Abstract
The ex vivo sensitivity of pediatric leukemia cells to the proteasome inhibitor bortezomib was compared to 3 next generation proteasome inhibitors: the epoxyketone-based irreversible proteasome inhibitors carfilzomib, its orally bio-available analog ONX 0912, and the immunoproteasome inhibitor ONX 0914. LC50 values were determined by MTT cytotoxicity assays for 29 childhood acute lymphoblastic leukemia and 12 acute myeloid leukemia patient samples and correlated with protein expression levels of the constitutive proteasome subunits (β5, β1, β2) and their immunoproteasome counterparts (β5i, β1i, β2i). Acute lymphoblastic leukemia cells were up to 5.5-fold more sensitive to proteasome inhibitors than acute myeloid leukemia cells (P<0.001) and the combination of bortezomib and dexamethasone proved additive/synergistic in the majority of patient specimens. Although total proteasome levels in acute lymphoblastic leukemia and acute myeloid leukemia cells did not differ significantly, the ratio of immuno/constitutive proteasome was markedly higher in acute lymphoblastic leukemia cells over acute myeloid leukemia cells. In both acute lymphoblastic leukemia and acute myeloid leukemia, increased ratios of β5i/β5, β1i/β1 and β2i/β2 correlated with increased sensitivity to proteasome inhibitors. Together, differential expression levels of constitutive and immunoproteasomes in pediatric acute lymphoblastic leukemia and acute myeloid leukemia constitute an underlying mechanism of sensitivity to bortezomib and new generation proteasome inhibitors, which may further benefit from synergistic combination therapy with drugs including glucocorticoids.
Introduction
Recently, the crystal structures of constitutive and immunoproteasomes in the absence or presence of the novel epoxyketone-based irreversible proteasome inhibitor ONX 0914 were solved.1 Structural evidence for selective inhibitory potential of ONX 0914 for the immunoproteasome was provided by the ability to induce conformational changes in the S1 binding pocket of the immunoproteasome subunit β5i but not in the constitutive proteasome subunit β5. Conceptually, immunoproteasome inhibitors may have dual impact on both cell proliferation and cytokine production2,3 and could thus serve a valuable alternative to the first generation clinically active proteasome inhibitors, e.g. bortezomib, which targets both constitutive and immunoproteasome subunits.4 Clinical evaluation of immunoproteasome inhibitors is still in an early phase. However, given the established efficacy of bortezomib in the treatment of various hematologic malignancies,4,5 adult and pediatric acute leukemias are valid candidates for further pre-clinical and clinical investigation.
Although over the last decades the treatment of children with acute leukemia has continued to improve, 20–40% of children still relapse following initial therapy and this is associated with a worse prognosis.6 For the survival of children with acute lymphoblastic leukemia (ALL) a good initial response to glucocorticoids is of favorable prognostic value.7 Hence, glucocorticoid-resistant and relapsed ALL patients may benefit from novel and glucocorticoid-sensitizing strategies. Based on its good pre-clinical activity in other hematologic malignancies,5 bortezomib was selected as a novel anti-leukemic drug in pediatric leukemias.8–10 Bortezomib is a reversible inhibitor of the 26S proteasome, a large intracellular protease expressed in all cell types.11 The proteasome consists of seven α-subunits and seven β-subunits, three of which harbor the catalytic activities of the proteasome, chymotrypsin-like, caspase-like, and trypsin-like, encoded by β5 (PSMB5), β1 (PSMB6), and β2 (PSMB7) subunits, respectively. The catalytic activities of the proteasome are responsible for the degradation of all poly-ubiquitinated proteins.11 Cells of the immune system express a distinct type of proteasome, the interferon-γ inducible immunoproteasome, in which all three catalytic constitutive subunits are exchanged for the immunosubunits β5i (PSMB8), β1i (PSMB9), and β2i (PSMB10).1 Besides immunoproteasomes, two additional proteasome hybrid types (β1+β2+β5i and β1i+β2+β5i) were identified, each of which has the capacity to process different tumor antigens.12 Immunoproteasomes play a major role in the provision of peptides for antigen presentation, partly by facilitating efficient clearance of protein aggregates that arise upon interferon-induced oxidative stress.13 Increased immunoproteasome expression has been noted in B-cell malignancies.14,15
In leukemic cell lines, bortezomib was shown to interact in an additive or synergistic way when combined with traditional drugs, including glucocorticoids.16 In pre-clinical TALL mouse models, bortezomib showed modest single-agent activity17 while almost no monotherapy activity was observed in phase I studies in children and adults.8,9 However, phase I or phase II studies in which bortezomib was combined with conventional chemotherapeutics showed promising clinical activity in adult AML patients18 and pediatric ALL patients.10,19
Despite the encouraging results of bortezomib in several hematologic malignancies, emergence of bortezomib resistance, as well as side effects, are factors that limit its long-term efficacy.20–22 To overcome these issues, several irreversible proteasome inhibitors have been developed.23 Carfilzomib is more selective for the proteasomal chymotrypsin-like activity and is a more effective anti-leukemic drug at low concentrations than bortezomib.24 Subsequently, an orally bioavailable analog of carfilzomib was developed, ONX 0912, which elicited anti-tumor activity by inhibiting chymotrypsin-like activity in Waldenstrom macroglobulinemia,15 and MM.25 Upon recognition of the important role of immunoproteasomes, ONX 0914 was developed as the first β5i selective proteasome inhibitor.1,3 Alternatively, to overcome bortezomib-resistance, proteasome inhibitors that target other non-catalytic parts of the proteasome may be attractive. In this context, the non-competitive proteasome inhibitor 5-amino-8-hydroxyquinoline (5AHQ) may serve as a prototypical drug that binds the structural α7 subunit of the proteasome and induces cell death of in vitro established bortezomib-resistant hematologic cell lines.26
The aim of the current study was to examine the ex vivo sensitivity of pediatric leukemia cells (ALL and AML) to: i) bortezomib as a single agent and in combination with dexamethasone; and ii) next generation epoxyketone-based irreversible proteasome inhibitors designed to overcome bortezomib resistance. To identify novel parameters that may predict proteasome inhibitor response, we explored whether or not their cytotoxic activity correlated with protein expression levels of the constitutive subunits β5, β1, β2, and α7, and the immunoproteasome subunits β5i, β1i and β2i. We show that higher ratios of immune versus constitutive proteasome level represent a novel indicator of sensitivity of pediatric acute leukemia cells to bortezomib and epoxyketone-based proteasome inhibitors.
Methods
Leukemic patient samples
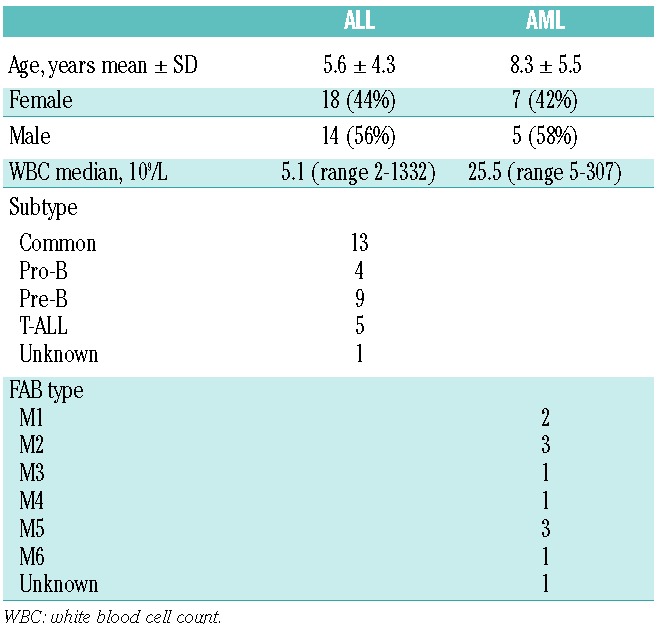
Forty-four pediatric leukemia samples (12 AML and 32 ALL) were included in this study. Table 1 gives an overview of patients’ characteristics. After thawing the vials, viable cells were counted and blast percentage was determined after May-Grunwald/Giemsa cytospin stainings. Inclusion criteria for the MTT assay were that more than 80% blasts were present in the leukemic samples. These non-proliferating cells were immediately used for MTT analysis, and the remaining cells were snap-frozen for proteasome subunit protein expression..
Table 1.
Patients’ characteristics.
MTT cytotoxicity assay
Cytotoxicity of bortezomib, dexamethasone, as well as their combination, and carfilzomib, ONX 0912, ONX 0914, and 5AHQ was determined using the MTT colorimetric dye reduction assay.27 For the drug combination study, CalcuSyn (Version 1.1.1 1996, Biosoft, Cambridge, UK) software was used to calculate a combination index (CI) based on the median-effect principle, for each drug combination tested.28 More detailed information is available in the Online Supplementary Appendix.
Protein expression
Western blotting: protein expression was determined by Western blot analysis, as previously described.20 Protein bands were quantified by Odyssey software, corrected for background, and normalized with β-actin to correct for any loading differences. To compare between gels, subunit expression of the patient samples were normalized using subunit expression in the leukemic T-ALL cell line CCRF-CEM.20 Since each antibody has different binding characteristics, it is not possible to determine ratios between the expression of each different subunit using Western blot analysis.
ProCISE analysis: a previously described ELISA-based method (ProCISE) was used to confirm the Western blot data and to accurately quantify the fraction of constitutive and immunoproteasome subunits per patient.29 Details on validation and statistical evaluation are described in the Online Supplementary Appendix.
Statistical analysis
Statistical significance of the differences between subunit expression and differences in drug sensitivity for AML and ALL patients was determined using the Mann-Whitney U test. Correlations of subunit expression with drug LC50 concentrations were calculated by Spearman’s correlation. P<0.05 was considered statistically significant. All statistical analyses were performed using SPSS (version 20.0).
This study has been approved by the Local Ethics Committee VUmc (Date of approval: December 5, 2000. Approval file number: TJFS/bz2568a).
Results
Constitutive proteasome and immunoproteasome composition in childhood ALL and AML cells
Quantitative assessment of total proteasome, constitutive- and immunoproteasome by ProCISE analysis was performed on 19 childhood ALL and 6 childhood AML samples (Figure 1). We refer to immunoproteasomes as forms composed of β5i, β1i and β2i subunits, although ProCISE assay could also identify β5i and β1i subunits being incorporated in hybrid proteasome forms.12 Total proteasome and constitutive proteasome subunit levels did not differ significantly between ALL and AML; however, immunoproteasome expression was significantly higher in ALL over AML (19 vs. 12.8 ng/μg total protein; P=0.036). Notably, total immunoproteasome levels outweigh constitutive proteasome levels by at least 2–3 fold (Figure 1A), which is also manifested in expression levels of individual immunoproteasomes and constitutive proteasomes (Online Supplementary Figure S1 and Online Supplementary Table S1).
Figure 1.
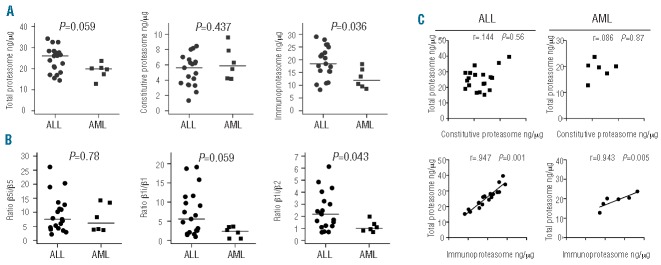
Proteasome expression of ALL and AML patient samples determined by ProCISE assay. (A) Total proteasome levels and subdivision of constitutive- and immunoproteasome levels (ng/μg total protein) in childhood ALL and AML cells. The total value of constitutive proteasome and immunoproteasome expression for each patient is shown and depicts the variation between patients. (B) The ratio of paired subunits (between β5i and β5 and between β1i and β1) is shown within each patient sample. The line denotes the mean for ALL (n=19) and AML (n=6). (C) Correlation plots of total proteasome as a function of constitutive proteasome, and total proteasome as a function of immunoproteasome levels. The individual subunit data are shown in Online Supplementary Figure S1.
Next, we determined the individual ratios of immuno-subunit over constitutive subunit expression. For ALL and AML the highest ratio was observed for β5i/β5 and lowest ratio for β2i/β2. Ratios of β2i/β2 were significantly increased in ALL versus AML samples (P=0.043) whereas a trend for higher β1i/β1 was noted (P=0.059) (Figure 1B). Furthermore, we examined the impact of variations in constitutive- and/or immunoproteasome expression for total cellular proteasome expression levels (Figure 1C). In contrast to constitutive subunit expression, increasing immunoproteasome subunit expression was significantly correlated with increased total proteasome levels in both ALL and AML cells.
The above-described analyses were further extended by comparing subgroups of ALL (BCP-ALL and T-ALL) for constitutive- and immunoproteasome expression and ratios of individual immunoproteasome over constitutive proteasome (Online Supplementary Figure S2). Although the sample size was limited, BCP-ALL, T-ALL and AML displayed similar levels of constitutive proteasome, but total immunoproteasome levels were higher in BCP-ALL (4-fold), AML (2-fold) and T-ALL (2-fold) compared to constitutive proteasome levels (Online Supplementary Figure S2A). Further dissection of the ratios of β5i/β5, β1i/β1 and β2i/β2 expression in ALL subgroups revealed high ratio of β5i/β5 the BCP-ALL subgroup (ratio 7.1) and AML (ratio: 7.9) compared to T-ALL (3-fold) (Online Supplementary Figure S2B). β1i/β1 ratios were high for BCP-ALL (ratio 7.4) compared to T-ALL and AML (2-fold) and β2i/β2 ratios were consistently the lowest among all leukemia subgroups. Further dissection of BCP-ALL in pro-B-ALL, pre-B-ALL and common-ALL revealed high ratios of β5i/β5 in all subgroups, with β1i/β1 ratios being particularly low in pro-B-ALL, T-ALL, and AML (Online Supplementary Figure S2C). Due to small subgroup-size, no statistics were applied on these subgroup comparisons. For comparison, Online Supplementary Figure S2C shows values of ratios for established in vitro cell line models of human T-ALL (CCRF-CEM) and AML (THP1).
The preliminary account shown in Online Supplementary Figure S1 suggested that differences in immuno/constitutive proteasome ratios between ALL and AML (Figure 1B) were associated with increased constitutive proteasome levels and decreased immunoproteasome levels in AML versus ALL cells. These observations were confirmed in a large group of ALL and AML patient samples (n=29 and n=12, respectively) by Western blot analysis of relative levels of immunoproteasomes and constitutive proteasomes, normalized on housekeeping gene β-actin and cell line CEM. Please note that these data depict relative quantifications of subunit expression, whereas ProCISE analysis provides absolute quantification of subunits. Figure 2 shows significantly increased levels of constitutive β5, β1 and β2 subunit levels in AML versus ALL samples, whereas AML cells had significantly lower levels of β1i and a tendency towards lowered β5i levels compared to ALL cells. No significant differences in expression level of the non-catalytic α7 subunit were observed. Upon classification of ALL samples into subgroups, pro-B ALL (n=4) and T-ALL (n=4) samples expressed relatively higher β5, β1, and β2 constitutive subunit expression levels than both pre-B ALL (n=7) and common-ALL (n=10), whereas there was a trend for the reverse with respect to β5i and β1i expression (Online Supplementary Figure S3). Taken altogether, the results demonstrate that proteasome composition in pediatric ALL and AML cells is largely represented (>70%) by immunoproteasome subunits and to a minor extent (<30%) by constitutive subunits. Beyond this, ratios of individual immuno/constitutive proteasomes vary considerably, mostly in the order of β5i/β5>β1i/βi>β2i/β2, of which the impact on response to constitutive- and immunoproteasome inhibitors warrants further investigation.
Figure 2.
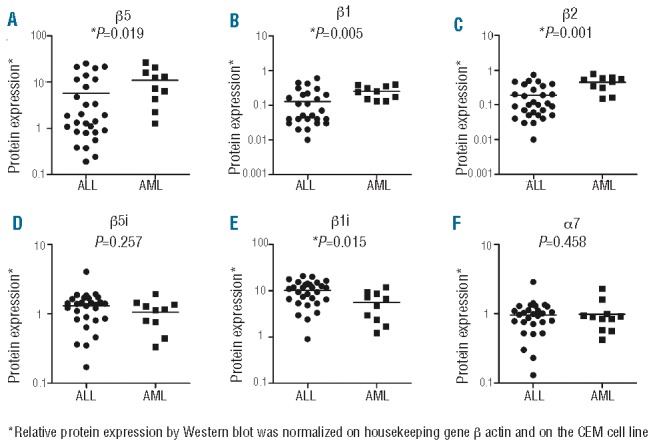
Proteasome protein subunit expression in ALL versus AML determined by Western blotting. Comparison of proteasome subunit expression of constitutive subunits; (A) β5, (B) β1, (C) β2, and immunoproteasome subunits; (D) β5i and (E) β1i, and (F) structural subunit α7 in ALL and AML patient samples. Protein expression determined by Western blotting was normalized on β-actin as loading control and to subunit expression of CCRF-CEM cell line as control between blots. Note that these data depict relative quantifications of subunit expression, whereas ProCISE analysis provides absolute quantification of subunits. The lines represent the mean.
Growth inhibitory effect of proteasome inhibitors against primary pediatric ALL and AML cells
Ex vivo sensitivity of pediatric leukemia patient cells towards different proteasome inhibitors was assessed in 4-day cytotoxicity assays (Figure 3). Apart from sensitivity to bortezomib, drug sensitivity was also determined for 3 epoxyketone-based irreversible proteasome inhibitors (carfilzomib, ONX 0912 and ONX 0914) and 5AHQ. With a median LC50 of 14.0 nM (range 10.1–61.0 nM), AML samples were significantly (P<0.001) less sensitive to bortezomib than ALL cells (median LC50: 6.0 nM, range 3.0–46.1 nM) (Figure 3A). Figure 3B and E and Online Supplementary Table S1 summarize the sensitivity to the individual proteasome inhibitors as median LC50 values. Statistical comparisons between proteasome inhibitor sensitivity of AML and ALL samples consistently revealed that ALL cells were the most drug sensitive (P<0.001). Notably, ALL samples were markedly sensitive to carfilzomib (Figure 3B) with a median LC50 of 4.1 nM, hence carfilzomib being 1.5-fold more potent than bortezomib (LC50: 6 nM). Again, AML samples were significantly more resistant to carfilzomib (median LC50: 20.8 nM) compared to ALL. Sensitivity for ONX 0912, an oral analog of carfilzomib, remained in the low nanomolar range for ALL (median LC50: 19.2 nM) and 4-fold lower than for AML cells (median LC50: 93.7 nM) (Figure 3C). The largest difference (5.6-fold) between ALL and AML cells was observed for the immunoproteasome inhibitor ONX 0914 (median LC50: 44.6 nM vs. 248.0 nM) (Figure 3D). Finally, ALL samples displayed 2.5-fold greater sensitivity for 5AHQ (median LC50: 20.1 μM vs. 53.8 μM) compared to the AML samples (Figure 3E). As reported previously,30 the latter cells were also the least sensitive to dexamethasone (Figure 3F). Within the ALL subgroups, pre-B-ALL and common-ALL samples appeared to be most sensitive for carfilzomib, ONX 0912, ONX 0914 and 5AHQ as compared to pro-B cells (Online Supplementary Figure S4).
Figure 3.
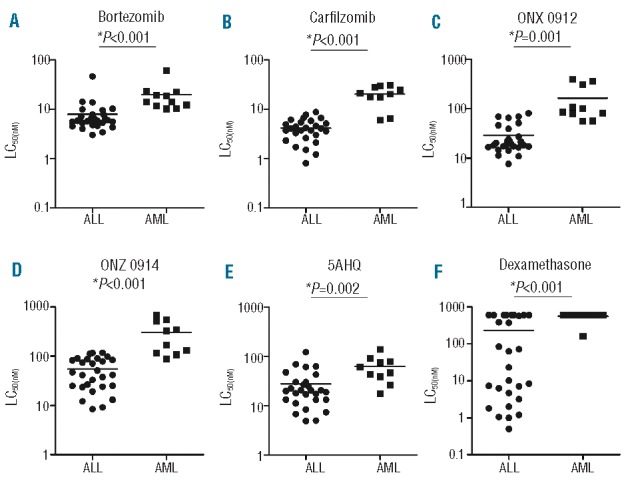
Sensitivity of pediatric ALL and AML patient samples to proteasome inhibitors and dexamethasone. Comparison of LC50 values obtained by MTT cytotoxicity assay after 96 h exposure to; (A) bortezomib, (B) carfilzomib, (C) ONX 0912, (D) ONX 0914, (E) 5AHQ, and (F) dexamethasone in ALL and AML patient samples. The lines represent the mean and the y-axis is depicted as a logarithmic scale.
Combination effects of bortezomib and dexamethasone
To investigate possible synergistic effects of combination therapy, dexamethasone was combined with bortezomib. Towards this end, pediatric leukemia samples were first tested for dexamethasone as single agent in MTT assays (Figure 3F). As expected, and consistent with previous studies,31 ALL samples were markedly more sensitive to dexamethasone than AML samples (median LC50: 62.4 nM vs. >600 nM; P<0.001). Within the group of AML samples, 10 of 11 (91%) had an LC50 >6 μM, which is the case in only 31% of ALL samples. Based on the dose-response curves of bortezomib and dexamethasone alone, 4 different concentrations of bortezomib were selected (range 2.4–20 nM) in combination with 5 concentrations of dexamethasone (range 0.18–750 nM). Nineteen ALL and 7 AML samples could be used for further calculation of a CI based on the median-drug effect analysis.28 Notably, the highest synergy was found for combinations with dexamethasone in the low bortezomib concentration range (2.4–11.8 nM) (Figure 4A). Furthermore, all 5 dexamethasone-resistant (defined as less than 50% cell kill) ALL patients displayed sensitivity for bortezomib as well as synergism in combination with dexamethasone. The only ALL patient with ex vivo bortezomib-resistance was sensitive for dexamethasone and to the bortezomib-dexamethasone combinations. Remarkably, bortezomib sensitized all 6 dexamethasone-resistant AML patients for dexamethasone. Lastly, the combination of dexamethasone and bortezomib was synergistic in the samples of 4 AML patients being resistant to both dexamethasone and to bortezomib. Regardless of the dexamethasone concentration used, 11.8 nM bortezomib established the most synergistic combinations and the highest fraction affected for AML. Overall, bortezomib was most sensitizing in dexamethasone-resistant cells, which may be of potential clinical relevance since these patients have a dismal prognosis.
Figure 4.
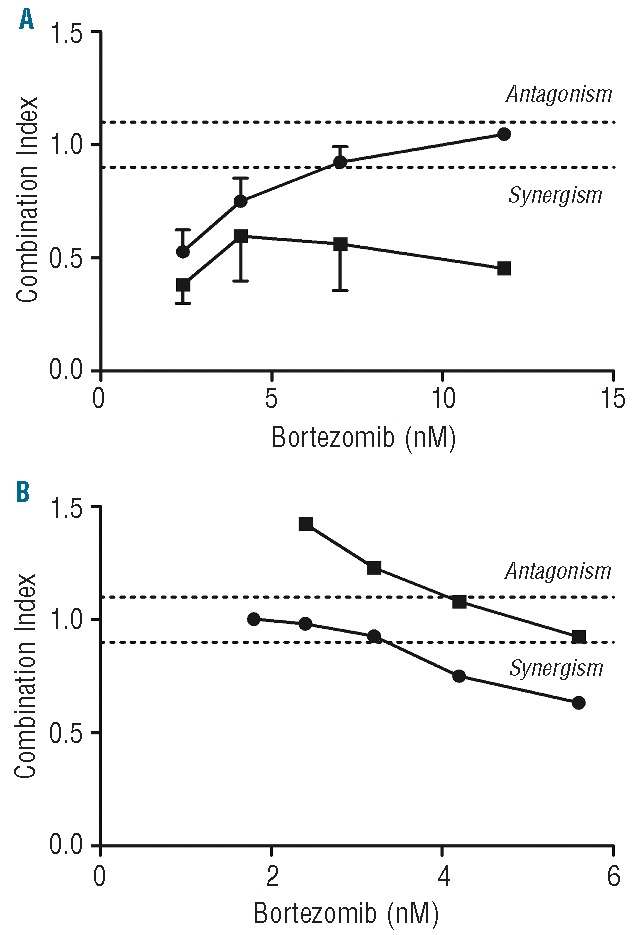
Combination Indices of bortezomib-dexamethasone combinations. (A) Mean CI of 3 representative dexamethasone concentrations per bortezomib concentration in patient samples. Symbols represent ALL (•) and AML (▪). For ALL (n=17), 1.5 nM, 11.7 nM, and 93.8 nM dexamethasone were selected, and for AML (n=6), 11.7 nM, 93.8 nM, and 750 nM dexamethasone were used. Antagonism (CI > 1.1) additivity (CI > 0.9 < 1.1), and synergism (CI < 0.9). Error bars represent standard error of the mean of 3 separate experiments. (B) Combination indices of T-ALL cell line CEM exposed to combinations of bortezomib and dexamethasone. Symbols represent (•) CEM pulsed for 1 h with 1 μM bortezomib prior to the combination assay and (▪) CEM control, not pulsed prior to the combination assay. Experiments were performed twice in triplicate. Results of the mean of these experiments are presented.
As a comparison, the T-ALL cell line CCRF-CEM was exposed to combinations of bortezomib and dexamethasone for four days, either with or without prior pulse exposure to 1 μM bortezomib for 1 h to mimic in vivo peak plasma pharmacokinetic concentrations of bortezomib followed by steady state plasma levels of 10–20 nM bortezomib. CEM cells that were pulse-treated with bortezomib showed increased sensitivity to dexamethasone (IC50: 18 nM) compared to CEM cells that were not pulse-treated with bortezomib (IC50: 24 nM). Furthermore, combination experiments in the clinically achievable concentration range of bortezomib and dexamethasone showed three additive (CI: 0.97±0.04) and two synergistic combination indices (CI: 0.69±0.08) in the pulse-treated T-ALL CEM cells (Figure 4B).
Correlates of proteasome subunit expression and sensitivity to proteasome inhibition
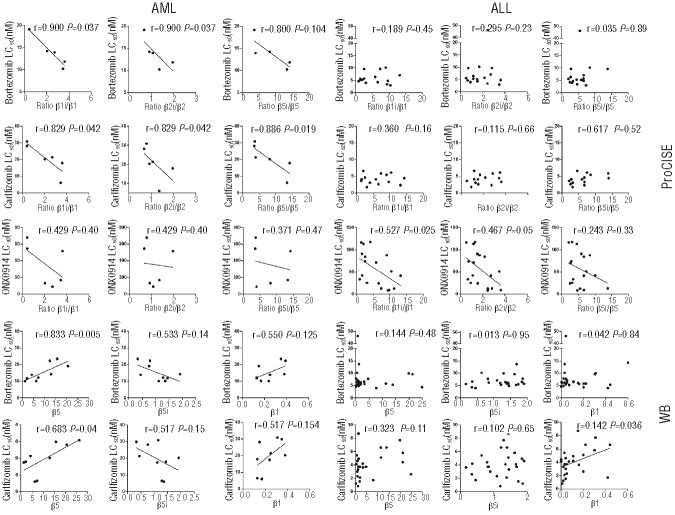
Recently, in vitro studies indicated that upregulation of the constitutive subunits was associated with decreased bortezomib sensitivity.20,22,32,33 Here we investigated whether differential expression levels of constitutive and/or immunoproteasome levels may underlie responsiveness to bortezomib and other proteasome inhibitors. Interestingly, ratios of immune/constitutive proteasome revealed most significant correlations or consistent trends such that increased ratios correlated with increased sensitivity to bortezomib and carfilzomib in AML, and ONX 0914 in ALL patients. This is illustrated in Figure 5, where sensitivity for bortezomib in AML inversely correlated with the ratios β1i/β1 and β2i/β2 (r=−0.900; P=0.037) whereas a trend was noted for β5i/β5 (r=−0.800; P=0.104). In addition, all three ratios correlated significantly with sensitivity for carfilzomib. For ALL, no significant correlations were revealed with bortezomib- or carfilzomib-sensitivity; however, β1i/β1 ratio correlated significantly with sensitivity for ONX 0914 (r=−0.527; P=0.025) and a trend for correlation with β2i/β2 was seen (r=−0.467; P=0.05). Overall, Online Supplementary Table S2 summarizes that, for AML samples, all proteasome inhibitors except 5AHQ displayed lower LC50 when immuno/constitutive proteasome ratio was higher. In ALL samples this was true for 11 of 15 proteasome inhibitors and immuno/constitutive proteasome ratio combinations. Other selected examples of correlates that were observed based on Western blot analysis of constitutive- and immunoproteasome levels in AML and ALL samples are shown in Figure 5. Carfilzomib and bortezomib LC50 values for AML correlated positively with β5 and β1 expression and inversely with β5i. For the same drugs in ALL samples, no such correlations were found.
Figure 5.
Representative correlations of proteasome inhibitor sensitivity and constitutive and immunoproteasome subunit expression in pediatric ALL and AML. Correlations of ratios (β1i/β1, β2i/β2 and β5i/β5) determined by ProCISE with LC50 values (bortezomib, carfilzomib, ONX 0914) for AML and ALL patient samples. Correlations of single subunits with LC50 values of bortezomib and carfilzomib for AML and ALL patient samples with β5, β5i, and β1.
Collectively, these results indicate that immuno/constitutive proteasome ratios, but not the mere expression of constitutive levels per se, is a novel correlative factor in response to proteasome inhibitors in pediatric ALL and AML cells, with increased ratios being predictive of increased sensitivity to proteasome inhibitors.
Effect of PSMB8 silencing on proteasome inhibitor sensitivity in THP1 cells
To study the role of the immunoproteasome subunits in the sensitivity to proteasome inhibitors at a mechanistic level, THP1 cells were pre-exposed to PSMB8 siRNA or PSMB9 siRNA. After 24 h of siRNA (mean knockdown of 3 experiments: 76%) (Online Supplementary Figure S5A), 4-day MTT assays were performed with bortezomib and ONX 0914 to assess cell growth inhibitory effects. PSMB8 siRNA had no significant impact on the intrinsically high bortezomib-sensitive phenotype of THP1 cells (IC50 3.2±0.5) compared to non-target siRNA (IC50 3.6±0.3). For ONX 0914, however, siRNA PSMB8-silencing was accompanied with a 2.1-fold loss of sensitivity (IC50: 71.4±5 nM) compared to non-target siRNA controls (IC50: 34.2±7 nM). Silencing PSMB9 had no effect on the sensitivity to both proteasome inhibitors (Online Supplementary Figure S5B).
Discussion
Proteasome inhibitors, particularly bortezomib, are being explored in the clinical setting in childhood acute leukemias.10 Unfortunately, however, chemoresistance to bortezomib may occur that could hinder its pharmacological activity. In this respect, several in vitro studies with human leukemia cell lines identified mechanisms of acquired bortezomib resistance, most frequently due to upregulation of the proteasomal β5 subunit20,22,32–34 as well as acquisition of β5 subunit mutations that decrease bortezomib binding.20,22,34,35 The current research is the first to address these proteasome-based drug sensitivity and resistance phenomena in a relatively large series of childhood acute leukemia patient specimens and in particular studying the immunoproteasome as a novel drug target. Although the patient samples evaluated in the current study displayed differential bortezomib sensitivity, this observation was not associated with mutations in the β5 subunit of the proteasome20 (data not shown). This may have been anticipated as all of the samples were initial diagnosis leukemia samples, and β5 mutations found in human leukemia cell lines were typically acquired after prolonged bortezomib exposure. In a clinical setting, though studies are limited, no PSMB5-associated mutations were found in patients treated with bortezomib.36,37 Rather, these and other studies20,33 point to upregulation of β5 subunits expression as a primary response mechanism to bortezomib, which may set the stage for acquisition of mutations following prolonged bortezomib exposure. The present study established that ALL cells were significantly more sensitive to bortezomib as a single agent than AML cells. Interestingly, even the prognostically unfavorable subgroups pro-B ALL and T-ALL, which are resistant to most chemotherapeutic drugs,38 were equally sensitive to bortezomib as pre-B/common ALL samples. This is underscored in a study by Szczepanek et al.39 who showed that bortezomib was even more potent in T-ALL patient samples compared to pre-B/common ALL. In this study, bortezomib was found to be a potent drug for this therapy-resistant subgroup of ALL although the 2 T-ALL patients treated in a phase II clinical trial with bortezomib did not reach complete remission.10 Clearly, larger clinical studies with bortezomib in this hematologic malignancy are warranted. Consistent with sensitivity patterns for other anti-leukemic drugs, ALL cells were also more sensitive to dexamethasone than AML cells. Sensitivity to glucocorticoids is an important prognostic marker for therapy response of ALL patients,7 hence the synergistic combination of dexamethasone and bortezomib may improve therapy response and decrease disease recurrence rates. Although the molecular basis for glucocorticoid resistance in AML patients remains elusive,40 our data showed that bortezomib can sensitize cells for dexamethasone-induced growth inhibition. In particular, the specimens of 4 AML patients resistant to bortezomib and dexamethasone were sensitive to the combination of these drugs (mean CI: 0.51). For ALL, bortezomib and dexamethasone combinations displayed a lesser synergistic effect probably due to the fact that ALL samples are intrinsically more sensitive to either bortezomib or dexamethasone alone. Consistent with this notion, dexamethasone resistant ALL samples could indeed be further sensitized by bortezomib/dexamethasone combinations as previously described by others41 for pre-clinical and clinical studies with MM cells. In fact, low-dose dexamethasone and bortezomib combinations were synergistic in 88% of ALL patients, and in all 5 AML patients. From a mechanistic perspective, the sensitization of dexamethasone-resistant AML cells by bortezomib might be possibly achieved via suppression of constitutively active NF-κB in AML cells,42 thereby triggering dexamethasone-mediated apoptosis. Consistently, combination experiments in T-ALL CEM cells also showed additive/synergistic effects at low nanomolar combinations of bortezomib and dexamethasone, even with short pre-exposures to high bortezomib concentrations mimicking peak plasma pharmacokinetic concentrations.
Given the emergence of bortezomib-resistance phenomena and untoward toxicity after chronic bortezomib administration, next generation irreversible proteasome inhibitors were developed to overcome these problems.4,23 In this respect, we tested a series of novel proteasome inhibitors in comparison with bortezomib to determine their efficacy in leukemic cell kill. ALL cells were more sensitive to these epoxyketone-based proteasome inhibitors than AML cells. For ALL patient samples, carfilzomib was most potent, followed by bortezomib, ONX 0912, ONX 0914 and 5AHQ. Of note, this drug-sensitivity pattern of ALL patient samples actually follows that of the human T-ALL cell line CCRF-CEM.20
The differential sensitivity of ALL versus AML cells for both bortezomib and epoxyketone-based proteasome inhibitors may, in part, be due to common mechanisms mediating drug resistance. Constitutively active NF-κB in AML cells42 can attenuate the induction of apoptosis, but also up-regulate the expression of the dominant multidrug efflux transporter MDR1/P-glycoprotein (Pgp/ABCB1).43 High Pgp expression has been associated with poor outcome in acute leukemia.44 Although bortezomib is considered to be a poor substrate for Pgp,22,45 the expoxyketone-based proteasome inhibitors carfilzomib, ONX 0912 and ONX 0914 were found to be bona fide substrates of Pgp.45 This notion, together with the fact that MDR1/Pgp expression is increased in AML cells over ALL cells,46 may result in a reduced sensitivity of AML cells to epoxyketone-based proteasome inhibitors.
Both ALL and AML patient cells displayed the lowest sensitivity to the immunoproteasome inhibitor ONX 0914, although effective drug concentrations still fall within the nanomolar range. This may be counterintuitive as pediatric acute leukemia cells do express appreciable levels of immunoproteasomes including β5i, the main target of ONX 0914. Hence, this result appears to be consistent with the outcome of a study by Parlati et al.14 indicating that beyond the inhibition of the β5i subunit, inhibition of additional subunits including β5 and β1, is required to elicit a superior anti-leukemic response.
Several attempts have been made in tumor cell line models to correlate differential bortezomib sensitivity to proteasome expression levels. Busse et al.47 showed that bortezomib-resistant solid tumor cell lines expressed lower levels of immunoproteasome than the more sensitive hematologic B-cell lines. Moreover, all cell lines classified as most bortezomib-resistant showed low β1i and/or β2i mRNA expression compared to bortezomib-sensitive cell lines. Hematologic cell lines with acquired resistance to bortezomib were also characterized with increased levels of constitutive proteasome levels and reduced levels of immunoproteasome.20,22 In two clinical studies, Matondo et al.48 suggested a relationship between 20S proteasome levels and sensitivity to proteasome inhibitors in AML patient samples, although no distinction between constitutive- and immunoproteasome subunit expression was made. Shuqing et al.37 described mRNA overexpression of β5 in a single MM patient (without a mutation in PSMB5) with clinical resistance to bortezomib-containing therapy compared to 3 sensitive patients. Together, these findings would imply that leukemic patients harboring higher constitutive (and lower immunoproteasome) subunit expression before bortezomib treatment would display a poorer response to bortezomib-based treatment than patients who have lower constitutive proteasome subunit expression. In the present study, ProCISE and Western blot analysis allowed the correlation of ex vivo drug-sensitivity to constitutive- and immunoproteasome subunit expression in ALL and AML cells.
Although total proteasome levels did not differ significantly between ALL and AML samples, we found that ALL cells had significantly lower expression of constitutive proteasome subunits and higher β1i expression than AML cells. Consistent with the above concept, increased bortezomib- and carfilzomib-sensitivity of AML cells correlated with lower β5 expression and with higher β5i expression. These correlations were further corroborated for ONX 0914 by demonstrating that sensitivity to this drug was observed with increasing ratios of immuno/constitutive proteasome in ALL. Since the ALL samples were all relatively sensitive for bortezomib and carfilzomib, no significant correlations were revealed with these drugs. The ex vivo part of this study has, however, some limitations such as relatively low numbers of patients per subgroup while performing many correlation analyses and the lack of mechanistic work due to the limited number of cells available. Notwithstanding these facts, several mechanistic data support the notion that modulation of expression levels of individual β5i and β5 subunits has an impact on immunoproteasome inhibitor sensitivity, e.g. β5i knockdown in THP1 cells abrogates sensitivity to ONX 0914 (Online Supplementary Figure S5) whereas interferon-γ induced upregulation of β5i sensitized for ONX 0914,49 and β5 knockdown in THP1 cells increased bortezomib sensitivity.22
The establishment of whether the immuno/constitutive proteasome ratio represents an additional contributing factor in proteasome inhibitor response would deserve further investigations in larger patient cohorts. It will be critical to further decipher the mechanisms regulating proteasome homeostasis, and in particular the equilibrium between the assembly of the immunoproteasome versus constitutive proteasome in AML and ALL (subgroups). Furthermore, it is important to mention that in this study we refer to the classical definition of immunoproteasomes as those composed of β5i+β1i+β2i subunits, but acknowledge that an unknown fraction of β1i and β5i subunits can be assembled in hybrid proteasome forms.12 It will be interesting to explore how these proteasome variants contribute to inhibition profiles by proteasome inhibitors. In this context, it is interesting to note that immunoproteasome rather than constitutive proteasome levels were dictating total proteasome levels in AML and ALL cells, which may provide a mechanistic rationale50 for the targeting of immunoproteasomes in order to disrupt proteasome homeostasis and elicit an anti-leukemic response.
In conclusion, bortezomib displayed potent cytotoxic effects against pediatric ALL and AML cells. This pharmacological efficacy was further enhanced in combination with dexamethasone, eliciting additive or synergistic effects. ALL cells were intrinsically more sensitive to proteasome inhibitors than AML cells, for which higher ratios of immuno/constitutive proteasome was an accountable factor. Thus, for next generation proteasome inhibitors including immunoproteasome inhibitors, these findings may hold promise in the future treatment of pediatric leukemia by avoiding toxicity of bortezomib, circumvention of bortezomib resistance and further assessment of their synergistic effect when combined with other drugs including glucocorticoids.
Footnotes
The Online version of this paper has a Supplementary Appendix.
Funding
YG Assaraf is recipient of a visiting professor fellowship of the Royal Netherlands Academy of Arts and Sciences. This study was supported (in part) by research funding from the Netherlands Organization for Scientific Research (YGA), KiKa (Children cancer-free) (GJKL and YGA) and ZonMW (NEF).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148(4): 727–38 [DOI] [PubMed] [Google Scholar]
- 2.Kuhn DJ, Orlowski RZ. The immunoproteasome as a target in hematologic malignancies. Semin Hematol. 2012;49(3): 258–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15(7): 781–7 [DOI] [PubMed] [Google Scholar]
- 4.Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14(6): 1649–57 [DOI] [PubMed] [Google Scholar]
- 5.Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A, et al. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120(5): 947–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pui CH, Mullighan CG, Evans WE, Relling MV. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120(6): 1165–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaspers GJ, Veerman AJ, Pieters R, Van Zantwijk CH, Smets LA, Van Wering ER, et al. In vitro cellular drug resistance and prognosis in newly diagnosed childhood acute lymphoblastic leukemia. Blood. 1997;90(7): 2723–9 [PubMed] [Google Scholar]
- 8.Cortes J, Thomas D, Koller C, Giles F, Estey E, Faderl S, et al. Phase I study of bortezomib in refractory or relapsed acute leukemias. Clin Cancer Res. 2004;10(10): 3371–6 [DOI] [PubMed] [Google Scholar]
- 9.Horton TM, Pati D, Plon SE, Thompson PA, Bomgaars LR, Adamson PC, et al. A phase 1 study of the proteasome inhibitor bortezomib in pediatric patients with refractory leukemia: a Children’s Oncology Group study. Clin Cancer Res. 2007;13(5): 1516–22 [DOI] [PubMed] [Google Scholar]
- 10.Messinger YH, Gaynon PS, Sposto R, Van der GJ, Eckroth E, Malvar J, et al. Bortezomib with chemotherapy is highly active in advanced B-precursor acute lymphoblastic leukemia: Therapeutic Advances in Childhood Leukemia & Lymphoma (TACL) Study. Blood. 2012;120(2): 285–90 [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6(1): 79–87 [DOI] [PubMed] [Google Scholar]
- 12.Vigneron N, Van den Eynde BJ. Proteasome subtypes and the processing of tumor antigens: increasing antigenic diversity. Curr Opin Hematol. 2012;24(1): 84–91 [DOI] [PubMed] [Google Scholar]
- 13.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142(4): 613–24 [DOI] [PubMed] [Google Scholar]
- 14.Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB, et al. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114(16): 3439–47 [DOI] [PubMed] [Google Scholar]
- 15.Roccaro AM, Sacco A, Aujay M, Ngo HT, Azab AK, Azab F, et al. Selective inhibition of chymotrypsin-like activity of the immunoproteasome and constitutive proteasome in Waldenstrom macroglobulinemia. Blood. 2010;115(20): 4051–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton TM, Gannavarapu A, Blaney SM, D’Argenio DZ, Plon SE, Berg SL. Bortezomib interactions with chemotherapy agents in acute leukemia in vitro. Cancer Chemother Pharmacol. 2006;58(1): 13–23 [DOI] [PubMed] [Google Scholar]
- 17.Houghton PJ, Morton CL, Kolb EA, Lock R, Carol H, Reynolds CP, et al. Initial testing (stage 1) of the proteasome inhibitor bortezomib by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50(1): 37–45 [DOI] [PubMed] [Google Scholar]
- 18.Attar EC, De Angelo DJ, Supko JG, D’Amato F, Zahrieh D, Sirulnik A, et al. Phase I and pharmacokinetic study of bortezomib in combination with idarubicin and cytarabine in patients with acute myelogenous leukemia. Clin Cancer Res. 2008;14(5): 1446–54 [DOI] [PubMed] [Google Scholar]
- 19.Niewerth D, Dingjan I, Cloos J, Jansen G, Kaspers G. Proteasome inhibitors in acute leukemia. Expert Rev Anticancer Ther. 2013;13(3): 327–37 [DOI] [PubMed] [Google Scholar]
- 20.Franke NE, Niewerth D, Assaraf YG, Van Meerloo J, Vojtekova K, Van Zantwijk CH, et al. Impaired bortezomib binding to mutant beta5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cells. Leukemia. 2012;26 (4): 757–68 [DOI] [PubMed] [Google Scholar]
- 21.Kale AJ, Moore BS. Molecular Mechanisms of Acquired Proteasome Inhibitor Resistance. J Med Chem. 2012;55(23): 10317–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oerlemans R, Franke NE, Assaraf YG, Cloos J, Van Zantwijk I, Berkers CR, et al. Molecular basis of bortezomib resistance: proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112(6): 2489–99 [DOI] [PubMed] [Google Scholar]
- 23.Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103(13): 1007–17 [DOI] [PubMed] [Google Scholar]
- 24.Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67(13): 6383–91 [DOI] [PubMed] [Google Scholar]
- 25.Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8(5): 407–19 [DOI] [PubMed] [Google Scholar]
- 26.Li X, Wood TE, Sprangers R, Jansen G, Franke NE, Mao X, et al. Effect of noncompetitive proteasome inhibition on bortezomib resistance. J Natl Cancer Inst. 2010;102(14): 1069–82 [DOI] [PubMed] [Google Scholar]
- 27.Van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol. 2011;731:237–45 [DOI] [PubMed] [Google Scholar]
- 28.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55 [DOI] [PubMed] [Google Scholar]
- 29.Suzuki E, Demo S, Deu E, Keats J, Rastu-Kapur S, Bergsagel PL, et al. Molecular mechanisms of bortezomib resistant adeno-carcinoma cells. PLoS One. 2011;6(12): e27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieters R, Den Boer ML, Durian M, Janka G, Schmiegelow K, Kaspers GJ, et al. Relation between age, immunophenotype and in vitro drug resistance in 395 children with acute lymphoblastic leukemia--implications for treatment of infants. Leukemia. 1998;12 (9): 1344–8 [DOI] [PubMed] [Google Scholar]
- 31.Kaspers GJ, Kardos G, Pieters R, Van Zantwijk CH, Klumper E, Hahlen K, et al. Different cellular drug resistance profiles in childhood lymphoblastic and non-lymphoblastic leukemia: a preliminary report. Leukemia. 1994;8(7): 1224–9 [PubMed] [Google Scholar]
- 32.Balsas P, Galan-Malo P, Marzo I, Naval J. Bortezomib resistance in a myeloma cell line is associated to PSM 5 overexpression and polyploidy. Leuk Res. 2012;36(2): 212–8 [DOI] [PubMed] [Google Scholar]
- 33.Ruckrich T, Kraus M, Gogel J, Beck A, Ovaa H, Verdoes M, et al. Characterization of the ubiquitin-proteasome system in bortezomib-adapted cells. Leukemia. 2009;23(6): 1098–105 [DOI] [PubMed] [Google Scholar]
- 34.Ri M, Iida S, Nakashima T, Miyazaki H, Mori F, Ito A, et al. Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia. 2010;24(8): 1506–12 [DOI] [PubMed] [Google Scholar]
- 35.Lu S, Yang J, Song X, Gong S, Zhou H, Guo L, et al. Point mutation of the proteasome beta5 subunit gene is an important mechanism of bortezomib resistance in bortezomib-selected variants of Jurkat T cell lymphoblastic lymphoma/leukemia line. J Pharmacol Exp Ther. 2008;326(2): 423–31 [DOI] [PubMed] [Google Scholar]
- 36.Lichter DI, Danaee H, Pickard MD, Tayber O, Sintchak M, Shi H, et al. Sequence analysis of -subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood. 2012;120(23): 4513–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuqing L, Jianmin Y, Chongmei H, Hui C, Wang J. Upregulated expression of the PSMB5 gene may contribute to drug resistance in patient with multiple myeloma when treated with bortezomib-based regimen. Exp Hematol. 2011;39(12): 1117–8 [DOI] [PubMed] [Google Scholar]
- 38.Kaspers GJ, Veerman AJ, Pieters R, Van Zantwijk I, Hahlen K, Van Wering ER. Drug combination testing in acute lymphoblastic leukemia using the MTT assay. Leuk Res. 1995;19(3): 175–81 [DOI] [PubMed] [Google Scholar]
- 39.Szczepanek J, Pogorzala M, Konatkowska B, Juraszewska E, Badowska W, Olejnik I, et al. Differential ex vivo activity of bortezomib in newly diagnosed paediatric acute lymphoblastic and myeloblastic leukaemia. Anticancer Res. 2010;30(6): 2119–24 [PubMed] [Google Scholar]
- 40.Kaspers GJ, Pieters R, Klumper E, De Waal FC, Veerman AJ. Glucocorticoid resistance in childhood leukemia. Leuk Lymphoma. 1994;13(3–4): 187–201 [DOI] [PubMed] [Google Scholar]
- 41.Ma MH, Yang HH, Parker K, Manyak S, Friedman JM, Altamirano C, et al. The proteasome inhibitor PS-341 markedly enhances sensitivity of multiple myeloma tumor cells to chemotherapeutic agents. Clin Cancer Res. 2003;9(3): 1136–44 [PubMed] [Google Scholar]
- 42.Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98(8): 2301–7 [DOI] [PubMed] [Google Scholar]
- 43.Bentires-Alj M, Barbu V, Fillet M, Chariot A, Relic B, Jacobs N, et al. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene. 2003;22(1): 90–7 [DOI] [PubMed] [Google Scholar]
- 44.Wood P, Burgess R, MacGregor A, Yin JA. P-glycoprotein expression on acute myeloid leukaemia blast cells at diagnosis predicts response to chemotherapy and survival. Br J Haematol. 1994;87(3): 509–14 [DOI] [PubMed] [Google Scholar]
- 45.Verbrugge SE, Assaraf YG, Dijkmans BA, Scheffer GL, Al M, den Uyl D, et al. Inactivating PSMB5 mutations and P-glycoprotein (multidrug resistance-associated protein/ATP-binding cassette B1) mediate resistance to proteasome inhibitors: ex vivo efficacy of (immuno)proteasome inhibitors in mononuclear blood cells from patients with. J Pharmacol Exp Ther. 2012;341(1): 174–82 [DOI] [PubMed] [Google Scholar]
- 46.Chauhan PS, Bhushan B, Singh LC, Mishra AK, Saluja S, Mittal V, et al. Expression of genes related to multiple drug resistance and apoptosis in acute leukemia: response to induction chemotherapy. Exp Mol Pathol. 2012;92(1): 44–9 [DOI] [PubMed] [Google Scholar]
- 47.Busse A, Kraus M, Na IK, Rietz A, Scheibenbogen C, Driessen C, et al. Sensitivity of tumor cells to proteasome inhibitors is associated with expression levels and composition of proteasome subunits. Cancer. 2008;112(3): 659–70 [DOI] [PubMed] [Google Scholar]
- 48.Matondo M, Bousquet-Dubouch MP, Gallay N, Uttenweiler-Joseph S, Recher C, Payrastre B, et al. Proteasome inhibitor-induced apoptosis in acute myeloid leukemia: a correlation with the proteasome status. Leuk Res. 2010;34(4): 498–506 [DOI] [PubMed] [Google Scholar]
- 49.Niewerth D, Franke N, Jansen G, Assaraf Y, Van Meerloo J, Kirk CJ, et al. Interferon-y-induced upregulation of immunoproteasome subunit assembly overcomes bortezomib resistance of leukemia cell lines harbouring bortezomib-induced mutations in constitutive PSMB5. ASH Annual Meeting Abstracts. 2012;120(21): 1346 [Google Scholar]
- 50.Heink S, Ludwig D, Kloetzel PM, Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci USA. 2005;102(26): 9241–6 [DOI] [PMC free article] [PubMed] [Google Scholar]