Abstract
Nodal marginal zone lymphoma is a poorly defined entity in the World Health Organization classification, based largely on criteria of exclusion and the diagnosis often remains subjective. Follicular lymphoma lacking t(14;18) has similar characteristics which results in a major potential diagnostic overlap which this study aims to dissect. Four subgroups of lymphoma samples (n=56) were analyzed with high-resolution array comparative genome hybridization: nodal marginal zone lymphoma, t(14;18)-negative follicular lymphoma, localized t(14:18)-positive follicular lymphoma and disseminated t(14;18)-positive follicular lymphoma. Gains on chromosomes 7, 8 and 12 were observed in all subgroups. The mean number of aberrations was higher in disseminated t(14;18)-positive follicular lymphoma than in localized t(14:18)-positive follicular lymphoma (P<0.01) and the majority of alterations in localized t(14:18)-positive follicular lymphoma were also found in disseminated t(14;18)-positive follicular lymphoma. Nodal marginal zone lymphoma was marked by 3q gains with amplifications of four genes. A different overall pattern of aberrations was seen in t(14;18)-negative follicular lymphoma compared to t(14;18)-positive follicular lymphoma. t(14;18)-negative follicular lymphoma is characterized by specific (focal) gains on chromosome 3, as observed in nodal marginal zone lymphoma. Our results support the notion that localized t(14:18)-positive follicular lymphoma represents an early phase of disseminated t(14;18)-positive follicular lymphoma. t(14;18)-negative follicular lymphoma bears aberrations that are more like those in nodal marginal zone lymphoma, suggesting a relation between these groups.
Introduction
Correct disease classification is essential for good care of patients. In lymphoma, unraveling the molecular mechanisms of lymphomagenesis has resulted in a highly refined system, based on objective, measurable, mostly genomic and immunophenotypic aberrations. Refined classification of disease entities now allows a more reliable prediction of clinical course, prognosis and more focused treatment for the majority of the lymphoma entities. Follicular lymphoma (FL) is one of the more common types of indolent B-cell lymphomas with well-defined characteristics in the World Health Organization (WHO) classification. FL commonly presents with a t(14;18) translocation in addition to a nodular growth pattern of neoplastic follicles of CD10+, BCL6+, BCL2+ B cells, hypermutated immunoglobulin genes and typically disseminated nodal disease in elderly patients.
However, not all lymphomas have been characterized to this extent. Nodal marginal zone lymphoma (NMZL), lymphoplasmacytic lymphoma and FL that lack the characteristic t(14;18) translocation pose classification problems.
The classical morphology of NMZL covers the full spectrum from small B cells to plasma cells. The NMZL immunophenotype is mostly defined by negative features such as no expression of CD10, BCL6, CD5 or cyclin D1 and variable expression of CD23.1 Colonization of pre-existing germinal centers is often seen and may reflect the behavior of the putative normal B-cell counterpart, but is not exclusive to NMZL. Several studies have described absence of known translocations in NMZL. As part of a large series of 218 samples [mucosa-associated lymphoid tissue (MALT) lymphoma, splenic marginal zone lymphoma (MZL) and NMZL], Rinaldi et al. described molecular features of 25 cases of NMZL.2 This study underpinned the notion that the genetic background of MZL, MALT-type and splenic type are relatively distinct. For NMZL however, no single recurrent characteristic structural chromosomal aberration could be identified. Only aberrations that are also commonly found in other types of indolent B-cell lymphoma were observed.
Translocation t(14;18)-negative FL is one of the diseases that potentially overlap with NMZL to a large extent. In a study of 10 such cases, Leich et al. showed that various characteristics of t(14;18)-positive FL, including amplification of 18q, were not present in the translocation-negative cases.3 In addition, no recurrent characteristic structural chromosomal aberrations could be identified in t(14;18)-negative FL, despite large differences at the gene expression level. A variant of t(14;18)-negative FL with a diffuse growth pattern, presenting as stage I disease preferentially with a large mass in the groin, seems to be characterized by 1p36 deletion and may be a distinct subtype.4
Upon review of patients treated within the EORTC phase III study on the role of low-dose total body irradiation and involved-field radiotherapy in patients with localized, stages I and II, low grade non-Hodgkin’s lymphoma (E20971), we observed a very high number of remarkable cases with morphological features of FL, but lacking a classical t(14;18) translocation. Moreover, frequent cases with features of both FL and NMZL were found, precluding an evidence-based choice between these classes. This series formed the basis of the present explorative study aimed at dissecting the overlapping area between follicular lymphoma and nodal marginal zone B-cell lymphoma using DNA copy number analysis with high-resolution array comparative genome hybridization (array CGH).5
Methods
Selection of patients
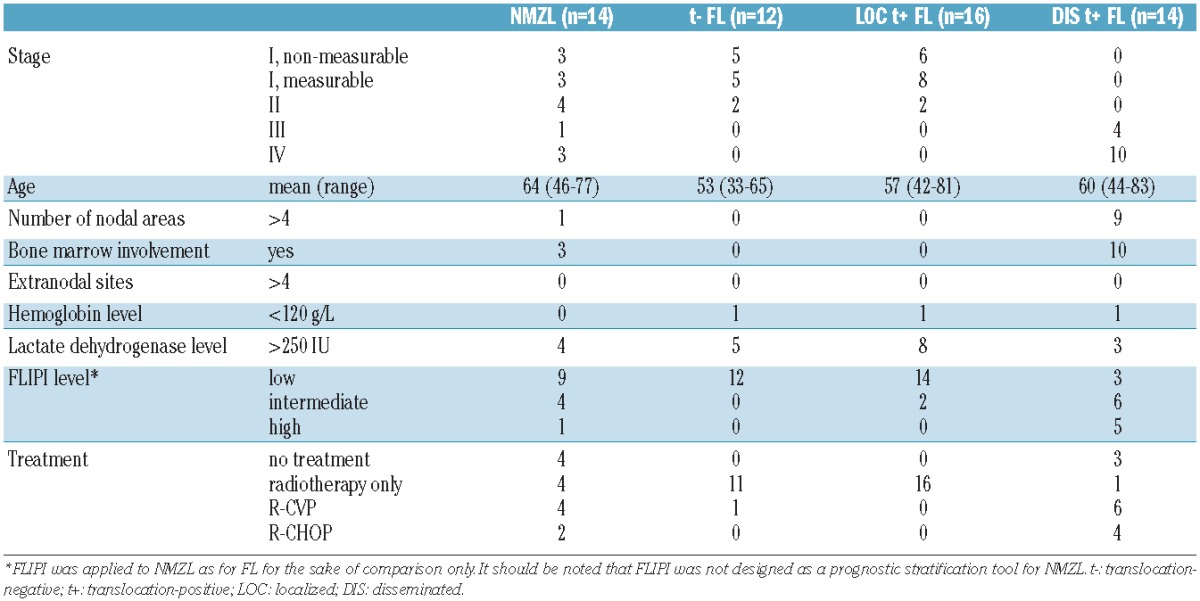
From a series of histological formalin-fixed, paraffin-embedded biopsy samples from patients treated in a randomized phase III EORTC trial (E20971) for localized indolent B-cell lymphoma and from the files of the Comprehensive Cancer Center Amsterdam Lymphoma Consult Panel, four sets were selected for this study: (i) morphologically and immunophenotypically classical NMZL (n=14); (ii) indolent B-cell lymphoma with features intermediate between FL and NMZL (translocation-negative FL) (n=12); (iii) morphologically and immunophenotypically classical FL bearing a t(14;18) translocation presenting as stage 1 or limited stage 2 disease (n=16); and (iv) morphologically and immunophenotypically classical FL bearing a t(14;18) translocation presenting as extensive disease (n=14).
All protocols for obtaining and studying human archived tissues and patients’ data were approved within the local ethical procedures at the Netherlands Cancer Institute and complied with the Code for Proper Secondary Use of Human Tissue in The Netherlands (http://www.fmwv.nl and www.federa.org).
Pathological review
All biopsy material was reviewed by two hematopathologists (DdJ, OBP) and was immunostained for at least CD20, CD79a, CD3, CD5, BCL2, BCL6, CD10, CD23, CD21, and MIB1.6 An alternative BCL2 antibody specific for another epitope was used in selected cases (SantaCruz C2, Santa Cruz Europe, Heidelberg, Germany) according to standard procedures,8 In addition, BCL2 translocation status was studied by fluorescence in situ hybridization (FISH) with BCL2 break-apart probes (DAKO, Glostrup, Denmark)7 or BCL2-IgH polymerase chain reaction (PCR). The following criteria were used to classify FL: (i) at least a section of the lymph node with unequivocal features of FL (nodular, centrocytes/centroblasts, no starry sky pattern and no macrophages in lymphoma nodes, uniformly CD10+, preferably supported by BCL6 expression); (ii) marginal zone differentiation could be present at the periphery of nodes; (iii) an interfollicular CD10+ population was considered as supportive evidence. The classification criteria for NMZL were as follows: (i) no areas of unequivocal FL as described above; (ii) the presence of reactive germinal centers or remnants thereof or neoplastic monomorphic nodes without reactive germinal centers negative for CD10 and IgD; (iii) presence of a follicular colonization pattern as distinguished by staining patterns of CD10, BCL6, CD21, CD23 and MIB1; (iv) monomorphic marginal zone B cells (“monocytoid B cells”) and plasmacytoid differentiation could be present; and (v) no t(14;18) or BCL2 break demonstrable by PCR or FISH.
Chromosomal copy number aberrations as measured by array comparative genome hybridization
Genomic DNA was isolated from formalin-fixed, paraffin-embedded tumor samples using the QIAamps DNA extraction kit (cat. 51306) as described by Beers et al.9 DNA from 68 samples was hybridized to the NimbleGen Human CGH 12×135K Whole-Genome Tilling v3.0 platform containing 134,937 in situ synthesized oligonucleotides (Roche NimbleGen, Madison, USA). Labeling was performed as previously described by Buffart et al.10
The samples were pre-processed as described by Wiel et al. using circular binary segmentation and the R-package CGHcall.11,12 To remove common germ-line copy number variants from the dataset all focal DNA copy number differences smaller than 3 Mb were compared with copy number variants in the healthy population as archived in the database of genomic variants (http://projects.tcag.ca/variation/).13–17
Further details of data pre-processing and the statistical analysis are given in the Online Supplementary Data. Raw data of all arrays are publicly available in the GEO database (accession number GSE40641).
Results
Morphological and clinical features of the selected lymphomas cases
The morphological, immunophenotypic and translocation data are summarized in Online Supplementary Table S1. All cases of NMZL and translocation-positive grade 1 and 2 FL were selected as prototypic examples, as outlined in the Methods section. NMZL showed marginal zone differentiation and a nodular pattern with uninvolved germinal centers, were negative for BCL2 (7/14), and had germinal center colonization as determined by the CD10, CD21 and BCL2 markers (9/14) and supported by additional markers such as BCL6 and MIB1. BCL2 breaks were absent in all cases. All FL that presented as disseminated disease had a nodular architecture and showed uniform expression of CD20, CD10, BCL6 and BCL2. Translocation-positive FL presenting as localized disease showed limited features of germinal center colonization in 2/16 cases, of which one case with morphological marginal zone differentiation. BCL2 breaks were not present in any case. One case of localized translocation-positive FL was negative for BCL2 protein when tested with the DAKO clone 124 antibody, but stained with the alternative antibody specific for another epitope.
The remaining cases were considered equivocal according to the pre-set criteria with discrepancies between morphological criteria such as CD10 expression and translocation status. In detail, all cases showed a uniform nodular staining pattern for CD10 and BCL2, of which 2/12 cases with the BCL2 alternative antibody only, reminiscent of FL. A varying interfollicular CD10+ component was noted (3/13). A “moth-eaten” pattern for CD10 (Online Supplementary Figure S1 and Online Supplementary Table S1) was noted in some follicles next to uniformly positive nodes in 3/12 cases, reminiscent of NMZL and follicular colonization by a CD10 weakly positive or negative population. Moreover, minor foci negative for BCL2 were seen in these germinal centers in two cases. A similar pattern was, however, seen in a case of translocation-positive FL with localized disease. By definition, a BCL2/IgH translocation could not be demonstrated using PCR in any of the 12 cases. Absence of a BCL2 break was confirmed in 8/12 cases, while FISH data could not be obtained in four cases because of insufficient availability (n=2) or quality (n=2) of the samples. Based on these combined features, these cases were classified as translocation-negative FL.
Translocation-positive FL with localized disease presented as stage I in 14/16 patients, of whom six showed no residual disease after the diagnostic lymph node excision. Translocation-negative FL with localized disease presented as stage I disease in 10/12 patients, of whom five had no residual disease after biopsy. NMZL presented as localized disease in 10/14 patients and as disseminated disease in 4/14 patients. The presenting features and distribution of FLIPI parameters were similar in NMZL and disseminated translocation-positive FL. Translocation-negative FL and localized translocation-positive FL were also relatively similar with regards to these aspects. The clinical and morphological data for all groups are summarized in Table 1.
Table 1.
Clinical data.
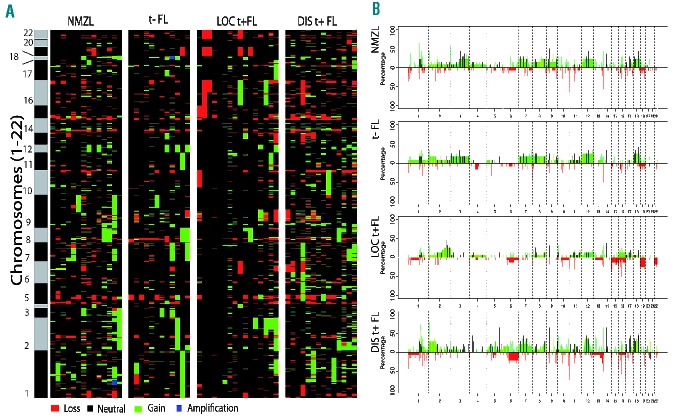
Identification of recurrent chromosomal copy number aberrations
DNA of 68 samples was hybridized to array CGH. Twelve samples did not meet the quality criteria leaving 56 samples for further analysis: NMZL (n=14), t(14;18)-negative FL (n=12), localized t(14;18)-positive FL (n=16) and disseminated t(14;18)-positive FL III/IV (n=14).
DNA copy number aberrations were observed in all groups (Figure 1). The median percentage of aberrant probes on the array was 16.3 (range, 8.9–32.5) for NMZL, 13.7 (range, 3.2–24.3) for translocation-negative FL, 13.6 (range, 0–39.9) for localized translocation-positive FL and 20.3 (range, 8.3–40.2) for disseminated translocation-positive FL, with the rate being significantly higher rate (P=0.009) in this last group compared to all other groups. A similar distribution was seen for focal chromosomal aberrations.
Figure 1.
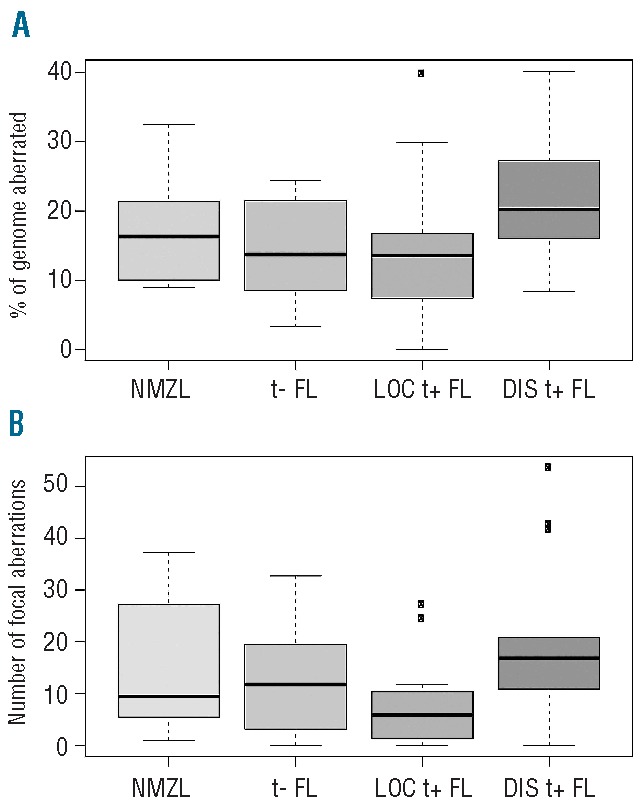
Number of aberrations in each group. (A) Percentage of the genome gained or lost and (B) number of somatic focal aberrations (<3 Mb) per group. t−: translocation-negative; t+: translocation-positive; LOC: localized; DIS: disseminated.
The frequency of chromosomal aberrations per sample and per group are illustrated in Figure 2 and listed in Table 2. Aberrations found in all groups were (partial) gains of chromosomes 8, 12 and 18. NMZL was marked additionally by gains of 3q (3/14). The fewest aberrations were detected in localized translocation-positive FL but these showed a gain at chromosome 2q in 6/16 cases with specific amplification at 2q33.2 - 2q34 (~17MB) only found sporadically in the other groups. This is a large and very gene dense genomic region, which makes it challenging to pinpoint driver genes.
Figure 2.
Heatmap (A) and frequency plot (B) of gains and losses of all analyzed samples. (A) Heatmap of aberrations of all analyzed tumors per group. Green represents a gain, red a loss. Black/gray bar represents the chromosomes. (B) Frequency plots for NMZL, translocation-negative (t−) FL, localized translocation-positive (LOC t+) FL and disseminated translocation-positive (DISt+) FL. The y-axis represents the percentage of aberrations and the x-axis shows the genomic location. Focal aberrations are indicated in black.
Table 2.
Most common aberrations per subgroup. Only aberrations found in at least 20% of the samples are shown.
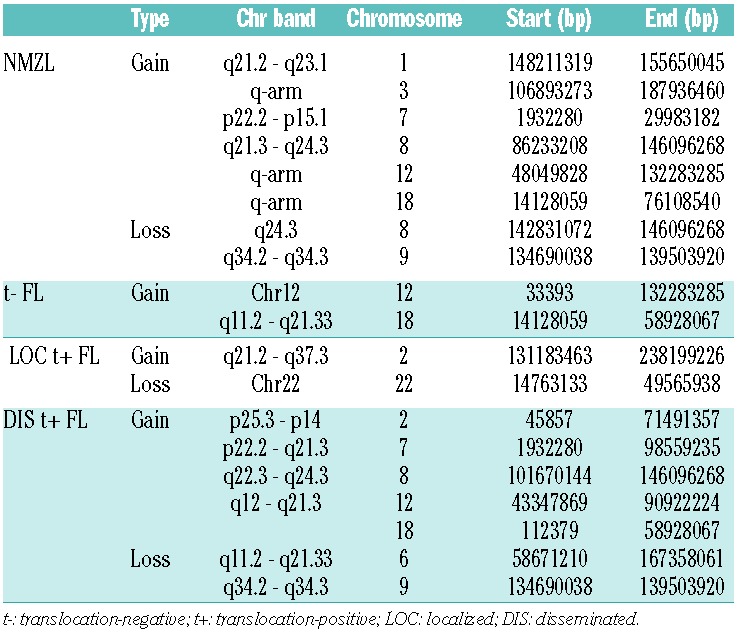
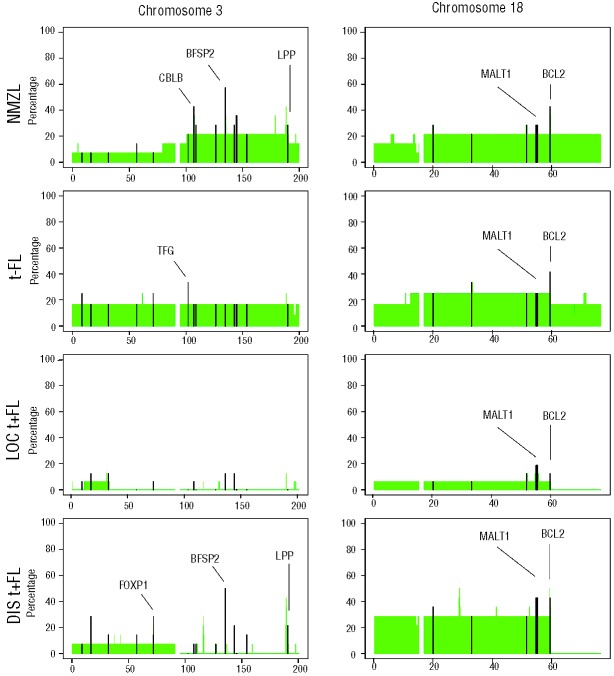
Focal chromosomal aberrations were significantly enriched for genes described in the Cancer Census list (P=0.0001, http://www.sanger.ac.uk/genetics/CGP/) and included ARHGEF12, BCL2, CBLB, CDK6, EBF1, FOXP1, IKZF1, LPP, MALT1, MSI2, RAD51L1, RANBP17, TFG, USP6 and WHSC1L1. The most frequent focal chromosomal aberrations found in all lymphoma groups were located at 14q24.1 (RAD51L1), 9p13.2 (ZCCHC7), 3q22.1 (BFSP2) and 18q21.33 (BCL2) (Table 3 and Online Supplementary Table S2). Focal chromosomal aberrations were significantly more frequently observed in disseminated translocation-positive FL (P=0.046), while the frequency in localized translocation-positive FL was significant lower (P=0.046). Chromosome 3 (more specifically 3q) was gained in 3/14 NMZL cases and in 2/12 translocation-negative FL cases, whereas no gains were observed in disseminated and localized translocation-positive FL (Figure 3). In addition, 15 focal chromosomal gains were observed at chromosome 3 of which gains on q22.1 (BFSP2), q23 (RASA2, ZBTB38), q24 (SLC9A9, CHST2) and q13.11 (CBLB) were most frequent.
Table 3.
Most common somatic focal aberrations. All focal aberrations found in at least 20% of the samples (in all subgroups combined). Gene names in bold indicate genes described in the Cancer Census List16.
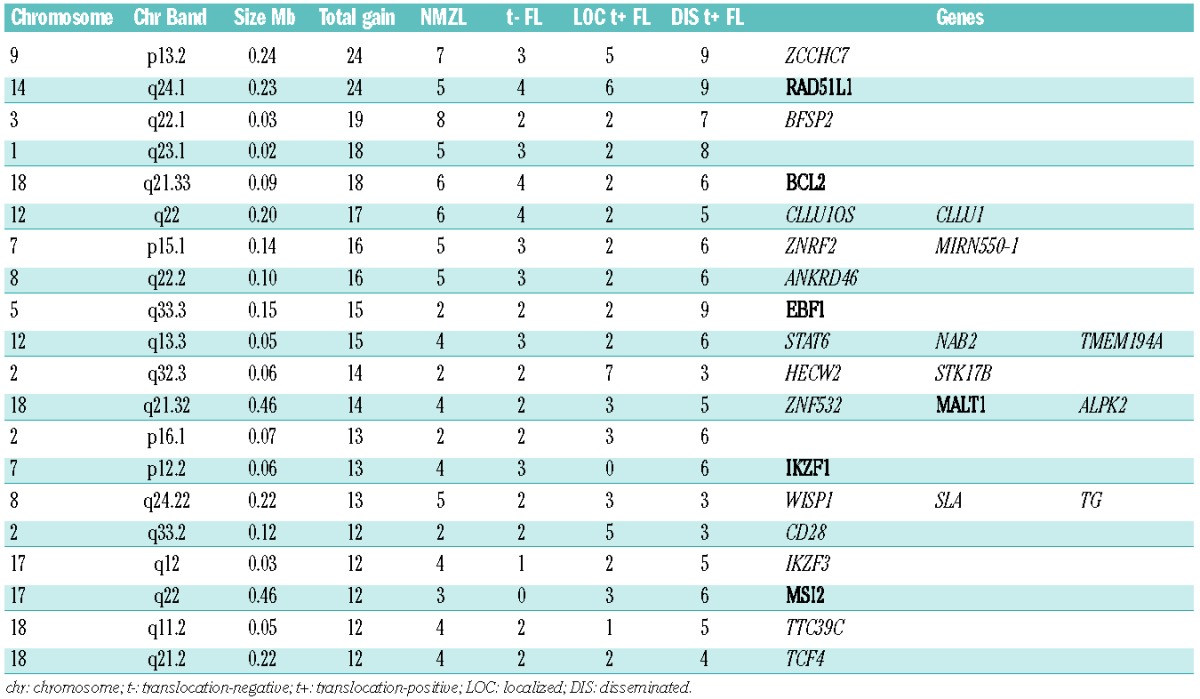
Figure 3.
Frequency plot of gains on chromosome 3 (left panel) and 18 (right panel) in NMZL, translocation-negative (t−) FL, localized translocation-positive (LOC t+) FL and disseminated translocation-positive (DIS t+) FL. The focal aberrations are indicated in black. Genes that were found altered in focal aberrations with a high frequency are added in the plot.
On chromosome 18, a recurrent breakpoint site was observed, flanking the BCL2 locus in translocation-positive FL, while in the other classes the complete chromosome 18 was gained or amplified.
At 1p36, a distinct deletion hotspot of 1.7Mb that included PRDM16, but not TNFRSF14, was observed in all classes (Online Supplementary Figure S2).
Translocation-positive localized and disseminated disease share common aberrations
There were significantly fewer aberrations in translocation-positive localized disease than in translocation-positive disseminated FL, both with regards to focal chromosomal aberrations (P=0.014) and larger deletions and amplifications (P=0.011) (Figure 1). Specific aberrations are illustrated in Figure 2. Common changes were seen at 1q23 as a gain in a small segment in localized FL (3/16 cases), while larger segments of 1q were amplified in disseminated disease (10/14 cases). A gain at 14q11 was also identified at a relatively high frequency in both localized and disseminated FL (4/16 and 9/14 patients, respectively). Gains at 6p21 were also found relatively frequently (3/16 with localized disease and 9/14 with disseminated disease). Gains at 2p23 and 17q22 were also found as a common change, albeit in only 3/16 with localized disease versus 6/14 in those with disseminated disease. The BCL2 region at 18q21 was amplified frequently in disseminated FL (6/14 cases), but in only one case of localized FL. Gains at chromosomes 2p, 3q13 and 3q27-28, 4p15-16, 5q, extensive changes on 7, 9, 10, 12q, 13q and 20q13 were (virtually) unique to disseminated FL. A remarkable difference was observed with gain of 2q in localized disease (4/16) in the absence of major aberrations in disseminated FL.
The global copy number aberrations odds ratio test (GAORT) was used to determine whether disseminated translocation-positive FL originates from localized translocation-positive FL based on the odds of aberrations. There was a significant, systematic increase in chromosomal aberrations in the disseminated form compared to the localized form (P=0.004).
Furthermore, it was noted that the relative level of numerical gains and losses of chromosomal regions differed within single cases, with more complexity of levels in disseminated FL than in localized FL, suggestive of more extensive genetic heterogeneity (Online Supplementary Figure S3).
Genes located in focal aberrations were MALT1, ALPK2, ZNF532, and RAD51L1, which were identified as genes predominantly involved in early phases of lymphomagenesis. Focal chromosomal aberrations with the genes EBF1, IKZF1, LBH, LCLAT1, CDK6, TTC39C, NCOA3, ZMYND8, and RPL35AP were predominantly detected in disseminated disease and can, therefore, be identified as genes related to later phases. Three focal aberrations (2q32.3, 2q33.2, 2q34) were found on chromosome 2q within a genomic region of 17 Mb and contained five genes, HECW2, STK17B, CD28, SPAG16 and IKZF2, which were remarkably more frequent in localized FL. Overall, these results underpin the notion that localized translocation-positive FL can be considered a precursor stage of disseminated translocation-positive FL.
Nodal marginal zone lymphoma shows a distinct spectrum of aberrations
NMZL is characterized by gains of the q-arm of chromosome 3. These characteristics gains occur in a landscape of aberrations common to all groups (Figure 2). More prominent differences between NMZL and the other groups were seen for focal chromosomal aberrations with dominant gain of CBLB (3q13.11), BBX (3q13.12), and SLC12A8 and HEG1 (3q21.2) in NMZL. Aberrations present in translocation-positive FL but not in NMZL (or only at a very low frequency) were RFTN1 and OXNAD1 (3p24.3), FOXP1 (3p14.1), EBF1 (5q33.3), NCOA3, ZMYND8 and RPL35AP (20q13.12) and ANKRD16, GD12 and C10ORF18 (10p15.1) (Online Supplementary Table S2 and Figure 3). This finding underlines that NMZL and translocation-positive FL have distinctly different tumor genetic backgrounds, but also indicates that NMZL has very few recurrent and characteristic numerical aberrations. Focal chromosomal aberrations at 3q13 may contain the most dominant candidate genes for this lymphoma.
Translocation-negative follicular lymphoma is more like nodal marginal zone lymphoma than translocation-positive follicular lymphoma
The pattern of aberrations in translocation-negative FL was characterized by complete chromosomal gains of chromosomes 3, 7, 8, 12 and 18 and gains at 1q without specific losses (Figure 2). Additional focal aberrations within these chromosomal gains were very heterogeneous. Focal aberrations most prominently indicate a gain at the BCL2 locus (5/12). The characteristic aberration seen in translocation-positive FL, including loss of 6q, specific gains at 2p, 3q13 and 3q27-28, 4p15-16, 5q as well as focal aberrations of, for example, NCOA3, ZMYND8, and RPL35AP, were virtually absent in the translocation-negative group (Figure 2). In contrast to aberrations characteristic of NMZL, various markers at chromosome 3 seemed to be specifically gained in translocation-negative FL, including SLC9A9, CHST2 and CBLB. On the other hand, genes more characteristically identified in translocation-positive FL, including FOXP1, were also seen in the translocation-negative group (Figure 3). Cluster analysis of translocation-negative FL, NMZL and disseminated translocation-positive FL yielded two groups that were significantly correlated with the subgroups (P=0.030, Figure 4). Group A contained 9/14 (64%) translocation-positive disseminated FL whereas group B contained 10/12 (83%) and 10/14 (71%) of translocation-negative FL and NMZL, respectively. The spectrum of characteristic aberrations, both larger than 3 Mb and focal, and the cluster analysis all support the notion that translocation-negative FL is more like NMZL than translocation-positive FL.
Figure 4.
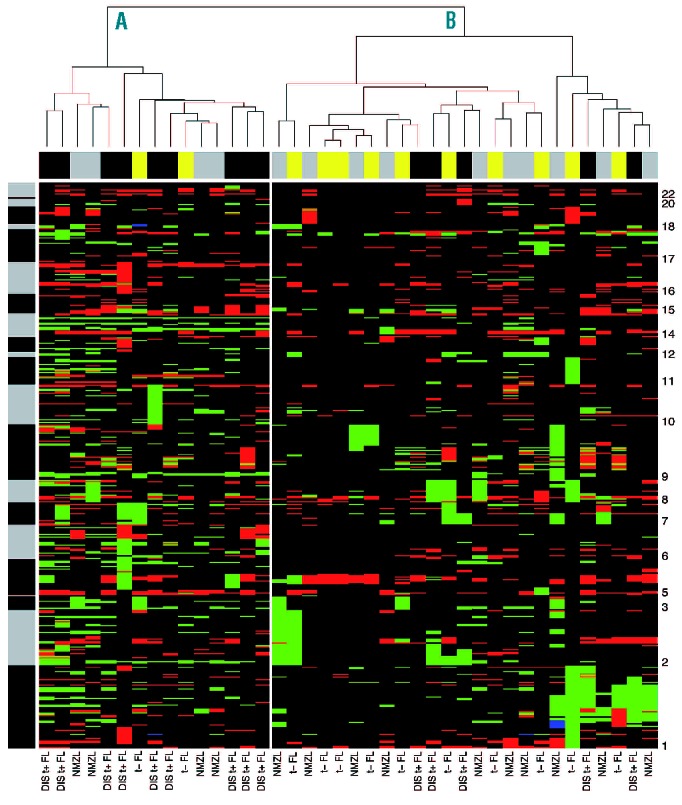
Cluster analysis of translocation-negative (t−) FL, NMZL and disseminated translocation-positive (DIS t+) FL. Hierarchical cluster analysis yielded two clusters significantly correlated with tumor types (P=0.03). Columns represent the different tumors and rows represent the different chromosomal regions, with chromosome 1 at the bottom and chromosome 22 at the top of the heatmap. DNA copy number gains and losses are indicated in green and red, respectively. The black and gray bar next to the cluster represents the chromosome separation. The bar above the heatmap represents the different subgroups, t− FL in yellow, NMZL in gray and DIS t+ FL in black.
It should be noted that the characteristic breakpoint site flanking the BCL2 locus in translocation-positive FL was observed in one translocation-negative case, suggesting misclassification, possibly due to technical reasons or an alternative translocation partner (PCR results were negative while FISH results were inconclusive).
Discussion
The WHO lymphoma classification system is based on “disease entities” that are defined by morphological, immunophenotypic, genetic as well as clinical criteria. Although FL is a prime example of a well-defined entity, it does have various undefined borders. In this study, we explored the borderlands to NMZL as well as the genetic make-up of the clinical exception of localized disease in FL using array CGH.
FL generally presents as disseminated disease and localized disease is seen in only 10–15% of patients at presentation.18 To define the genetic aberrations within the clinical spectrum of FL, a series of t(14;18)-positive FL with localized and disseminated disease was studied. The mean number of copy number aberrations was significantly higher in disseminated disease than in localized disease and the majority of specific aberrations present in localized disease were also found in disseminated disease, supporting the notion that localized disease might represent an early phase of the same entity. Specifically, gains at 1q23, 14q11, 6p21 were relatively frequent and gains at the BCL2 locus were somewhat less frequently seen, all suggestive of early aberrations in the clonal evolution of FL. Previous studies by Hoglund et al. and d’Amore et al. using computational modeling have also implicated gains at 1q and at the BCL2 locus (+der18) and dup6p as early events, supporting this view.19,20 However, our data do not support gains of chromosomes 7 and 8 as being common early events. A deletion at 1p36 was detected as a frequent, but not FL-specific aberration, and surprisingly did not include the previously reported TNFRSF14.21 This specific gene is a focus of uniparental disomy, which cannot be detected with the currently used technique.21–23 A 17 Mb amplification of 2q33.2-2q34 seems relatively specific for localized disease. This is a very gene-dense area, which makes driver gene identification challenging. Various specific focal chromosomal amplifications were found in this region, of which the hematopoietic-specific transcription factor IKZF2 may be the most interesting.
To define the genetic spectrum of NMZL, “prototypic” cases were carefully selected, fulfilling criteria of: classical marginal zone morphology, lack of CD10 and BCL6 expression and no class-specific translocations. Furthermore, cases with splenomegaly or MALT-type extranodal localizations were excluded. Alongside frequent aberrations shared with FL, such as gains of chromosomes 7, 8, 12 and 18 and loss of 9q, NMZL was characterized by more specific gains at 3q. Specific amplifications were seen for CBLB (3q13.11), BBX (3q13.12) and SLC12A8 and HEG1 (3q21.2). Of these, the proto-oncogene CBLB may be of interest as RING-type E3 ubiquitin ligase functions as a negative regulator of T-cell activation and has been described incidentally in hematologic malignancies.24BCL6 at 3q was not included in a specific focal aberration. In FL, gains on chromosome 18 were characterized by a sharp break at the BCL2 locus as is typically seen at translocation sites (Figure 2). In contrast, no such pattern on chromosome 18 was present in NMZL, consistent with the absence of a BCL2 translocation in this disease and neither was this seen at any other chromosome region.
In an integrative study, Arribas et al. showed that the NMZL gene-expression profile was dominated by microenvironment-related immunological factors, supporting the notion that antigen stimulation and immunological growth support rather than primary genetic aberrations may play an important role in the oncogenesis of NMZL, as in FL.25–28 Indeed, integrated analysis of copy-number and gene-expression analysis indicated very few, if any, potentially driving genes.
Neither Rinaldi et al. in a series of 25 NMZL nor Braggio et al. in a series of 20 NMZL could identify any dominant, reproducible aberrations in these lymphomas, in contrast to the situation in MALT-type lymphoma and splenic MZL. Only chromosome 3 gains were identified at high frequencies (24% and 15%), validating the observations in our study. Mostly, this involved the complete chromosome with a minimum region of 3q and no major differences were seen between NMZL and other marginal zone B-cell-related malignancies. In our series, specific deletion of TNFAIP3 (A20, 6q21–25) could not be confirmed, as this loss was present in only one case of NMZL, but also in five cases of (translocation-positive) FL.2,29 Braggio et al. also confirmed that loss of TNFAIP3 is not specific to NMZL since it is also found in splenic and MALT-type MZL.33 Selection of cases considered to be NMZL remains very difficult and often subjective, which may explain discrepancies in study results.
Translocation-negative FL may be considered as the major subjectively overlapping class with NMZL. Using strict pre-set definitions, we selected a series of cases that lacked a BCL2 break, but showed the morphological and immunophenotypic characteristics of FL. Interestingly, in a few cases, signs of possible germinal center colonization, reminiscent of NMZL, were seen. This phenomenon was also observed in one case of translocation-positive FL. Since these cases all concerned localized disease, the colonization pattern may be related to this form. In our series, translocation-negative FL showed a rather different overall pattern of large and focal aberrations as compared to the translocation-positive group. In comparison to translocation-positive FL, the characteristic translocation-related break of the amplification on 18q was not present and other aberrations were not class-specific, such as whole chromosomal gains of chromosomes 7, 8 and 12, which are the most common aberrations in all classes of B-cell lymphoma. The dominant aberration on chromosome 3 is, however, quite typical of the group of MZL. More specific focal gains related to NMZL, as seen in our series or as reported by others, were noted with a distinct amplification of BCL2 in 50% of the cases, three complete chromosomal gains, two focal gains as well as characteristic, similar gains at SLC9A9, CHST2, CBLB and FOXP1. These amplifications, together with the common aberrations on chromosome 3 may be an argument for a relation between translocation-negative FL and NMZL. Amplification of BCL2 has been reported at different rates by others, again most likely highlighting the influence of case selection, despite identical selection criteria.3,30 It should be noted that our series did not include any cases of inguinal, diffuse-type FL, described by Katzenberger et al. or cases with an aberrant phenotype (CD10−/BCL2−/ MUM1+) and/or a diffuse growth pattern or cases of FL grade 3B, which have all been suggested to bear different and possibly specific alterations.4,30–32,34 Indeed, in a study by Tagawa et al., the CD10−/BCL2−/MUM1+ cases with predominantly blastic morphology (grades 3A and 3B) were considered translocation-negative FL and were characterized by a high frequency of trisomy 3.34 This is a different selection from ours, which was restricted to CD10+ FL grade 1–2.
In conclusion, classical, translocation-positive FL is characterized by a relatively specific spectrum of genetic aberrations that are also reflected in early stage disease. So-called translocation-negative FL bears aberrations that are reminiscent of NMZL rather than FL. This is also reflected in cluster analysis, in which translocation-positive FL clusters predominantly together with NMZL. The spectrum of aberrations of NMZL is less specific and diagnostic criteria between NMZL and translocation-negative FL are relatively subjective, precluding full comparison between various studies and preventing definitive conclusions from being drawn.
Acknowledgments
OK is supported by the VUMC-CCA (VU University Medical Center, Cancer Center Amsterdam).
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (ed 4th). Lyon, France: IARC Press; 2008 [Google Scholar]
- 2.Rinaldi A, Mian M, Chigrinova E, Arcaini L, Bhagat G, Novak U, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood. 2011;117(5): 1595–604 [DOI] [PubMed] [Google Scholar]
- 3.Leich E, Salaverria I, Bea S, Zettl A, Wright G, Moreno V, et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood. 2009;114(4): 826–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katzenberger T, Kalla J, Leich E, Stöcklein H, Hartmann E, Barnickel S, et al. A distinctive subtype of t(14;18)-negative nodal follicular non-Hodgkin lymphoma characterized by a predominantly diffuse growth pattern and deletions in the chromosomal region 1p36. Blood. 2009;113(5): 1053–61 [DOI] [PubMed] [Google Scholar]
- 5.Costa JL, Meijer G, Ylstra B, Caldas C. Array comparative genomic hybridization copy number profiling: a new tool for translational research in solid malignancies. Semin Radiat Oncol. 2008;18(2): 98–104 [DOI] [PubMed] [Google Scholar]
- 6.Fenton JA, Vaandrager JW, Aarts WM, Bende RJ, Heering K, van Dijk M, et al. Follicular lymphoma with a novel t(14;18) breakpoint involving the immunoglobulin heavy chainswitch mu region indicates an origin from germinal center B cells. Blood. 2002;99(2): 716–8 [DOI] [PubMed] [Google Scholar]
- 7.Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8(2): 141–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schraders M, de Jong D, Kluin P, Groenen P, van Krieken H. Lack of Bcl-2 expression in follicular lymphoma may be caused by mutations in the BCL2 gene or by absence of the t(14;18) translocation. J Pathol. 2005;205(3): 329–35 [DOI] [PubMed] [Google Scholar]
- 9.van Beers EH, Joosse SA, Ligtenberg MJ, Fles R, Hogervorst FB, Verhoef S, et al. A multiplex PCR predictor for aCGH success of FFPE samples. Br J Cancer. 2006;94(2): 333–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buffart TE, Israeli D, Tijssen M, Vosse SJ, Mrsić A, Meijer GA, et al. Across array comparative genomic hybridization: a strategy to reduce reference channel hybridizations. Genes Chromosomes Cancer. 2008;47(11): 994–1004 [DOI] [PubMed] [Google Scholar]
- 11.van de Wiel MA, Picard F, van Wieringen WN, Ylstra B. Preprocessing and downstream analysis of microarray DNA copy number profiles. Brief Bioinform. 2011;12 (1): 10–21 [DOI] [PubMed] [Google Scholar]
- 12.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23(6): 657–63 [DOI] [PubMed] [Google Scholar]
- 13.Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105(42): 16224–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bierkens M, Krijgsman O, Wilting SM, Bosch L, Jaspers A, Meijer GA, et al. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes Chromosomes Cancer. 2013;52(1): 56–68 [DOI] [PubMed] [Google Scholar]
- 15.Brosens RP, Haan JC, Carvalho B, Rustenburg F, Grabsch H, Quirke P, et al. Candidate driver genes in focal chromosomal aberrations of stage II colon cancer. J Pathol. 2010;221(4): 411–24 [DOI] [PubMed] [Google Scholar]
- 16.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3): 177–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464 (7289): 704–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiddemann W, Buske C, Dreyling M, Weigert O, Lenz G, Forstpointner R, et al. Treatment strategies in follicular lymphomas: current status and future perspectives. J Clin Oncol. 2005;23(26): 6394–9 [DOI] [PubMed] [Google Scholar]
- 19.Höglund M, Sehn L, Connors JM, Gascoyne RD, Siebert R, Säll T, et al. Identification of cytogenetic subgroups and karyotypic pathways of clonal evolution in follicular lymphomas. Genes Chromosomes Cancer. 2004;39(3): 195–204 [DOI] [PubMed] [Google Scholar]
- 20.d’Amore F, Chan E, Iqbal J, Geng H, Young K, Xiao L, et al. Clonal evolution in t(14;18)-positive follicular lymphoma, evidence for multiple common pathways, and frequent parallel clonal evolution. Clin Cancer Res. 2008;14(22): 7180–7 [DOI] [PubMed] [Google Scholar]
- 21.Cheung KJ, Johnson NA, Affleck JG, Severson T, Steidl C, Ben-Neriah S, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70(22): 9166–74 [DOI] [PubMed] [Google Scholar]
- 22.O’Shea D, O’Riain C, Gupta M, Waters R, Yang Y, Wrench D, et al. Regions of acquired uniparental disomy at diagnosis of follicular lymphoma are associated with both overall survival and risk of transformation. Blood. 2009;113(10): 2298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13(16): 4777–85 [DOI] [PubMed] [Google Scholar]
- 24.Makishima H, Jankowska AM, McDevitt MA, O’Keefe C, Dujardin S, Cazzolli H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117(21): e198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arribas AJ, Campos-Martín Y, Gómez-Abad C, Algara P, Sánchez-Beato M, Rodriguez-Pinilla MS, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119(3): e9–e21 [DOI] [PubMed] [Google Scholar]
- 26.Glas AM, Kersten MJ, Delahaye LJ, Witteveen AT, Kibbelaar RE, Velds A, et al. Gene expression profiling in follicular lymphoma to assess clinical aggressiveness and to guide the choice of treatment. Blood. 2005;105(1): 301–7 [DOI] [PubMed] [Google Scholar]
- 27.Glas AM, Knoops L, Delahaye L, Kersten MJ, Kibbelaar RE, Wessels LA, et al. Gene-expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007;25(4): 390–8 [DOI] [PubMed] [Google Scholar]
- 28.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21): 2159–69 [DOI] [PubMed] [Google Scholar]
- 29.Novak AJ, Slager SL, Fredericksen ZS, Wang AH, Manske MM, Ziesmer S, et al. Genetic variation in B-cell-activating factor is associated with an increased risk of developing B-cell non-Hodgkin lymphoma. Cancer Res. 2009;69(10): 4217–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horsman DE, Okamoto I, Ludkovski O, Le N, Harder L, Gesk S, et al. Follicular lymphoma lacking the t(14;18)(q32;q21): identification of two disease subtypes. Br J Haematol. 2003;120(3): 424–33 [DOI] [PubMed] [Google Scholar]
- 31.Jardin F, Gaulard P, Contentin N, Lepretre S, Lenanin P, Stamatoullas A, et al. Follicular lymphoma without t(14;18) and with BCL6 rearrangements: a lymphoma subtype with distinct pathological, molecular and clinical characteristics. Leukemia. 2002;16(11): 2309–17 [DOI] [PubMed] [Google Scholar]
- 32.Karube K, Guo Y, Suzymiya Y, Nomura Y, Yamamoto K, Shimizu K, et al. CD10-MUM1+ follicular lymphoma lacks BCL2 gene translcation and shows characteristic biologic and clinical features. Blood. 2007;109(7): 3076–9 [DOI] [PubMed] [Google Scholar]
- 33.Braggio E, Dogan A, Keats JJ, Chng WJ, Huang G, Matthews JM, et al. Genomic analysis of marginal zone and lymphoplasmacytic lymphomas identified common and disease-specific abnormalities. Mod Pathol. 2012;25(5): 651–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagawa H, Karube K, Guo Y, Takeshita M, Kikuchi M, Morishima Y, et al. Trisomy 3 is a specific genomic aberration of t(14;18) negative follicular lymphoma. Leukemia 2007;21:2549–51 [DOI] [PubMed] [Google Scholar]