Abstract
Cancer-associated fibroblasts (CAFs) have been reported to support tumor progression by a variety of mechanisms. However, their role in the progression of non-small cell lung cancer (NSCLC) remains poorly defined. In addition, the extent to which specific proteins secreted by CAFs contribute directly to tumor growth is unclear. To study the role of CAFs in NSCLC, a cross-species functional characterization of mouse and human lung CAFs was performed. CAFs supported the growth of lung cancer cells in vivo by secretion of soluble factors that directly stimulate the growth of tumor cells. Gene expression analysis comparing normal mouse lung fibroblasts (NFs) and mouse lung CAFs identified multiple genes that correlate with the CAF phenotype. A gene signature of secreted genes upregulated in CAFs was an independent marker of poor survival in NSCLC patients. This secreted gene signature was upregulated in NFs after long-term exposure to tumor cells, demonstrating that NFs are “educated” by tumor cells to acquire a CAF-like phenotype. Functional studies identified important roles for CLCF1-CNTFR and IL6-IL6R signaling, in promoting growth of NSCLC cells. This study identifies novel soluble factors contributing to the CAF protumorigenic phenotype in NSCLC and suggests new avenues for the development of therapeutic strategies.
Keywords: Carcinoma-associated fibroblasts, lung cancer, cytokines, il6, clcf1
Introduction
Cancer is initiated and progresses within a microenvironment that is itself altered as a consequence of the tumorigenic process. Stromal cells in contact with cancer cells secrete growth factors and cytokines that may act directly by signaling to tumor cells or indirectly by recruiting other stromal components to promote tumor progression (1-3). An important aspect of this process is the expansion of cancer-associated fibroblasts (CAFs) (4-6). CAFs are a diverse population of stromal cells with distinct characteristics in different tumors and tissues. To date, they have been best characterized in human breast and prostate cancer (7, 8) as well as in several mouse cancer models (9-11). However, the exact origins of these cells and how they acquire their tumor-promoting capacities remains poorly understood. Several studies have identified key CAF secreted molecules that play a role in tumor progression. For example, in human breast cancer, CAFs recruit endothelial cells and signal directly to tumor cells via the cytokine SDF-1 (12). In mouse dermal, cervical and islet cell cancers CAFs secrete a proinflammatory array of cytokines in response to NFκB activation (11). CAFs have also been reported to modulate the self-renewal and proliferation of cancer stem cells in vitro and in vivo (13, 14). These observations link CAFs to the regulation of multiple processes that control tumor development. Thus, CAF-tumor cell interactions play an important role in cancer progression and represent relatively unexplored target for cancer therapy. Consequently, understanding the precise molecular mechanisms that lead to tumor promotion by CAFs is an important goal in cancer biology.
Lung cancer is one of the leading causes of cancer death in the world. Non-small cell lung cancer (NSCLC) accounts for most lung cancer cases and the overall 5-year survival of patients with this disease remains approximately 15% (15). Most NSCLC tumors are characterized by a significant desmoplastic/fibrotic component at the time of diagnosis (16, 17). Several studies have suggested a role for lung fibroblasts in NSCLC using in vitro models (18-20). Recently, CAF-secreted Vegf has been suggested to promote tumor invasion (21). Whether other secreted proteins also play a role in lung CAF-induced tumor proliferation has not been reported. Here we report a systematic, cross-species study of the molecular phenotype of NFs and CAFs and identify novel mediators of tumor-stroma cross-talk in NSCLC.
Oncogenic Kras-driven mouse models of lung cancer recapitulate many features of human NSCLC (22), including oncogene-specific gene signatures (23). In this study, mouse NFs and CAFs isolated from a Kras-driven lung adenocarcinoma model were used to identify individual genes and gene signatures relevant to the pathogenesis of lung cancer. While both NFs and CAFs contribute to tumor progression, lung CAFs fostered tumor growth in vivo more efficiently than NFs. A gene signature specific to lung CAFs including multiple secreted proteins and genes related to inflammatory processes was identified and demonstrated to be a marker of poor survival in NSCLC patients. Mouse and human NFs upregulated genes in this gene signature after exposure to tumor cells, strongly suggesting that CAFs derive from normal resident lung fibroblasts. Finally, a critical and non-redundant functional role for one member of the CAF secreted gene signature, CLCF1, was confirmed using both knockdown and overexpression studies. While IL6 also contributes to tumor growth, signaling through the IL6 receptor is dispensable for tumor growth. In contrast, blockade of CLCF1-CNTFR paracrine signaling significantly decreases the growth of tumor cells in vivo. Thus these studies underscore a previously uncharacterized role for CLCF1 in paracrine signaling in NSCLC.
Material and methods
Reagents
ORFs for lacZ and clcf1 were obtained from Open Biosystems (ThermoFisher, USA). Il6 cDNA was synthesized from mouse CAF RNA with the DyNAmo cDNA synthesis kit (New England Biolabs). All ORFs were PCR-amplified using reverse primers that lacked the stop codon for each gene, cloned into an entry D-TOPO vector, and transferred to a pL6/V5-DEST lentiviral vector using Gateway® technology (Invitrogen, USA). Mouse-raised antibodies to α-smooth-muscle actin, vimentin, cytokeratin, β-actin (Sigma, St. Louis, MO, USA), and V5 (Invitrogen), and rabbit-raised antibodies to β-actin (LiCOR) were used.
Cell lines
LKR10 and LKR13 cells are mouse lung cancer cell lines derived by serial passage of minced lung adenocarcinoma tissues from two tumors isolated from separate lobes of the same KrasLA1 mouse (24). These cells were a gift of Dr. Julien Sage. A549 and H1299 cells were from ATCC (Manassas, VA, USA). LSZ2 cells were derived through xenograft passages from KrasLSLG12D mice and established as cell lines in vitro. All cell lines were grown in DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin. MLE12 cells were purchased from ATCC and grown according to vendor's specifications. These cells were originally isolated by Wikenheiser et al. (25).
Isolation of NFs and CAFs
Mouse fibroblasts were isolated as described (26). Cells could be grown in vitro for 4-6 passages before replicative senescence became apparent under these culture conditions. Human NFs and CAFs were isolated from normal lung or lung adenocarcinomas obtained with approval from the Stanford Institutional Review Board. Tissues were digested to obtain single cells and grown in 96-well culture plates before expansion for different analyses. For immortalization of NFs, Cells were transfected using a 1:10 ratio of pEmpty puro and pSV40Tag vectors using lipofectamine (Invitrogen) and following manufacturer's specifications. The vectors were a gift of Dr. Steve Artandi.
Retro and lentiviral infections
Virus was produced by transfection into 293FT cells of a retroviral vector expressing eGFP (27) using Fugene (Roche) according to manufacturer's recommendations. A pLKO.1 vector expressing mCherry was constructed by replacing the puromycin cassette with mCherry cDNA (Clontech, USA). pLKO and pL6 vectors were used to produce lentivirus by transfection into 293FT cells as previously described (28). Viruses were filtered and applied directly to cells for infection at a MOI <1.
Cell proliferation assay
Cells were plated into 96-well plates and treated with (3-[4,5-dimethlythiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT). Experiments were read on the indicated days according to manufacturer's instructions (Cell proliferation Kit I, Roche). Data were normalized to day 0 of experiment. 3T3 assays were as described (29) except that for NFs and CAFs 200,000 cells were plated and counted every 3 days.
Coinjection experiments
Xenograft experiments were done in accordance with Stanford Institutional Animal Care and Use Committee (IACUC) guidelines. For experiments with mouse lung cancer cells, 5×103 LKR10, LKR13 or LSZ2 were resuspended, alone or in combination with 5×104 NFs or CAFs (ratio 1:10), in 100 ml of serum free DMEM and injected subcutaneously into the two lower flanks of athymic Balb/cnu/nu mice (Charles River). Beginning one week after injection, tumor dimensions were measured every 3-4 days, and tumor volume was calculated using Volume= π/6 × (length) × (width)2. For human experiments, 5×105 A549 or H1299 cells were injected alone or in combination with 5×106 NFs or iCAFs.
Conditioned media and coculture (CM) experiments
CM from NFs/CAFs: 1×106 LFs and CAFs were plated into 10cm and media was changed on the following day. Fresh media (DMEM, 10% FBS and 1% Penicillin-Streptomycin) was added and cells were grown for another 2 days. Media was then removed and concentrated using Amicon Ultra columns (Milipore). Total protein was determined using the BCA kit (Pierce, USA) and indicated concentrations were used to treat LKR10 or LKR13 cells. CM from LKR10: 2×106 cells were plated and grown in fresh media for 5 days, and CM was harvested. 1:1 ratio of filtered CM plus fresh media was used to treat NFs for 36 hours.
For coculture, mCherry-NFs and LKR10 cells were mixed in a 1:2 ratio for 5 or 15 days. Then mCherry-positive cells were sorted out for gene expression analysis. NFs were cocultured with CAFs for 5 or 15 days, and mCherry-negative cells sorted for mRNA analysis. Human NFs were cocultured with GFP-expressing A549 cells at a 1:2 ratio for 12 and 24 days, followed by sorting of GFP-negative cells for gene expression analysis. G10 and G13 cells were cocultured with NFs overexpressing clcf1, il6 and control cells at a 1:5 ratio for 7, 14 and 21 days.
Western blotting
Cells were scraped, washed and lysed in NP40 buffer supplemented with protease inhibitor cocktail (Roche), 25mM sodium fluoride and 1mM sodium orthovanadate (Sigma). Protein samples were resolved by SDS PAGE and transferred to Immobilon-FL membranes (Millipore) and incubated in blocking buffer (Li-COR Biosciences) for 1 h prior to addition of primary antibodies. Antibody incubations and washing steps were done according to Li-COR protocols. Membranes were scanned using an Odyssey® Imager (Li-COR Biosciences).
Immunofluorescence and trichrome staining
Cells were trypsinized and grown on glass slides overnight. Cells were fixed using 4% paraformaldehyde, processed and stained with indicated primary antibodies. AlexaFluor secondary antibodies were from Invitrogen. Bioquant software was used to montage the entire xenograft sections and calculate the percentage of trichrome staining (Bioquant Inc, Nashville, TN). Images were taken using an Eclipse E800 microscope (Nikon).
Quantitative RT-PCR (qRT-PCR)
RNA was isolated using TRIzol reagent (Invitrogen) following manufacturer's specifications and cleaned-up using Qiagen miniRNA columns (Qiagen). cDNA was synthesized with a DyNAmo cDNA synthesis kit (New England Biolabs) and qRT-PCR was performed using SYBRGreen (Applied Biosystems) (see Supplementary information for primer sequences).
Gene Expression Analysis
The Robust Multichip Average (RMA) Express 1.0.4 program (30) was used for background adjustment and quantile RMA normalization of the 45,101 probe sets encoding mouse genome transcripts. Linear Models for Microarray data (LIMMA) (31) was used for comparison between the groups on RMA normalized signal intensities. After preprocessing steps a moderated Student t-test was used to select differentially expressed genes between groups (CAFs and NFs). P-values were adjusted for multiple testing using the Benjamini–Hochberg method. Fold change (Fc) was calculated by comparing gene expression levels between CAFs and NFs samples and expressed as the ratio between the averages of normalized intensities of the two groups. Only genes with fold change threshold greater than 2 and adjusted p-value <0.075 were considered as differentially expressed. The overexpressed genes identified from LIMMA were analyzed using Ingenuity Pathway Analysis (IPA) software (32). For network analysis, we chose to include only direct interactions in IPA. We used Onto-Express Ensemble (33, 34) for pathway analysis and Gene Ontology (GO) studies of the overexpressed genes from LIMMA. P-values were computed using the hypergeometric distribution and adjusted for multiple hypotheses using Benjamini-Hochberg correction.
Meta-Analysis
We perform meta-analysis of five human non-small cell lung cancer data sets obtained using DNA microarrays. Samples included tumor and stroma. Details of the meta-analysis procedure will be described elsewhere (Khatri et al., manuscript in preparation). Briefly, we estimated the effect size for each gene in each data set as Hedges' adjusted g (35). The study-specific effect sizes for each gene were then combined into a single meta effect-size using a linear combination of study-specific effect sizes, fi, where each study-specific effect size was weighted by inverse of the variance in the corresponding study (Eq. 1). After computing meta-effect size, significant genes were identified using Z-statistic, and p-values were corrected for multiple hypotheses testing using Benjamini-Hochberg false discovery rate (FDR) correction. Equation 1:
Survival analysis
Survival analysis was performed on both gene sets and individual genes. Four different datasets were used (36). Data was generated from RNA samples that included both tumor and stromal populations. Only samples at N0 and N1 stage and with time period less than 60 months for the study were analyzed. Each data set was normalized independently and standardization (mean centering) and scaling (dividing the centered value by the standard deviation) was performed prior to uni and multivariate analysis. High and low expressed groups were determined by taking the average of the standardized values across all samples for a specific gene. For gene sets a summation of all the genes for a particular sample was calculated and the average of the sum values was used to determine the high and low groups. Samples which have a value less than the calculated average were assigned as the low risk group and the rest to high risk group. Cox hazard ratios were calculated for both uni and multivariate analysis to estimate the relative risk along with confidence intervals and log rank tests.
Statistical analyses
A two-tailed t-test was used for comparisons of groups. Error bars correspond to either standard deviation (s.d) or standard error of the mean (s.e.m). Significant p values in the text correspond to <0.05 (*), <0.01 (**) or <0.001 (***).
Results
CAFs promote lung tumor development in vivo
To compare the effect of normal lung fibroblasts (NFs) and lung carcinoma-associated fibroblasts (CAFs) on lung cancer progression, we used cells derived from the KrasLA1 mouse model of lung adenocarcinoma (24). KrasLA1 lung tumors contained a significant fibrotic component, suggesting a role for CAFs in the progression of these tumors (Fig. 1A). CAFs isolated from tumors in this model and NFs from littermate wild-type mice were expanded in vitro for approximately 7-10 days before gene expression analysis or in vivo experiments (see methods). Expression of fibroblast specific protein-1 (fsp-1) (37) was detected in both NFs and CAFs whereas markers of other cells types were very low or undetectable (Supplementary Fig. 1A). Both NFs and CAFs showed expression of mesenchymal markers (α-smooth muscle actin –α-sma– and vimentin) by immunofluorescence (Fig. 1B) whereas they did not express cytokeratins (Supplementary Fig. S1B). As expected (12, 21), expression levels of α-sma were higher in CAFs than in NFs. It has been suggested that CAFs may be derived from epithelial cells that have undergone an epithelial-to-mesenchymal transition (38). However, KrasLA1 CAFs did not demonstrate recombination of the Kras locus, which occurs in epithelial tumors in this model (Supplementary Fig. S1C), demonstrating that they are not of tumor origin. Finally, neither NFs nor CAFs formed subcutaneous tumors when injected alone (data not shown).
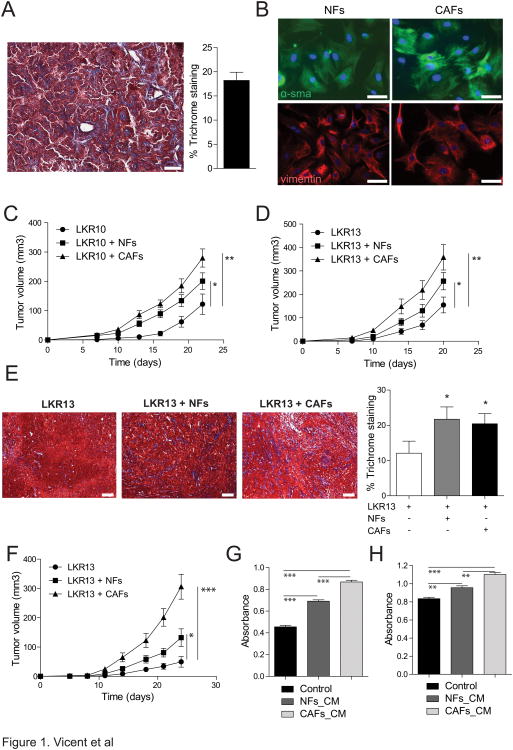
Figure 1.
Lung CAFs stimulate lung cancer growth in vivo. A. Representative image of a lung adenocarcinoma section from a KrasLA1 mouse stained with Masson's trichrome. Scale bars are 300μm. Bar graph represents the percentage of the stromal component in lung adenocarcinomas (n=4). B. Immunofluorescence on LFs and CAFs using antibodies against smooth-muscle actin and vimentin. Scale bars are 200μm. C and D. LKR10 and LKR13 xenografts injected alone or with either NFs or CAFs. Endpoint is 22 days (LKR10) and 20 days (LKR13). Graphs show average tumor volume at indicated time points (n=10 per group). Error bars indicate ± s.e.m. E. Representative images of Masson's trichrome staining of xenografts derived from tumor cells alone (LKR13), or tumor cells plus NFs or CAFs. Bar graph shows percentage of stromal component for each type of tumor (n=4 tumors per group). Scale bars are 300μm. F. Xenograft experiment of LKR13 injected alone or either with NFs or CAFs that have been passaged in vitro for >2 weeks. Endpoint is 24 days. Graphs show averaged tumor volume at indicated time points (n=10 injections per group). Error bars indicate ± s.e.m. G and H. Tumor cells (G-LKR10) and (H-LKR13) were treated with 2.5 μg/ml of conditioned media from NFs and CAFs and cell viability was analyzed by MTT 72 hours after treatment. Results are representative of 3 independent experiments using conditioned media from 3 different NF and CAF lines. All p values are for a two-tailed t-test.
Next, mouse lung cancer cell lines derived from the same KrasLA1 model (LKR10 and LKR13) were grafted alone, or coinjected with either NFs or CAFs (ratio 1:10) into immunodeficient mice. Tumors harboring CAFs were significantly larger than tumors harboring only tumor cells. Interestingly, tumors harboring NFs were also larger than tumors lacking fibroblasts, although significantly smaller than those harboring CAFs (Fig. 1C and D). NFs and CAFs had similar proliferative capacity in vitro (Supplementary Fig. S1D). In addition, the fibrotic component was similar in tumors that incorporated NFs or CAFs (Fig. 1E) and closely recapitulated that of the KrasLA1 tumors (Fig. 1A). Thus, the differential contribution to tumor growth of CAFs and NLFs is not likely to be due to a difference in their proliferative potential in vivo. The ability of NFs and CAFs to influence tumor growth in vivo was maintained after two weeks in culture (Fig. 1F), demonstrating that the tumor promoting capacity of NFs and CAFs is stable over time and independent of ongoing contact with tumor cells, at least after 2 weeks in culture. Therefore, resident lung fibroblasts contribute to tumorigenesis in vivo and CAFs provide a further advantage to tumor growth.
CAFs have been proposed to promote tumor growth through direct interaction with tumor cells or by recruitment of other cells to the tumor microenvironment (11, 12). To determine whether CAFs from KrasLA1 mice directly stimulate the growth of tumor cells through secreted factors, LKR10 and LKR13 cells were treated in vitro with conditioned media (CM) from NFs or CAFs. Proliferation of both lung cancer cell lines was significantly increased by conditioned media from CAFs when compared to that of NFs or control medium (Fig. 1G and H). This suggests that CAFs act at least in part by secreting soluble ligands that directly stimulate the growth of tumor cells.
CAFs are characterized by a secretory and inflammatory expression profile
Gene expression microarrays were used to identify changes between NFs and CAFs that may contribute to their different tumor-enhancing capacity. Unsupervised hierarchical clustering separated NF and CAF gene expression into two distinct clusters (Supplementary Fig. S2A). One hundred and sixty-four genes were upregulated in CAFs compared to NFs (CAF-HIGH), whereas only eleven genes were downregulated in CAFs (CAF-LOW) (fold change >2 or <0.5 and FDR <0.075) (Supplementary Fig. S2B and Supplementary Table S1). Interrogation of the molecular pathways represented in CAF-HIGH genes using Onto-Express (34) identified cytokine-cytokine receptor interaction (p-value=5.04×10−4), apoptosis (p-value=0.0086) and Jak-STAT signaling (p-value=0.0087) as pathways enriched in CAF-HIGH genes (Supplementary Table S2). In addition, genes coding for proteins secreted into the extracellular space were significantly enriched in the CAF-HIGH gene list (corrected p-values= 0.0026 and 0.0058 respectively) as were membrane proteins (corrected p-value= 5.8×10−4) (Supplementary Table S3). Thus, extracellular proteins secreted by CAFs may represent a key component of their phenotype. A heat map for these 15 secretome genes is shown in Fig. 2A. Differential expression of most of these secreted genes in CAFs was validated by qRT-PCR (Fig. 2B).
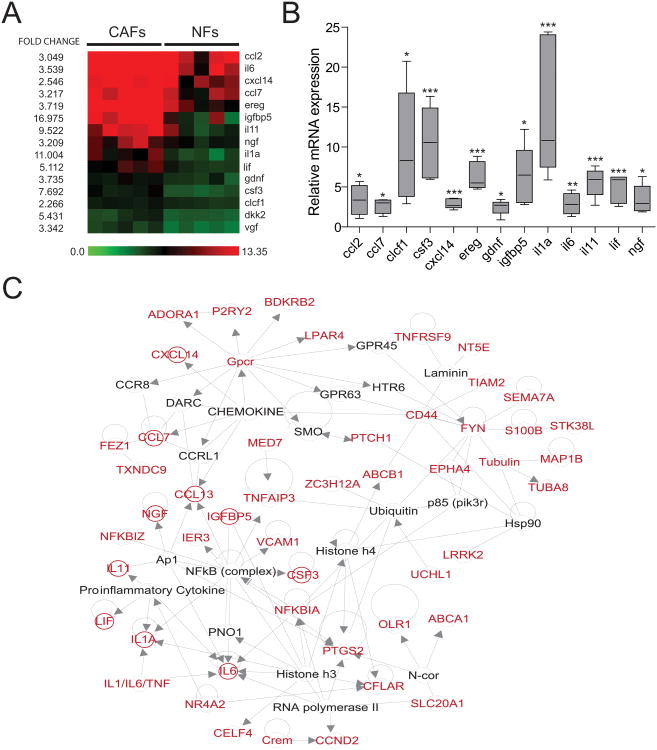
Figure 2.
A secretory and inflammatory gene signature specific to CAFs. A. Heat map of soluble ligands identified in the CAF gene signature. B. mRNA expression analysis of the 15 genes identified in A. Results are shown as CAF expression (n=5 samples) relative to average NF expression (n=5 samples) for each gene indicated in the X axis. C. 2D representation of network 1 obtained using Ingenuity Pathway Analysis (IPA) software. Genes in red correspond to those with a fold change >2 in CAFs. Secreted proteins are highlighted by red circles. Lines indicate interaction between two genes and arrows show direction of a gene interaction when information is publicly available.
To identify whether the gene signature of CAFs was centered around specific gene networks, we used a database of curated information regarding gene interactions (see methods). This identified four major gene networks based on the human orthologues of the CAF-HIGH genes (Supplementary Table S4). These networks were enriched for molecular pathways of inflammatory response and disease, cell death, cellular growth and development, and cell-to-cell signaling and interaction (p-value<0.0001) (Supplementary Table S5). Interestingly, the inflammatory response pathway was represented in 2 networks, highlighting the biological relevance of this cellular function in CAFs. The 15 secretome genes were particularly enriched in “network 1” which contained 10 of these secreted proteins (Fig. 2C). Thus, an inflammatory expression profile involving multiple secreted proteins is characteristic of lung CAFs. Collectively, these results identify secreted proteins that are part of an inflammatory phenotype in lung CAFs that might contribute to tumor development.
A CAF secreted-gene signature is a marker of poor survival in human NSCLC
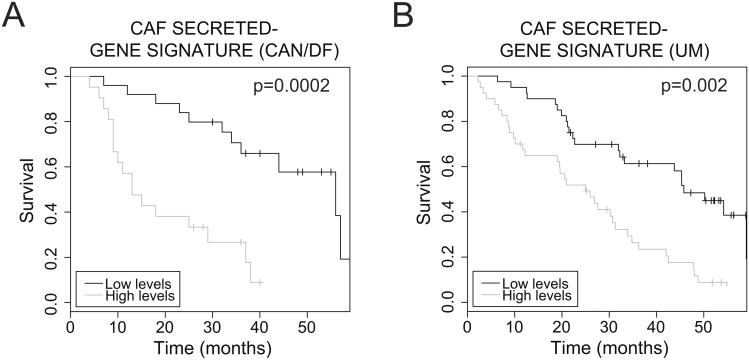
To probe the relevance of these findings to human NSCLC, we performed cross-species gene-expression analysis using publicly available human NSCLC gene expression data sets. As NSCLC samples often contain a significant percentage of tumor stroma, we reasoned that expression profile analyses from NSCLC biopsy samples could identify gene expression changes due to overexpression of stromal genes rather than the tumor cells themselves. First, gene expression of the human orthologues of the mouse-defined secreted-gene signature was assessed in five independent microarray data sets from normal human lung and NSCLC samples (36, 39-42). Several of these genes (CCL7, CCL13, CLCF1, EREG, IGFBP5, IL11, LIF and NGF) were upregulated in NSCLC compared to normal lung (Supplementary Fig. 3A and Supplementary Table S6). Next, the prognostic potential of the CAF gene signature was queried in a large expression profile data set of human NSCLC (36). In two independent data sets, univariate analysis revealed that high levels of the secreted CAF-gene signature was associated with poor survival in NSCLC patients (CAN/DF: logrank test p=0.0002, hazard ratio= 3.16±0.26; and UM: longrank test p= 0.002, hazard ratio= 2.93±0.14) (Fig. 3A and B). Survival prediction using this gene set was independent of stage and gender in a multivariate analysis (CAN/DF: logrank test p= 0.001, hazard ratio= 3.12±0.26; and UM: longrank test p= 0.007, hazard ratio= 2.91±0.14). These observations demonstrate that the secreted gene signature identified in mouse CAFs is highly relevant to human NSCLC and is a strong predictor of patient survival.
Figure 3.
Survival prognosis of a mouse CAF-secretory gene signature in human NSCLC. A and B. Kaplan Meier plots of NSCLC patients from two different data sets, Dana Farber Cancer Institute (CAN/DF) and University of Michigan (UM), using high and low levels of a secretory gene signature corresponding to 15 secretome genes identified in mouse CAFs.
Tumor-stroma cross-talk directly alters the phenotype of resident lung fibroblasts to become cancer-associated fibroblasts
The mechanism by which NFs are activated to become CAFs is poorly understood. For example, it is unclear whether tumor cells can directly activate NFs or whether other stromal components are first recruited into the tumor microenvironment leading to an indirect mechanism for NF activation. Furthermore, it is unclear whether NFs are altered to become CAFs, or whether CAFs perhaps arise from a rare subpopulation of resident either resident cells or cells mobilized from bone-marrow derived mesenchymal stem cells (37, 43-46). Therefore, we tested whether tumor cells could induce changes via secreted proteins that would directly lead to upregulation of CAF secreted genes in NFs. CM from mouse tumor cells was used to treat immortalized NFs (see methods and supplementary Fig. S4A), and the expression of the secretory gene signature was assessed by qRT-PCR 36 hours later. NFs subjected to tumor cell CM displayed increased expression (>1.5 fold) of mRNAs for 11 out of 15 CAF genes identified by microarray analysis (Fig. 4A). Similar findings were observed in an independent NF line (Supplementary Fig. S4B). The upregulation of the secretory gene signature in NFs after a short exposure to CAF CM suggests that CAFs may arise directly from reprogramming of resident NFs rather than requiring expansion of a rare subpopulation of cells within the stromal compartment. However, expression of these secreted proteins in NFs returned to basal levels after 72h of treatment (data not shown), suggesting that CM provides a transient effect that fails to permanently reprogram NFs towards a CAF phenotype.
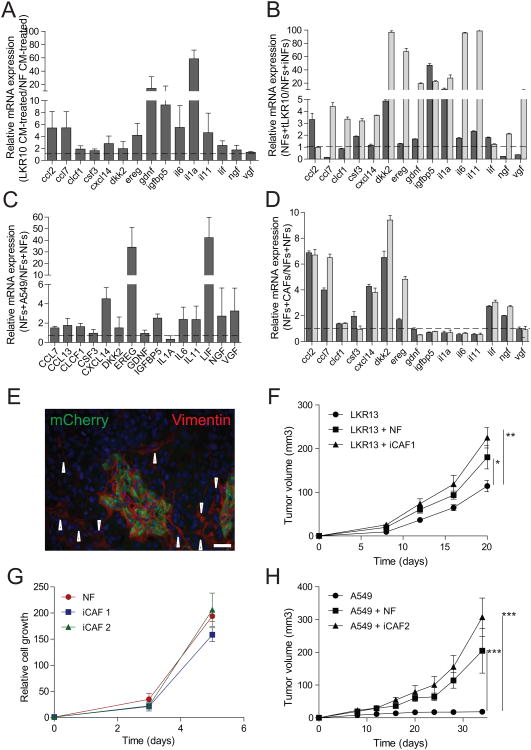
Figure 4.
Lung tumor cells and CAFs induce a of CAF-like phenotype in NFs. A. NFs were grown in the presence of conditioned media from NFs or LKR10 cells for 24 hours, and the expression levels of CAF soluble ligands was assessed by qRT-PCR. Conditioned media was harvested from cells after 5 days in culture. Results are relative to conditioned media from NFs and representative of 3 experiments. B. mCherry-NFs were cocultured in vitro with NFs or LKR10 cells for 5 (dark grey) or 15 (light grey) days, FACS sorted and studied by qRT-PCR for the expression levels of CAF secreted proteins. Experiments were done in triplicate and repeated at least twice. Results are relative to coculture with NFs. C. Human NFs (hNFs) were cocultured with A549 cells expressing GFP for 24 days. NFs were then sorted out for expression analysis of indicated genes. Results are relative to hNFs cocultured with GFP-positive hNFs and representative of two independendent experiments. D. NFs were cocultured in vitro with mCherry-NFs or mCherry-CAFs (ratio 1:2) for 5 (dark grey) and 15 (light grey) days and the 15 secretome genes were studied by qRT-PCR. E. Immunostaining of xenografts using anti-mCherry and anti-vimentin antibodies. Arrow heads point at endogenous, host fibroblasts. Scale bars are 200μm. F. Averaged tumors from LKR13 cells injected alone, with NFs, or NFs passaged in vivo with G13 cells (induced CAFs or iCAFs; n=8 mice per group). Endpoint is 20 days. G. Cell proliferation assay of NFs and iCAFs measured by MTT analysis. H. Same experiment as in B using human NSCLC cell lines (A549) and one iCAF line (iCAF2). Endpoint is 23 days 34 days (A549). Error bars indicate ± s.e.m. All p values are for a two-tailed t-test.
To determine whether direct cell-to-cell contact or continuous stimulation by tumor cells would provide stable upregulation of the CAF secretory gene signature, NFs and tumor cells were cocultured in vitro and the expression profile of the secreted gene signature was studied in these “educated” NFs. NFs were modified to express a fluorescent reporter (mCherry) to allow isolation by FACS after coculture (Supplementary Fig. S4C). An upregulation of 9 out of 15 secreted proteins, similar to what was observed after CM treatment, was observed after 5 days of coculture. A further increase in the expression level of the majority of secreted proteins (13/15 genes) was observed after 15 days (Fig. 4B). These results were confirmed using a coculture system incorporating human NFs (hNFs) and GFP-positive NSCLC cell lines (A549), indicating the relevance of tumor-NF interactions to human lung cancer. Exposure of hNFs to NSCLC cells for 12 days enhanced the expression of 9 genes of the secretory gene signature >1.5 fold (data not shown). Upregulation of 12 out of 15 members of the secretory gene signature was further increased after 24 days of coculture in vitro (Fig. 4C). These studies demonstrate the existence of functional cross-talk between tumor cells and NFs, and suggest that 1) the CAF phenotype is the consequence of direct and chronic stimulation of NFs by tumor cells, and 2) CAFs can be derived directly from resident lung fibroblasts and do not necessarily arise from a distinct population of mesenchymal cells recruited from a distant site.
While the above results suggest that tumor cells directly contribute to NF-to-CAF transition, CAFs may also establish homotypic interactions with resident NFs and contribute to further recruitment of fibroblasts towards a CAF phenotype. Treatment of NFs with CAF CM did not upregulate expression of secretory genes (data not shown) indicating that, unlike tumor cell CM, CAF CM is not sufficient to induce gene expression changes in NFs. However, coculture of NFs and CAFs led to upregulation of expression of 8 out of 15 CAF genes in NFs, (Fig. 4D and Supplementary Fig. S4D). Taken together, these results suggest that tumor cells initiate a transition of NFs to a proinflammatory CAF state that is further propagated to other NFs by already activated CAFs.
To analyze the relevance of these findings in vivo and to determine whether NFs could be stably reprogrammed to a CAF-like phenotype, fluorescently labeled NFs (mCherry) and tumor cells (GFP) were co-injected to form xenografts (Supplementary Fig. S4E). Tumors from these coinjections yielded similar volumes to those observed with primary NFs (Supplementary Fig. S4F) demonstrating that manipulation of NFs and tumor cells with reporter constructs did not result in significantly phenotypic changes. Double staining with anti-mCherry and anti-vimentin antibodies demonstrated the presence of exogenous, donor-derived NFs within expanding tumors (Fig. 4E). Thus, NFs expand within xenografts and are an active and sustained component of developing tumors in vivo. The in vitro data above suggested that tumor cells can progressively drive the NF-to-CAF transition and that this transition is further promoted by CAFs acting directly on NFs. To study whether NFs could acquire a stable CAF-like phenotype after in vivo passage with tumor cells, NFs were sorted from tumors and expanded in vitro for approximately 7-10 days. Next, tumor cells were injected alone, with parental NFs or with NFs isolated from tumor xenografts (here called induced CAFs or iCAFs). Secondary xenografts incorporating iCAFs were larger than those including “uneducated” primary NFs and the difference closely paralleled that between primary NFs and CAFs (Fig. 4F). These differences were not due to variations in the proliferation rate between iCAFs and NFs (Fig. 4G). iCAFs isolated from secondary xenografts (siCAFs) also recapitulated the tumor-promoting abilities shown by iCAFs from primary xenografts (Supplementary Fig. S4G). These results indicate that NFs can be “educated” to display characteristics of CAFs in vivo, and that the long-term interaction of NFs with tumor cells provides a point of no return from a CAF-like phenotype even after in vitro expansion for >1 week.
Next, we tested the relevance of the CAF phenotype established in mouse fibroblasts to human lung cancer oncogenesis. The NFs and iCAFs described above were mixed with the human NSCLC cell lines A549 or H1299. A549 and H1299 cells grew significantly better when mixed with iCAFs (Fig. 4H and Supplementary Fig. S4H). Thus, the acquisition of a CAF-like phenotype in resident lung fibroblasts leads to the promotion of oncogenesis in vivo via the direct stimulation of tumor cells proliferation by secreted proteins. These findings support the relevance of studies of mouse-derived lung CAFs to human lung cancer.
The cytokines clcf1 and il6 contribute to tumor progression in vivo
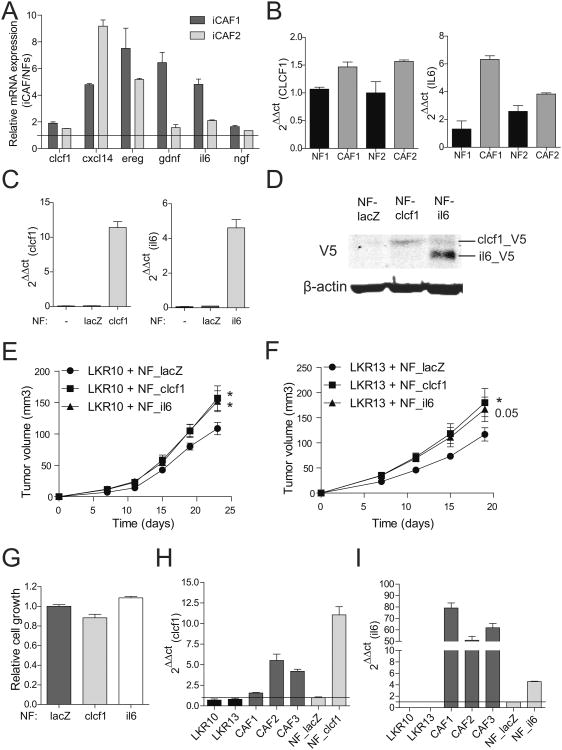
As NFs can transition to iCAFs that are phenotypically similar to CAFs isolated directly from tumors, the most functionally relevant secreted genes that drive the CAF phenotype should be common to both iCAFs and CAFs. To identify these genes, the expression profile of the CAF secretory gene signature was queried in NFs compared to iCAFs. Six genes (clcf1, cxcl14, ereg, gdnf, il6 and ngf) in the original CAF signature were consistently upregulated by at least 1.5 fold in iCAFs when compared to NFs (Fig. 5A and Supplementary Fig. S5A). Thus, this subset of the secreted gene signature may be the most critical for the CAF phenotype. As a further functional test, NFs and CAFs were isolated from primary human lung adenocarcinomas (Supplementary Fig. S5B). Two pairs of matched primary human NFs and CAFs were analyzed for the expression of these 5 genes. Notably, only CLCF1 and IL6 were upregulated in human CAFs compared to their matched NFs (Fig. 5B, and Supplementary Fig. S5C), suggesting that these two genes may play a central role in driving the protumorigenic phenotype of CAFs.
Figure 5.
Clcf1- and il6-expressing NFs enhance tumor growth in vivo. A. mRNA expression levels of clcf1, cxcl14, ereg, gdnf, il6 and ngf analyzed in two iCAF lines analyzed by qRT-PCR. Results are normalized against NF gene expression levels. B. Expression levels of CLCF1 and IL6 mRNA in human NFs and CAFs were assessed by qRT-PCR. C. mRNA expression levels analysis in NFs overexpressing clcf1 and il6. D. V5-tag protein detection by Western blot of cells in C. Cells were treated with 10μg/ml brefeldin A and 5μg/ml monensin for 5h to inhibit the secretory pathway and facilitate protein detection by Western blot. E and F. Xenograft experiment of LKR10 and LKR13 injected with control lacZ-, clcf1-, or il6-overexpressing NFs. Graphs show averaged tumor volume at indicated time points (n=10 injections per group). Endpoints are 23 days (LKR10) and 19 days (LKR13). Error bars indicate ± s.e.m. All p values are for a two-tailed t-test. G. Cell proliferation assay of NFs overexpressing lacZ, clcf1 and il6 at day 3 of experiment, as measured by MTT. H and I. mRNA expression levels of tumor cells (LKR10 and LKR13), primary CAFs, control NFs and NFs overexpressing either clcf1 or il6 assessed by qRT-PCR. Results are relative to NFs_lacZ (black line).
To test the functional role of clcf1 and il6 in lung cancer in vivo, NFs were transduced with lentiviral vectors containing cDNAs coding for either of these two mRNAs (Fig. 5C and D). NFs over-expressing clcf1, il6 or control (lacZ) were coinjected with tumor cells. Coinjection with NFs overexpressing clcf1 and il6 yielded significantly larger tumors (Fig. 5E and F). Similar results were obtained using an independent lung tumor cell line derived from the KrasLSLG12D mouse model (47)(Supplementary Fig. S5D). Importantly, overexpression of clcf1 or il6 did not alter the proliferation rate of the corresponding NFs (Fig. 5G). Thus, clcf1 and il6 secreted by CAFs play a direct role in stimulating tumor growth in vivo. This effect is likely due to direct stimulation of tumor cells by fibroblasts, as both clcf1- and il6-overexpressors increased the relative cell number of cancer cells when compared to control NFs when cocultured in vitro (Supplementary Fig. S5E and F). No expression of either clcf1 or il6 was detected in tumor cells (Fig. 5H and I), suggesting that the primary source of both cytokines is derived from CAFs which establish a paracrine signaling axis to promote tumor cell growth.
Suppression of clcf1-cntfr paracrine signaling axis decreases lung cancer in vivo
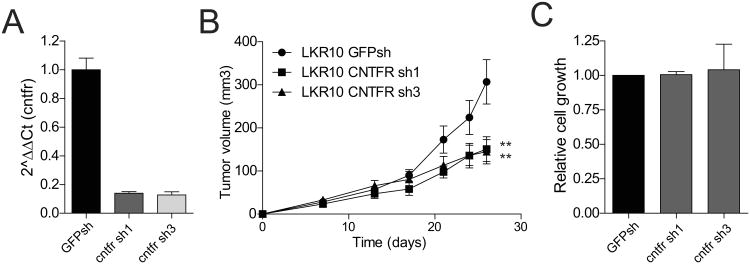
Clcf1 and il6 act through a receptor complex containing specific α-subunit receptors, cntfr and il6r, which recognize and bind clcf1 and il6 respectively. These distinct α-subunits act together with the common β-subunit receptor gp130 to activate JAK/STAT signaling (48). Abrogating the ligand-receptor interaction via decreased expression of cntfr and il6r in tumor cells would thus be expected to halt tumor growth in vivo. To test this hypothesis, two independent shRNAs to cntfr and il6r were validated and used to infect LKR10 cells (Fig. 6A and Supplementary Fig. S6A). We coinjected CAFs along with cntfr-and il6r-deficient LKR10 cells or controls, and assessed tumor volume. Tumor cells carrying single shRNAs directed against cntfr were significantly smaller than controls (Fig. 6B). A similar trend was observed with two independent il6r shRNAs, however the differences with regard to control cells were not statistically significant for both hairpins (Supplementary Fig. S6B). Importantly loss of cntfr or il6r did not alter tumor cell growth in vitro, suggesting that the in vivo effect is likely due to an important role of this signaling pathway in mediating paracrine effects in vivo but not intrinsic tumor proliferation (Fig. 6C and Supplementary Fig. S6C). Taken together, the results of the overexpression and knockdown studies indicate that while both clcf1 and il6 stimulate tumor growth, il6-il6r signaling may be dispensable whereas loss of the clcf1-cntfr axis causes a consistent decrease in tumor formation, suggesting that the two signaling pathways are distinct and yet together constitute an important paracrine signaling network between CAFs and tumor cells in NSCLC.
Figure 6.
Decrease response to CAF-derived clcf1 in cntfr-depleted tumor cells. A. qRT-PCR analysis of LKR10 cells transduced with two independent shRNAs to cntfr or a control shRNA. B. Tumor volume analysis of xenografts including CAFs and cntfr-depleted LKR10 cells. Error bars indicate ± s.e.m. All p values are for a two-tailed t-test. Endpoint is 26 days. C. Cell viability analysis of LKR10 cells expressing cntfr shRNAs assessed by MTT.
Discussion
Accumulating evidence points to a critical role for CAFs in tumor progression (4-6). As increased stromal proportion is a marker of poor survival in NSCLC patients (16, 17, 49), CAFs are likely to play an important role in lung cancer progression. We utilized a cross-species approach to identify a secretory gene signature in CAFs that promotes lung cancer growth and to query individual components of this signature for relevance to the phenotype of CAFs and to human NSCLC. These studies link secretion of CLCF1 and IL6 from CAFs to tumor progression in vivo. CLCF1 and IL6 are members of the IL6 family (50). CLCF1 (cardiotrophin-like cytokine factor 1, also known as NNT-1 or BSF-3) is a cytokine that has been implicated in the pathogenesis of multiple myeloma (51) but has not been reported to play a role in NSCLC. While autocrine secretion of IL6 has been proposed to occur in cell lines carrying EGFR mutations (52), our results suggest that IL6 is produced primarily by CAFs. Indeed, the functional analysis presented here suggests that CLCF1 and IL6 secretion by CAFs play a significant role in the contribution of this component of the tumor microenvironment to tumor growth.
CLCF1 and IL6 are cytokines that signal through multimeric complexes that include the gp130 receptor/β-subunit and ultimately trigger a signaling cascade that is mediated by the Jak-STAT and MAPK pathways (50). The specificity of the cellular responses elicited by these cytokines is governed by the expression patterns of different α-subunit receptors, CNTFR for CLCF1, and IL6R for IL6, which can be activated by each cytokine independently (50). Interestingly, overexpression of another IL6 family member upregulated in mouse CAFs, IL11, which signals through an alternative α-subunit receptor (IL11RA), does not induce tumor growth in vivo in our model system (data not shown), suggesting that distinct receptor complexes may be activated in specific tumor types. Our loss-of-function experiments show a more prominent role of the CLCF1-CNTFR axis in our model, and suggest that this paracrine network is less redundant and more relevant to NSCLC progression. Thus, these results provide rationale for the development of new therapies targeting CAFs, and suggests that inhibition of the CLCF1-CNTFR axis may be a novel strategy.
In vitro and in vivo experiments in models of breast and skin carcinoma have reported that tumor cells can induce a CAF-like phenotype in normal fibroblasts, suggesting that direct tumor cell-fibroblast cross talk plays an important role in tumor development (11, 45). In this study, we demonstrate that transient upregulation of some CAF-specific genes in NFs can be induced by tumor-derived secreted cues. However, the ability of NFs to fully upregulate a secretory CAF-gene signature is only achieved through long-term, direct exposure to tumor cells. Our results suggest that while NFs are responsive to tumor-derived signals, the generation of NF populations stably expressing a CAF-like gene signature (and thus, recapitulating the CAF tumor-promoting phenotype) likely involves genetic and/or epigenetic changes resulting from chronic tumor cell-to-NF interactions (53, 54). The fact that NFs passaged through a xenograft (iCAFs) are phenotypically indistinguishable from primary CAFs after 3-4 weeks of in vivo growth (despite intermediate expansion in vitro for >1 week) supports the notion that long-term interaction between tumor cells and NFs leads to the acquisition of a stable CAF phenotype. In addition, in vitro experiments described here also demonstrate a role for human NSCLC cells in the reprogramming of hNFs. Thus, tumor cells are critical in regulating the reprogramming of NFs to CAFs in both mouse and human lung cancer. Further work will be required to elucidate the molecular mechanisms that induce this transition.
Cross-species gene-expression analysis has been used successfully to identify mouse gene signatures relevant to human NSCLC (23). By analyzing mouse stromal cells we identified a gene signature predictive of survival in human NSCLC, supporting the relevance of studies of stromal compartments in mice to the pathogenesis of human cancer. Previous work identified an 11-gene signature of human CAFs in NSCLC samples with prognostic relevance (55). Surprisingly, we found no significant overlap between our mouse CAF-HIGH gene set and the upregulated CAF genes uncovered in the former study (data not shown). This discrepancy is likely due to the heterogeneous genetic background of the different human samples and highlights the ongoing value of cross-species comparison as a tool for functional genomics studies.
In summary, the data presented in this work describe a novel CAF gene signature with significant relevance in human NSCLC. We demonstrate that CAF-derived CLCF1 and IL6 exert specific paracrine effects to promote tumor growth in vivo and may be particularly important components of the CAF secretome. In addition, we establish that direct interaction between tumor cells and lung fibroblasts is sufficient to alter the phenotype of the later to become CAFs. These observations highlight a prominent role for CAFs in NSCLC and provide a framework for further mechanistic studies to identify relevant therapeutic targets. Lastly, these studies underscore the need to further elucidate the mechanisms that lead to the conversion of normal NFs to CAFs as this may provide a way to inhibit the “education” of NFs to a pro-tumorigenic CAF state.
Supplementary Material
Acknowledgments
We thank Julien Sage, Laura Attardi, Steve Artandi, Monte Winslow and all the members of the Sweet-Cordero laboratory for helpful discussions. We thank Oihana Murillo for technical assistance with flow cytometry analyses.
Grant support: This work was supported by the Alfonso Martín Escudero Foundation and the Tobacco-Related Disease Research Program (TRDRP) of California (UCAEF SPO#44443) (to S.V.), the National Cancer Institute (1R01CA138256-01) (to P.K. and A.B.) the Center for Cancer Systems Biology (CCSB) at Stanford (funded by a grant from the National Cancer Instiute (U54 CA149145)) (to O.G. and S.P.), the Fund for Scientific Research Flanders (FWO-Vlaanderen) (to O.G.), and an American Cancer Society Research Scholar Award (to E.A.S-C.).
S.V. was funded by the Alfonso Martín Escudero Foundation and the Tobacco-Related Disease Research Program (TRDRP) of California (UCAEF SPO#44443). P.K. and A.J.B. and were funded by the US National Cancer Institute (1R01CA138256-01). O.G. and S.P. were supported by the Center for Cancer Systems Biology (CCSB) at Stanford (U54 CA149145). O.G. is a fellow of the Fund for Scientific Research Flanders (FWO-Vlaanderen). E.A.S-C. was funded by a Research Scholar Grant from the American Cancer Society.
Footnotes
Declaration of conflict of interest: the authors declare no conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 4.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 5.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–7. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirri P, Chiarugi P. Cancer associated fibroblasts: the dark side of the coin. Am J Cancer Res. 2011;1:482–97. [PMC free article] [PubMed] [Google Scholar]
- 7.Augsten M, Hagglof C, Olsson E, Stolz C, Tsagozis P, Levchenko T, et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc Natl Acad Sci U S A. 2009;106:3414–9. doi: 10.1073/pnas.0813144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–51. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 10.Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–91. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–47. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 13.Liao CP, Adisetiyo H, Liang M, Roy-Burman P. Cancer-associated fibroblasts enhance the gland-forming capability of prostate cancer stem cells. Cancer Res. 2010;70:7294–303. doi: 10.1158/0008-5472.CAN-09-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannoni E, Bianchini F, Masieri L, Serni S, Torre E, Calorini L, et al. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer Res. 2010;70:6945–56. doi: 10.1158/0008-5472.CAN-10-0785. [DOI] [PubMed] [Google Scholar]
- 15.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimosato Y, Suzuki A, Hashimoto T, Nishiwaki Y, Kodama T, Yoneyama T, et al. Prognostic implications of fibrotic focus (scar) in small peripheral lung cancers. Am J Surg Pathol. 1980;4:365–73. doi: 10.1097/00000478-198008000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Maeshima AM, Niki T, Maeshima A, Yamada T, Kondo H, Matsuno Y. Modified scar grade: a prognostic indicator in small peripheral lung adenocarcinoma. Cancer. 2002;95:2546–54. doi: 10.1002/cncr.11006. [DOI] [PubMed] [Google Scholar]
- 18.Zhong L, Roybal J, Chaerkady R, Zhang W, Choi K, Alvarez CA, et al. Identification of secreted proteins that mediate cell-cell interactions in an in vitro model of the lung cancer microenvironment. Cancer Res. 2008;68:7237–45. doi: 10.1158/0008-5472.CAN-08-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson IC, Mari SE, Broderick RJ, Mari BP, Shipp MA. The angiogenic factor interleukin 8 is induced in non-small cell lung cancer/pulmonary fibroblast cocultures. Cancer Res. 2000;60:269–72. [PubMed] [Google Scholar]
- 20.Nazareth MR, Broderick L, Simpson-Abelson MR, Kelleher RJ, Jr, Yokota SJ, Bankert RB. Characterization of human lung tumor-associated fibroblasts and their ability to modulate the activation of tumor-associated T cells. J Immunol. 2007;178:5552–62. doi: 10.4049/jimmunol.178.9.5552. [DOI] [PubMed] [Google Scholar]
- 21.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, et al. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim CF, Jackson EL, Kirsch DG, Grimm J, Shaw AT, Lane K, et al. Mouse models of human non-small-cell lung cancer: raising the bar. Cold Spring Harb Symp Quant Biol. 2005;70:241–50. doi: 10.1101/sqb.2005.70.037. [DOI] [PubMed] [Google Scholar]
- 23.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 24.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 25.Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, et al. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci U S A. 1993;90:11029–33. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar RK, O'Grady R, Li W, Smith LW, Rhodes GC. Primary culture of adult mouse lung fibroblasts in serum-free medium: responses to growth factors. Exp Cell Res. 1991;193:398–404. doi: 10.1016/0014-4827(91)90112-8. [DOI] [PubMed] [Google Scholar]
- 27.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–51. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 28.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 29.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–50. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 32.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 33.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–70. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 34.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 35.Hedges LV, Olkin I. Statistical methods for meta-analysis. Orlando: Academic Press; 1985. [Google Scholar]
- 36.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 38.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–84. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, Lubin JH, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–75. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishii G, Sangai T, Oda T, Aoyagi Y, Hasebe T, Kanomata N, et al. Bone-marrow-derived myofibroblasts contribute to the cancer-induced stromal reaction. Biochem Biophys Res Commun. 2003;309:232–40. doi: 10.1016/s0006-291x(03)01544-4. [DOI] [PubMed] [Google Scholar]
- 44.Sangai T, Ishii G, Kodama K, Miyamoto S, Aoyagi Y, Ito T, et al. Effect of differences in cancer cells and tumor growth sites on recruiting bone marrow-derived endothelial cells and myofibroblasts in cancer-induced stroma. Int J Cancer. 2005;115:885–92. doi: 10.1002/ijc.20969. [DOI] [PubMed] [Google Scholar]
- 45.Mishra PJ, Humeniuk R, Medina DJ, Alexe G, Mesirov JP, Ganesan S, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–9. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haviv I, Polyak K, Qiu W, Hu M, Campbell I. Origin of carcinoma associated fibroblasts. Cell Cycle. 2009;8:589–95. doi: 10.4161/cc.8.4.7669. [DOI] [PubMed] [Google Scholar]
- 47.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. The Journal of clinical investigation. 2011;121:3375–83. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito M, Ishii G, Nagai K, Maeda R, Nakano Y, Ochiai A. Prognostic impact of cancer-associated stromal cells in stage I lung adenocarcinoma patients. Chest. 2012 doi: 10.1378/chest.11-2458. [DOI] [PubMed] [Google Scholar]
- 50.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burger R, Bakker F, Guenther A, Baum W, Schmidt-Arras D, Hideshima T, et al. Functional significance of novel neurotrophin-1/B cell-stimulating factor-3 (cardiotrophin-like cytokine) for human myeloma cell growth and survival. Br J Haematol. 2003;123:869–78. doi: 10.1046/j.1365-2141.2003.04686.x. [DOI] [PubMed] [Google Scholar]
- 52.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–56. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 54.Fiegl H, Millinger S, Goebel G, Muller-Holzner E, Marth C, Laird PW, et al. Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res. 2006;66:29–33. doi: 10.1158/0008-5472.CAN-05-2508. [DOI] [PubMed] [Google Scholar]
- 55.Navab R, Strumpf D, Bandarchi B, Zhu CQ, Pintilie M, Ramnarine VR, et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2011;108:7160–5. doi: 10.1073/pnas.1014506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.