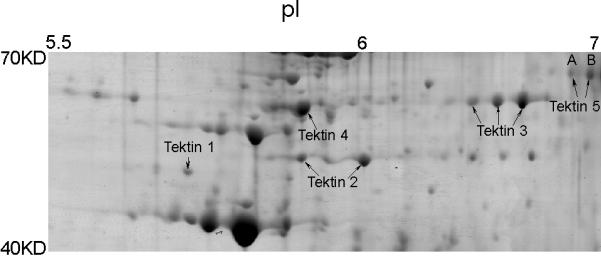
Five tektin members present in flagellar accessory structures. Sperm were processed by 1% S-EDTA, homogenized. Detached head and tail remnants were then separated by sucrose gradient centrifugation. The enriched sperm tail fraction (flagellar accessory structures) was further solubilized in 2D sample buffer. The protein sample (~900 μg) was separated by IEF on a non-linear pH 3–11 IPG strip, followed by a second dimension of SDS-PAGE. The gel was stained with colloidal Coomassie Blue. Gel pieces containing individual protein spots were excised and treated with trypsin. The resulting peptides were analyzed by mass spectrometry for protein identification. All identified tektins are labeled. Based on the two-dimensional gel data, the molecular weights of the two TEKT5 spots (A, B) were around 62,000 Mr, and the isoelectric points were near 7, which corresponded to the respective values of 62,730 and 8.83 predicted by the MacVector computer program. The molecular weights of the tektins were estimated by comparison to the theoretical molecular weights of other proteins identified in this analysis.
Table 2.
Peptides identified from Spots A and B (TEKT5)
| Start | End | Peptide sequence | Spot A | Spot B |
|---|---|---|---|---|
| 47 | 58 | YLNAWRPSVFHK | + | + |
| 59 | 73 | IATSQTIPEECSGIR | + | + |
| 74 | 83 | RPPTILPSLR | + | + |
| 94 | 104 | DWDRSNDLQIR | + | |
| 123 | 140 | IMQDKDQLIHQMQEGTSR | + | |
| 146 | 153 | LSDLGFWK | + | + |
| 154 | 162 | SELCYELDR | + | + |
| 201 | 213 | RIGIDLVHDNVEK | + | + |
| 202 | 213 | IGIDLVHDNVEK | + | |
| 224 | 230 | CCQDQMR | + | |
| 235 | 241 | RIDFQIR | + | + |
| 245 | 252 | DAQHSLER | + | |
| 253 | 271 | DIEDKSSAQYIDENCFNL | + | |
| 258 | 271 | SSAQYIDENCFNLR | + | + |
| 272 | 285 | STSDSISFFHGVEK | + | + |
| 286 | 298 | FDGTVSIPETWAK | + | + |
| 299 | 311 | FSNDNIRHAQNMR | + | |
| 335 | 346 | QFTNTNLAFNAR | + | + |
| 364 | 380 | ILQEIFQAENTIMLLER | + | + |
| 391 | 399 | MAQTMLACR | + | |
| 400 | 409 | TRRPNVELCR | + | |
| 402 | 409 | RPNVELCR | + | + |
| 416 | 431 | LVNEVFTIDDTLQTLK | + | + |
| 434 | 448 | LRETQDTLQLLVMTK | + | |
| 449 | 458 | SRLEHELAIK | + | + |
| 451 | 466 | LEHELAIKANTLCIDK | + | |
| 474 | 481 | KSFPSTPR | + | |
| 482 | 501 | LTGYTCSAIGSGPYANHAPR | + | + |
Figure 2.
Determination of the Tekt5 cDNA sequence. (A): TEKT5 peptide sequences determined by mass spectrometry (Table 2) were used to search the NCBI nucleotide database using the blastP algorithm. The four corresponding nucleotide sequences are listed, schematically showing the comparison of four possible nucleotide sequences. The same color denotes the same DNA sequence. The arrows indicate the approximate positions of primers used for cloning the testicular Tekt5 cDNA. The primer sequences are listed in Table 1. (B): Primers derived from peptide sequence of TEKT5 successfully amplified the expected size product and the amplicons were sequenced. (C): Northern blot, using the previous amplicon as probe, showed the size of Tekt5 in mixed germ cell (MGC) and round spermatid (RS) mRNA is around 2.3 kb. AKAP4 was used as a loading control.
Cloning of Tekt5
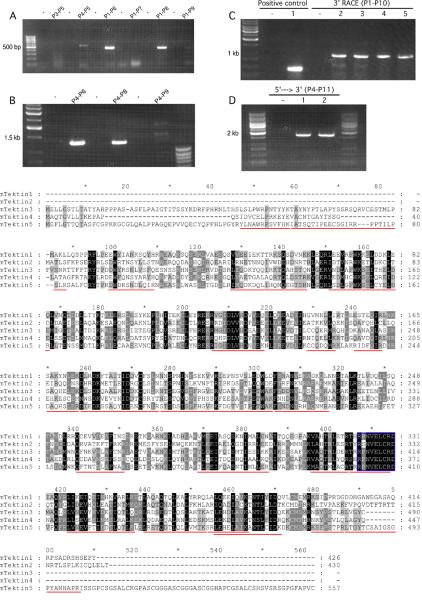
Based on the peptide sequences identified by mass spectrometry and cDNA sequences available in NCBI, primers (P1 and P2) were designed and RT-PCR was successfully performed. The amplicon was sequenced (Fig. 2B). Northern blotting was conducted to determine the size of Tekt5 mRNA using the PCR product as a probe, revealing the transcript to be about 2.3 kb (Fig. 2C). To clone the full-length cDNA, regular RT-PCR and 3'-RACE were performed. First, three 5'-primers (P1, P3, and P4) were designed based on the predicted Tekt5 cDNA and genomic sequences. Also, a number of 3'-primers specific to the different purported Tekt5 sequences in the NCBI database were designed (Fig. 2A). A pilot experiment testing for small amplicons was done to determine which primer pairs (P3–P5, P4–P5, P1–P6, P1–P7, P1–P8, and P1–P9) would work and the PCR results showed that P4–P5, P1–P6, and P1–P8 successfully yielded amplicons (Fig. 3A). Furthermore, larger products could be amplified using the 5'-primer P4 with the 3'-primers P6 and P8 whereas the P4–P9 primer combination was unsuccessful (Fig. 3B). It should be noted that the primer P8 sequence (TCA CCT GAA CTG TGG CAC ATC) is based on the 3'end of the cDNA BC115969, which terminates with a stop codon (TGA); the remaining 18 bases of the primer are identical to the corresponding regions of two other purported Tekt5 sequences (XM_908332 and XM_980627). To determine the 3' sequence of the Tekt5 mRNA expressed in male germ cells, 3'-RACE with the P1 primer and the P10 primer of the commercial kit was conducted, which yielded an approximately 1 kbp band (Fig. 3C). Further analysis demonstrated that the 3' sequence is identical to XM_908332. In addition, we identified a complete polyadenylation signal (AATAAA) and a poly-A tail. The cDNA sequences of the P4–P6 PCR product and the 3'-RACE product overlapped by 418 base pairs and were spliced together in silico by Sequencher 4.5 software, providing the complete sequence of male germ cell Tekt5 mRNA (Supplemental Figure 2). To confirm the above finding, we performed long-range RT-PCR of male germ cell mRNA to amplify the complete cDNA product from 5'-UTR to 3'-UTR. The expected sized PCR product (1747 bp) was seen and sequenced from both directions (Fig. 3D). The complete Tekt5 mRNA sequence, which contains an in-frame stop codon upstream from the start codon as well as the polyadenylation signal and poly-A tail, was submitted to GenBank (accession number GQ292766). The total cDNA sequence was 1785 bp, which differed from the 2.0–2.3 kb length of the mRNA as determined by northern blotting analysis (Fig. 2C). The discrepancy between the lengths of the experimentally determined cDNA sequence and the mRNA detected by northern blotting is probably due to the incompletely defined 5'-UTR and a more extended poly A region of the 3'-UTR of the mRNA (our 5'-UTR was 36 bp and the poly-A tail was only 11 bp). The 1785 bp cDNA reported here is 508 bp longer than the cDNA available at NCBI (Accession BC115969), which terminates at amino acid 415 of TEKT5.
Figure 3.
Utilization of PCR and 3'-RACE to clone the testicular Tekt5 cDNA. The primers used in the molecular cloning procedure are labeled in Fig. 2A, and listed in Table 1. (A): Primer P4 was designed from the 5'-UTR region described for Tekt5 sequences present in the NCBI database. Primer P3 was based upon genomic sequence upstream from this region. P4 produced the expected size of product with P5 and was verified by DNA sequencing. P6, P7, P8, P9 are 3' corresponding primers for XM_908332, XM_980627, BC115969, and XM_156349, respectively. P6 and P8 produced the expected sizes of PCR products with P1. (B): Attempts to amplify longer PCR products with P4–P6 and P4–P8 were successful. The identities of the PCR products were verified by sequencing. (C): The 3'-UTR of Tekt5 was characterized by 3'-RACE. A positive control encompassing part of the Tekt5 open reading frame, (lane 1), was included to confirm that the RACE reverse transcription was successful. Gradient PCR reactions at different annealing temperatures (lanes 2–5 are 55°C, 57.5°C, 60°C, and 62.5°C, respectively) were used to optimize the RACE conditions. A predominant PCR product was seen at about 1 kb. (D): P11 was located in the 3'-UTR of Tekt5 RACE results. P4–P11 amplified the Tekt5 cDNA sequence from 5'-UTR to 3'-UTR (lanes 1–2 are 49°C, 52°C). The sequencing results showed that our Tekt5 cDNA sequence is similar to the computer-predicted sequence XM_908332, but that our Tekt5 cDNA sequence includes a complete polyadenylation signal (AATAAA) and a poly-A tail. Note: All PCR products were verified by nucleotide sequencing. The no-template control PCR reaction used water in place of the sample. (E): alignment of encoded Tektin 5 with other known Tektins. The peptide sequences identified by mass spectroscopy are underlined in red and the canonical tektin domain is outlined with a blue box.
Features of the TEKT5 protein
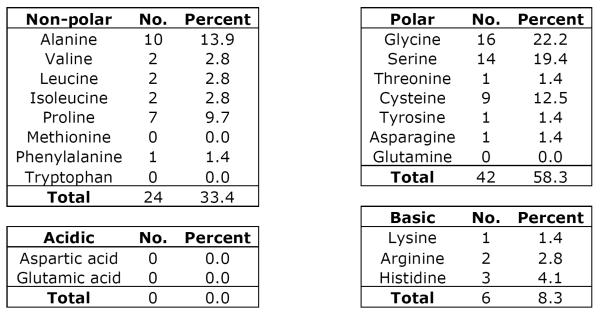
TEKT5 has a deduced molecular weight of 62,730, which is 107 Daltons smaller than the rat sequence (Murayama et al, 2008). The identical amino acids between the orthologues of mouse and rat account for 89%. Among all identified tektins, TEKT5 has the highest homology to TEKT3 (Fig. 3E). Using bl2seq (NCBI/BLAST), the identity ratio is 48% and the positive ratio is 67%. The closer homology between TEKT3 (56,669 Da) and TEKT5 partly results from the property that both of these tektins possess extended N-termini. Like other tektins, TEKT5 possesses the tektin motif, RPNVELCRD, which is located at amino acids 402–410 and a coiled coil region (amino acids 345–385). The tektin motif in most tektins is located close to the C-terminus (<100 amino acid from the end of the protein), whereas the tektin domain of TEKT5 is 147 amino acids away from the C-terminus as the result of an unusually long C-terminal tail (Fig. 3E). By analyzing the unique C-terminal tail between amino acids 486–557, we found there is a low complexity region CSGSALCKGPASCGGGASCGGGASCGGHAPCGSAL (amino acids 507–541) (supplemental figure 3). Analyses of amino acid composition also revealed that there are no aspartates or glutamates in the region of amino acids 485–557 but basic amino acids account for 8.3%. That is probably the reason why TEKT5 is the most basic tektin (Fig 1). Also this region has a high abundance of small amino acids like alanine (13.9%) and glycine (22.2%), as well as serine (19.4%), cysteine 12.5% and proline 9.7% (Table 3).
Table 3.
Amino Acid Composition of TEKT5486-557
|
Tekt5 mRNA expression is up-regulated during late spermiogenesis
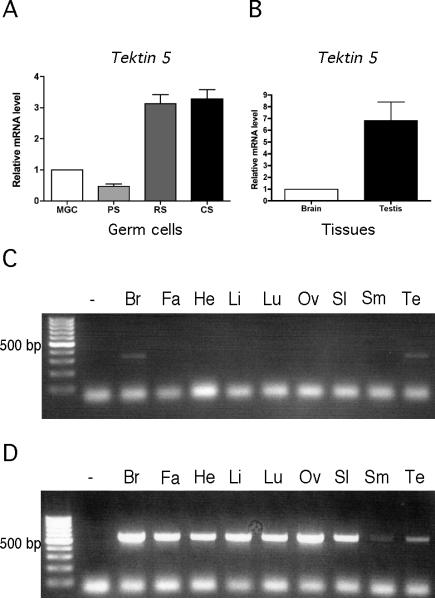
To examine the expression pattern of Tekt5 mRNA in male germ cells, quantitative real-time RT-PCR was conducted (Fig. 4A). Tekt5 mRNA was present in pachytene spermatocytes, round spermatids, and condensing spermatids. On a per microgram of RNA basis, the level was highest in condensing spermatids and was greater than 6~7-fold higher than that found in pachytene spermatocytes. Tekt5 mRNA expression in several somatic tissues was also examined. The results showed that Tekt5 mRNA was specifically expressed in testes and brain (Fig. 4C). Beta-actin served as a loading control (Fig. 4D). By quantitative real-time RT-PCR, the expression level of Tekt5 mRNA in the testis was about 6.8-fold higher than that in the brain (Fig. 4B).
Figure 4.
Tekt5 mRNA levels increase substantially in spermiogenesis. (A): Quantitative RT-PCR of Tekt5 from mixed germ cell (MGC), pachytene spermatocytes (PS), round spermatids (RS), and condensing spermatids (CS). Results were normalized to mRNA corresponding to ribosomal protein S16. (B): Tekt5 mRNA was detected in testis and brain by RT-PCR. (C): RT-PCR analysis of Tekt5 mRNA from brain (Br), fat (Fa), heart (He), liver (Li), lung (Lu), oviduct (Ov), spleen (Sl), smooth muscle (Sm), and testis (Te). (D): Beta-actin was used as a loading control for various tissue samples.
TEKT5 protein is enriched in flagellar accessory structures
Recombinant fusion protein GST-TEKT5486-557 was expressed in bacteria and purified by chromatography on a GSTrap™ FF column. The antiserum against GST-TEKT5486-557 was characterized using immunoblotting. The results showed that the antiserum recognized the GST-TEKT5486-557 fusion protein, while preimmune serum did not exhibit any signal (Fig. 5A). The antiserum also recognized a ~62,000 Mr protein in sperm, which is consistent with the deduced mass of the translation product of Tekt5 transcript (62,730 Da). Of note, a minor immunoreactive band at 43,000 Mr was detectable in whole sperm. This band was not seen in flagellar accessory structures (Fig. 5C). Antiserum preabsorbed with the GST-TEKT5486-557 fusion protein did not recognize the ~62,000 Mr TEKT5, but still reacted with the 43,000 Mr protein, indicating that the detection of this smaller protein resulted from a non-specific interaction with the antiserum (Fig. 5B). When equivalent amounts of protein were examined, a stronger TEKT5 signal was detected in flagellar accessory structure preparations relative to whole sperm extracts, indicating that TEKT5 was enriched in these cytoskeletal elements (Fig. 5C–b). The purity of the flagellar accessory structures was assessed biochemically by immunoblotting with antibody against α-tubulin (Fig. 5C–a), which confirmed that this sperm fraction lacked microtubules and demonstrated that TEKT5 was more likely associated with a flagellar cytoskeletal element other than the axoneme. Further experiments showed that TEKT5 was resistant to 1% S-EDTA extraction, whereas the 43,000 Mr protein was not (Fig. 5D). The brain extract did not contain the immunoreactive band at 62,000 Mr (Fig. 5E).
Figure 5.
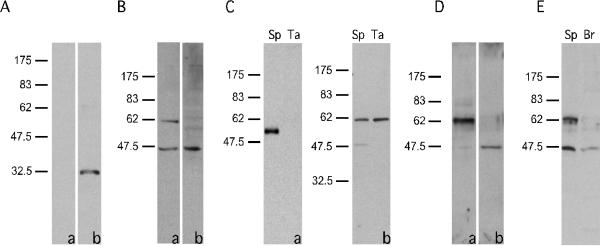
TEKT5 protein was present in sperm (Sp) and was particularly enriched in sperm flagellar accessory structures (Ta). (A): Preimmune serum (a) did not recognize the recombinant GST-TEKT5486-557 but immune serum (b) did. (B): Anti-GST-TEKT5486-557 recognized a ~62,000 Mr protein present in sperm, as well as a 43,000 Mr band (a). Reactivity of the ~62,000 Mr protein with the antibody was blocked by purified antigen GST-TEKT5486-557, while the 43,000 Mr signal was not, demonstrating the non-specific nature of this band (b). (C): The flagellar accessory structures lacked axonemal components as demonstrated by immunoblotting of equal amounts of sperm (Sp) and flagellar accessory structure protein (Ta). The axonemal protein, α–tubulin, was present in extracts of whole sperm but was absent in the flagellar accessory structure protein preparation (a). After 1% S-EDTA treatment, TEKT5 remained associated with the flagellar accessory structures, whereas the 43,000 Mr protein, which non-specifically bound to the anti-GST-TEKT5486-557 antibody, was solubilized. (b). (D): Equal amounts of protein from the 1% S-EDTA soluble supernatant or insoluble pellet (flagellar accessory structures) were analyzed by immunoblotting with the anti-GST-TEKT5486-557 antibody. This treatment effectively partitions the ~62,000 Mr protein into the S-EDTA insoluble fraction (a), whereas the non-specific 43,000 Mr signal was extracted into the supernatant (b). (E): The ~62,000 Mr, signal was detectable in sperm protein (Sp) but was absent in protein extracts of brain (Br).
TEKT5 is present in the sperm tail
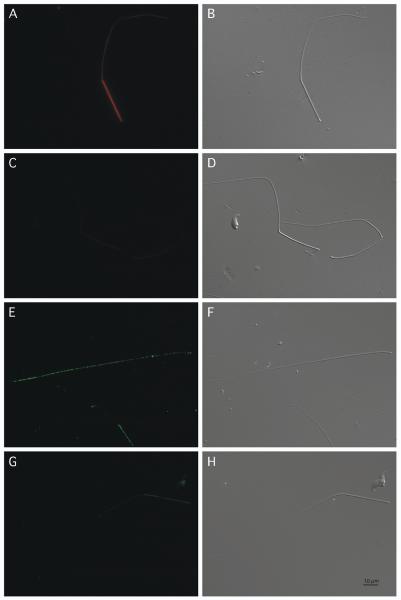
We were concerned that the 43,000 Mr band recognized by anti-GST-TEKT5486-557 might confound any results seen in whole sperm, so immunolocalization was performed using flagellar accessory structures, which do not contain this non-specific protein (Fig. 5C and 5D). The immunofluorescence images demonstrate that the TEKT5 signal stained the full length of flagellar accessory structures (note: the SDS-processed sperm tail structures lack axonemes and thus do not contain end piece, please see the Supplemental Figure 1), which further indicated that TEKT5 was present in the midpiece and principal piece (Fig. 6A, B). This suggests that TEKT5, like TEKT4 (Iida et al, 2006), was associated with outer dense fibers since these structures are common to both the midpiece and principal piece. The pre-absorbed antibody exhibited a much weaker signal (Fig. 6C, D), indicating the signal caused by anti-GST-TEKT5486-557 antibody was specific. The immunostaining of ODF2 was also present in the full length of flagellar accessory structures, but the staining pattern was different from the TEKT5. Although the TEKT5 staining was present throughout the flagellar accessory structures, the antibody signal was much more intense in the midpiece than ODF2.
Figure 6.
As determined by indirect immunofluorescence, TEKT5 was present along the full length of flagellar accessory structures with much stronger staining in the midpiece. Paired indirect immunofluorescence and Nomarski differential interference contrast images of sperm probed with anti-GST-TEKT5486-557 antibody (A and B) or anti-GST-TEKT5486-557 antibody preabsorbed with GST-TEKT5486-557 peptide (C and D). Paired indirect immunofluorescence and Nomarski differential interference contrast images of sperm probed with anti-ODF2 antibody (E and F) or normal goat IgG (G and H), Bar = 10 μm.
Discussion
Axonemes are highly structurally conserved from unicellular algae to mammals. This core structure of eukaryotic cilia and flagella is responsible for generating the motive force required for ciliary beating and sperm motility and is composed of a characteristic “9 + 2” array of microtubules and associated structures (Supplemental Figure 1). Tektins were first isolated from sea urchin sperm tail axonemes (Linck and Stephens, 1987) but the locations and functions of the multitude of tektin forms in mammalian sperm flagella remain unclear. In sea urchin flagella, tektins were proposed to form protofilaments near the site where the B tubules attach to A tubules in the outer doublets (Nojima et al, 1995). Also, electron microscopy has revealed a longitudinally continuous thin filament, which might bind to the protofilaments between the A-and B-tubules and support microtubule sliding and bending (Sui and Downing, 2006). Sea urchin tektins A1 and B1 heterodimers co-assemble with tektin C1 dimers to produce filaments with overall repeats of multiples of 8 nm and form the basis for the complex spatial arrangements of axonemal components (Norrander et al, 1996).
Compared to the sea urchin, mammalian sperm flagella are more complex. Echinoderm sperm contain a very small mitochondrial midpiece at the head-tail junction. Mammalian sperm possess a more developed midpiece with an extensive mitochondria sheath surrounding nine outer dense fibers that emanate from the nine outer doublets of the axoneme. At the annulus, the mitochondrial sheath terminates and two of the outer dense fibers (numbers 3 and 8) are replaced with the longitudinal columns of the fibrous sheath, which run the length of the principal piece and are bridged by circumferential ribs. The tektin involvement in mammals appears to be complicated as well. To date, five tektin genes have been cloned in rodents. Mouse TEKT1 is found in the centrosomes of round spermatids and gradually disappears during spermiogenesis (Larsson et al, 2000). The actual function of TEKT1 is poorly understood. Human TEKT2 (also named as Tektin-t) is present in the principal piece (Tanaka et al, 2004). Disruption of the corresponding gene in the mouse causes male infertility, which might result from impaired inner dynein arm function. The location of TEKT3 in mouse sperm in unclear but mice deficient in TEKT3 display reduced sperm motility and forward progression (Roy et al, 2009). Mouse TEKT4 is present in outer dense fibers and deletion of the Tekt4 gene causes drastically reduced forward motility resulting from poor coordinated beating of the flagellum (Roy et al, 2007). Rat TEKT5 is localized between the mitochondria and the outer dense fibers (Murayama et al, 2008). Functional studies have not been reported for TEKT5. However, it is clear that tektins are intimately involved with spermiogenesis and dysfunction of tektins can severely impair sperm motility, leading to male infertility.
Flagellar accessory structures were once considered as passive supportive structures (Supplemental Figure 1). However, the discovery of the protein kinase A-anchoring property of AKAP4 of the sperm fibrous sheath suggested that these structures could have more active roles than people originally imagined (Carrera et al, 1994, Miki and Eddy, 1999). Proteomic studies conducted by our group identified protein components of flagellar accessory structures, including many glycolytic and metabolic proteins, anti-oxidant enzymes, all five tektins, as well as other proteins (Cao et al, 2006a). We were surprised to find tektins in the flagellar accessory structures and confirmed that these samples were not contaminated by axonemal proteins such as tubulin (Fig. 5C). Additional studies are required to establish the function of TEKT5 in the flagellar accessory structures.
Prior to this study, a cDNA encoding mouse Tekt5 cDNA had not been cloned. There are several sequences for Tekt5 cDNA in the NCBI database but these in silico predictions are either computer deduced sequences (Accession Numbers XM_908332 and XM_980627), or identified from global transcriptome analyses, such as Accession Number BC115969 (Carninci et al, 2005, Strausberg et al, 2002). These high-throughput procedures provide much valuable information, but the data must be interpreted with caution because sequence errors are occasionally introduced by the methodology. In 2005, our proteomic study of flagellar accessory structures identified TEKT5 peptides that correspond to nucleotide sequences XM_156349, which was assigned the Tekt5 gene nomenclature. As the genome annotation progressed, the representative sequence became known as BC115969 in the NCBI database. This predicts a protein of 48,000 Da, but this size does not correspond to our two-dimensional gel electrophoresis experimental results (Fig. 1). In our study with RNA from mixed germ cells, we characterized a transcript possessing a complete open reading frame, the size of which was consistent with our Northern blot result (Fig. 2C). Furthermore, this transcript encoded a protein whose deduced molecular weight and pI were consistent with our two-dimensional gel result (Fig. 1). In addition, the peptides we identified by mass spectrometry spanned the full length of the amino acid sequence deduced from our cDNA, including the extended C-terminal region unique to TEKT5 (Table 2). Sequence analyses of our Tekt5 cDNA and peptides identified by mass spectrometry demonstrated that TEKT5 contains the characteristic C-terminal signature tektin motif “R(P/S)NVELCRD” (Fig. 3E) and coiled-coil secondary structure (Supplemental Figure 3) characteristic of tektin family members (Norrander et al, 1996). Although we cannot completely exclude the possible existence of an mRNA corresponding to the shorter reported transcript BC115969, the majority of transcripts should resemble GQ292766, as determined by northern blotting and 3'-RACE. If transcripts similar to BC115969 were highly abundant, a different PCR product would have been detected by northern blotting (Fig. 2C) and 3'-RACE (Fig. 3C). Since a shorter transcript was not detected, we suspect that BC115969 represents an incomplete cDNA. The cloned mouse Tekt5 cDNA sequence reported in our study establishes a solid foundation for further functional studies using methodology such as genetic manipulation.
The deduced mouse TEKT5 is 89% identical to rat TEKT5, which is a higher homology than is found for any of the other mouse tektins, suggesting that each type of tektin has a specific role. In comparison to other tektins, one distinguishing feature of TEKT5 is its long C-terminal tail composed of over 70 extra amino acids (Fig. 3E). Use of the Simple Modular Architecture Research Tool (SMART) demonstrated that this region exhibits extremely low complexity (Letunic et al, 2009, Schultz et al, 1998) (Supplemental Figure 3). In particular, this tail is rich in glycine (22.2%), serine (19.4%), alanine (13.9%), cysteines (12.5%), and proline (9.7%) with six repeats of the hexpeptide pattern C-[GSK]-G-[GSPH]-A-[SLP] from amino acids 507–541. There are no acidic amino acids in this region and the few basic amino acids confer a predicted isoelectric point of 11.23 to this domain. The biological significance of this region is unclear but the sequence and composition suggest that this domain may possess the ability to form multiple disulfide cross-links and/or interact with acidic regions of other proteins. Low complexity regions are widely used by signaling proteins or to mediate protein-protein interactions (Dunker et al, 2001, Kay et al, 2000, Michelitsch and Weissman, 2000). Coincidently, the rat sperm outer dense fiber proteins have high cysteine (as high as 11–12%) and proline content (Olson and Sammons, 1980). The C-terminus of ODF1 contains a Cys-Gly-Pro repeat (Morales et al, 1994). By yeast two-hybrid screening, ODF1 interacts with axonemal proteins Spag4 and Spag5 (Shao et al, 1999, Shao et al, 2001). Therefore, ODF1 mediates interactions between the outer dense fibers and the axoneme. The structural similarity between ODF and TEKT5 (low complexity region with repeats high in cysteine and proline) suggests to us that this C-terminal extension may be important for axonemal or outer dense fiber protein interactions.
We compared the expression pattern of mouseTekt5 mRNA to that of rat Tekt5 (Murayama et al, 2008). Murayama et al reported that Tekt5 expression in the rat testis was first detectable by RT-PCR at 5 weeks after birth. We found that Tekt5 was expressed at a low level at the pachytene spermatocyte stage. During spermiogenesis when flagella biogenesis occurs in round and condensing spermatids, the level of Tekt5 expression increased about 6–7 fold (Fig. 4A). When we examined extracts of cilia-rich tissues, Tekt5 mRNA was identified by RT-PCR in extracts of the brain but was undetectable in the lung. This finding differs from the reported lack of mRNA expression in the rat brain (Murayama et al, 2008). However, TEKT5 protein was not detected in immunoblots of mouse brain (Fig. 5E). The biological significance of the Tekt5 mRNA expression in the brain remains to be elucidated.
Consistent with our previous proteomic study, mouse sperm TEKT5 was insoluble in 1% S-EDTA, suggesting that this protein was localized in the flagellar accessory structures (Fig. 5D). The lower molecular weight band of 43,000 Mr detected by immunoblotting posed some challenges for our studies of the localization of TEKT5 in sperm (Fig. 5C). The extra band was due to non-specific binding since the immunoreactivity of the specific band at 62,000 Mr could be blocked by preabsorption with the recombinant protein antigen (Fig. 5B). Fortunately for our immunolocalization experiments, the non-specific protein was removed by extraction with 1% S-EDTA (Fig. 5D), allowing us to localize TEKT5 in flagellar accessory structures. The results showed that mouse TEKT5 was present throughout the full length of accessory structures with a much stronger signal in the midpiece (Fig. 6A). However, using indirect immunofluorescence and immunoelectron microscopy of autoclaved cells, Murayama et al. (2008) concluded that rat TEKT5 is restricted to the sperm midpiece. Our ability to detect lower levels of TEKT5 in the mouse sperm principal piece might be due to our immunohistochemical protocol or a difference in species. We localized TEKT5 in mouse sperm flagellar accessory structures that had been prepared in 1% S-EDTA and fixed in 4% paraformaldehyde. On the other hand, Murayama et al (2008) were unable to preserve immunoreactivity with their antibody following extraction with the detergents Triton X-100 or 1% SDS, and they autoclaved the rat sperm samples as part of their immunolocalization protocol. We repeatedly tried to localize TEKT5 in mouse sperm by immunoelectron microscopy without success.
In summary, we cloned Tekt5 cDNA through RACE and RT-PCR. It contained a complete coding region with polyadenylation signal and poly-A tail. The cDNA sequence encodes an appropriately sized TEKT5 protein compared to the smaller size predicted from the NCBI and MGI database representative sequence. The TEKT5 protein was highly expressed in mouse spermatids and was localized in the flagellar accessory structures. The stable association of tektins with the flagellar accessory structures raises many questions concerning their roles in sperm. The N-terminal extension of TEKT5 and its unusually long C terminus with a strange amino acid composition containing six repeats of the hexpeptide pattern C-[GSK]-G-[GSPH]-A-[SLP] is intriguing and worthy of more extensive study. Genetic manipulation of the Tekt5 gene may yield additional important information concerning the functions of this flagellar protein.
Supplementary Material
The diagram of sperm structures and accessory structures. After treatment with 1% S-EDTA and homogenization, the sperm flagellum was reduced to the mitochondrial sheath, fibrous sheath, and outer dense fibers.
The nucleotide sequences of 5′ PCR and 3′-RACE products overlapped 700 by 418 bp. The merging of the PCR and 3′-RACE products was performed using Sequencher 701 4.5.
The coiled-coil analysis of mouse TEKT5 amino acid sequence using SMART. Coiled-coil regions (green, 345–386) were determined by the Coils2 program (Lupas, 1996), whereas, segments of low compositional complexity (pink, 515–540) were determined by the SEG program (Wootton and Federhen, 1996). Intron positions are indicated with vertical lines showing the intron phase and exact positions in the amino acid sequence.
Acknowledgments
We thank Drs. Bayard T. Storey and Mariano G. Buffone for constructive criticisms. This work was supported by NIH grants P01-HD006274 and R01-HD057144 to GLG and T32-HD007305 to TWI.
Supported in part by NIH grants P01-HD006274 and R01-057144 to GLG and NIH Training Grant T32-HD007305 to TWI.
References
- Bellvé AR, Millette CF, Bhatnagar YM, O'Brien DA. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977;25:480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- Cao W, Gerton GL, Moss SB. Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol Cell Proteomics. 2006a;5:801–10. doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]
- Cao W, Haig-Ladewig L, Gerton GL, Moss SB. Adenylate kinases 1 and 2 are part of the accessory structures in the mouse sperm flagellum. Biol Reprod. 2006b;75:492–500. doi: 10.1095/biolreprod.106.053512. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrera A, Gerton GL, Moss SB. The major fibrous sheath polypeptide of mouse sperm: Structural and functional similarities to the A-kinase anchoring proteins. Dev Biol. 1994;165:272–284. doi: 10.1006/dbio.1994.1252. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Godeas C, Tramer F, Micali F, Soranzo M, Sandri G, Panfili E. Distribution and possible novel role of phospholipid hydroperoxide glutathione peroxidase in rat epididymal spermatozoa. Biol Reprod. 1997;57:1502–8. doi: 10.1095/biolreprod57.6.1502. [DOI] [PubMed] [Google Scholar]
- Iida H, Honda Y, Matsuyama T, Shibata Y, Inai T. Tektin 4 is located on outer dense fibers, not associated with axonemal tubulins of flagella in rodent spermatozoa. Mol Reprod Dev. 2006;73:929–36. doi: 10.1002/mrd.20486. [DOI] [PubMed] [Google Scholar]
- Jeong YJ, Choi HW, Shin HS, Cui XS, Kim NH, Gerton GL, Jun JH. Optimization of real time RT-PCR methods for the analysis of gene expression in mouse eggs and preimplantation embryos. Mol Reprod Dev. 2005;71:284–9. doi: 10.1002/mrd.20269. [DOI] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–41. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson M, Norrander J, Graslund S, Brundell E, Linck R, Stahl S, Hoog C. The spatial and temporal expression of Tekt1, a mouse tektin C homologue, during spermatogenesis suggest that it is involved in the development of the sperm tail basal body and axoneme. Eur J Cell Biol. 2000;79:718–25. doi: 10.1078/0171-9335-00097. [DOI] [PubMed] [Google Scholar]
- Linck RW, Stephens RE. Biochemical characterization of tektins from sperm flagellar doublet microtubules. J Cell Biol. 1987;104:1069–1075. doi: 10.1083/jcb.104.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–25. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–5. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K, Eddy EM. Single amino acids determine specificity of binding of protein kinase A regulatory subunits by protein kinase A anchoring proteins. J Biol Chem. 1999;274:29057–62. doi: 10.1074/jbc.274.41.29057. [DOI] [PubMed] [Google Scholar]
- Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci U S A. 2004;101:16501–6. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales CR, Oko R, Clermont Y. Molecular cloning and developmental expression of an mRNA encoding the 27 kDa outer dense fiber protein of rat spermatozoa. Mol Reprod Dev. 1994;37:229–240. doi: 10.1002/mrd.1080370215. [DOI] [PubMed] [Google Scholar]
- Murayama E, Yamamoto E, Kaneko T, Shibata Y, Inai T, Iida H. Tektin5, a new Tektin family member, is a component of the middle piece of flagella in rat spermatozoa. Mol Reprod Dev. 2008;75:650–658. doi: 10.1002/mrd.20804. [DOI] [PubMed] [Google Scholar]
- Nojima D, Linck RW, Egelman EH. At least one of the protofilaments in flagellar microtubules is not composed of tubulin. Curr Biol. 1995;5:158–67. doi: 10.1016/s0960-9822(95)00037-6. [DOI] [PubMed] [Google Scholar]
- Norrander JM, Perrone CA, Amos LA, Linck RW. Structural comparison of tektins and evidence for their determination of complex spacings in flagellar microtubules. J Mol Biol. 1996;257:385–97. doi: 10.1006/jmbi.1996.0170. [DOI] [PubMed] [Google Scholar]
- O'Brien DA, Bellvé AR. Protein constituents of the mouse spermatozoon. I. An electrophoretic characterization. Dev Biol. 1980;75:386–404. doi: 10.1016/0012-1606(80)90171-2. [DOI] [PubMed] [Google Scholar]
- Olson GE, Sammons DW. Structural chemistry of outer dense fibers of rat sperm. Biol Reprod. 1980;22:319–322. doi: 10.1095/biolreprod22.2.319. [DOI] [PubMed] [Google Scholar]
- Pirner MA, Linck RW. Tektins are heterodimeric polymers in flagellar microtubules with axial periodicities matching the tubulin lattice. J Biol Chem. 1994;269:31800–6. [PubMed] [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: 2007. [Google Scholar]
- Romrell LJ, Bellvé AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. Faseb J. 2007;21:1013–25. doi: 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Tektin 3 is required for progressive sperm motility in mice. Mol Reprod Dev. 2009;76:453–9. doi: 10.1002/mrd.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Yan W, Burns KH, Matzuk MM. Tektin3 encodes an evolutionarily conserved putative testicular microtubules-related protein expressed preferentially in male germ cells. Mol Reprod Dev. 2004;67:295–302. doi: 10.1002/mrd.20025. [DOI] [PubMed] [Google Scholar]
- Shao X, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. Spag4, a Novel Sperm Protein, Binds Outer Dense-Fiber Protein Odf1 and Localizes to Microtubules of Manchette and Axoneme. Dev Biol. 1999;211:109–123. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- Shao X, Xue J, van der Hoorn FA. Testicular protein Spag5 has similarity to mitotic spindle protein Deepest and binds outer dense fiber protein Odf1. Mol Reprod Dev. 2001;59:410–6. doi: 10.1002/mrd.1047. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 2006;442:475–8. doi: 10.1038/nature04816. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004;24:7958–64. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–71. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The diagram of sperm structures and accessory structures. After treatment with 1% S-EDTA and homogenization, the sperm flagellum was reduced to the mitochondrial sheath, fibrous sheath, and outer dense fibers.
The nucleotide sequences of 5′ PCR and 3′-RACE products overlapped 700 by 418 bp. The merging of the PCR and 3′-RACE products was performed using Sequencher 701 4.5.
The coiled-coil analysis of mouse TEKT5 amino acid sequence using SMART. Coiled-coil regions (green, 345–386) were determined by the Coils2 program (Lupas, 1996), whereas, segments of low compositional complexity (pink, 515–540) were determined by the SEG program (Wootton and Federhen, 1996). Intron positions are indicated with vertical lines showing the intron phase and exact positions in the amino acid sequence.