Abstract
Loss of skeletal muscle mass occurs frequently in clinical settings in response to joint immobilization and bed rest, and is induced by a combination of unloading and inactivity. Disuse-induced atrophy will likely affect every person in his or her lifetime, and can be debilitating especially in the elderly. Currently there are no good therapies to treat disuse-induced muscle atrophy, in part, due to a lack of understanding of the cellular and molecular mechanisms responsible for the induction and maintenance of muscle atrophy. Our current understanding of disuse atrophy comes from the investigation of a variety of models (joint immobilization, hindlimb unloading, bed rest, spinal cord injury) in both animals and humans. Under conditions of unloading, it is widely accepted that there is a decrease in protein synthesis, however, the role of protein degradation, especially in humans, is debated. This review will examine the current understanding of the molecular and cellular mechanisms regulating muscle loss under disuse conditions, discussing the similarities and areas of dispute between the animal and human literature.
Keywords: Unloading, Protein synthesis, Protein degradation, Ubiquitin ligases, Reloading
1. Introduction
Skeletal muscle atrophy occurs in response to a variety of stressors including decreases in external loading and neural activation, increases in inflammatory cytokines and glucocorticoids, and malnutrition. A combination of unloading and reduced neural activity, commonly referred to as “disuse”, occurs frequently in clinical settings following limb immobilization, bed rest, spinal cord injury and partial/complete peripheral nerve damage, resulting in significant loss of muscle mass and force production. The extent of muscle atrophy under disuse conditions is variable and dependent on a variety of factors including age, the physiological function and fiber type composition of the muscle, and the degree of unloading and inactivity. A number of disuse models, having variable degrees of unloading and inactivity, such as ankle immobilization, hindlimb unloading, and spinal cord isolation have been studied in rodents and other mammals, providing valuable information on the extent of morphological and functional alterations, as well as insights into the molecular and cellular mechanisms responsible for disuse-induced muscle loss. While disuse muscle atrophy is a common clinical problem there are no good therapies to prevent the loss of muscle mass and tension. Moreover, there are no good pharmaceutical options to enhance the recovery of muscle mass following an atrophy-inducing event. Currently, only resistance exercise can be used to promote recovery of mass/strength following disuse atrophy, and many patients are unable or unwilling to exercise at a sufficient intensity to promote muscle growth.
2. Loss of muscle mass following disuse
The loss of skeletal muscle mass and fiber cross-sectional area is well documented in rodent models of ankle-joint immobilization, hindlimb unloading via tail suspension, and neural inactivity as the result of spinal cord injury (Booth and Gollnick, 1983; Ohira et al., 2002; Roy et al., 1991; Thomason and Booth, 1990). In general, under conditions of disuse in rodents, where the nerve is intact, there is rapid loss of mass within the first one-two weeks of unloading followed by a slowing of the rate of muscle loss until muscle mass reaches a nadir and maintains a new lower steady state. Muscle mass will remain at this new steady state until an anabolic stimuli, such as an increase in external loading, stimulates muscle regrowth (see Fig. 1). The rate and amount of muscle mass lost is dependent on both the muscle type and the degree of unloading and inactivity. In rodent models of immobilization and hindlimb unloading, muscle loss is generally greater in the extensor muscles of the ankle (i.e., soleus and gastrocnemius) than the flexor muscles (i.e., tibialis anterior and extensor digitorum longus) (Ohira et al., 2002; Roy et al., 1991; Zhong et al., 2005; Adams et al., 2003). Further, within a particular muscle, the loss of fiber cross-sectional area is first seen in the slow, type I fibers followed by the fast type IIa and then fast type IIx and IIb fibers (Ohira et al., 2002, Thomason and Booth, 1990). For some muscles (e.g., medial gastrocnemius and tibialis anterior) the amount of muscle loss is often greater under conditions of joint immobilization than hindlimb unloading or bed rest (Bodine et al., 2001a; Clark, 2009). Muscle atrophy under conditions of immobilization varies significantly depending on how completely the joint is restricted from movement (e.g., casting versus pinning) and the angle at which the joint is fixed; muscle atrophy being greatest when a muscle is immobilized in a shortened versus a neutral or lengthened position (Spector et al., 1982; Goldspink, 1977).
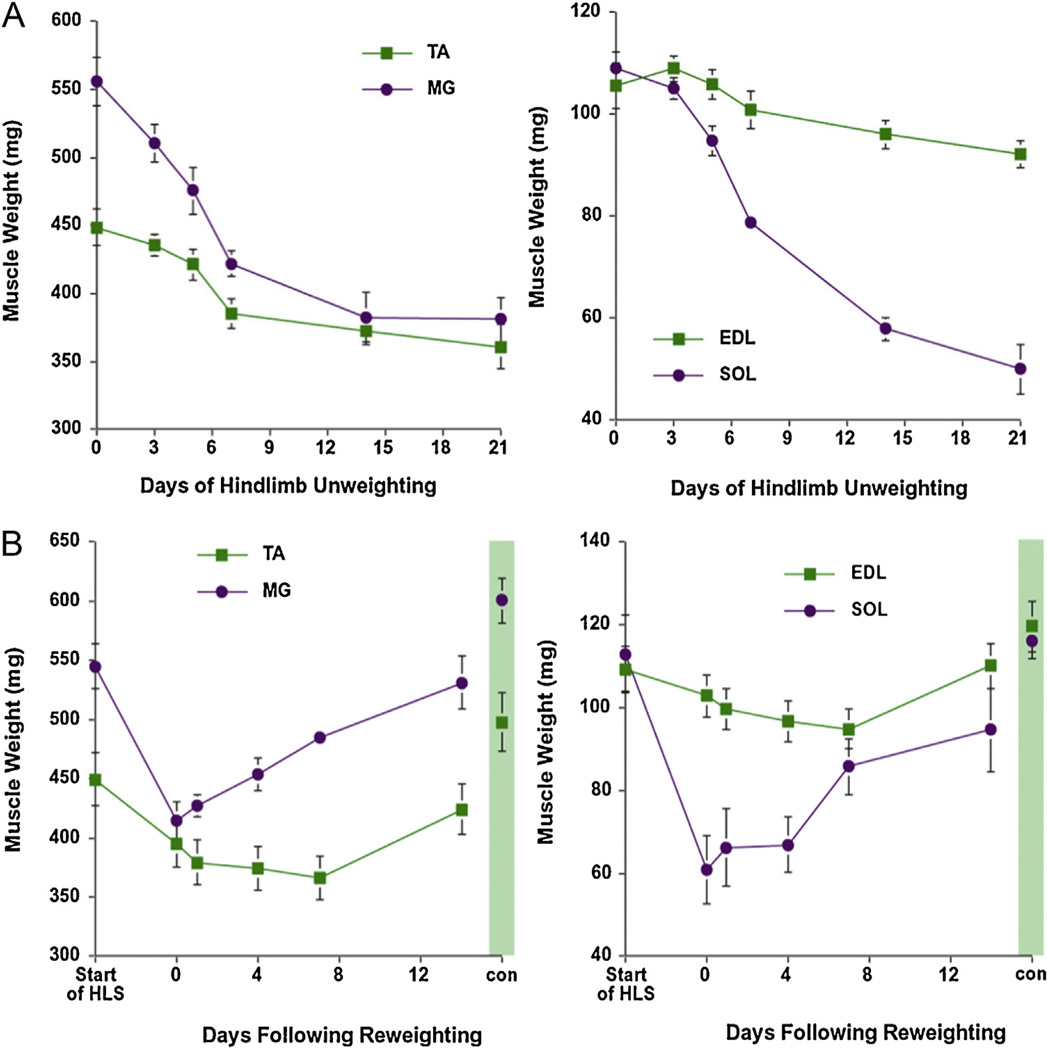
Fig. 1.
Effect of hindlimb unloading and reloading on muscle mass. Muscle wet weight of the soleus (Sol), medial gastrocnemius (MG), tibialis anterior (TA), and extensor digitorum longus (EDL) of female Sprague Dawley rats following hindlimb unloading for 21 days (A) and hindlimb unloading for 14 days followed by 14 days of reloading (B). Data points are mean ± SD (n = 10/time point). A separate cohort of controls was taken at the start (start of HLS) and end (shaded area) of the experiment to assess normal growth over the 28 day experiment. The control rats were allowed food and water ad libitum over the 28 day period.
In humans, differential atrophy across muscles and fiber types can be observed in response to disuse. Following prolonged (~180 days) spaceflight Fitts et al. (2010) found significant loss of fiber size and force in the soleus and gastrocnemius muscles with the hierarchy of effect being soleus type I > soleus type II >gastrocnemius type I >gastrocnemius type II. The same fiber type response was found after 35 days of bed rest in the vastus lateralis (VL) muscle where the percent loss of fiber cross-sectional was greater in the type I than the type II fibers (no difference being observed between the IIa and IIx) (Brocca et al., 2012). While these studies show differential atrophy related to muscle fiber type (i.e., slow versus fast myosin heavy chain), not all human studies have observed this relationship (Hvid et al., 2010; Bamman et al., 1998; Trappe et al., 2007). Those studies that have been unable to detect differential atrophy have generally been for short durations (≤14 days) of immobilization and restricted to one muscle (usually the VL). One weakness of the human studies is that the majority of data on fiber cross-sectional area and atrophy is based on a small biopsy sample taken from a single site in periphery of the muscle belly, with the majority of biopsies taken from the VL muscle (a knee extensor), with occasional examination of the soleus or medial gastrocnemius muscles (ankle extensors).
Human studies have also utilized magnetic resonance imaging (MRI) to examine volume and cross-sectional area changes in multiple muscles during disuse atrophy. A recent study used longitudinal MRI to examine atrophy in muscles of the lower limb for 43 days following ankle immobilization due to ankle fracture (Psatha et al., 2012). In this study, the greatest rate and amount of atrophy occurred in the soleus and medial gastrocnemius muscle, followed by the lateral gastrocnemius and tibialis anterior, which is consistent with the recruitment patterns of these muscles during locomotion. Another recent study examined the loss of cross-sectional area along the entire muscle length in nineteen lower limb muscles following 27 and 60-days of head-down tilt bed rest, and showed differential atrophy across muscles and between muscles of similar physiological function (i.e., synergist) (Miokovic et al., 2012). For example the posterior calf muscles had the fastest rates of atrophy relative to other limb muscles, with the soleus and medial gastrocnemius having a faster rate of atrophy than their synergist, the lateral gastrocnemius. Overall the posterior calf muscles had faster rates and greater amounts of atrophy than the knee extensors (vasti, rectus femoris) and ankle flexors (tibialis anterior, extensor digitorum longus). Another important finding was that many muscles (e.g., VL and MG) do not atrophy uniformly along the length of the muscle, which has significant implications for the interpretation of cross-sectional areas taken from a single biopsy site. Comparison of disuse atrophy in rodents and humans suggest that there are considerable similarities, and that the biggest difference appears to be in the rate at which atrophy occurs, with loss of mass in rodents being considerably faster than that observed in humans (Phillips et al., 2009; Rennie et al., 2010).
3. Mechanisms of muscle atrophy following disuse: alterations in protein synthesis
The maintenance of muscle mass is dependent on the balance of two processes: the rate of protein synthesis and protein degradation. Under atrophy conditions there is a shift in the balance of these two processes such that there is a net loss of muscle proteins. There is strong evidence in humans (Ferrando et al., 1996; Glover et al., 2008; Paddon-Jones et al., 2006; Biolo et al., 2004) and rodent models of disuse (Kelleher et al., 2013; Booth and Seider, 1979; Lang et al., 2012; Goldspink, 1977) that the rate of basal protein synthesis declines immediately after unloading and stays at a suppressed level for the duration of the disuse. Short-term immobilization in humans (≤14 days), with no other complicating factors, produces a reduction in VL muscle mass of 5–10% (de Boer et al., 2007; Jones et al., 2004; Suetta et al., 2009); and it is argued that this amount of atrophy can be solely accounted for by decreases in basal protein synthesis with no changes in protein degradation (Phillips et al., 2009; Rennie et al., 2010). It should be pointed out, however, that MuRF1 and/or MAFbx were shown to be upregulated within the first 14 days of immobilization (de Boer et al., 2007; Jones et al., 2004) and could have contributed to the loss of mass through an increase in the degradation of specific substrates. As will be discussed later in this review, measurement of protein degradation in humans has been difficult. Further, the exact cellular pathways affected by MuRF1 and MAFbx upregulation during atrophy are still unclear.
In contrast in rat models of immobilization or hindlimb unloading, there is much greater muscle loss, e.g., 20–40% in the medial gastrocnemius in 7–14 days, which cannot be accounted for solely by decreases in protein synthesis and thus require some increase, at least transiently, in protein degradation (Bodine et al., 2001b; Lang et al., 2012; Ohira et al., 2006). The soleus, a predominately slow twitch muscle in the rat, can atrophy as much as 50% following 14–21 days of hindlimb unloading (Thomason and Booth, 1990). Thomason and Booth (1990) predicted a model where loss of mass in the unloaded soleus is induced by an immediate and sustained decrease in the rate of protein synthesis, followed by a delayed increase in protein degradation that peaks at around 14 days and then returns to basal levels. Direct measurement of both the rate of protein synthesis and degradation has not been performed for multiple muscle types at multiple time points over an extended period of unloading, however, over a short duration of immobilization (~7 days) protein synthesis has been shown to be suppressed and degradation increased in the MG muscle (Lang et al., 2012; Magne et al., 2012)
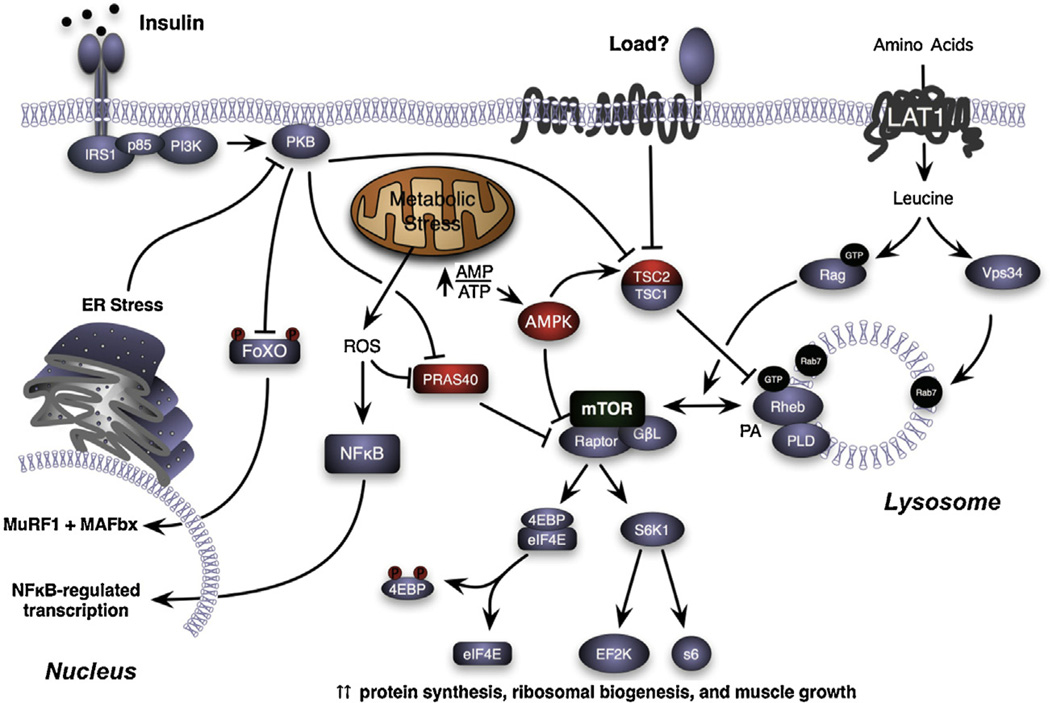
While decreases in the rate of protein synthesis have been measured following unloading, the cellular mechanisms responsible for the decline are poorly understood. Over the past decade, it has been demonstrated that adult skeletal muscle growth in response to increases in loading is critically regulated by the activation of the serine/threonine kinase, the mammalian Target Of Rapamycin (mTOR), resulting in increases in protein translation initiation and ribosome biogenesis (Bodine et al., 2001b; Goodman et al., 2011; Hornberger et al., 2004; von Walden et al., 2012). Given the importance of mTORC1 activation (the complex comprised of mTOR, raptor, and GβL (also known as mLST8)) in muscle growth, it has been hypothesized that under conditions of unloading decreases in protein synthesis are the result of inhibition of mTORC1 (Fig. 2). In rodent models of immobilization and hindlimb unloading decreases in the activation of Akt and mTORC1 (as measured by phosphorylation of S6K1 and 4E–BP1 and eIF4E•4E–BP1 binding) have been reported in the soleus and medial gastrocnemius muscles (Bodine et al., 2001b; Kelleher et al., 2013; Haddad et al., 2006; Hornberger et al., 2001). In contrast, in human immobilization studies, decreases in Akt/mTORC1 signaling have not been observed, even though decreases in protein synthesis have been measured (de Boer et al., 2007; Marimuthu et al., 2011; Glover et al., 2008; Greenhaff et al., 2008). The explanation for the disparate results in rodents and humans is not clear, but could be related to a number of factors including maturation state of the subjects, time points examined, muscle under study, or the immobilization procedure. In the majority of rodent studies, disuse is induced in juvenile animals that are still growing and where mTORC1 activity is elevated relative to an adult, weight stable animal. In mature rodents, short-term treatment (≤7 days) with the mTORC1 inhibitor, rapamycin, does not induce skeletal muscle loss (Bodine, 2006; Bodine et al., 2001b). Thus, the lack of a suppression of the mTORC1 pathway in human studies may reflect the fact that the subjects are mature and that basal mTORC1 activity is already low (Rennie et al., 2010).
Fig. 2.
Regulation of mTORC1 activation. Schematic diagram illustrating potential signaling pathways that could contribute to the inhibition of mTORC1 (mTOR, raptor, GβL (also known as mLST8)), and contribute to muscle loss following unloading and muscle regrowth following reloading. The proteins depicted in RED are known inhibitors of mTORC1.
The data are quite strong that in response to unloading the rate of protein synthesis declines, however, the question remains as to what cellular pathways are responsible for the immediate and sustained depression of protein synthesis. One pathway that has received limited investigation is the phosphorylation of glycogen synthase kinase-3β (GSK3β), which leads to its inhibition and an increase in global protein synthesis through an increase in the activity of eukaryotic initiation factor 2B (eIF2B) (Kimball et al., 2002). eIF2B is a guanine nucleotide exchange factor that mediates the exchange of GDP for GTP on eukaryotic initiation factor 2 (eIF2). The active eIF2-GTP binds to met-tRNAi, which then binds to the 40S ribosomal sub-unit. A decrease in the phosphorylation GSK3β, and thus an increase in its activity, has been measured following hindlimb unloading in rats (Stevenson et al., 2003; Childs et al., 2003), and following 48 h of immobilization in humans (Urso et al., 2006). Another potential pathway that may be modified following unloading is elongation factor 2. Elongation factor 2 (eEF2) is regulated through the mTOR pathway and by cellular energy stress status (Browne and Proud, 2002), and mediates the translocation step of elongation. Phosphorylation of eEF2 at Thr 56 inhibits its activity by preventing binding to the ribosome. Following hindlimb suspension, eEF2 phosphorylation increased immediately (day 1) and remained elevated relative to control for at least 7 days (Stevenson et al., 2003, unpublished data). Phosphorylation of eEF2 is catalyzed by eEF2 kinase, the activity of which is also inhibited by phosphorylation. Phosphorylation of eEF2 kinase is controlled by p70s6k and p90RSK1, suggesting control by not only mTORC1 pathways, but also MAP kinase pathways. Finally, protein translation is dependent on ribosomes and a decrease in the number of ribosome available for translation could affect the rate of protein synthesis and the maintenance of muscle mass under unloading conditions. It has been demonstrated that ribosome biogenesis is critical for adaptive hypertrophy of skeletal muscle (von Walden et al., 2012), but little is known of the effect of unloading or inactivity on ribosome biogenesis.
4. Mechanisms of muscle atrophy following disuse: alterations in protein degradation
There is considerable debate regarding the role of protein degradation in disuse atrophy (immobilization and bed rest) in humans, while the data in rodent disuse models (immobilization and hindlimb unloading) suggest a significant increase in proteolysis. One major difference between the human and rodent disuse models is the rate of atrophy, with atrophy occurring at a much greater rate in rodents than in humans (Phillips et al., 2009; Rennie et al., 2010). Protein degradation may play a major role in disuse-induced atrophy (with no nerve damage), but it appears that it is increased for a relatively short period after the start of unloading, and may be greater under conditions with significant decreases in neural activity and other complications associated with fractures and illness such as elevated glucocorticoids or cytokines (Ochala et al., 2011). The evidence supporting a role for protein degradation during disuse atrophy is limited in humans, in part, because of the difficulty in measuring in vivo rates of protein degradation. Few studies have actually measured in vivo protein degradation during disuse, however, an increase in muscle protein degradation in the VL following 72 h of unilateral lower limb suspension was demonstrated through the measurement of intramuscular 3-methylhistidine levels by means of microdialysis (Tesch et al., 2008). The majority of studies have inferred an increase in protein degradation through the measurement of genes associated with specific protein degradation pathways.
The major proteolytic pathways in skeletal muscle include the lysosomal system (i.e., cathepsins), Ca2+-dependent proteases (i.e., calpains), caspases, and the ubiquitin proteasome pathway (UPP) (Glickman and Ciechanover, 2002; Ventadour and Attaix, 2006; Sandri, 2011). While increases in the expression and activities of the calpains (Andrianjafiniony et al., 2010; Salazar et al., 2010; Talbert et al., 2013) and cathepsins (Taillandier et al., 1996; Ferreira et al., 2009) have been noted with unloading, the primary degradation pathway is thought to be the ubiquitin proteasome pathway (Solomon and Goldberg, 1996; Taillandier et al., 1996). The ubiquitin proteasome pathway (UPP) is the major nonlysosomal pathway for intracellular protein degradation. Ubiquitination consists of a three-step sequential cascade that involves the E1-activating, E2-conjugating, and E3 ligating enzymes. The E1 enzyme activates ubiquitin via an ATP dependent thiol-ester linkage with the carboxy-terminal glycine. The activated ubiquitin is transferred to an E2 conjugating enzyme before covalent linkage of the amino-end of a lysine residue to a targeted protein substrate that is directed by an E3 ubiquitin protein ligase. There are two main families of E3 ubiquitin ligases: HECT-domain E3s and RING and RING-like E3s (Metzger et al., 2012). The RING/RING-like E3s function as scaffolds bringing the substrate and an ubiquitin charged E2 enzyme into close proximity enabling the ubiquitin transfer, whereas the HECT E3s are thought to function to directly catalyze the covalent attachment of the ubiquitin to the substrate. The specificity of the ubiquitylation process is determined by the E3 ligases which have substrate specificity, while the length of the ubiquitin chain and the type of ubiquitin linkage added to the substrate is determined by the specific E2–E3 pairing (Napolitano et al., 2011). It is estimated that the human genome contains ~600 E3 and 35–40 E2 proteins (Li et al., 2008). The precise pairing of specific E3s with E2s during inactivity and unloading conditions is presently unknown, and is an important topic for additional study.
The 26S proteasome is made up of two protein complexes: a core 20S proteasome complex that is either connected to one or flanked by two regulatory 19S complexes. The binding of the 19S complex to the 20S proteasome is considered to lead to the activation of the proteasome. The 20S proteasome is a cylindrical and barrel-shaped that contains four stacked rings, each containing seven subunits. The inner two rings are made up of the β-subunits, designated β1-β7, while the outer two rings consist of the α1-α7 subunits. While research on the roles of the various β-subunits continues, it is believed that the β1, β2, and β5 subunits are the most important in the proteasome, as they possess caspase-like, trypsin-like, and chymotrypsin-like catalytic activity, respectively (Orlowski and Wilk, 2000). A few studies have actually measured an increase in the 20S and/or 26S proteasome following unloading (Lang et al., 2012; Magne et al., 2012; Slimani et al., 2012). However, the most frequent observation is an increase in the amount of ubiquitinated protein conjugates and in the expression of selected genes associated with the UPP (Brocca et al., 2012; Slimani et al., 2012; Stevenson et al., 2003; Koncarevic et al., 2007; Ferreira et al., 2009).
5. The role of MuRF1 and MAFbx in disuse atrophy
In 2001, the mRNA expression of two E3 ubiquitin ligases was found to increase in response to atrophy associated with immobilization, hindlimb unloading and denervation (Bodine et al., 2001a). These two genes are known as MuRF1 (for Muscle Ring Finger 1) and MAFbx/atrogin-1 (for Muscle Atrophy F-box), which are selectively expressed in striated muscle at relatively low levels under resting conditions, but, become quickly induced under conditions of unloading and inactivity. The expression pattern of these genes under disuse conditions shows a rapid increase upon unloading with peak expression occurring around seven days followed by a decline to baseline levels at around fourteen days (Bodine et al., 2001a). The fact that the rapid rise in expression of these two E3 ubiquitin ligases coincides with the rapid loss of muscle mass in rodent models has lead to the hypothesis that MuRF1 and MAFbx expression regulates proteasome-mediated degradation of muscle proteins during atrophy. Both MuRF1 and MAFbx have ubiquitin ligase activity and putative substrates have been identified for each E3 ligase based on binding studies and in vitro ubiquitin ligase assays (Glass, 2010). It has been suggested that MAFbx controls protein synthesis through its regulation of the translation initiation factor, eIF3f (Lagirand-Cantaloube et al., 2008), however, muscles from mice with a null deletion of MAFbx are not larger than normal and expression of eIF3f is not increased in MAFbx null mice (unpublished observations). MuRF1 has been reported to selectively bind and facilitate the ubiquitinylation of thick filament proteins such as myosin binding C, myosin light chain, and myosin heavy chain during muscle atrophy, thereby increasing their degradation by the 26S proteasome (Clarke et al., 2007; Cohen et al., 2009). More recently, it has been reported that MuRF1 can bind actin, a thin filament protein, and target it for ubiquitinylation and degradation in vitro (Polge et al., 2011). Thus, the question still remains as to whether MuRF1 selectively targets thick filament proteins for degradation during muscle atrophy in vivo. In our recent studies, we have found no evidence of selective sparing of thick versus thin filament proteins in the MuRF1 null mice following denervation or dexamethasone induced atrophy (Baehr et al., 2011; Gomes et al., 2012). Thus further in vivo validation of the putative substrates of MuRF1 is a necessity, and the list of in vivo substrates for MuRF1 and MAFbx is likely incomplete and could differ under different atrophy conditions depending on which E2 ligases are expressed. Currently, we have no knowledge of which E2s are associated with MuRF1 or MAFbx in vivo during atrophy.
The role of MuRF1 and MAFbx in disuse-induced atrophy in humans continues to be debated. Following knee immobilization, MuRF1 and MAFbx expression in the VL was not elevated after 48 h (Urso et al., 2006), but was significantly elevated after 10 (de Boer et al., 2007) and 14 (Jones et al., 2004) days followed by a return to baseline after 21 days of unloading (de Boer et al., 2007). These data show that the expression of MuRF1 and MAFbx can be increased following unloading in humans, but the increase in transcription may be delayed relative to rodents, which may reflect the muscle under study and/or the degree of unloading and inactivity. For example, the expression of both MuRF1 and MAFbx are significantly increased in humans two days after spinal cord injury (Urso et al., 2007). Even though an increase in MuRF1 and MAFbx expression has been measured during disuse-induced atrophy, their relative importance has been questioned because of the lack of an associated increase in protein degradation. Atrophy-associated increases in MuRF1 and MAFbx expression without concomitant increases in proteasome activities have been reported in rodents following dexamethasone treatment and alcohol intoxication (Baehr et al., 2011; Vary et al., 2008). Thus, while MuRF1 and MAFbx appear to be excellent markers of muscle atrophy, their substrates and role in regulating protein degradation and turnover requires further study.
6. Attenuation of disuse-induced atrophy
The examination of muscle atrophy in mice with null deletions of MuRF1 (Trim63) or MAFbx (Fbxo32) has provided evidence that these E3 ligases play an important role in the regulation of muscle atrophy under disparate conditions; deletion of MuRF1 spares muscle mass following hindlimb unloading (Labeit et al., 2010) and denervation (Bodine et al., 2001a; Gomes et al., 2012), while deletion of MAFbx spares muscle mass following denervation (Bodine et al., 2001a) and immobilization (unpublished data). However, examination of muscle sparing following extended denervation (28 days) revealed that functional sparing occurred in the MuRF1 null mice, but not the MAFbx null mice (Gomes et al., 2012). Measurement of both 20S and 26S proteasome activities in WT and MuRF1 null mice following 3 and 14 days of denervation revealed that proteasome activity increases in both WT and MuRF1 null mice, and in fact the increase in 20S and 26S proteasome activity is significantly greater in the MuRF1 null mice than the WT mice (Gomes et al., 2012); an observation that is in direct contradiction to the prediction that deletion of MuRF1 would result in a decrease in proteasome-mediated degradation. It is not entirely surprising that overall proteasome activity is not suppressed in the MuRF1 null mice given that E3 ligases are selective to a small number of proteins. The fact that deletion of MuRF1 produces muscle sparing that appears to be functional under a variety of catabolic conditions is impressive and suggests that the targets of MuRF1 may be important regulators of key signaling pathways. One could argue that absence of MuRF1 could be partially compensated by the other family members, MuRF2 and MuRF3, but we have not seen increased expression of either MuRF2 or MuRF3 in the MuRF1 null mice following inactivity (unpublished observation).
While deletion of MuRF1 did not inhibit proteasome activity, it did suppress the expression of certain components of the UPP such as Nedd4, HSP19, USP14, UCHL1 and others following denervation, but did not suppress proteasome activity or other degradation pathways such as calcium-dependent calpain I and II proteases or the lysosomal enzyme, cathepsin L. Interestingly, muscle sparing in the MuRF1 null mice following denervation occurred even though MAFbx expression was maintained at elevated levels for 14 days, showing that inhibition of both ligases is not required to achieve muscle sparing. These data suggest that MuRF1, and not MAFbx, is the better drug target for inhibiting the loss of muscle mass in response to unloading and inactivity, and suggest that some level of MAFbx expression is required for proper turnover of modified proteins.
MuRF1 and MAFbx expression is regulated by a number of transcription factors including the glucocorticoid receptor (GR), the forkhead transcription factors (FOXO1, FOXO3a) and NFκB transcription factors (p50 and Bcl-3) (Sacheck et al., 2007; Sandri et al., 2004; Wu et al., 2011; Waddell et al., 2008). Glucocorticoids have been shown to increase the expression of MuRF1 and MAFbx, however, they are likely not responsible for the increase in MuRF1 and MAFbx expression during unloading and inactivity. We recently reported that mice with a muscle specific deletion of GR have reduced MuRF1 and MAFbx expression relative to wild-type mice and muscle sparing in response to treatment with a synthetic glucocorticoid, however, in response to denervation MuRF1 and MAFbx expression increases to levels comparable to wild-type mice and there is no muscle sparing (Watson et al., 2012). Thus, while MuRF1 and MAFbx expression can be regulated by glucocorticoids, activation of the GR is not required to increase the transcription of these genes under conditions of unloading and inactivity.
Increased expression of the class O type of forkhead transcription factors (FOX01 and FOXO3a) is observed under a variety of atrophy-inducing conditions, including disuse (Senf et al., 2008; Lecker et al., 2004; Sacheck et al., 2007); and has been shown to regulate the transcription of a number of key atrophy associated genes, including MuRF1 and MAFbx (Waddell et al., 2008; Sandri et al., 2004; Stitt et al., 2004). Increased activation and nuclear translocation of the FOXO transcription factors following unloading is thought to occur due to a decrease in Akt/PKB signaling (Sandri et al., 2004; Latres et al., 2005; Brunet et al., 1999) leading to a decrease in their phosphorylation level. However, the activity of the FOXO transcription factors can be regulated by other posttranslational modifications, such as acetylation, and recently it was demonstrated that increased expression of the p300 histone acetyltransferase could inhibit FOXO activity in response to limb immobilization in rats, and decrease the expression of FOXO-responsive genes, such as MuRF1 and MAFbx (Senf et al., 2011). Interestingly, elevated HAT activity suppressed transcriptional up-regulation of FOXO1, but not FOXO3a, in response to immobilization highlighting the fact that FOXO1 and FOXO3a should not be treated as the same molecule. It is clear that FOXO transcription factors (especially FOXO1 and FOXO3a) play an important role in the induction of muscle atrophy under disuse conditions, in part, through the increased expression of MuRF1 and MAFbx, but also through the activation of other gene pathways (Senf et al., 2010). Moreover, direct (Senf et al., 2010) and indirect (Senf et al., 2008) inhibition of FOXO transcription activity has been shown to attenuate disuse-induced muscle loss. The exact mechanism by which the FOXO transcription factors are activated is unclear since the FOXO transcription factors are not always activated by a decrease in Akt/PKB activity (Reynolds et al., 2012), and Akt/PKB activity is not always decreased in response to unloading (Marimuthu et al., 2011).
Additional transcription factors have been shown to be upregulated in response to unloading and include NFkB transcription factors (Bcl3 and p50 (Jackman et al., 2013)), activating transcription factor 4 (ATF4 (Ebert et al., 2012)) and growth arrest and DNA damage-inducible 45a (Gadd45a (Ebert et al., 2012)). Deletion of each of these transcription factors has been shown to result in some degree of muscle sparing following unloading. Mice lacking the Nfkb1 gene (that encodes the NFkB transcription factor, p50) or Bcl-3 have significant muscle sparing following hindlimb unloading, which could be related in part to the reduced up-regulation of both MuRF1 and MAFbx (Hunter and Kandarian, 2004). Recently, mice lacking the ATF4 gene were protected from immobilization-induced atrophy after 3 days, but not 7 days of immobilization (Ebert et al., 2012). In this same study, inhibition of Gadd45a was shown to protect type II, but not type I fibers, from immobilization and denervation-induced atrophy. Interestingly, neither ATF4 nor Gadd45a control the transcription of MuRF1 or MAFbx, thus the muscle sparing is through some other mechanism. In general, attenuation of muscle atrophy, or muscle sparing, accomplished as the result of inhibition of a single transcription factor is rarely complete and can be limited to particular phases of the atrophy process (e.g., ATF4 spares early muscle loss and MuRF1 spares late muscle loss). Near complete sparing of disuse-induced muscle atrophy has usually been observed when multiple transcription factors or pathways are affected. For example, over-expression of HSP70 was shown to spare muscle mass in response to seven days of immobilization (Senf et al., 2008), and did so through the suppression of MuRF1 and MAFbx expression in addition to suppression of the transcriptional activities of FOXO3a and NF-κB. Muscle sparing during unloading has also been achieved through the activation of mTORC1 by using the β2 adrenergic agonist, clenbuterol (Kline et al., 2007) or constitutively active Akt/PKB (Bodine et al., 2001b). Thus, while inhibition of the mTORC1 pathways may not be responsible for suppressing protein synthesis and inducing muscle atrophy following unloading, activation of mTORC1 and its downstream targets may be a good strategy to increase protein synthesis and attenuate disuse-induced atrophy. Interestingly, mTORC1 activation by nutrients appears to be inhibited in response to immobilization and bed rest (Kelleher et al., 2013; Glover et al., 2008).
7. Recovery of muscle mass following disuse atrophy
The loss of skeletal muscle due to unloading and inactivity can be reversed upon the return of normal weight bearing (or reloading) of the limbs (see Fig. 1). The recovery of muscle mass following disuse atrophy is usually complete in young healthy adults, but is delayed and often incomplete in older individuals (Magne et al., 2012; Suetta et al., 2009). In young rodents, recovery of muscle mass upon reloading is dependent on activation of protein synthesis pathways, and in particular activation of mTORC1 (Bodine et al., 2001b). The importance of mTORC1 for recovery was also shown by (Lang et al., 2012) who examined recovery of mass following limb immobilization in mice heterozygous (+/−) for mTOR, having a 50% reduction in total mTOR protein in skeletal muscle. In response to ankle joint immobilization in plantar-flexion, the medial gastrocnemius atrophied rapidly and to a similar amount (~15% loss of mass) in WT and mTOR+/− mice. Upon cast removal and reloading, MG mass returned to pre-casting levels by 10 days, however, the MG in mTOR−/+ mice showed only limited growth (<5%) within this time period. The failure to recover mass in the mTOR−/+ mice was associated with a lower rate of protein synthesis relative to WT mice. The mTOR−/+ mice ultimately showed a complete recovery after 20 days of reloading. Examination of the Akt/mTORC1 signaling pathways showed that the amount of eIF4E bound to eIF4G was reduced in the mTOR−/+ mice relative to WT after ten days of reloading. In addition, there was a decrease in the amount of raptor-4E•BP1 binding and an increase in the amount of raptor–deptor binding in the mTOR−/+ mice.
In both rodents (Hwee and Bodine, 2009; Magne et al., 2012) and humans (Suetta et al., 2009; Hvid et al., 2010), impaired recovery from disuse-induced atrophy has been observed as a consequence of aging. Impaired growth in response to increased loading has been associated with a decrease in activation of the Akt/mTORC1 pathway (Parkington et al., 2004; Thomson and Gordon, 2006). Interestingly, a decrease in eIF4E•eIF4G association with increased loading has been observed in aging muscle, similar to what was reported in the mTOR−/+ mice (Hwee and Bodine, 2009). The mechanisms responsible for the inhibition of mTORC1 signaling in aging muscle in response to loading are unknown. In addition to suppression of anabolic pathways in aging muscle, there may be activation of proteolysis in some muscles upon reloading. There is some evidence of an increase in UPP mediated proteolysis immediately upon reloading in the soleus and medial gastrocnemius, however, it appears to be suppressed within one to seven days of reloading (Lang et al., 2012; Taillandier et al., 2003). In addition, there is a transient upregulation of MuRF1 and MAFbx (Slimani et al., 2012). In some muscles, such as the tibialis anterior, there appears to be an injury response upon reloading with a loss of muscle mass for up to 7 days following reloading (Slimani et al., 2012) (Fig. 1B). This loss of muscle mass is correlated with a prolonged elevation in proteasome activity and apoptosome-associated caspase-9 activity. Interestingly, with aging there is incomplete recovery in the extensor muscles upon reloading from disuse atrophy, and what appears to be increased muscle injury and lack of recovery in the flexor muscles (TA and EDL (Hwee and Bodine, 2009)). Additional research is needed to understand the response of different muscle types to reloading following disuse atrophy, and the effect of aging on the recovery of muscle mass.
8. Future directions
Understanding of the cellular and molecular mechanisms regulating muscle loss in response to unloading and inactivity has increased substantially over the past decade; however, it remains incomplete. While there is general agreement in animals and humans that unloading leads to a decrease in protein synthesis, there is considerable debate as to whether increases in protein degradation contribute to the loss of muscle mass. Additional research is needed to determine the exact cell signaling pathways involved in the suppression of protein synthesis in response to unloading and inactivity in the young, mature, and aging individual. While suppression of mTORC1 signaling may contribute to atrophy in young, growing individuals it may not be the primary driving force in adult individuals, and thus other mechanisms involved in the control of protein translation, such as elongation and ribosome biogenesis, should be examined. Strategies for inhibiting the loss of muscle mass under conditions of inactivity and unloading should consider targeting both the inhibition of protein degradation and/or stimulation of protein synthesis. In response to unloading, the transcription of MuRF1 and MAFbx and a number of transcription factors increases. While it is accepted that MuRF1 and MAFbx have ubiquitin ligase activity, their in vivo substrates and the pathways affected by the ubiquitination of their substrates remain unclear. In the same way, much remains to be learned about the pathways affected by the activation of the FOXO and NFκB transcription factors following unloading and inactivity. Further studies are required to understand how the activation of MuRF1, MAFbx, FOXO1 and other genes affect both protein degradation and protein synthesis. In addition, other cellular pathways, such as the endoplasmic reticulum and oxidative stress, could affect protein turnover during both acute and chronic conditions of disuse and should be examined.
Acknowledgments
Grant support
This research was supported by grants from the Muscular Dystrophy Association and NIH/NIDDK (DK75801). Additional support for students and postdoctoral fellow came from HHMI basic science research training program and a Kirchstein National Research Service Award.
References
- Adams GR, Caiozzo VJ, Baldwin KM. Skeletal muscle unweighting: spaceflight and ground-based models. Journal of Applied Physiology. 2003;95:2185–2201. doi: 10.1152/japplphysiol.00346.2003. [DOI] [PubMed] [Google Scholar]
- Andrianjafiniony T, Dupre-Aucouturier S, Letexier D, Couchoux H, Desplanches D. Oxidative stress, apoptosis, and proteolysis in skeletal muscle repair after unloading. American Journal of Physiology – Cell Physiology. 2010;299:C307–C315. doi: 10.1152/ajpcell.00069.2010. [DOI] [PubMed] [Google Scholar]
- Baehr LM, Furlow JD, Bodine SC. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. Journal of Physiology. 2011;589:4759–4776. doi: 10.1113/jphysiol.2011.212845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamman MM, Clarke MS, Feeback DL, Talmadge RJ, Stevens BR, Lieberman SA, et al. Impact of resistance exercise during bed rest on skeletal muscle sarcopenia and myosin isoform distribution. Journal of Applied Physiology. 1998;84:157–163. doi: 10.1152/jappl.1998.84.1.157. [DOI] [PubMed] [Google Scholar]
- Biolo G, Ciocchi B, Lebenstedt M, Barazzoni R, Zanetti M, Platen P, et al. Short-term bed rest impairs amino acid-induced protein anabolism in humans. Journal of Physiology. 2004;558:381–388. doi: 10.1113/jphysiol.2004.066365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC. mTOR signaling and the molecular adaptation to resistance exercise. Medicine and Science in Sports and Exercise. 2006;38:1950–1957. doi: 10.1249/01.mss.0000233797.24035.35. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001a;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature Cell Biology. 2001b;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Booth FW, Gollnick PD. Effects of disuse on the structure and function of skeletal muscle. Medicine and Science in Sports and Exercise. 1983;15:415–420. [PubMed] [Google Scholar]
- Booth FW, Seider MJ. Early change in skeletal muscle protein synthesis after limb immobilization of rats. Journal of Applied Physiology. 1979;47:974–977. doi: 10.1152/jappl.1979.47.5.974. [DOI] [PubMed] [Google Scholar]
- Brocca L, Cannavino J, Coletto L, Biolo G, Sandri M, Bottinelli R, et al. The time course of the adaptations of human muscle proteome to bed rest and the underlying mechanisms. Journal of Physiology. 2012;590:5211–5230. doi: 10.1113/jphysiol.2012.240267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. European Journal of Biochemistry. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Childs TE, Spangenburg EE, Vyas DR, Booth FW. Temporal alterations in protein signaling cascades during recovery from muscle atrophy. American Journal of Physiology – Cell Physiology. 2003;285:C391–C398. doi: 10.1152/ajpcell.00478.2002. [DOI] [PubMed] [Google Scholar]
- Clark BC. In vivo alterations in skeletal muscle form and function after disuse atrophy. Medicine and Science in Sports and Exercise. 2009;41:1869–1875. doi: 10.1249/MSS.0b013e3181a645a6. [DOI] [PubMed] [Google Scholar]
- Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metabolism. 2007;6:376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. Journal of Cell Biology. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer MD, Selby A, Atherton P, Smith K, Seynnes OR, Maganaris CN, et al. The temporal responses of protein synthesis, gene expression and cell signalling in human quadriceps muscle and patellar tendon to disuse. Journal of Physiology. 2007;585:241–251. doi: 10.1113/jphysiol.2007.142828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, et al. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. Journal of Biological Chemistry. 2012;287:27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Lane HW, Stuart CA, Davis-Street J, Wolfe RR. Prolonged bed rest decreases skeletal muscle and whole body protein synthesis. American Journal of Physiology. 1996;270:E627–E633. doi: 10.1152/ajpendo.1996.270.4.E627. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Vitorino R, Neuparth MJ, Appell HJ, Duarte JA, Amado F. Proteolysis activation and proteome alterations in murine skeletal muscle submitted to 1 week of hindlimb suspension. European Journal of Applied Physiology. 2009;107:553–563. doi: 10.1007/s00421-009-1151-1. [DOI] [PubMed] [Google Scholar]
- Fitts RH, Trappe SW, Costill DL, Gallagher PM, Creer AC, Colloton PA, et al. Prolonged space flight-induced alterations in the structure and function of human skeletal muscle fibres. Journal of Physiology. 2010;588:3567–3592. doi: 10.1113/jphysiol.2010.188508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ. Signaling pathways perturbing muscle mass. Current Opinion in Clinical Nutrition & Metabolic Care. 2010;13:225–229. doi: 10.1097/mco.0b013e32833862df. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiological Reviews. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glover EI, Phillips SM, Oates BR, Tang JE, Tarnopolsky MA, Selby A, et al. Immobilization induces anabolic resistance in human myofibrillar protein synthesis with low and high dose amino acid infusion. Journal of Physiology. 2008;586:6049–6061. doi: 10.1113/jphysiol.2008.160333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink DF. The influence of immobilization and stretch on protein turnover of rat skeletal muscle. Journal of Physiology. 1977;264:267–282. doi: 10.1113/jphysiol.1977.sp011667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AV, Waddell DS, Siu R, Stein M, Dewey S, Furlow JD, et al. Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2012;26:2986–2999. doi: 10.1096/fj.12-204495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhaff PL, Karagounis LG, Peirce N, Simpson EJ, Hazell M, Layfield R, et al. Disassociation between the effects of amino acids and insulin on signaling, ubiquitin ligases, and protein turnover in human muscle. American Journal of Physiology – Endocrinology and Metabolism. 2008;295:E595–E604. doi: 10.1152/ajpendo.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad F, Adams GR, Bodell PW, Baldwin KM. Isometric resistance exercise fails to counteract skeletal muscle atrophy processes during the initial stages of unloading. Journal of Applied Physiology. 2006;100:433–441. doi: 10.1152/japplphysiol.01203.2005. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Hunter RB, Kandarian SC, Esser KA. Regulation of translation factors during hindlimb unloading and denervation of skeletal muscle in rats. American Journal of Physiology – Cell Physiology. 2001;281:C179–C187. doi: 10.1152/ajpcell.2001.281.1.C179. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Stuppard R, Conley KE, Fedele MJ, Fiorotto ML, Chin ER, et al. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochemical Journal. 2004;380:795–804. doi: 10.1042/BJ20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RB, Kandarian SC. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. Journal of Clinical Investigation. 2004;114:1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvid L, Aagaard P, Justesen L, Bayer ML, Andersen JL, Ortenblad N, et al. Effects of aging on muscle mechanical function and muscle fiber morphology during short-term immobilization and subsequent retraining. Journal of Applied Physiology. 2010;109:1628–1634. doi: 10.1152/japplphysiol.00637.2010. [DOI] [PubMed] [Google Scholar]
- Hwee DT, Bodine SC. Age-related deficit in load-induced skeletal muscle growth. Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2009;64:618–628. doi: 10.1093/gerona/glp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman RW, Cornwell EW, Wu CL, Kandarian SC. Nuclear factor-kappaB signalling and transcriptional regulation in skeletal muscle atrophy. Experimental Physiology. 2013;98:19–24. doi: 10.1113/expphysiol.2011.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SW, Hill RJ, Krasney PA, O’Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- Kelleher AR, Kimball SR, Dennis MD, Schilder RJ, Jefferson LS. The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. American Journal of Physiology – Endocrinology and Metabolism. 2013;304:E229–E236. doi: 10.1152/ajpendo.00409.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Farrell PA, Jefferson LS. Invited review: role of insulin in translational control of protein synthesis in skeletal muscle by amino acids or exercise. Journal of Applied Physiology. 2002;93:1168–1180. doi: 10.1152/japplphysiol.00221.2002. [DOI] [PubMed] [Google Scholar]
- Kline WO, Panaro FJ, Yang H, Bodine SC. Rapamycin inhibits the growth and muscle-sparing effects of clenbuterol. Journal of Applied Physiology. 2007;102:740–747. doi: 10.1152/japplphysiol.00873.2006. [DOI] [PubMed] [Google Scholar]
- Koncarevic A, Jackman RW, Kandarian SC. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2007;21:427–437. doi: 10.1096/fj.06-6665com. [DOI] [PubMed] [Google Scholar]
- Labeit S, Kohl CH, Witt CC, Labeit D, Jung J, Granzier H. Modulation of muscle atrophy, fatigue and MLC phosphorylation by MuRF1 as indicated by hindlimb suspension studies on MuRF1-KO mice. Journal of Biomedicine and Biotechnology. 2010;2010:693741. doi: 10.1155/2010/693741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, et al. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO Journal. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang SM, Kazi AA, Hong-Brown L, Lang CH. Delayed recovery of skeletal muscle mass following hindlimb immobilization in mTOR heterozygous mice. PLoS ONE. 2012;7:e38910. doi: 10.1371/journal.pone.0038910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, et al. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. Journal of Biological Chemistry. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne H, Savary-Auzeloux I, Migne C, Peyron MA, Combaret L, Remond D, et al. Contrarily to whey and high protein diets, dietary free leucine supplementation cannot reverse the lack of recovery of muscle mass after prolonged immobilization during ageing. Journal of Physiology. 2012;590:2035–2049. doi: 10.1113/jphysiol.2011.226266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimuthu K, Murton AJ, Greenhaff PL. Mechanisms regulating muscle mass during disuse atrophy and rehabilitation in humans. Journal of Applied Physiology. 2011;110:555–560. doi: 10.1152/japplphysiol.00962.2010. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. Journal of Cell Science. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miokovic T, Armbrecht G, Felsenberg D, Belavy DL. Heterogeneous atrophy occurs within individual lower limb muscles during 60 days of bed rest. Journal of Applied Physiology. 2012;113:1545–1559. doi: 10.1152/japplphysiol.00611.2012. [DOI] [PubMed] [Google Scholar]
- Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochemical Journal. 2011;434:309–319. doi: 10.1042/BJ20101487. [DOI] [PubMed] [Google Scholar]
- Ochala J, Gustafson AM, Diez ML, Renaud G, Li M, Aare S, et al. Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. Journal of Physiology. 2011;589:2007–2026. doi: 10.1113/jphysiol.2010.202044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira Y, Yoshinaga T, Nomura T, Kawano F, Ishihara A, Nonaka I, et al. Gravitational unloading effects on muscle fiber size, phenotype and myonuclear number. Advances in Space Research. 2002;30:777–781. doi: 10.1016/s0273-1177(02)00395-2. [DOI] [PubMed] [Google Scholar]
- Ohira Y, Yoshinaga T, Ohara M, Kawano F, Wang XD, Higo Y, et al. The role of neural and mechanical influences in maintaining normal fast and slow muscle properties. Cells Tissues Organs. 2006;182:129–142. doi: 10.1159/000093963. [DOI] [PubMed] [Google Scholar]
- Orlowski M, Wilk S. Catalytic activities of the 20 S proteasome, a multicatalytic proteinase complex. Archives of Biochemistry and Biophysics. 2000;383:1–16. doi: 10.1006/abbi.2000.2036. [DOI] [PubMed] [Google Scholar]
- Paddon-Jones D, Sheffield-Moore M, Cree MG, Hewlings SJ, Aarsland A, Wolfe RR, et al. Atrophy and impaired muscle protein synthesis during prolonged inactivity and stress. Journal of Clinical Endocrinology & Metabolism. 2006;91:4836–4841. doi: 10.1210/jc.2006-0651. [DOI] [PubMed] [Google Scholar]
- Parkington JD, LeBrasseur NK, Siebert AP, Fielding RA. Contraction-mediated mTOR, p70S6k, and ERK1/2 phosphorylation in aged skeletal muscle. Journal of Applied Physiology. 2004;97:243–248. doi: 10.1152/japplphysiol.01383.2003. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Glover EI, Rennie MJ. Alterations of protein turnover underlying disuse atrophy in human skeletal muscle. Journal of Applied Physiology. 2009;107:645–654. doi: 10.1152/japplphysiol.00452.2009. [DOI] [PubMed] [Google Scholar]
- Polge C, Heng AE, Jarzaguet M, Ventadour S, Claustre A, Combaret L, et al. Muscle actin is polyubiquitinylated in vitro and in vivo and targeted for breakdown by the E3 ligase MuRF1. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2011;25:3790–3802. doi: 10.1096/fj.11-180968. [DOI] [PubMed] [Google Scholar]
- Psatha M, Wu Z, Gammie FM, Ratkevicius A, Wackerhage H, Lee JH, et al. A longitudinal MRI study of muscle atrophy during lower leg immobilization following ankle fracture. Journal of Magnetic Resonance Imaging. 2012;35:686–695. doi: 10.1002/jmri.22864. [DOI] [PubMed] [Google Scholar]
- Rennie MJ, Selby A, Atherton P, Smith K, Kumar V, Glover EL, et al. Facts, noise and wishful thinking: muscle protein turnover in aging and human disuse atrophy. Scandinavian Journal of Medicine & Science in Sports. 2010;20:5–9. doi: 10.1111/j.1600-0838.2009.00967.x. [DOI] [PubMed] [Google Scholar]
- Reynolds THt, Merrell E, Cinquino N, Gaugler M, Ng L. Disassociation of insulin action and Akt/FOXO signaling in skeletal muscle of older Akt-deficient mice. American Journal of Physiology – Regulatory, Integrative and Comparative Physiology. 2012;303:R1186–R1194. doi: 10.1152/ajpregu.00358.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy RR, Baldwin KM, Edgerton VR. The plasticity of skeletal muscle: effects of neuromuscular activity. Exercise and Sport Sciences Reviews. 1991;19:269–312. [PubMed] [Google Scholar]
- Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- Salazar JJ, Michele DE, Brooks SV. Inhibition of calpain prevents muscle weakness and disruption of sarcomere structure during hindlimb suspension. Journal of Applied Physiology. 2010;108:120–127. doi: 10.1152/japplphysiol.01080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M. New findings of lysosomal proteolysis in skeletal muscle. Current Opinion in Clinical Nutrition & Metabolic Care. 2011;14:223–229. doi: 10.1097/MCO.0b013e3283457a75. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senf SM, Dodd SL, Judge AR. FOXO signaling is required for disuse muscle atrophy and is directly regulated by Hsp70. American Journal of Physiology – Cell Physiology. 2010;298:C38–C45. doi: 10.1152/ajpcell.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senf SM, Dodd SL, McClung JM, Judge AR. Hsp70 overexpression inhibits NF-kappaB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2008;22:3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senf SM, Sandesara PB, Reed SA, Judge AR. p300 acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. American Journal of Physiology – Cell Physiology. 2011;300:C1490–C1501. doi: 10.1152/ajpcell.00255.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slimani L, Micol D, Amat J, Delcros G, Meunier B, Taillandier D, et al. The worsening of tibialis anterior muscle atrophy during recovery post-immobilization correlates with enhanced connective tissue area, proteolysis, and apoptosis. American Journal of Physiology – Endocrinology and Metabolism. 2012;303:E1335–E1347. doi: 10.1152/ajpendo.00379.2012. [DOI] [PubMed] [Google Scholar]
- Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. Journal of Biological Chemistry. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- Spector SA, Simard CP, Fournier M, Sternlicht E, Edgerton VR. Architectural alterations of rat hind-limb skeletal muscles immobilized at different lengths. Experimental Neurology. 1982;76:94–110. doi: 10.1016/0014-4886(82)90104-2. [DOI] [PubMed] [Google Scholar]
- Stevenson EJ, Giresi PG, Koncarevic A, Kandarian SC. Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. Journal of Physiology. 2003;551:33–48. doi: 10.1113/jphysiol.2003.044701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Molecular Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Suetta C, Hvid LG, Justesen L, Christensen U, Neergaard K, Simonsen L, et al. Effects of aging on human skeletal muscle after immobilization and retraining. Journal of Applied Physiology. 2009;107:1172–1180. doi: 10.1152/japplphysiol.00290.2009. [DOI] [PubMed] [Google Scholar]
- Taillandier D, Aurousseau E, Combaret L, Guezennec CY, Attaix D. Regulation of proteolysis during reloading of the unweighted soleus muscle. International Journal of Biochemistry & Cell Biology. 2003;35:665–675. doi: 10.1016/s1357-2725(03)00004-9. [DOI] [PubMed] [Google Scholar]
- Taillandier D, Aurousseau E, Meynial-Denis D, Bechet D, Ferrara M, Cottin P, et al. Coordinate activation of lysosomal, Ca2+-activated and ATP–ubiquitin-dependent proteinases in the unweighted rat soleus muscle. Biochemical Journal. 1996;316(Pt 1):65–72. doi: 10.1042/bj3160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert EE, Smuder AJ, Min K, Kwon OS, Powers SK. Calpain and capase-3 play required roles in immobilization-induced limb muscle atrophy. Journal of Applied Physiology. 2013;114:1482–1489. doi: 10.1152/japplphysiol.00925.2012. [DOI] [PubMed] [Google Scholar]
- Tesch PA, von Walden F, Gustafsson T, Linnehan RM, Trappe TA. Skeletal muscle proteolysis in response to short-term unloading in humans. Journal of Applied Physiology. 2008;105:902–906. doi: 10.1152/japplphysiol.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason DB, Booth FW. Atrophy of the soleus muscle by hindlimb unweighting. Journal of Applied Physiology. 1990;68:1–12. doi: 10.1152/jappl.1990.68.1.1. [DOI] [PubMed] [Google Scholar]
- Thomson DM, Gordon SE. Impaired overload-induced muscle growth is associated with diminished translational signalling in aged rat fast-twitch skeletal muscle. Journal of Physiology. 2006;574:291–305. doi: 10.1113/jphysiol.2006.107490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappe S, Creer A, Slivka D, Minchev K, Trappe T. Single muscle fiber function with concurrent exercise or nutrition countermeasures during 60 days of bed rest in women. Journal of Applied Physiology. 2007;103:1242–1250. doi: 10.1152/japplphysiol.00560.2007. [DOI] [PubMed] [Google Scholar]
- Urso ML, Chen YW, Scrimgeour AG, Lee PC, Lee KF, Clarkson PM. Alterations in mRNA expression and protein products following spinal cord injury in humans. Journal of Physiology. 2007;579:877–892. doi: 10.1113/jphysiol.2006.118042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urso ML, Scrimgeour AG, Chen YW, Thompson PD, Clarkson PM. Analysis of human skeletal muscle after 48 h immobilization reveals alterations in mRNA and protein for extracellular matrix components. Journal of Applied Physiology. 2006;101:1136–1148. doi: 10.1152/japplphysiol.00180.2006. [DOI] [PubMed] [Google Scholar]
- Vary TC, Frost RA, Lang CH. Acute alcohol intoxication increases atrogin-1 and MuRF1 mRNA without increasing proteolysis in skeletal muscle. American Journal of Physiology – Regulatory, Integrative and Comparative Physiology. 2008;294:R1777–R1789. doi: 10.1152/ajpregu.00056.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventadour S, Attaix D. Mechanisms of skeletal muscle atrophy. Current Opinion in Rheumatology. 2006;18:631–635. doi: 10.1097/01.bor.0000245731.25383.de. [DOI] [PubMed] [Google Scholar]
- von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. American Journal of Physiology – Cell Physiology. 2012;302:C1523–C1530. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, et al. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. American Journal of Physiology – Endocrinology and Metabolism. 2008;295:E785–E797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML, Baehr LM, Reichardt HM, Tuckermann JP, Bodine SC, Furlow JD. A cell-autonomous role for the glucocorticoid receptor in skeletal muscle atrophy induced by systemic glucocorticoid exposure. American Journal of Physiology-Endocrinology and Metabolism. 2012;302:E1210–E1220. doi: 10.1152/ajpendo.00512.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Kandarian SC, Jackman RW. Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS ONE. 2011;6:e16171. doi: 10.1371/journal.pone.0016171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Roy RR, Siengthai B, Edgerton VR. Effects of inactivity on fiber size and myonuclear number in rat soleus muscle. Journal of Applied Physiology. 2005;99:1494–1499. doi: 10.1152/japplphysiol.00394.2005. [DOI] [PubMed] [Google Scholar]