SUMMARY
Protein synthesis is critical for circadian clock function, but little is known on how translational regulation controls the master pacemaker in mammals, the suprachiasmatic nucleus (SCN). Here we demonstrate that the pivotal translational repressor, the eukaryotic translational initiation factor 4E binding protein 1(4E-BP1) is rhythmically regulated via the mechanistic target of rapamycin (mTOR) signaling in the SCN and preferentially represses vasoactive intestinal peptide (Vip) mRNA translation. Knockout (KO) of Eif4ebp1 (gene encoding 4E-BP1) leads to upregulation of VIP and higher amplitude of molecular rhythms in the SCN. Consequently, the 4E-BP1 null mice exhibit accelerated re-entrainment to a shifted light/dark cycle and are more resistant to the rhythm-disruptive effects of constant light. Conversely, in Mtor+/− mice VIP expression is decreased and susceptibility to the effects of constant light is increased. These results reveal a novel role for mTOR/4E-BP1-mediated translational control in regulating entrainment and synchrony of the master clock.
Keywords: circadian clock, SCN, entrainment, synchrony, mTOR, 4E-BP1, VIP, mRNA translation
INTRODUCTION
Circadian rhythmicity is a fundamental biological property that orchestrates various behavioral, physiological, and metabolic processes in a wide range of organisms (Rosbash, 2009). In mammals, the master circadian clock is located in the suprachiasmatic nucleus (SCN) of the hypothalamus (Reppert and Weaver, 2002). The cellular clockwork is driven by interconnected transcriptional and post-transcriptional feedback loops (Rosbash et al., 2007; Takahashi et al., 2008). In a major negative feedback loop, the transcription factors CLOCK and BMAL1 form heterodimers and activate transcription of Period (Per) and Cryptochrome (Cry) genes. In turn, PER and CRY proteins associate with CLOCK/BMAL1 heterodimers and repress their own gene transcription.
SCN neurons are heterogeneous in their oscillatory activities, neuropeptide expression and responses to light (Welsh et al., 1995; Herzog et al., 1998; Antle and Silver, 2005). Cellular oscillators in the SCN are coupled to form a coherent and stable oscillator network (Aton and Herzog, 2005; Welsh et al., 2010). Intercellular synchronization confers robustness and accuracy to SCN-generated rhythms and distinguishes SCN from peripheral oscillators, where coupling is weak (Yamazaki et al., 2000; Yamaguchi et al., 2003; Liu et al., 2007a). Although the mechanisms of such synchrony are not fully understood, recent evidence points to an essential role for vasoactive intestinal peptide (VIP) (Shen et al., 2000; Harmar et al., 2002; Colwell et al., 2003; Anton et al., 2005; Maywood et al., 2006). VIP is a 28-amino-acid neuropeptide, which is cleaved from the precursor protein prepro-VIP encoded by the Vip gene (Gozes and Brenneman, 1989). In the SCN, Vip is expressed by a subset of ventromedial SCN neurons (Abrahamson and Moore, 2001). However, the molecular mechanisms regulating prepro-VIP synthesis are not understood.
Protein synthesis is controlled primarily at the step of mRNA translation initiation (Sonenberg and Hinnebusch, 2009). A critical event in this process is the association of the eukaryotic translation initiation factor 4E (eIF4E) with the mRNA 5′ m7GpppN (where N is any nucleotide) cap structure. eIF4E binding to the cap structure is controlled by the eIF4E-binding proteins (4E-BPs). Binding of 4E-BPs to eIF4E causes inhibition of cap-dependent translation initiation and is relieved by 4E-BP phosphorylation through the mechanistic target of rapamycin (mTOR) signaling (Gingras et al., 1999). mTOR is an evolutionarily conserved Ser/Thr kinase that forms two different multiprotein complexes, mTOR complex 1 (TORC1) and mTORC2, which couple extracellular and intracellular signals (growth factors, energy status, nutrient availability, and stress) with cellular metabolic resources to balance anabolic and catabolic processes (Laplante and Sabatini, 2012). In the developing brain, mTOR controls neuronal survival and differentiation, neurite growth, and synaptogenesis (Cao et al., 2009). In the adult brain, mTOR mediates various forms of synaptic plasticity and plays an important role in learning and memory (Costa-Mattioli et al., 2009). In the hypothalamus, mTOR functions as a homeostatic sensor to control food intake and body weight (Cota et al., 2006).
Recent work has begun to reveal key roles for mTOR signaling in circadian clocks. Notably, mTOR activity exhibits robust circadian rhythms in the SCN (Cao et al., 2011), and light exposure activates mTOR signaling in a phase-dependent manner (Cao et al., 2008). Pharmacological inhibition of mTOR activation decreases light-induced PER protein expression and modulates behavioral phase shifts in animals (Cao et al., 2010a). In Drosophila, elevation of mTOR activity by genetic manipulation lengthens the circadian period (Zheng and Sehgal, 2010). One of the best-studied roles of mTORC1 is control of protein synthesis via phosphorylation of its major targets, 4E-BPs and S6 kinases (Topisirovic and Sonenberg, 2011). Interestingly, 4E-BPs are strongly phosphorylated in the SCN, in striking contrast with other brain regions (Cao et al., 2010b). Furthermore, 4E-BP phosphorylation in the SCN is stimulated by light in an mTOR-dependent manner, suggesting the involvement of 4E-BPs in the SCN clock physiology (Cao et al., 2008). Here, using a combination of behavioral, biochemical, and molecular approaches we investigated the functions of 4E-BP1 in the mammalian circadian clock. We show that 4E-BP1 regulates entrainment and synchrony of the SCN clock by repressing Vip mRNA translation, thus demonstrating a key role for mTOR/4E-BP1-mediated translational control in the master pacemaker.
RESULTS
Circadian Regulation of 4E-BP1 Phosphorylation in SCN
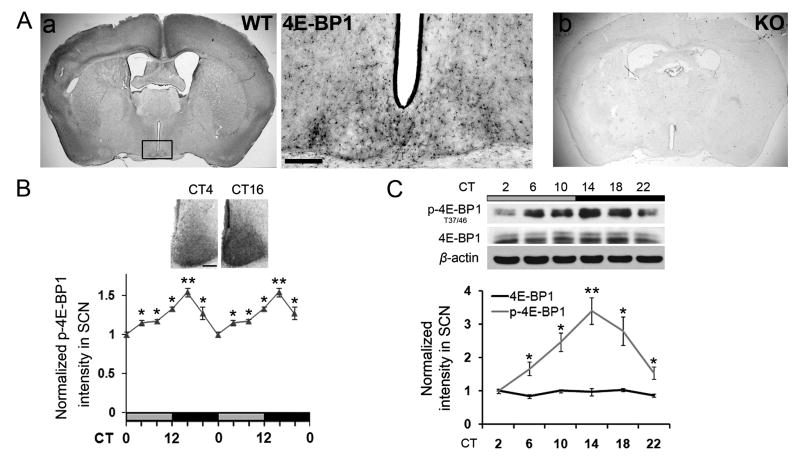
Abundant expression of 4E-BP1 was detected in the SCN, as compared to other hypothalamic brain regions (Figure 1A left). High magnification imaging revealed that 4E-BP1 was extensively expressed in the SCN (Figure 1A middle). To confirm the specificity of the 4E-BP1 immunostaining, we performed parallel staining on brain sections from Eif4ebp1 (gene encoding 4E-BP1) knockout (KO) mice (see below). No signal was detected in the KO brain (Figure 1A right).
Figure 1. Expression and Circadian Phosphorylation of 4E-BP1 in SCN (see also Figure S1).
A. Bright field microscopy images showing 4E-BP1 expression in the brain of WT (a) and Eif4ebp1 KO (b) mice. In sub-panel (a), the framed area is magnified to the right. Scale bar: 100 μm.
B. Circadian 4E-BP1 phosphorylation (at Thr37/46) in the SCN. Double-plotted line graph shows a curve of normalized p-4E-BP1 staining intensity in the SCN. The values are presented as the mean ± SEM. Representative microscopic images at CT4 and CT16 are shown above. *p<0.05 vs. CT0, **p<0.01 vs. CT0. Scale bar: 100 μm.
C. Circadian phosphorylation of 4E-BP1 (at Thr37/46) in the SCN. 8% polyacrylamide gel was used. Note that phosphorylation of 4E-BP1 exhibited different levels with circadian time, while 4E-BP1 level did not change. β-actin was used as a protein loading control. Quantitative data are from three biological replicates. The values are presented as the mean ± SEM. *p<0.05 vs. CT0, **p<0.01 vs. CT0. In each experiment, SCN tissue from five mice was pooled for each time point. Half of the lysate was used for detecting phosphorylation of 4E-BP1 and the other half was used for 4E-BP1 and β-actin blotting.
Next, we examined 4E-BP1 phosphorylation in the SCN over a 24 h period when mice were kept under constant dark (DD). mTOR-dependent phosphorylation of 4E-BP1 at Thr37 and Thr46 primes 4E-BP1 for subsequent phosphorylation at Ser65 and Thr70 and, therefore, is an indicator of 4E-BP1 activity (Gingras et al., 1999). Strong 4E-BP1 phosphorylation(at Thr37/46) was detected in the SCN by immunostaining, with highest level at circadian time (CT) 16 and lowest level at CT0 (CT4, 8, 12, and 20 vs. CT0, p<0.05; CT16 vs. CT0, p<0.01, analysis of variance (ANOVA), Figure 1B). Importantly, 4E-BP1 phosphorylation is mTOR-dependent, as rapamycin decreased the signal (Supplemental Figure 1A). In contrast to SCN, other brain regions exhibited weak 4E-BP1 phosphorylation (Supplemental Figure 1A), consistent with low 4E-BP1 expression in these regions. Consistent with the immunostaining results, Western blotting revealed that 4E-BP1 phosphorylation was highest at around CT14 and lowest at around CT2 (CT6, 10, 18, and 22 vs. CT2, p<0.05; CT14 vs. CT2, p<0.01, ANOVA, Figure 1C and Supplemental Figure 1B). Total 4E-BP1 and Eif4ebp1 mRNA level did not oscillate in the SCN (Supplemental Figure 1C). ERK/MAPK contributes to circadian mTOR activity in the SCN (Cao et al., 2011). As expected, MEK inhibitor U0126 decreased 4E-BP1 phosphorylation in the SCN (Supplemental Figure 1D). Together, these findings indicate that 4E-BP1 activity is controlled by the circadian clock via mTOR signaling in the SCN.
Accelerated Re-entrainment of Circadian Behavior in Eif4ebp1 KO Mice
To investigate the potential roles of 4E-BP1 in the circadian clock, we utilized an Eif4ebp1 KO mouse strain (Tsukiyama-Kohara et al., 2001). Confocal microscopic examination of DRAQ5 (a nuclear stain)-labeled sections revealed no difference in the histological features of SCN tissues between wild-type (WT) and KO mice (Supplemental Figure 2A). To study the effects of Eif4ebp1 gene deletion on circadian behavior, mice were kept in a 12 h/12 h light/dark (LD) cycle for 10 d and then released into DD for 9 d. The KO mice entrained normally to the LD cycle and displayed robust free-running rhythms of locomotor activities in DD (Supplemental Figure 2B). However, the circadian period (tau) of KO mice was slightly decreased compared to the WT mice (KO vs WT, 23.67 ± 0.06, n=12 vs.23.81 ± 0.03, n=12, p<0.05, Student’s t-test, Supplemental Figure 2C). Dark pulses applied during the light phase did not induce significant wheel-running activities in the mice and no difference was noted between WT and KO mice (Supplemental Figure 2D).
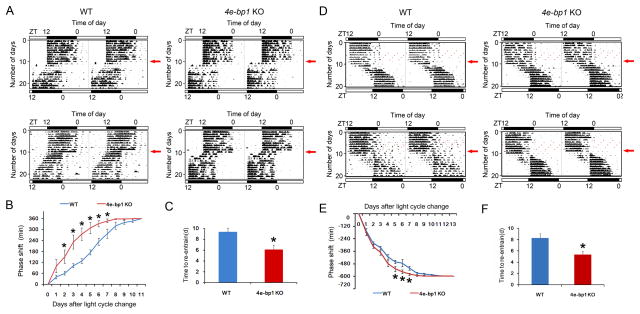
Next, to further characterize the circadian behavior of the Eif4ebp1 KO mouse, we used a “jet lag” model to study clock entrainment. For this purpose, mice were kept in a 12 h/12 h LD cycle for 10 d, followed by an abrupt 6-h phase advance of the LD cycle, with light on at zeitgeber time (ZT) 18. During re-entrainment, the mice displayed increased nighttime rest and daytime running and gradually re-entrained to the new LD cycle (Figure 2A). Notably, the KO mice re-entrained more quickly than the WT mice (Figure 2A). From day 2 to day 7 following the LD cycle shift, the KO mice exhibited a larger phase advance than the WT mice (KO vs. WT, p<0.05, ANOVA, Figure 2B), and the time to re-entrain was ~40% shorter for the KO mice (KO vs. WT, p<0.05, Student’s t-test, Figure 2C). In the second experiment, after the mice were entrained to a 12 h/12 h LD cycle for 10 d, the LD cycle was abruptly delayed by 10 h (light off at ZT22). Similar to the phase advancing experiment, during re-entrainment, both the WT and the KO mice displayed increased daytime running and nighttime rest (Figure 2D). However, the KO mice re-entrained more quickly to the delayed LD cycle (Figure 2D). From day 5 to day 7 following the shift, the KO mice exhibited a larger phase delay than the WT mice (KO vs. WT, p<0.05, ANOVA, Figure 2E), and the time to re-entrain was ~40% shorter for the KO mice (KO vs. WT, p<0.05, Student’s t-test, Figure 2F).
Figure 2. Accelerated Circadian Behavioral Re-entrainment in Eif4ebp1 KO Mice (see also Figure S2).
A and D. Representative double-plotted actograms of wheel-running activities from two WT (left) and two Eif4ebp1 KO mice (right). The x-axis (top) indicates the Zeitgeber time (ZT) of the day. The y-axis (left) indicates the number of days during the experiment. For these experiments, the animals were entrained to a 12 h/12 h light/dark (LD) cycle for 10 days, and on the 11th day, the LD cycle was either advanced for 6 h (light on at previous ZT18)(A) or delayed for 10 h (light off at previous ZT22) (D). Red arrows (right) indicate the day when the light cycle was shifted.
B and E. Line graphs showing the daily phase advance (B) or delay (E) of wheel-running activities following a 6 h advancing LD cycle shift (B) or a 10 h delaying LD cycle shift (E). Values are presented as the mean ± SEM. Each data point is averaged from nine mice. *p<0.05 KO vs. WT by ANOVA.
C and F. Histograms showing time (in days) needed for the mice to re-entrain to a 6 h LD cycle advance (C) or a 10 h LD cycle delay (F). Values are presented as the mean ± SEM. Each data point is averaged from nine mice. *p<0.05 KO vs. WT by Student’s t-test
Because dark pulses did not induce significant wheel-running activities in the KO mice (see Supplemental Figure 1D), the accelerated re-entrainment is unlikely due to enhanced negative masking. ERK phosphorylation and Jun expression are sensitive and reliable markers of photic stimulation of the SCN clock (Kornhauser et al., 1992; Obrietan et al., 1998). Light-induced ERK phosphorylation (at Thr202/Tyr204) and c-Jun expression in the SCN were not different in the KO mice (Supplemental Figure 3A and B; KO vs. WT, p>0.05, ANOVA). Further, as a core component of the clock feedback loop, light-pulse induced PER1 was not altered in the core region of SCN of the KO mice (Supplemental Figure 3C and D). Basal PER levels were not changed in the brain of the KO mice (Supplemental Figure 3E), indicating that 4E-BP1 does not regulate cellular PER expression. Taken together, these results demonstrate that although masking behavior and photic entrainment pathway are intact, re-entrainment of circadian behavior is accelerated in Eif4ebp1 KO mice.
Accelerated Resynchronization of PER Rhythms in Eif4ebp1KO Mice
When animals are stably entrained to the 12 h/12 h LD cycle, PER (including PER1 and PER2) rhythms in different regions of the SCN are synchronized and the overall PER levels in the SCN peak at around the light/dark transition (ZT12) (Hastings et al., 1999; Field et al., 2000). When the LD cycle is abruptly shifted, PER rhythms within different regions of the SCN become desynchronized due to their different re-entraining speeds (Reddy et al., 2002; Nagano et al., 2003; Albus et al., 2005; Nakamura et al., 2005; Davidson et al., 2009). Consequently, PER levels of the entire SCN are lower at ZT12 than when the clock is well entrained. As SCN cells become resynchronized, the PER levels at ZT12 recover to the original levels prior to the LD cycle shift (Amir et al., 2004). Thus, to investigate the molecular mechanisms responsible for the accelerated re-entrainment of circadian behavior in the Eif4ebp1 KO mice, we measured PER levels in the SCN at ZT12 before and after the LD cycle shift.
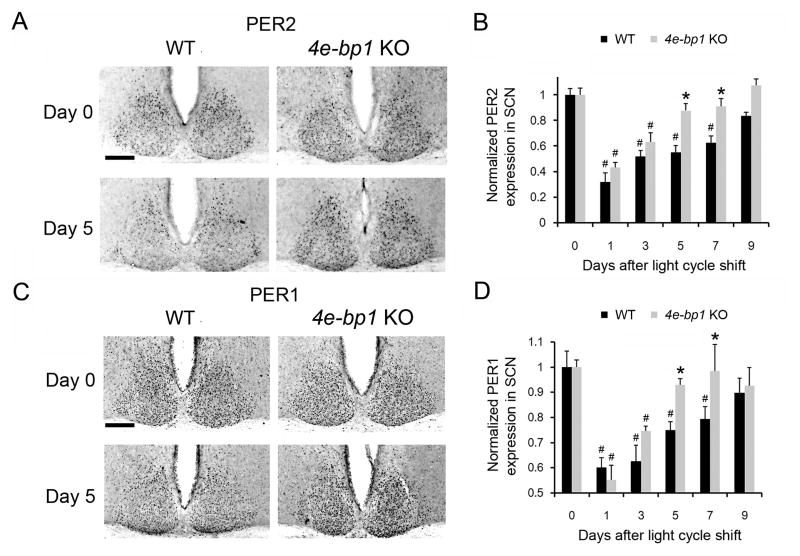
Before the LD cycle shift, PER1 and PER2 levels in the KO mice were not different from those in the WT animals (day 0, Figure 3A and C). In contrast, on day 5 after the 6h advancing LD cycle shift, PER1 and PER2 levels were higher in the SCN of the KO mice, suggesting better resynchronization of cellular clocks by day 5 (Figure 3A and C). Quantitation of PER levels before and after the light cycle shift is presented in Figure 3B and D. One day after the LD cycle shift, PER levels at ZT12 were dramatically decreased in the WT mice. They increased with time and reached pre-shifted control (day 0) levels 9 days after the light cycle shift (days 1, 3, 5, and 7 vs. day 0, p<0.05; day 9 vs. day 0, p>0.05, ANOVA, Figure 3B and D). In the KO mice, PER1/2 at ZT12 decreased to levels similar to those in WT mice following the light cycle shift, indicating a similar degree of desynchronization. Significantly, however, in the SCN of KO mice PER1/2 reached the pre-shifted levels 5 days after the light cycle shift, ~40% faster than in the WT mice (days 1 and 3 vs. day 0, p<0.05; days 5, 7, and 9 vs. day 0, p>0.05, ANOVA). Thus, on days 5 and 7 following the light cycle shift, PER levels in the SCN of the KO mice were significantly higher than in the WT mice (days 5 and 7, KO vs. WT, p<0.05, ANOVA, Figure 3B and D).
Figure 3. Accelerated PERIOD Rhythm Resynchronization in Eif4ebp1 KO Mice (see also Figure S3).
A and C. Representative microscopic images of PER2 (A) and PER1(C) immunostaining in the SCN. WT and Eif4ebp1 KO animals were entrained in a 12 h/12 h LD cycle for 10 days, and on the 11th day, the LD cycle was advanced for 6 h (light on at previous ZT18). Mice were sacrificed at ZT12 before and different days after the LD cycle shift. Representative images are shown before (Day 0) and 5 days after the LD cycle shift (Day 5). Scale bars: 100 μm. The quantitative data for all time points are shown in B (for PER2) and D (for PER1). PER levels at ZT12 on Day 0 are normalized to be “1” in WT and KO SCN, respectively. Values are presented as the mean ± SEM. Each data point is averaged from eight mice. #p<0.05 vs. Day 0; *p<0.05 vs. WT by ANOVA.
Notably, the PER staining data are remarkably consistent with the behavioral entrainment data (see Figure 2), showing that the WT mice re-entrained to a shifted light cycle in approximately 9 days, whereas the KO mice reach a new steady phase in approximately 5 days. Taken together, these results support the idea that KO mice re-entrain more quickly because cellular clocks in the SCN of these mice resynchronize faster to the shifted LD cycle.
Resistance to Constant Light (LL)-induced Clock Desynchrony in Eif4ebp1 KO Mice
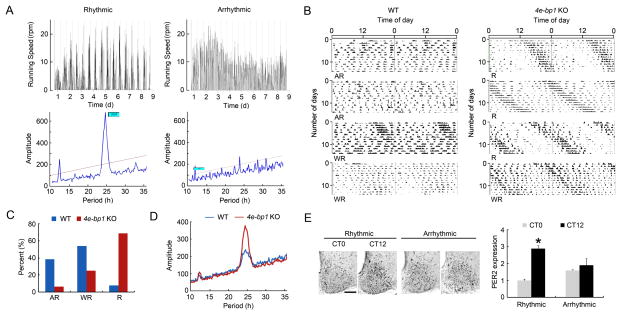
Prolonged exposure to LL extends endogenous circadian period and induces arrhythmic behavior in a sizable percentage of animals, depending on the light intensity and animal species (Daan and Pittendrigh, 1976). In the arrhythmic animals, LL disrupts the coupling among individual SCN neurons without affecting intracellular clock function (Ohta et al., 2005). To study the effect of LL on circadian behavior and PER2 expression in the SCN, Eif4ebp1 KO and WT mice were first housed in regular colony cages in LL (200 lx at cage level) for 14 d. Subsequently, the animals were transferred to individual cages equipped with running wheels in LL (55 lx at cage level) and their circadian behavior was recorded for 14d. The wheel-running behavior in LL was classified into three types based on the amplitude of the main peak of the periodograms: 1) rhythmic (R, amplitude >200, Figure 4A, left); 2) arrhythmic (AR, amplitude≤ 50 or no main peak, Figure 4A, right) and 3) weakly rhythmic (WR, 50<amplitude≤ 200).
Figure 4. Resistance to Constant Light-Induced Clock Desynchnony in Eif4ebp1 KO Mice A.
Constant light (LL)-induced arrhythmic wheel-running behavior in mice. Representative wheel-running speed line graphs (top) and periodograms (bottom) from a rhythmic (left) and an arrhythmic (right) mouse in LL. Note that in the periodogram of the rhythmic animal, a main peak is seen at around 25 h, indicating lengthened circadian period under LL. In the arrhythmic periodogram, there is no main peak.
B. Representative wheel-running actograms of mice in LL. Actograms from four WT (left) and four KO (right) mice are shown. R, rhythmic; AR, arrhythmic; WR, weakly rhythmic. The percentages of three types of behavior are shown in C.
D. Pooled periodograms from all mice used in the experiment. Note that the overall rhythmicity of KO mice was stronger compared to the WT mice, as indicated by a higher amplitude of the main peak of the periodogram. Thirteen WT and sixteen KO mice were used in the experiment.
E. PER2 expression in the SCN. Left: representative microscopic images of PER2 staining in the SCN. Right: Quantitation of PER2 levels in the SCN. The values are presented as the mean ± SEM. *p<0.05 vs. CT0. Note that in the arrhythmic mice, PER2 levels were not different at the two time points separated by 12 h. For this experiment, both WT and KO mice were used. PER2 levels were not different between WT and KO mice. Scale bar: 100 μm.
LL induced all three types of behavior in both WT and Eif4ebp1 KO animals. Most WT mice were either arrhythmic (AR) or weakly rhythmic (WR), while most KO mice were rhythmic (R) in LL (Figure 4B). Distribution of the three types of behavior (AR, WR and R) in both genotypes is quantified in Figure 4C. Strikingly, a smaller percentage of KO mice (6.3%, 1/16) exhibited arrhythmic behavior than did WT mice (38.5%, 5/13) (KO vs. WT, p<0.05, X2 test). The pooled periodograms from all the mice used in the experiment are shown in Figure 4D. The main peak of the periodogram is higher in the KO mice than in the WT mice, demonstrating stronger rhythmicity in the KO mice in LL. To verify that the rhythms of clock protein expression are disrupted in behaviorally arrhythmic mice, PER2 was immunostained in the SCN at CT0 and CT12 for the rhythmic mice and at two random time points 12 h apart for the arrhythmic mice. CT12 was defined as the onset time of the active phase, and CT0 was defined as the time point 12 h apart from CT12. As expected, PER2 was not rhythmic in the SCN of behaviorally arrhythmic mice (KO or WT), as compared to the rhythmic mice (Figure 4E). Thus, these data show that Eif4ebp1 KO mice are more resistant to LL-induced disruption of circadian behavioral and PER2 rhythms, consistent with enhanced synchrony in the SCN cells.
Regulation of Vip mRNA Translation by mTOR/4E-BP1 Signaling
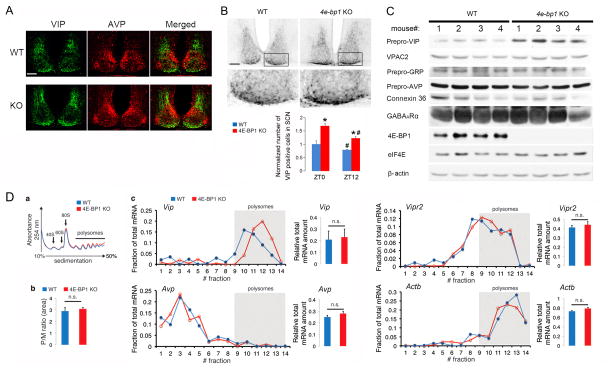
VIP plays a critical role in mediating synchrony in SCN cells. To investigate the mechanisms of enhanced re-entrainment and synchrony of the SCN clock in Eif4ebp1 KO mice, we first studied VIP expression in these animals. Using double immunofluorescent labeling, we first examined the expression pattern of VIP and arginine vasopressin (AVP) in the SCN. AVP is generally used as a neuropeptide marker for the dorsolateral SCN (Abrahamson and Moore, 2001). Confocal microscopic imaging revealed that VIP was expressed in a subset of ventromedial (core) SCN neurons, while AVP was expressed in some cells in the dorsal and lateral (shell) SCN (Figure 5A). The spatial distribution of VIP and AVP was similar in the SCN of KO and WT animals. Immunohistochemical staining also revealed robust VIP expression in the SCN (Figure 5B and Supplemental Figure 4A). In both the WT and the KO mice, expression of VIP at ZT12 was decreased compared to ZT0 (ZT12 vs. ZT0, p<0.05, ANOVA), which is consistent with a previous report (Takahashi et al., 1989). Interestingly, VIP level was increased by ~2 fold in the Eif4ebp1 KO mice at both ZT0 and ZT12 (KO vs. WT, p<0.05, ANOVA) (Figure 5B), suggesting constitutive repression of VIP expression by 4E-BP1.
Figure 5. Regulation of VIP expression by 4E-BP1 (see also Figure S4).
A. Confocal microscopic images showing expression of VIP (green) and AVP (red) in the SCN. For these experiments, the mice were entrained and sacrificed at ZT0. Note that VIP and AVP expression pattern in the KO SCN was similar to that in the WT SCN. Scale bar: 100 μm.
B. Top: Representative bright-field microscopy images showing VIP immunostaining in the SCN. For these experiments, mice were sacrificed at ZT0. Note that number of VIP positive cells was increased in the Eif4ebp1 KO SCN. Scale bars: 100 μm. Framed regions are magnified to below. Bottom: Quantitation of number of VIP-positive cells in the SCN. The values are presented as the mean ± SEM. *p<0.05 vs WT;# p<0.05 vs ZT0 by ANOVA, Four animals were used for each group.
C. Western blots of whole-forebrain lysates. For these experiments, animals were sacrificed at ZT0. The numbers 1–4 indicate four different animals for each group. Note that prepro-VIP, but not VPAC2, prepro-GRP, prepro-AVP, Connexin 42 or GABAARα was significantly increased in the brain of Eif4ebp1 KO mice. eIF4E and β-actin were used as loading controls. See Results section for quantitation of the blots.
D. Brain polysome profiling assay. (a) Polysome profiles from whole-brain lysates. The positions of the 40S, 60S, and 80S ribosome peaks and polysomes are indicated. (b) Polysome to monosome ratio. No difference was found between WT and Eif4ebp1 KO profiles (n=3, p>0.05, Student’s t-test). (c) Left figures show qRT-PCR results of Vip, Avp, Vipr2 and Actinb on RNA extracted from whole-brain polysome fractions. qRT-PCR results on total mRNA extracted from brain lysates are shown to the right. Note that for Vip mRNA distribution, a shift to the right (heavier gradient fractions) indicates increased translation initiation (n=3). No significant shifts were detected on distribution profiles of Avp, Vipr2 or Actinb mRNAs. No differences were found in Vip, Avp, Vipr2 and Actb transcription between WT and Eif4ebp1 KO (n=3, p>0.05 by Student’s t-test).
To investigate the mechanisms underlying VIP increase in Eif4ebp1 KO mice, we examined the expression of the VIP precursor protein, prepro-VIP, in the brain. We used whole brain lysate for Western blotting analysis, because the available antibody was not sensitive enough to detect prepro-VIP in the small amount of SCN lysate, and because VIP is extensively expressed in all brain regions (Gozes and Brenneman, 1989). Prepro-VIP level was increased significantly in the brain of Eif4ebp1 KO mice (normalized band intensities: KO vs. WT, 3.88±0.36 vs. 1±0.18, p<0.05, Student’s t-test) (Figure 5C). In contrast, expression of VPAC2 (the VIP receptor expressed in the SCN), and of the precursor proteins of other neuropeptides implicated in the SCN synchrony (Piggins et al., 1995; Maywood et al., 2011), including prepro-GRP and prepro-AVP, was not changed (Figure 5C). In addition to the neuropeptides, we examined other proteins involved in SCN synchrony, including GABAa receptor (Liu and Reppert, 2000; Colwell et al., 2003; Albus et al., 2005) and gap junction protein Connexin 36 (Long et al., 2005). The levels of Connexin 36 and the GABAa receptor α subunit were not altered in the Eif4ebp1 KO brain (Figure 5C). Furthermore, the expression of the 4E-BP1 binding partner, eIF4E, was not changed (Figure 5C). These results demonstrate specific regulation of prepro-VIP by 4E-BP1.
To complement the in vivo data, we studied prepro-VIP expression in mouse Neuro2A and human SHEP neuroblastoma cells (Waschek et al., 1988). Treatment of Neuro2A cells with the specific mTOR active-site inhibitor, PP242, resulted in reduced prepro-VIP levels and in dephosphorylation of 4E-BP1 after 3 h (Supplemental Figure 4B). To determine whether the effect of mTOR inhibition on prepro-VIP expression is dependent on 4E-BP1, we knocked down 4E-BP1 in SHEP cells using lentivirus (Supplemental Figure 4C). Prepro-VIP was increased by ~1 fold in 4E-BP1 knockdown cells. Rapamycin decreased 4E-BP1 phosphorylation and inhibited prepro-VIP expression in control cells (scrambled), but not in 4E-BP1 knockdown cells (sh4e-bp1) (Supplemental Figure 4C). Serum stimulation induced strong prepro-VIP expression in control cells, but to a lesser extent in 4E-BP1 knockdown cells (Supplemental Figure 4D), indicating that inducible prepro-VIP expression is at least partially dependent on 4E-BP1. Consistent with these data, overexpression of 4E-BP1 led to a reduction in prepro-VIP (Supplemental Figure 4E). Overexpression of WT eIF4E, but not the W56A mutant, which cannot bind to the mRNA cap (Gingras et al. 1999), increased prepro-VIP (Supplemental Figure 4F), demonstrating that prepro-VIP synthesis is dependent on eIF4E and cap-dependent translation in neuroblastoma cells. Expression of Vip 5′UTR-RLuc mRNA, but not RLuc or Grp 5′UTR-RLuc mRNA was enhanced in Eif4ebp1 KO (~ 2-fold) as compared to WT mouse embryonic fibroblasts (Supplemental Figure 4G; p<0.05, ANOVA). Grp mRNA 5′UTR has a similar length but lesser secondary structure than Vip mRNA 5′UTR. Thus, these results demonstrate that Vip mRNA translation is preferentially enhanced in 4E-BP1 KO cells.
Because 4E-BP1 inhibits translation initiation it was anticipated that prepro-VIP upregulation in the Eif4ebp1 KO brain is at the mRNA translation initiation step. To demonstrate this, we studied Vip mRNA translation by polysome profiling. Brain tissue lysate from ZT0 was fractionated by sucrose density gradient centrifugation. mRNAs for which the translation initiation rate is fast are associated with multiple ribosomes and sediment at the heavy-density fractions (Figure 5D-a). The polysome/monosome ratio was not changed in the KO brain (Figure 5D-b), indicating that the translation of most mRNAs is not changed. Next, the abundance of Vip, Avp, Vipr2 (the gene encoding VPAC2) and Actb mRNAs in each fraction was quantified by qRT-PCR, and the distribution of the mRNAs was compared between the WT and KO mice (Figure 5D-c). In the KO brain, Vip mRNA was shifted towards the heavy-density fractions, but total Vip mRNA level was not changed (Figure 5D-c), demonstrating enhanced Vip mRNA translation initiation. This effect on Vip mRNA translation was highly specific, as Avp, Vipr2 and Actb mRNA distribution was not changed (Figure 5D-c). Taken together, these data demonstrate that 4E-BP1 inhibits VIP expression by specifically repressing Vip mRNA translation initiation.
Reversal of Circadian Phenotypes in Eif4ebp1 KO Mice by Antagonizing VIP Signaling
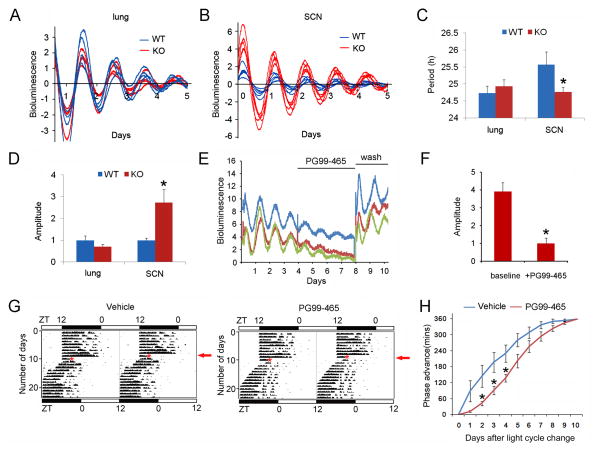
To study the dynamics of molecular rhythms in 4E-BP1 null mice, we made the Eif4ebp1−/−:mPER2:: LUC mice. We first examined the PER2::LUC bioluminescence expression patterns of tissue explants of the SCN, and as a representative of peripheral oscillators, the lung. No significant difference in period length and amplitude was observed between WT and KO lung explants (KO vs WT, p>0.05, Student’s t-test, Figure 6A, C and D), demonstrating that the circadian properties of peripheral oscillators are not changed in the KO mice. However, for the SCN rhythms, the KO explants displayed shorter period than the WT explants (WT, 25.57 h ± 0.39, n = 9; KO, 24.76 h ± 0.14, n = 8, KO vs. WT, p<0.05, Student’s t-test) (Figure 6B and C), consistent with animal behavioral data (see Supplemental Figure 2C). Strikingly, the amplitude of SCN rhythms was higher in the KO explants (WT, 1 ± 0.21, n = 9; KO, 2.74 ± 0.60, n = 8, KO vs. WT, p<0.05, Student’s t-test) (Figure 6B and D).
Figure 6. Reversal of Phenotypes in Eif4ebp1 KO Mice by Antagonizing VIP Signaling.
A and B. mPer2::Luc bioluminescence patterns of lung (A) and SCN (B) explants: representative plots of lung showing similar amplitudes and periods between WT and KO mice, but representative plots of SCN showing higher amplitudes and shorter periods in the KO animals. Each plot was from one SCN explant.
C and D. Histograms showing period (C) and amplitude (D) of mPER2::LUC bioluminescence rhythms from lung and SCN explants. The values are presented as the mean ± SEM. Eight to twelve animals were used for each group. *p<0.05 vs. WT
E. mPer2::Luc bioluminescence patterns of SCN explants from Eif4ebp1 KO Mice. Each plot was from one explant. Note that VPAC2 antagonist PG99-465 (50 nM) decreased amplitudes of mPer2::Luc rhythms in a reversible manner.
F. Histograms showing amplitudes of mPER2::LUC bioluminescence rhythms of SCN explants from Eif4ebp1 KO Mice before and after PG99-465 treatment. The values are presented as the mean ± SEM. Twelve SCN explants were used.
G. Rescue of circadian behavioral phenotypes in Eif4ebp1 KO Mice by PG99-465. Representative double-plotted actograms of wheel-running activities of a vehicle-infused (left) and a PG99-465-infused mouse (right). The Eif4ebp1 KO Mice animals were cannulated and entrained to a 12 h/12 h light/dark (LD) cycle for 10 days, and on the 11th day, mice were infused with 4 μL of saline or PG99-465 (100 μM) at ZT15 and the light cycle was advanced for 6 h (light on at ZT18). Red asterisks indicate the time of infusion and red arrows (right) indicate the day when the light cycle was shifted.
H. Line graphs showing the daily phase advance of wheel-running activities of the Eif4ebp1 KO Mice following a 6 h advancing light cycle shift. Values are presented as the mean ± SEM. Each data point is averaged from six mice. *p<0.05 PG99-465 vs. Vehicle by ANOVA.
The SCN pacemaker distinguishes from the peripheral oscillators in its neuronal network coupling capacity and the resulting system robustness (Liu et al., 2007a; Welsh et al., 2010). Experimental evidence and in silico modeling indicate that coupling strength (e.g. through VIP signaling) and phase relation between neurons can affect the amplitude of a multicellular oscillator (To et al., 2007; vander Leest et al., 2009; Abraham et al., 2010). Thus, our results are consistent with this notion and indicate that, whereas cellular oscillators that lack functional intercellular coupling (e.g., in the lung) function normally in the 4E-BP1 null mice, changes in intercellular coupling within the SCN network (e.g., elevated VIP signaling) can influence the properties of the SCN clock. To investigate whether VIP signaling is responsible for the increased amplitude in the KO mice, we applied VPAC2 antagonist PG99-465 (Cutler, et al., 2003) to the SCN explants from KO mice. Low concentration (50 nM) of PG99-465 reversibly decreased the amplitudes of KO explants to a level that is similar to WT explants (Figure 6E and F). These results indicate that the amplitude increase in KO explants is caused by VIP-dependent enhancement of coupling in SCN cells.
Next, to test whether the VPAC2 antagonist can reverse the faster entrainment behavioral phenotype in 4E-BP1 null mice, Eif4ebp1 KO mice were infused with PG99-465(100 μM, 4μL) or vehicle (physiological saline, 4μL) into the lateral ventricle at ZT15, before the light cycle was advanced for 6 h (light on at ZT18). Both groups of mice re-entrained to the new LD cycle. Notably, however, the mice infused with PG99-465 re-entrained more slowly than those infused with saline (Figure 6G). From day 2 to day 4 following the LD cycle shift, PG99-465-infused mice exhibited a smaller phase advance than control (PG99-465 vs. vehicle, p<0.05, ANOVA, Figure 6H). Together, these results demonstrate that VIP overexpression in the SCN underlies the phenotypes of Eif4ebp1 KO mice.
Decreased VIP and Increased Susceptibility to LL-induced Clock Desynchrony in Mtor+/− mice
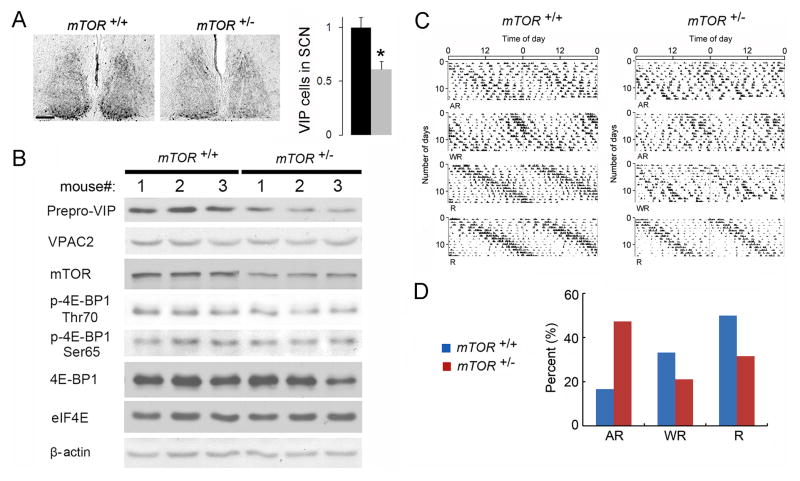
mTOR phosphorylates 4E-BP1 and decreases its translational inhibitory activity in the SCN (Cao et al., 2008). To corroborate the regulation of VIP and the clock function by 4E-BP1, we utilized an Mtor+/− mouse strain. In the Mtor+/− SCN, VIP was decreased by ~50% (Mtor+/− vs. Mtor+/+, p<0.05, Student’s t-test; Figure 7A). In the Mtor+/− brain, mTOR activity was decreased, as indicated by decreased phosphorylation of 4E-BP1 (normalized band intensities: Mtor+/− vs. Mtor+/+, at Thr70: 0.69±0.04 vs. 1±0.07; at Ser65: 0.67±0.07 vs. 1±0.19, p<0.05, Student’s t-test), and prepro-VIP was reduced (normalized band intensities: Mtor+/− vs. Mtor+/+, 0.43±0.08 vs. 1±0.08, p<0.05, Student’s t-test) (Figure 7B).
Figure 7. Decreased VIP Expression and Increased Susceptibility to Constant Light-Induced Clock Desynchnony in Mtor+/− mice.
A. Representative bright-field microscopy images showing VIP immunostaining in the SCN. Mice were entrained and sacrificed at ZT0. Quantitation of VIP expression in the SCN is shown to the right. The values are presented as the mean ± SEM. Five mice were used for each group. *p<0.05 by Student’s t-test. Note that number of VIP positive cells was decreased in the Mtor+/− SCN. Scale bars: 100 μm.
B. Western blots of whole-forebrain lysates. Animals were sacrificed at ZT0. The numbers 1–3 indicate three different animals for each group. Note that mTOR expression and 4E-BP1 phosphorylation was decreased in the brains of Mtor+/− animals. Accordingly, prepro-VIP, but not VPAC2, expression was decreased in these mice. eIF4E and β-actin were used as loading controls. See Results for quantitation of the blots.
C. Representative wheel-running actograms of mice in constant light. Actograms from four Mtor+/+ (left) and four Mtor+/− (right) mice are shown. R, rhythmic; AR, arrhythmic; WR, weakly rhythmic. Note that more Mtor+/− mice exhibited arrhythmic behavior compared to Mtor+/+ mice. The percentages of three types of behavior are shown in D.
To investigate the effects of lower VIP level on the circadian clock function, we monitored circadian behavior of the Mtor+/− mice in LL. Mice were housed in regular cages in LL (200 lx) for 14 d and then transferred to cages equipped with running wheels in LL (55 lx) to record their circadian behavior for 14 d. LL induced three types of behavior (R, AR and WR) in both Mtor+/− and Mtor+/+animals. A larger percentage of Mtor+/− mice (47.4%, 9/19) exhibited arrhythmic behavior than did Mtor+/+ mice (16.7%, 3/18) (p<0.05, X2 test, Figure 7C and D), indicating increased susceptibility to LL-induced clock desynchrony in Mtor+/− mice. Taken together, the results demonstrate that mTOR/4E-BP1 signaling bidirectionally regulates VIP level and susceptibility of the SCN clock to desynchronizing effect of LL.
DISCUSSION
In the present study, we found that mTOR signaling promotes Vip mRNA translation by repressing 4E-BP1. Consequently, in Eif4ebp1 KO mice, VIP is increased in the SCN, which is associated with larger amplitude of PER2 rhythms, accelerated circadian clock entrainment and enhanced synchrony. The phenotypes can be reversed by pharmacologically antagonizing VIP signaling and in Mtor+/− mice, in which VIP is decreased in the SCN. The findings demonstrate a key role for mTOR/4E-BP1-mediated translational control in the SCN circadian clock physiology.
Our findings indicate that entrainment and synchrony of the SCN clock are enhanced in Eif4ebp1 KO mice. This conclusion is based on three lines of evidence: First, Eif4ebp1 KO mice re-entrain faster to a shifted LD cycle than WT littermates. Photic entrainment of the SCN clock involves photic reception and resynchronization within the SCN cells. The photic input pathway appears to be normal in the KO mice. However, cellular PER rhythms resynchronize faster in their SCN. Importantly, the temporal profile of PER rhythm resynchronization is consistent with the progress of animal behavioral re-entrainment, suggesting that faster resynchronization of molecular rhythms in the SCN underlies accelerated behavioral re-entrainment. Second, Eif4ebp1KO mice are more resistant to forced clock desynchrony by constant light. Constant light disrupts intercellular synchrony, but does not affect individual cellular clocks in the SCN (Ohta et al., 2005). More resistance to constant light is consistent with enhanced synchrony among SCN cells in Eif4ebp1KO mice. Conversely, in Mtor+/− mice in which 4E-BP1 activity is enhanced, the SCN clock is more susceptible to the disruptive effects of constant light, consistent with compromised synchrony in the SCN of Mtor+/− mice. Third, SCN explants of Eif4ebp1KO mice display higher amplitudes of PER2::LUC rhythms. As there is no change in amplitude and period in peripheral oscillators, a plausible explanation is that coupling strength among SCN cells is increased in the Eif4ebp1KO mice and consequently the amplitude of circadian rhythms is increased at the tissue level.
Mounting evidence has established VIP as an essential mediator of SCN synchrony (Shen et al., 2000; Harmar et al., 2002; Colwell et al., 2003; Anton et al., 2005; Maywood et al., 2006). For example, microinjection of VIP induces phase shifts in the SCN circadian pacemaker, and VIP antagonists disrupt circadian function (Piggins et al., 1995; Gozes et al., 1995; Reed et al., 2001; Cutler, et al., 2003). VIP- (Colwell et al., 2003) and VPAC2-deficient mice (Harmar et al., 2002) show arrhythmic wheel-running behavior in constant darkness. Electrophysiological recordings show that SCN neurons in slices from Vip−/− and Vipr2−/− (encoding VPAC2) mice do not exhibit circadian rhythms of firing and lack inter-neuronal synchrony. Daily application of a VIP agonist to the Vip−/− SCN restores synchrony (Aton et al., 2005). Similarly, bioluminescence recordings from Vipr2−/− SCN slices also suggest that VIP signaling is necessary to synchronize individual SCN neurons as well as to maintain intracellular rhythms within these cells (Maywood et al., 2006, 2011).
4E-BP1 represses prepro-VIP synthesis by inhibiting Vip mRNA translation. In Eif4ebp1 KO mice, VIP expression is constitutively upregulated in the SCN due to overexpression of prepro-VIP. Compared with transcriptional control, translational regulation of Vip by 4E-BP1 does not significantly affect the phase of VIP daily rhythms (Takahashi et al., 1989); rather, it increases the abundance of VIP throughout the 24 h cycle. Of note, upregulating VIP signaling is sufficient to accelerate entrainment of the SCN circadian clock. For example, in Vipr2 transgenic mice, where VPAC2 is constitutively overexpressed, re-entrainment of circadian behavioral rhythms to a shifted LD cycle is accelerated and animals exhibit shorter circadian behavioral period (Shen et al., 2000). Moreover, pharmacological application of VIP facilitates behavioral re-entrainment to a shifted LD cycle and re-entrainment of PER rhythms in SCN slices to a changed temperature cycle (An, 2011a). To further link the phenotype of the Eif4ebp1 KO mice to VIP signaling, we applied the VPAC2 antagonist PG99-465 to the SCN and demonstrate that VIP antagonism can reverse the faster-entrainment phenotype of Eif4ebp1 KO mice and decrease the amplitude of PER2::LUC rhythms in the KO SCN explants.
In the SCN, VIP is rhythmically released by a subset of neurons in the core region that receives direct synaptic inputs from the retina (Welsh et al., 2010). Its receptor, VPAC2 is expressed in about 60% of SCN neurons, including half of the VIPergic neurons and almost all AVP-expressing neurons in the shell region (Abrahamson and Moore, 2001; An et al., 2012). VIP depolarizes SCN neurons by closing potassium channels and induces Per1 and Per2 expression via parallel changes in adenylate cyclase and phospholipase C activities (Nielsen et al. 2002; Meyer-Spasche and Piggins 2004; An et al., 2011b). Synaptic inputs from the core SCN synchronize neurons in the shell region, consistent with dense anatomical projections from the core to the shell but sparse reciprocal projections (Abrahamson and Moore, 2001). Resetting to a shifted LD cycle is initiated by phase shifts of a small group of core SCN neurons that are quickly synchronized (indicated by clock gene expression and firing rates) following the LD cycle shift (Nagano et al., 2003; Rohling et al., 2011). In turn, these neurons synchronize those in the shell region via GABAergic and neuropeptidergic synaptic transmission (Albus et al., 2005; Maywood et al., 2006, 2011).
Although VIPergic synaptic transmission is known to be essential for SCN synchrony in general, its role in core-shell synchronization during the SCN entrainment is not fully appreciated. The degree of core-shell synchronization contributes to the ability of the SCN pacemaker to reset (Rohling et al., 2011). Conceivably, increased VIP level would enhance the efficacy of synaptic transmission from the core to the shell region and thereby accelerate synchronization of the shell by the core. This model is supported by the close correlation between increased VIP expression, faster clock protein resynchronization and accelerated behavioral entrainment in the Eif4ebp1 KO mice. Under conditions of “forced clock desynchrony” such as a 22h LD cycle or constant light, the rhythms from the core and the shell can become out of phase and animals show split behavioral rhythms or become arrhythmic (de la Iglesia HO et al., 2004; Ohta et al., 2005). Similar to the role in entrainment, enhanced VIPergic synaptic transmission from the core to the shell would make the SCN ensemble more strongly coupled and thus more resistant to the desynchronizing effects of these conditions, whereas decreased VIP level would make the clock more susceptible to the clock-disruptive effects. This model is consistent with the opposite changes of susceptibility to constant light in Eif4ebp1 KO and Mtor+/−mice.
In addition to VIP, we examined other mediators of SCN synchrony such as GRP and GABA that may underlie the phenotypes of 4E-BP1 mutants. The expression of the relevant proteins is not altered in the 4E-BP1 KO mice, thus not supporting a role for these mediators in regulation of the clock by 4E-BP1. A previous study showed that mTOR signaling modulates photic entrainment of the SCN clock by facilitating PER1 and PER2 expression (Cao et al., 2010). Although 4E-BP1 is a downstream effector of mTOR, pharmacological disruption (i.e. using rapamycin) of mTOR signaling in vivo only transiently inhibits 4E-BP1 activity (up to a couple of hours, unpublished data), and thus cannot be used to study circadian functions of 4E-BP1. Here we show that 4E-BP1 does not regulate PER1 and PER2 expression. Thus, the effects of mTOR on PER expression are mediated through other mTOR downstream targets.
Besides its role in Vip regulation, 4E-BP1 may have other functions in circadian clocks. 4E-BP1 inhibits translational initiation by binding to eIF4E and impairing the formation of the translational preinitiation complex, which consists of eIF4E, eIF4A and eIF4G. Indeed, several studies have reported the roles of the eIF4E and its binding proteins in circadian clock physiology. For example, knockdown of the eIF4G homolog, NAT1, significantly reduces PER expression and lengthens the behavioral period in Drosophila (Bradley et al., 2012). Moreover, a recent study reported that the clock coordinates ribosomal biogenesis in the liver by rhythmic activation of the mTOR/4E-BP1/eIF4E pathway (Jouffe et al., 2013). Therefore, circadian rhythmicity of the mTOR/4E-BP1 signaling may be a general feature of circadian oscillators. Regulation of Vip mRNA translation is a SCN (or VIP-producing tissue)-specific function of the mTOR/4E-BP1 signaling. In the peripheral oscillators, where there is no obvious role for VIP or circadian coupling, the mTOR/4E-BP1 pathway may serve as an output signaling of the circadian clock to coordinate rhythmic mRNA translation.
EXPERIMENTAL PROCEDURES
Animals
Eif4ebp1 KO mice (Tsukiyama-Kohara et al., 2001) were backcrossed to WT C57BL/6 mice for over 10 generations to obtain a pure C57BL/6 background. mtor floxed mice on a C57BL/6 background (kindly provided by Dr. Sara C. Kozma, University of Cincinnati) were crossed to cytomegalovirus promoter (a ubiquitously expressed promoter)-Cre mice to generate mtor+/− mice (Sauer and Henderson, 1988). Eif4ebp1 KO mice were crossed with mPER2::LUC transgenic reporter mice (Yoo et al., 2004) to obtain Eif4ebp1−/−:mPER2::LUC mice. Animals were maintained in the animal facility at McGill University in accordance with institutional guidelines. All procedures were approved by the Institutional Animal Care and Use Committee.
Cannulation and Infusion
Mice were cannulated in the lateral ventricles using the techniques described by Cao et al. (2008). The coordinates (posterior, 0.34 mm from bregma; lateral, 0.90 mm from the midline; dorsoventral, −2.15 mm from bregma) were used to place the tip of a 24 gauge guide cannula into the lateral ventricle. To disrupt VIP signaling, PG99-465 (100 μM, 4 μl; Bachem, Switzerland) was infused through the cannula at ZT15. Control animals were infused with physiological saline (4μl).
Circadian Behavioral Assay
Eight- to 10-week-old male mice were individually housed in cages equipped with running wheels. Wheel rotation was recorded using the VitalView program (Mini Mitter, Bend, USA) (Hood et al., 2010). For the “jet lag” experiments, mice were entrained to a 12 h/12 h LD cycle (12 lx) for 10 d. On the 11th day the LD cycle was either advanced for 6 h or delayed for 10 h, and animal behavior was recorded for 10 d following the LD cycle shift. For the constant light (LL) experiments, mice were first kept in common cages under LL (200 lx) for 14 d and then transferred to running-wheel cages in LL (55 lx) to record their intrinsic rhythms for 14 d. The actograms of wheel-running activities were analyzed using the ActiView software (Mini Mitter, Bend, USA) and the ClockLab software (Actimetrics, Wilmette, USA).
Brain Tissue Processing, Immunostaining, and Microscopic Imaging Analysis
Under indicated conditions, mice were sacrificed and brain tissue was harvested. SCN sections were processed and immunostained for 4E-BP1, p-4E-BP1, PER1, PER2, AVP and VIP as previously reported (Cao et al., 2008, 2010, 2011). Bright-field microscopy images were captured using a digital camera mounted on an inverted Zeiss microscope (Oberkochen, Germany). Confocal microscopy images were captured using a Zeiss 510 Meta confocal microscope. See Supplemental Data for antibody information and image quantitation methods.
Protein Extraction and Western Blotting Analysis
The SCN tissue was excised using a 700 μm tissue punch and frozen on dry ice. SCN tissue was pooled from five animals per condition. Brain tissue was homogenized with a pestal grinder (Fisher Scientific Limited, Nepean, Canada) and lysed using a lysis buffer previously reported (Lee, 2007). Western blotting analysis was performed as described (Dowling et al., 2010).
Brain Polysome Profiling
Brain polysome profiling was performed as described (Gkogkas et. al. 2013). The polysome to monosome ratio was calculated as the area under the A254 absorbance curve, using the absorbance values processed with the definite integral command in MATLAB.
Real-Time Quantitative Reverse Transcription–PCR (qRT-PCR)
RNA extraction and qRT-PCR were performed as reported (Dowling et al., 2010). See Supplemental Data for primer information.
Explant Culture, Kinetic Bioluminescence Recording and Data Analysis
Explants of SCN and lung tissues from Eif4ebp1−/−:mPER2::LUC and mPER2::LUC mice were dissected and cultured as reported (Liu et al., 2007b). Real-time circadian reporter assays were performed using a Lumicycle luminometer (Actimetrics, Inc.) as previously described (Khan et al., 2012). Baseline-subtracted data (counts/sec) were plotted against time (days) in culture. For comparison, the first peak was aligned in the plotted data. The LumiCycle Analysis program (version2.31, Actimetrics, Inc.) was used to analyze rhythm parameters. For period length analysis, raw data were baseline fitted, and the baseline-subtracted data were fitted to a sine wave (damped), from which the period was determined. All samples showed persistent rhythms and goodness-of-fit of >90% was achieved. For amplitude analysis, baseline-subtracted data (polynomial order = 1; 3~6 days of recording data) were fitted to a sine wave, from which the amplitude was determined using Sin Fit.
Statistical Analysis
The values are presented as the mean ± standard error of the mean (SEM) or percentage (%). Statistical analysis was performed using SPSS software (SPSS Inc, Chicago, IL). Mean values from multiple groups were compared via one-way ANOVA, followed by the Student-Newman-Keuls test. Mean values from two groups were compared via Student’s t-test. Arrhythmia rates of WT and KO mice were compared via the X2 test. p < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
We thank Michael Rosbash, Isaac Edery, Erik Herzog and Jane Stewart for advice and critical reading of the manuscript and Maritza Jaramillo, Alex Gavrila, Annie Sylvestre and Isabelle Harvey for excellent technical assistance. We are indebted to Joseph Takahashi for his generous gift of the mPER2::LUC transgenic mice, Sara C. Kozma for the mtor floxed mice and Linda Penn and Manfred Schwab for the SHEP neuroblastoma cell line. This work was supported by Canadian Institute of Health Research (CIHR) Grants MOP 114994 to N.S. and MOP 13625 to S.A., National Science Foundation (NSF) Grant IOS-0920417 to A.C.L.. N.S. is a senior international research scholar of the Howard Hughes Medical Institute (HHMI). R.C. is a Fonds de recherche du Québec – Santé (FRQS) Postdoctoral Training Award recipient.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. Coupling governs entrainment range of circadian clocks. MolSystBiol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. CurrBiol. 2005;15:886–893. doi: 10.1016/j.cub.2005.03.051. [DOI] [PubMed] [Google Scholar]
- Amir S, Lamont EW, Robinson B, Stewart J. A circadian rhythm in the expression of PERIOD2 protein reveals a novel SCN-controlled oscillator in the oval nucleus of the bed nucleus of the stria terminalis. J Neurosci. 2004;24:781–790. doi: 10.1523/JNEUROSCI.4488-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S. Doctoral dissertation. Washington University; St. Louis: 2011a. The Roles of Vasoactive Intestinal Polypeptide in Circadian Entrainment of Suprachiasmatic Nucleus. ProQuest/UMI. (pqid:2342376571.) [Google Scholar]
- An S, Irwin RP, Allen CN, Tsai C, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011b;105:2289–2296. doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Tsai C, Ronecker J, Bayly A, Herzog ED. Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmaticnucleus. J Comp Neurol. 2012;520:2730–2741. doi: 10.1002/cne.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–151. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Herzog ED. Come together, right…now: synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley S, Narayanan S, Rosbash M. NAT1/DAP5/p97 and Atypical Translational Control in the Drosophila Circadian Oscillator. Genetics. 2012;192:943–957. doi: 10.1534/genetics.112.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79–88. doi: 10.1016/j.neuroscience.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol Cell Neurosci. 2008;38:312–324. doi: 10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Li A, Cho HY. mTOR signaling in epileptogenesis: too much of a good thing? J Neurosci. 2009;29:12372–12373. doi: 10.1523/JNEUROSCI.3486-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmaticcircadian clock. J Neurosci. 2010a;30:6302–6314. doi: 10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Obrietan K. mTOR Signaling and Entrainment of the Mammalian Circadian Clock. Mol Cell Pharmacol. 2010b;2:125–130. doi: 10.4255/mcpharmacol.10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler DJ, Haraura M, Reed HE, Shen S, Sheward WJ, Morrison CF, Marston HM, Harmar AJ, Piggins HD. The mouse VPAC2 receptor confers suprachiasmatic nuclei cellular rhythmicity and responsiveness to vasoactive intestinal polypeptide in vitro. Eur J Neurosci. 2003;17:197–204. doi: 10.1046/j.1460-9568.2003.02425.x. [DOI] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J PhysiolRegulIntegr Comp Physiol. 2003;285:R939–949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Daan S, Pittendrigh CS. A Functional Analysis of Circadian Pacemakers in Nocturnal Rodents III. Heavy Water and Constant Light: Homeostasis of Frequency? J Comp Physiol. 1976;106:253–266. [Google Scholar]
- Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur J Neurosci. 2009;29:171–180. doi: 10.1111/j.1460-9568.2008.06534.x. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O’Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozes I, Brenneman DE. VIP: molecular biology and neurobiological function. Mol Neurobiol. 1989;3:201–236. doi: 10.1007/BF02740606. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- Hood S, Cassidy P, Cossette MP, Weigl Y, Verwey M, Robinson B, Stewart J, Amir S. Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J Neurosci. 2010;30:14046–14058. doi: 10.1523/JNEUROSCI.2128-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoSBiol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SK, Xu H, Ukai-Tadenuma M, Burton B, Wang Y, Ueda HR, Liu AC. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem. 2012;287:25917–25926. doi: 10.1074/jbc.M112.368001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS. Regulation of jun-B messenger RNA and AP-1 activity by light and a circadian clock. Science. 1992;255:1581–1584. doi: 10.1126/science.1549784. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Protein extraction from mammalian tissues. Methods MolBiol. 2007;362:385–389. doi: 10.1007/978-1-59745-257-1_29. [DOI] [PubMed] [Google Scholar]
- Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007a;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007b;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–128. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Long MA, Jutras MJ, Connors BW, Burwell RD. Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat Neurosci. 2005;8:61–66. doi: 10.1038/nn1361. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A. 2011;108:14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Meyer-Spasche A, Piggins HD. Vasoactive intestinal polypeptide phase-advances the rat suprachiasmatic nuclei circadian pacemaker in vitro via protein kinase A and mitogen-activated protein kinase. Neurosci Lett. 2004;358:91–94. doi: 10.1016/j.neulet.2003.12.114. [DOI] [PubMed] [Google Scholar]
- Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, Sehgal A, Shigeyoshi Y. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci. 2003;23:6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25:5481–5487. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HS, Hannibal J, Fahrenkrug J. Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci. 2002;15:570–574. doi: 10.1046/j.0953-816x.2001.01882.x. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Field MD, Maywood ES, Hastings MH. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci. 2002;22:7326–7330. doi: 10.1523/JNEUROSCI.22-17-07326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed HE, Meyer-Spasche A, Cutler DJ, Coen CW, Piggins HD. Vasoactive intestinal polypeptide (VIP) phase-shifts the rat suprachiasmatic nucleus clock in vitro. Eur J Neurosci. 2001;13:839–843. doi: 10.1046/j.0953-816x.2000.01437.x. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Rohling JH, vanderLeest HT, Michel S, Vansteensel MJ, Meijer JH. Phase resetting of the mammalian circadian clock relies on a rapid shift of a small population of pacemaker neurons. PLoS One. 2011;6:e25437. doi: 10.1371/journal.pone.0025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M, Bradley S, Kadener S, Li Y, Luo W, Menet JS, Nagoshi E, Palm K, Schoer R, Shang Y, Tang CH. Transcriptional feedback and definition of the circadian pacemaker in Drosophila and animals. Cold Spring Harb Symp Quant Biol. 2007;72:75–83. doi: 10.1101/sqb.2007.72.062. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Spratt C, Sheward WJ, Kallo I, West K, Morrison CF, Coen CW, Marston HM, Harmar AJ. Overexpression of the human VPAC2 receptor in the suprachiasmatic nucleus alters the circadian phenotype of mice. Proc Natl Acad Sci U S A. 2000;97:11575–11580. doi: 10.1073/pnas.97.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Okamura H, Yanaihara N, Hamada S, Fujita S, Ibata Y. Vasoactive intestinal peptide immunoreactive neurons in the rat suprachiasmatic nucleus demonstrate diurnal variation. Brain Res. 1989;497:374–377. doi: 10.1016/0006-8993(89)90283-7. [DOI] [PubMed] [Google Scholar]
- To TL, Henson MA, Herzog ED, Doyle FJ., 3rd A molecular model for intercellular synchronization in the mammalian circadian clock. Biophys J. 2007;92:3792–3803. doi: 10.1529/biophysj.106.094086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb Symp Quant Biol. 2011;76:355–367. doi: 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- vanderLeest HT, Rohling JH, Michel S, Meijer JH. Phase shifting capacity of the circadian pacemaker determined by the SCN neuronal network organization. PLoS One. 2009;4:e4976. doi: 10.1371/journal.pone.0004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschek JA, Hsu CM, Eiden LE. Lineage-specific regulation of the vasoactive intestinal peptide gene in neuroblastoma cells is conferred by 5.2 kilobases of 5′-flanking sequence. ProcNatlAcadSci U S A. 1988;85:9547–9551. doi: 10.1073/pnas.85.24.9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.