Abstract
Clinical and genetic heterogeneity renders confirmation or exclusion of autoimmune lymphoproliferative syndrome difficult. To re-evaluate and improve the currently suggested diagnostic approach to patients with suspected FAS mutation, the most frequent cause of autoimmune lymphoproliferative syndrome, we prospectively determined 11 biomarkers in 163 patients with splenomegaly or lymphadenopathy and presumed or proven autoimmune cytopenia(s). Among 98 patients sequenced for FAS mutations in CD3+TCRα/β+CD4−CD8− “double negative” T cells, 32 had germline and six had somatic FAS mutations. The best a priori predictor of FAS mutations was the combination of vitamin B12 and soluble FAS ligand (cut-offs 1255 pg/mL and 559 pg/mL, respectively), which had a positive predictive value of 92% and a negative predictive value of 97%. We used these data to develop a web-based probability calculator for FAS mutations using the three most discriminatory biomarkers (vitamin B12, soluble FAS ligand, interleukin-10) of the 11 tested. Since more than 60% of patients with lymphoproliferation and autoimmune cytopenia(s) in our cohort did not harbor FAS mutations, 15% had somatic FAS mutations, and the predictive value of double-negative T-cell values was rather low (positive and negative predictive values of 61% and 77%, respectively), we argue that the previously suggested diagnostic algorithm based on determination of double-negative T cells and germline FAS sequencing, followed by biomarker analysis, is not efficient. We propose vitamin B12 and soluble FAS ligand assessment as the initial diagnostic step with subsequent decision on FAS sequencing supported by a probability-calculating tool.
Introduction
Splenomegaly and/or chronic lymphadenopathy (lymphoproliferation) and autoimmune cytopenias are common clinical problems in different immunodeficiencies and constitute a diagnostic challenge.1 A key differential diagnosis in patients with these clinical manifestations is autoimmune lymphoproliferative syndrome (ALPS),2 which is caused by defects in the extrinsic FAS-mediated apoptosis pathway.3,4 Due to advances in the immunological and genetic understanding of this disease,5–8 the diagnostic criteria and classification have recently been revised.9 A definitive diagnosis of ALPS requires chronic lymphoproliferation and raised levels of CD3+TCRα/β+CD4−CD8− double-negative T (DNT) cells plus one primary accessory criterion, i.e. either defective FAS-mediated T-cell apoptosis in vitro or a somatic or germline mutation in the genes encoding FAS (ALPS-FAS and ALPS-sFAS), FAS ligand (ALPS-FASLG) or caspase 10 (ALPS-CASP10). Patients who fulfill ALPS diagnostic criteria with defective in vitro FAS-induced apoptosis, in whom no causative mutation can be identified, are classified as ALPS-U. This leaves a significant proportion of patients with lymphoproliferation and autoimmunity unclassified (ALPS-phenotype: lymphoproliferation with or without autoimmune cytopenia(s), raised DNT but normal apoptosis). The latter two groups of patients may include rare cases with ALPS related diseases including caspase 8 deficiency state and RAS-associated autoimmune leukoproliferative disease.
Recently, a number of biomarkers including soluble FAS ligand (sFASL), interleukin-10 (IL-10) and vitamin B12 were shown to be specifically altered in ALPS patients10,11 with a high positive predictive value (PPV) for the presence of FAS mutations.12 Consequently, as secondary accessory diagnostic criteria for ALPS, these markers have become part of a suggested algorithm for the diagnostic work-up of patients with suspected ALPS. This algorithm recommends FAS germline sequencing in all patients with lymphoproliferation and elevated DNT, while further analysis for somatic FAS mutations depends on the bio-marker profile.9
Despite these important advances, a number of questions remain unresolved. In the study by Caminha et al. at the National Institutes of Health (NIH)12, the utility of bio-markers was retrospectively evaluated in a large cohort of ALPS-FAS, ALPS-sFAS, ALPS-U, ALPS-phenotype patients as well as mutation-positive and -negative healthy relatives. It remains to be shown whether prospective analysis of an independent cohort can confirm the high predictive value of the biomarkers. Patients were selected based on the criteria of chronic lymphoproliferation and raised DNT and thus the selection was biased by a priori inclusion of raised DNT as a biomarker. It therefore remains unclear how the biomarkers perform in the initial laboratory evaluation of unselected patients with lymphoproliferation and autoimmune cytopenia(s). Moreover, a number of additional biological parameters such as memory B cells, soluble CD25 (sCD25), expression of B220 on DNT and serum lipids show characteristic alterations in ALPS, but have so far not been evaluated. If their high discriminatory value can be confirmed, this may allow biomarkers to be used for initial screening with a subsequent biomarker-based decision on FAS sequencing. This could result in significant cost-saving.
We, therefore, initiated a prospective study to evaluate the previously assessed biomarkers as well as additional biomarkers for their ability to predict or exclude FAS mutations in a cohort of patients selected on the basis of their clinical presentation with splenomegaly and/or chronic lymphadenopathy and presumed or proven autoimmune cytopenia(s). We aimed to use the best identified biomarkers to develop a simple tool to calculate the individual probability of a FAS mutation prior to any other selection criteria such as DNT levels.
Methods
Patients
All patients presenting to one of the collaborating immunodeficiency centers with lymphoproliferation (defined as chronic non-malignant lymphadenopathy for >6 months at more than two sites and/or splenomegaly) and presumed or proven autoimmune cytopenia(s) were eligible for this study. Autoimmune cytopenia was defined as cell counts repeatedly below normal in one or more blood cell lineages with detectable autoantibodies. A number of patients with neutropenia or thrombocytopenia did not undergo autoantibody testing, but were diagnosed with presumed autoimmune cytopenia by the referring physician. The study was approved by the ethics committee of the University of Freiburg (protocol number 40/09).
Clinical chemistry, flow cytometry and apoptosis assay
Serum and blood collected into EDTA were sent to the coordinating study center in Freiburg by overnight delivery and the following 11 parameters were determined in a single reference laboratory in all patients: vitamin B12 (Eclia: Electrochemiluminescence Immunoassay, Roche Diagnostics), sCD25 (turbidimetry), high-density lipoprotein cholesterol (HDL; homogenous enzymatic color test), apolipoprotein A1 (APO-A1; turbidimetry), IgG (turbidimetry). sFASL and IL-10 were determined by an enzyme-linked immunosorbent assay as described elsewhere.13 CD3+TCRα/β+CD4−CD8− DNT cells, the percentage of DNT cells expressing B220 (B220) and marginal zone-like (CD19+CD27+IgD+IgM+, MZB) and class-switched memory B cells (CD19+CD27+IgD−IgM−; SMB) were determined by flow cytometry as described previously.13,14 FASL-induced apoptosis was analyzed as described elsewhere.13
Genetic analysis
In patients with abnormal apoptosis, exons and exon-intron boundaries of the FAS gene were sequenced using DNA from granulocytes or whole blood. FAS ligand (FASLG) and caspase 10 (CASP10) genes were sequenced in 60% of patients, and caspase 8 (CASP8) in 30%. If no mutations were found, DNA from sorted DNT cells was sequenced. In patients with normal apoptosis, sequencing was performed directly from sorted DNT cells. Somatic mutations were confirmed by sequencing of germline DNA. In some patients, we could not obtain further material for DNT sorting or could not reliably rule out somatic mutations because DNT numbers were too low. Only patients with completed genetic analysis were included in the calculation of predictive values.
Statistics
Differences in the distribution of the biomarkers across subgroups of patients were visualized using dot plots and their significance assessed by Kruskal-Wallis tests. The diagnostic values of the different markers in predicting the presence or absence of a FAS mutation were assessed in a first step using receiver operator characteristic (ROC) curves and corresponding area under the curve (AUC) values. Comparison of AUC values of different parameters were based on the non-parametric approach suggested by DeLong et al.15P-values for comparisons with published AUC values were based on inversion of confidence intervals. Biomarkers with an AUC value ≥0.7 were chosen for further evaluation. In a second step, the ability of each marker to define subgroups of clinically relevant size with positive or negative predictive values close to 100% was examined. We first considered cutoffs and combinations already suggested in the publication by Caminha et al.,12 and then continued using cut-offs values chosen in a way that the prevalence of positives tests corresponded to the prevalence of FAS mutations in our study population. These predictive values were compared with the published values using binomial tests.
All statistical analyses were done using STATA 12.16
Results
The majority of patients presenting with lymphoproliferation and autoimmune cytopenia(s) remain without a molecular diagnosis
One hundred and sixty-three patients from nine European countries (Online Supplementary Table S1) fulfilled the inclusion criteria of splenomegaly or chronic lymphadenopathy and presumed or proven autoimmune cytopenia(s). Autoantibodies against red blood cells, platelets or granulocytes were detectable in 57 (35%), the other patients were either not tested or negative, but diagnosed with probable autoimmune cytopenia according to clinical judgment. Apoptosis testing was performed in 157 patients and was abnormal in 39 (24%). Among these 39 patients, 28 had germline FAS mutations (ALPS-FAS) and 11 had no germline mutation in FAS, FASL, CASP8 or CASP10 (ALPS-U). Three of these 11 patients were investigated for a somatic FAS mutation, which was found in two of them. The other eight patients were not investigated for somatic mutations either because defective apoptosis was a familial trait (n=3) or because the patients’ material was not sufficient for sorting DNT (n=5). Among 113 patients with normal apoptosis, 53 were investigated for mutations in sorted DNT cells (n=46) or in germline DNA (n=7). Of these, four patients had a somatic FAS mutation (ALPS-sFAS), while 49 had no mutations in any of the four genes (designated “ALPS-phenotype sequenced”: ALPS-ph-s). Eleven of the 163 patients did not have interpretable apoptosis tests. Six of these 11 patients were sequenced for germline FAS mutations, which were detectable in four. Among the 65 patients who did not undergo genetic analysis (designated “ALPS-phenotype not sequenced”: ALPS-ph-ns), 11/61 (18%) had DNT cells <1%, precluding reliable exclusion of somatic mutations and for the remaining patients, the physicians pursued other diagnoses so that additional samples were not sent for DNT sorting.
Overall, in our cohort of 163 patients with splenomegaly or chronic lymphadenopathy and presumed or proven autoimmune cytopenia(s), 32 were diagnosed with ALPS-FAS and six with ALPS-sFAS. In seven patients, a molecular diagnosis other than ALPS was eventually established, including one patient each with DiGeorge syndrome, ICOS deficiency, LRBA deficiency, STIM-1 deficiency, X-linked lymphoproliferative syndrome-2, X-linked chronic granulomatous disease and RAS-associated autoimmune leukoproliferative disease. In addition, 19 of 36 patients with lymphoproliferation, autoimmune cytopenia(s) and hypogammaglobulinemia were clinically classified as having common variable immunodeficiency. No disease-causing mutations were found in CASP8 or CASP10 (Online Supplementary Figure S1).
Biomarker profiling in patients with lymphoproliferation and autoimmune cytopenia(s)
We then analyzed the distribution of values for 11 bio-markers in the five categories of patients (detailed information on missing values is provided in Online Supplementary Table S2). Similar to a previous retrospective report on a large cohort of patients with lymphoproliferation and more than 1% DNT cells,12 sFASL, IL-10, vitamin B12 and DNT levels were increased in ALPS-FAS and ALPS-sFAS patients when compared to ALPS-U and ALPS-phenotype patients (Figure 1 and Online Supplementary Figure S2). We also studied a number of additional bio-markers that were previously reported to be altered in ALPS. Their distribution between ALPS-FAS/sFAS and control groups is shown in Online Supplementary Figure S2. sCD25 was elevated in most ALPS-FAS patients, but also in all other groups of patients, indicating an association with lymphoproliferation and autoimmunity in general rather than with ALPS-FAS in particular. Serum HDL and APO-A1 were decreased in most ALPS-FAS patients, but also in many other patients. B220 expression on DNT cells was increased in only 12/20 of ALPS-FAS patients, 3/5 ALPS-sFAS patients but also in 2/6 of ALPS-U and 10/36 of sequenced and 5/40 of not sequenced ALPS-phenotype patients. When compared to age-related normal values, MZB and SMB were reduced in 90% and 86% of ALPS-FAS patients, respectively, and in 100% and 83% of ALPS-sFAS patients, compared to 56% and 78% of ALPS-U, 63% and 88% of sequenced and 70% and 89% of not sequenced ALPS phenotype patients. IgG levels were above normal in 15/27 (56%) and below normal in 3/27 (11%) of ALPS-FAS patients. Elevated IgG was also observed in 17% of sequenced ALPS-phenotype patients, while 27% of not sequenced ALPS-phenotype patients had IgG levels below the age-related normal range. Biomarker values in patients with defined molecular diagnoses other than ALPS are shown in Online Supplementary Table S3.
Figure 1.
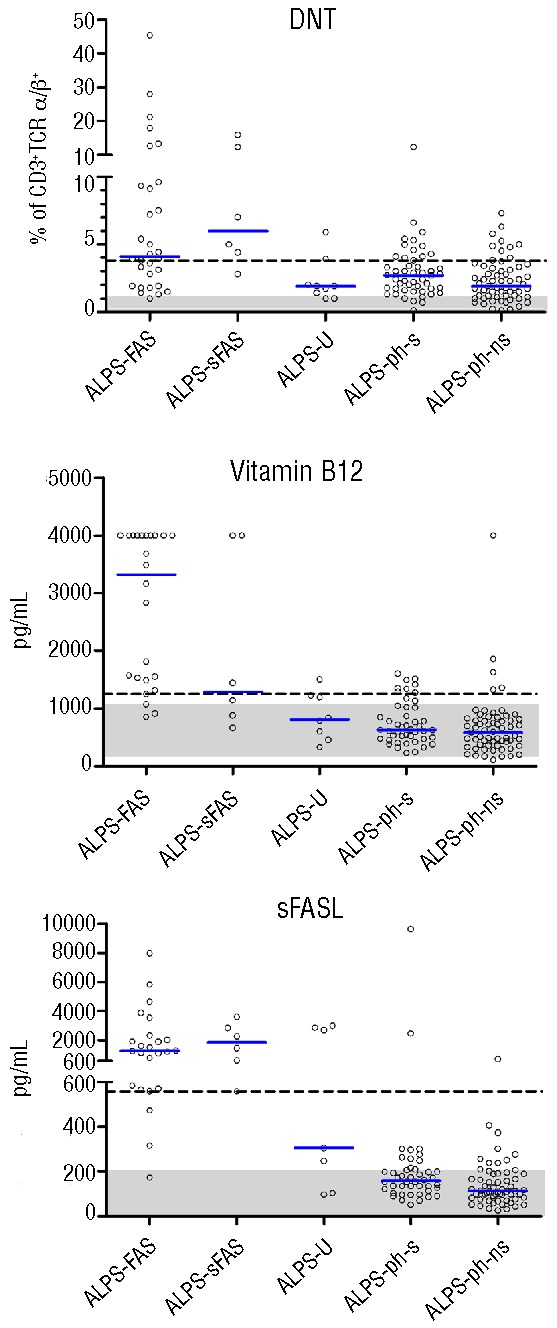
Biomarker values in the analyzed groups of patients. Values for DNT (CD4−CD8− of CD3+TCRα/β+ T cells), vitamin B12 and sFASL are shown. The vertical blue bars denote median values. Shaded areas represent normal ranges. In the case of known age dependency (vitamin B12), the variable normal ranges cannot be depicted for the whole cohort in one plot; the range shown represents normal values for the most prevalent age group in our cohort (10–18 years). The dashed lines indicate the cut-offs identified in this study based on the prevalence of FAS mutations in our cohort as described in the Methods section. The upper limit of detection for vitamin B12 was 4000 pg/mL. Differences between the ALPS-FAS/sFAS and the other groups, examined with the Kruskal Wallis test, reached high statistical significance for all three parameters (P<0.001).
Table 1.
Description and classification of patients included in our study.
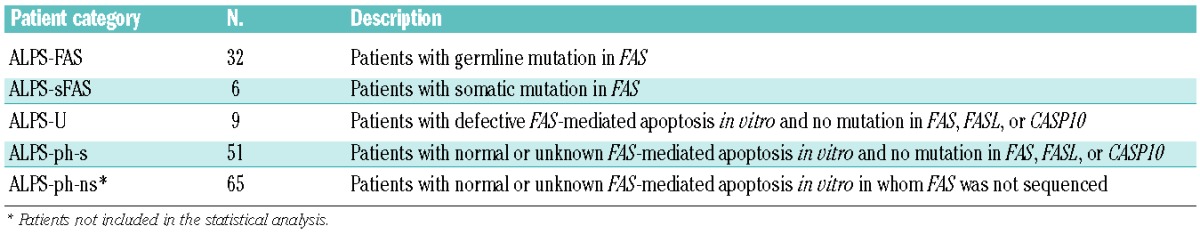
To investigate the potential of the single biomarkers to diagnose a FAS mutation, we determined AUC values for each biomarker (Table 2) and ROC curves (Online Supplementary Figure S3) for the 98 sequenced patients. The AUC was 0.92 for vitamin B12, 0.91 for sFASL and 0.86 for IL-10, which were the best discriminators. The value for DNT (0.74) was comparable to those for B220, IgG, sCD25, HDL, APO-A1 or MZB, all of which were in the range of 0.69–0.76. Thus, in our cohort of patients with splenomegaly or lymphadenopathy and autoimmune cytopenia(s), vitamin B12 and sFASL were the best predictors of a FAS mutation and significantly better than DNT levels (P=0.006 and P=0.012, respectively). Comparison to the available published AUC values in the NIH study showed that sFASL performed in a similar way to the NIH study, DNT slightly worse while vitamin B12 and IL-10 performed significantly better (P<0.01 and P=0.04, respectively).
Table 2.
Area under the curve (AUC) values for all tested biomarkers.
Positive and negative predictive values of biomarker combinations for the diagnosis of ALPS-FAS
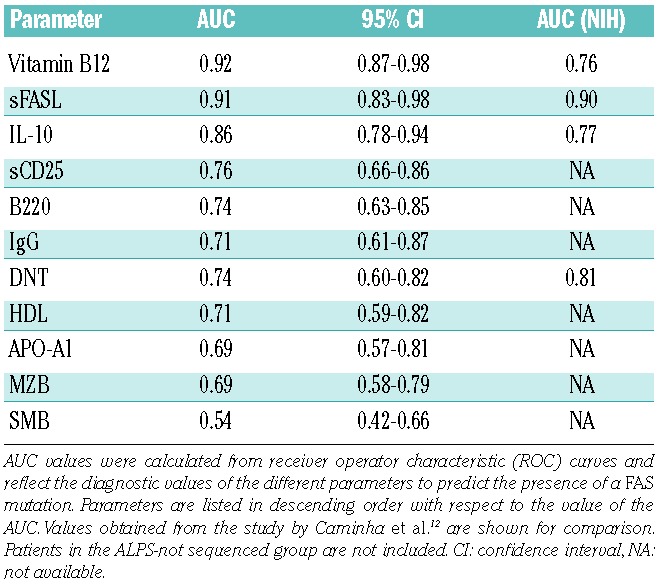
Subsequently, we analyzed the predictive values of the biomarkers studied. In order to facilitate comparison between the studies, we first used cut-off values and combinations suggested by Caminha et al.12 (Table 3A). In our cohort with a prevalence of FAS mutations of 39%, vitamin B12 (>1500 pg/mL), sFASL (>300 pg/mL) or IL-10 (>40 pg/mL) alone identified between 27%, 44% and 43% of the patients with predictive values of 87%, 81% and 73%, respectively. In contrast, DNT (>4%) only reached a PPV of 63%, although it identified a group of patients of similar size (34%). The PPV of DNT was significantly lower than that in the NIH study (P<0.001). Combination of vitamin B12 and sFASL led to a PPV of 95% and still identified a group of 25%. Combining DNT with vitamin B12 or sFASL also yielded PPV above 90%, but identified fewer patients (Online Supplementary Table S4A).
Table 3.
Positive predictive values (PPV) and negative predictive values (NPV) of selected biomarkers or their combinations for having a FAS mutation.
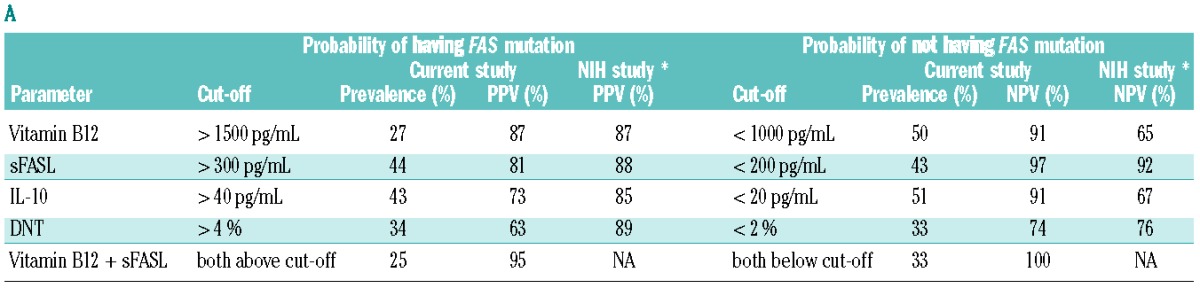
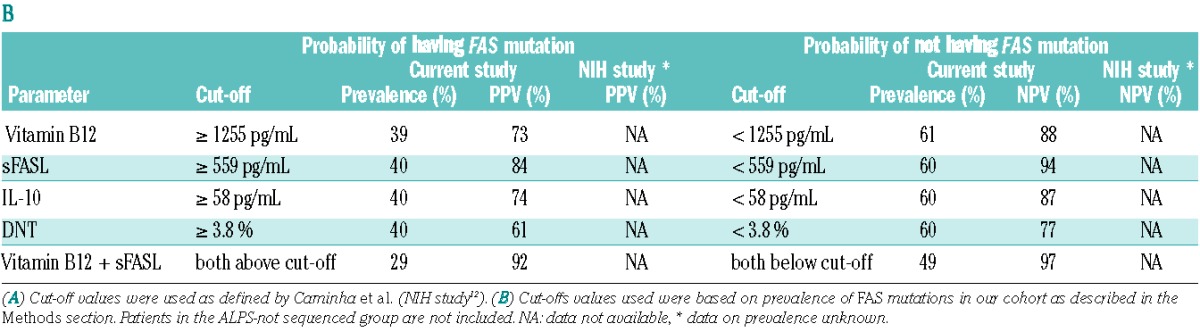
Looking at the prediction of the absence of a FAS mutation (Table 3A), sFASL (<200 pg/mL) had a negative predictive value (NPV) of 97% and identified 43% of all patients. Vitamin B12 (<1000 pg/mL) and IL-10 (<20 pg/mL) identified slightly larger groups and had NPV of 91%, which were significantly higher compared to those in the NIH study (P<0.001 for both). DNT (<2%) had a NPV above 70% and identified only a third of all patients. Combining sFASL and vitamin B12 identified a group of similar size and with a NPV of 100%. Combining DNT with sFASL and vitamin B12 identified fewer than 25% of the patients (Online Supplementary Table S4B).
One of the problems of the cut-off values defined by Caminha et al.12 was that they covered highly variable fractions of our population. For example, while 44% of our population had elevated sFASL levels above 300 pg/mL, only 27% had elevated vitamin B12 levels above 1500 pg/mL. In a second step, we therefore considered a different set of cut-off values that were chosen according to the prevalence of the FAS mutation in our cohort leading to identification of 40% of the patients as positive and 60% as negative (Table 3B). Using these cut-offs (sFASL 559 pg/mL, vitamin B12 1255 pg/mL, DNT 3.8%) the best bio-markers were sFASL (PPV 84% and NPV 94%) followed by vitamin B12 (PPV 73% and NPV 88%), while DNT performed much worse (PPV 61% and NPV 77%). Using bio-marker combinations, vitamin B12 and sFASL above these cut-offs identified 29% of our population with a PPV of 92%, while both values below these cut-offs identified 49% of the population with a NPV of 97%. Other bio-marker combinations did not perform better (Online Supplementary Tables S4C and S4D). Combining the more easily available biomarkers vitamin B12, HDL and APO-A1 yielded a PPV of 83% and identified 15% of our population. When these three values were below the cut-offs, 36% were identified with a NPV of 90%.
Using biomarker combinations to re-evaluate the results of genetic analysis
The high AUC values of the single markers and the high positive and negative predictive values observed for some combinations of markers suggested that these biomarkers can nearly separate the group of FAS mutation-positive from the group of FAS mutation-negative patients. This is illustrated by a scatter plot of vitamin B12 against sFASL with marking of the five considered categories of patients (Figure 2). ALPS-FAS/sFAS patients were well separated from patients with chronic lymphoproliferation and presumed or proven autoimmune cytopenia(s) of unknown molecular origin. Of note, in our original dataset there were five individuals among ALPS-U and ALPS-phenotype patients, who had a biomarker profile highly suggestive of a FAS mutation, with both vitamin B12 and sFASL values above the cut-offs of 1255 pg/mL and 559 pg/mL, respectively. We carefully re-evaluated the original genetic findings in these patients. In one patient, a misnaming of samples was uncovered and the patient was reclassified as having ALPS-sFAS. In another patient, two initial cell sortings did not yield sufficient numbers of DNT cells, and only when a third sample was sent, the genetic analysis could be completed revealing ALPS-sFAS. These two patients were reclassified prior to the analyses presented in this paper and are marked with arrows in Figure 2. One patient with familial ALPS-U had reduced FAS expression on T-cell subsets (not shown), suggesting a disturbed regulation of FAS expression. Two patients had their DNT reanalyzed but no mutation could be identified.
Figure 2.
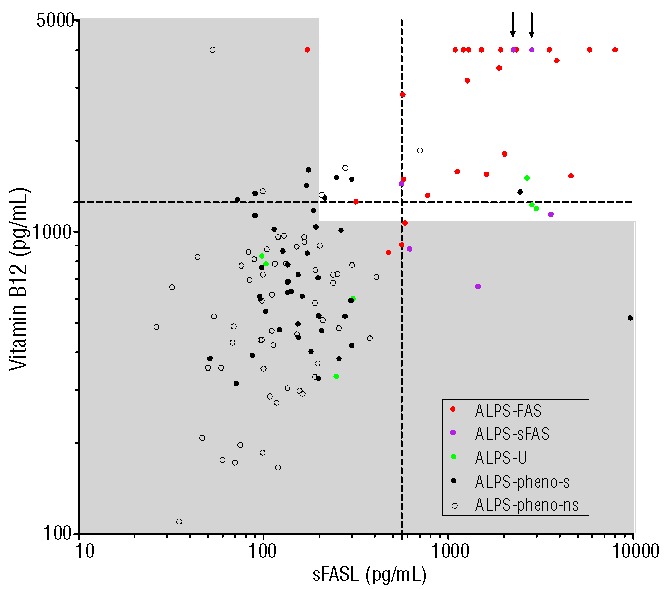
Scatter plot showing individual patients’ values of vitamin B12 against sFASL for all investigated groups. Shaded areas represent normal values, the dashed lines depict the prevalence matched cut-offs (vitamin B12: 1255 pg/mL, sFASL: 559 pg/mL). In patients marked with arrows, somatic FAS mutations were detected upon re-analysis (see text for details).
A biomarker-based probability calculator for selection or exclusion of patients with lymphoproliferation and autoimmunity to undergo FAS sequencing
As the best single markers were already highly predictive and the size of our population was limited, it was not possible to determine optimal scores combining several markers using methods such as logistic regression. To combine the information from single markers, we therefore used the naïve Bayes classification approach, which is based on the assumption of conditional independence of the markers given the FAS gene status, but is also known to be rather robust against violations of this assumption.17,18 This approach makes use only of the means and standard deviations of each marker in FAS mutation-positive and FAS mutation-negative patients and allows computation of the probability that a patient with lymphoproliferation and presumed or proven autoimmune cytopenia(s) carries a FAS mutation based on measurements for any set of markers. Based on this algorithm, we have created a web-based tool (Figure 3), in which clinicians can enter the values for vitamin B12, sFASL and IL-10 of their patients to estimate the probability of underlying FAS mutations (http://www.alps.uni-freiburg.de/). We included IL-10 to allow a more flexible use of the tool. In particular, it will enable retrospective validation of the tool with patients who did not have the two best biomarkers determined. It is important to note that calculated probabilities are only estimates which are clinically useful for selection of patients with high (>95%) versus low (<5%) probabilities of a FAS mutation. Values between 5% and 95% can only exclude high or low risk of a FAS mutation but do not represent reliable estimates of FAS mutation probability.
Figure 3.
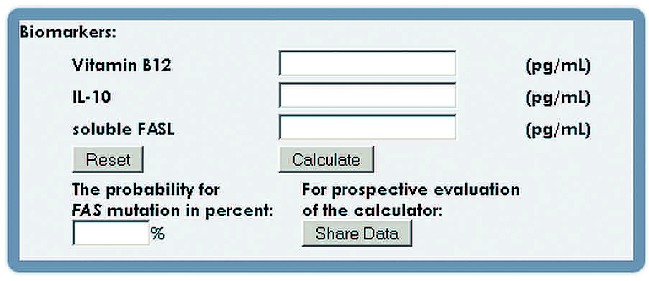
A web-based calculator of FAS mutation probability. Any number of the three most discriminative biomarkers, vitamin B12, sFASL and IL-10, can be entered to calculate the individual’s probability of having a FAS mutation. (http://www.alps.uni-freiburg.de)
Although validation of the tool and diagnostic path need to be carried out on different data sets in the future, we analyzed the distribution of the individual probabilities according to this tool in our population in order to check whether the general robustness of the naïve Bayes classifier held in our situation. If not, this should have become visible by such an analysis. ALPS-U patients were excluded from this evaluation as their biomarker values do not fit into either of the ALPS-FAS or ALPS-phenotype groups. When the probability of FAS mutation was calculated using the three strongest predictors (vitamin B12, sFASL and IL-10), we observed that 35% of the study population had a high probability (>95%), of whom 83% actually had the FAS mutation, and 56% had a low probability (<5%) with only 2% carrying a FAS mutation.
Discussion
In this prospective study on patients referred for a diagnostic evaluation of chronic lymphoproliferation and presumed or proven autoimmune cytopenia(s), we evaluated 11 biomarkers in the diagnosis of ALPS. Our results suggest determination of vitamin B12 and sFASL with a subsequent decision on FAS sequencing as the most efficient and cost-effective diagnostic pathway to establish a molecular diagnosis. The positive and negative predictive values determined in this study were integrated into a simple web-based probability calculating tool that can support the decision whether or not to perform FAS sequencing based on any combination of values for the best predictive biomarkers: IL-10, sFASL and vitamin B12.
Lymphoproliferation and autoimmune cytopenia(s) are common manifestations of impaired immune regulation and are frequently associated with primary immunodeficiencies, including, but not limited to ALPS. When conceiving our study, we reasoned that a prospective evaluation of diagnostic laboratory parameters could best be performed in an unselected cohort referred for a combination of clinical symptoms rather than in patients preselected for biological markers such as DNT cells. This approach should provide the most relevant disease population for this particular clinical situation. However, this implies that our study does not provide information on patients referred for the evaluation of lymphoproliferation or autoimmune cytopenia alone or in combination with other autoimmune manifestations or with DNT elevation. In fact, during the time of the study, we also evaluated 23 other patients who did not fulfill our clinical entry criteria and identified FAS mutations in 12 (11 germline, 1 somatic). It is important to be aware of this conceptual difference when discussing our findings in relation to the landmark studies of Magerus-Chatinet et al.10 and Caminha et al.12
Despite these differences in study concepts, we could confirm the high predictive value of vitamin B12, sFASL and IL-10 for the presence of FAS mutations in an independent, prospectively evaluated European cohort. Positive and negative predictive values were best for sFASL, IL-10 and vitamin B12 as single markers and best for vitamin B12 and sFASL as a biomarker combination. Obviously, positive and negative predictive values of biomarkers critically depend on the definition of cut-offs, but the cut-offs defined by Caminha et al.12 and the cut-offs generated in our cohort yielded similar results. Of note, single biomarkers performed better in exclusion of FAS mutation in our cohort than in the study by Caminha et al.12 Although the different sizes of the study populations may be a relevant factor, this is most probably related to the different structure of the cohorts of patients with important implications for the calculation of predictive values. Thus, the percentage of patients with FAS mutations among all symptomatic patients was much higher (171 of 263; 65%) than in our study (38 of 98; 39%), which may be related to the fact that the retrospective study by Caminha et al.12 was performed in a dedicated ALPS referral center. Moreover, the inclusion of healthy relatives in the control group of that study12 represents a significant difference to our approach of only considering symptomatic patients.
The most relevant difference between the two studies was observed in the diagnostic value of DNT cells. When we initiated the study, DNT were defined as CD4−CD8− of CD3+TCRα/β+ cells among peripheral blood mononuclear cells using a threshold >1%. New guidelines recommend whole blood staining and thresholds of CD4−CD8−TCRα/β+ of CD3+ cells >2.5% or CD4−CD8−TCRα/β+CD3+ of lymphocytes >1%. Following publication of these guidelines we performed both our initial as well as the recommended staining in all patients. DNT values determined using the suggested protocol9 were slightly higher than those determined with our protocol. This has a limited impact on the results because it affected patients and controls equally. Since we did not have full blood stains for all recruited patients, we used the data obtained with our initial protocol for this analysis. DNT of CD3+/TCRα/β+ were raised above 1% in 92% and above 2% in 69% of patients with disease control who did not carry FAS mutations. Since the proportion of disease controls was higher in our study, the predictive value of DNT determinations was much lower than in the study by Caminha et al.12 Only when greater than 8%, were DNT levels reliably associated with a FAS mutation. In patients with chronic lymphoproliferation and autoimmune cytopenia(s), DNT levels therefore appear no more useful than the panel of additional biomarkers that were tested in our study including sCD25, serum lipids, memory B cells13 and B220 expression on DNT cells.14 Although characteristic alterations of these markers were observed in most patients with FAS mutations, they were similarly elevated or reduced in significant proportions of disease control groups, reducing their predictive value. It is relevant to note that the combination of the more easily available markers, vitamin B12, HDL and APO-A1, could exclude FAS mutation with a probability of 90% in 36% of our cohort (Online Supplementary Table S4D). However, the addition of any one of the other nine markers considered in our analyses to sFASL and vitamin B12 did not improve the positive or negative predictive values.
Interestingly, as observed previously,12 some individuals among the ALPS-U and ALPS- phenotype patients had biomarker profiles highly suggestive of a FAS mutation. Given the high specificity of these markers, we hypothesized that they may have been misclassified or require more extensive genetic analysis. Indeed, two of these five patients were eventually reclassified as having ALPS-sFAS (Figure 2). In the other three, the characteristic biomarker profile could point to a more complex disturbance of FAS expression/function or to yet unknown defects in the FAS pathway; these patients are currently being analyzed for splice site and intronic FAS mutations, two have undergone whole-exome sequencing.
Another important issue in the diagnostic evaluation is the stability of biomarkers over time. It has been observed previously that sFASL, IL-10 and DNT cells decrease in response to immunosuppressive therapy.10 We therefore performed additional analyses of our cohort, separating untreated patients (n=58) from treated patients (n=32) (Online Supplementary Table S4). Overall, positive and negative predictive values were comparable in untreated and treated patients. However, a clear decrease of the PPV of DNT was seen in treated patients. The variability of biomarkers in the long-term course of the disease and in response to different treatment regimen after initial diagnosis as well as their potential predictive impact on clinical outcome are additional relevant issues that need to be addressed in continued diagnostic and follow-up studies.
The identification of biomarkers which are specifically raised in patients with FAS mutations by Magerus-Chatinet et al.10 and Caminha et al.12 have very usefully extended the ALPS diagnostic panel and have been integrated into a revised diagnostic algorithm. This revised algorithm proposes germline sequencing in all patients with lymphoproliferation and raised DNT and makes use of biomarkers for subsequent selection of patients who require additional DNT sorting in search for somatic FAS mutations. Our results suggest that for the diagnostic evaluation of patients presenting with chronic lymphoproliferation and autoimmune cytopenia(s), an alternative diagnostic pathway can be followed. We propose initial determination of vitamin B12 and sFASL in order to decide who will need FAS sequencing. We still include analysis of DNT in the initial screening, but mainly to quantify the surface FAS expression of these cells, which can be helpful to identify patients with somatic second-hit mutations.19,20 The combination of vitamin B12 and sFASL helps to identify patients with germline mutations as well as those with somatic FAS mutations and possibly also patients with as yet unknown mutations in the FAS pathway (‘ALPS by biomarkers’). Moreover, it can identify FAS mutations as highly unlikely in a large proportion of patients with lymphoproliferation and autoimmunity.
Based on these data, it is our current policy to enter vitamin B12 and sFASL values into the probability calculator and if the probability is <5% (low risk), we do not sequence FAS. If clinical suspicion remains in the subsequent clinical course, biomarkers are determined again, since they can vary. If the calculated probability is >95% (high risk), we proceed to germline FAS sequencing and, if this is negative, we sequence DNA from sorted DNT cells. In patients with a probability between 5% and 95%, we base the decision on FAS sequencing on DNT levels and apoptosis induction: if both are normal, we do not sequence FAS, but if either DNT levels are above 3.8% or apoptosis is defective or both we proceed to FAS sequencing from germline and/or DNT DNA. The results of applying this algorithm retrospectively to our cohort for estimation of robustness of the calculating tool are shown in Online Supplementary Figure S4. This sequential diagnostic approach must be validated prospectively in independent cohorts of patients. The online calculator tool developed in this study will contribute to this evaluation. Thus, our findings provide additional prospective information and can contribute to evolving consensus recommendations addressing diagnostic algorithms in patients with lymphoproliferation and autoimmune cytopenia(s).
The spectrum of other genetic diseases identified in our cohort reflects the broad differential diagnosis in patients with chronic lymphoproliferation and autoimmune cytopenia(s). To date the majority of these patients remain without a genetic diagnosis. One important challenge for future studies is to better define a rational diagnostic approach to these patients. This includes algorithms to select those in whom FASL, CASP8, CASP10 or K/N-RAS mutations must be looked for very thoroughly, both at germline and somatic levels.
Acknowledgments
We would like to thank the patients and their families for their participation. We also thank the technicians of the CCI advanced diagnostic unit and the study nurse Henrike Ritterbusch. We also gratefully acknowledge the assistance of Stefan Rusch, Daniel Fix and Gerhard Kindle in designing the database and Lukas Sättler and Dr. Martin Boeker in preparing the web-based calculator.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
This study was supported by the Bundesministerium für Bildung und Forschung (BMBF 01 EO 0803 and 01 GM 1111B). Dr. A. Janda is a recipient of an unrestricted fellowship grant from the European Society for Immunodeficiencies (ESID) provided by Baxter.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Notarangelo LD, Gambineri E, Badolato R. Immunodeficiencies with autoimmune consequences. Adv Immunol. 2006;89:321–70 [DOI] [PubMed] [Google Scholar]
- 2.Teachey DT. New advances in the diagnosis and treatment of autoimmune lymphoproliferative syndrome. Curr Opin Pediatr. 2012;24(1): 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, et al. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81(6): 935–46 [DOI] [PubMed] [Google Scholar]
- 4.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IA, Debatin KM, Fischer A, et al. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268(5215): 1347–9 [DOI] [PubMed] [Google Scholar]
- 5.Holzelova E, Vonarbourg C, Stolzenberg MC, Arkwright PD, Selz F, Prieur AM, et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N Engl J Med. 2004;351(14): 1409–18 [DOI] [PubMed] [Google Scholar]
- 6.Niemela JE, Lu L, Fleisher TA, Davis J, Caminha I, Natter M, et al. Somatic KRAS mutations associated with a human non-malignant syndrome of autoimmunity and abnormal leukocyte homeostasis. Blood. 2011;117(10): 2883–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira JB, Bidere N, Niemela JE, Zheng L, Sakai K, Nix CP, et al. NRAS mutation causes a human autoimmune lymphoproliferative syndrome. Proc Natl Acad Sci USA. 2007;104(21): 8953–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takagi M, Shinoda K, Piao J, Mitsuiki N, Matsuda K, Muramatsu H, et al. Autoimmune lymphoproliferative syndrome-like disease with somatic KRAS mutation. Blood. 2011;117(10): 2887–90 [DOI] [PubMed] [Google Scholar]
- 9.Oliveira JB, Bleesing JJ, Dianzani U, Fleisher TA, Jaffe ES, Lenardo MJ, et al. Revised diagnostic criteria and classification for the autoimmune lymphoproliferative syndrome (ALPS): report from the 2009 NIH International Workshop. Blood. 2010;116 (14): e35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magerus-Chatinet A, Stolzenberg MC, Loffredo MS, Neven B, Schaffner C, Ducrot N, et al. FAS-L, IL-10, and double-negative CD4- CD8- TCR alpha/beta+ T cells are reliable markers of autoimmune lymphoproliferative syndrome (ALPS) associated with FAS loss of function. Blood. 2009;113(13): 3027–30 [DOI] [PubMed] [Google Scholar]
- 11.Dowdell KC, Niemela JE, Price S, Davis J, Hornung RL, Oliveira JB, et al. Somatic FAS mutations are common in patients with genetically undefined autoimmune lymphoproliferative syndrome. Blood. 2010; 115(25): 5164–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caminha I, Fleisher TA, Hornung RL, Dale JK, Niemela JE, Price S, et al. Using biomarkers to predict the presence of FAS mutations in patients with features of the autoimmune lymphoproliferative syndrome. J Allergy Clin Immunol. 2010; 125(4): 946–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rensing-Ehl A, Warnatz K, Fuchs S, Schlesier M, Salzer U, Draeger R, et al. Clinical and immunological overlap between autoimmune lymphoproliferative syndrome and common variable immunodeficiency. Clin Immunol. 2010;137(3): 357–65 [DOI] [PubMed] [Google Scholar]
- 14.Bleesing JJ, Brown MR, Dale JK, Straus SE, Lenardo MJ, Puck JM, et al. TcR-alpha/beta(+) CD4(−)CD8(−) T cells in humans with the autoimmune lymphoproliferative syndrome express a novel CD45 isoform that is analogous to murine B220 and represents a marker of altered O-glycan biosynthesis. Clin Immunol. 2001;100(3): 314–24 [DOI] [PubMed] [Google Scholar]
- 15.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3): 837–45 [PubMed] [Google Scholar]
- 16.StataCorp Stata Statistical Software. College Station, TX: StataCorp LP; 2011 [Google Scholar]
- 17.Bickel PJ, Levina E. Some theory of Fisher’s linear discriminant function, ‘naive Bayes’, and some alternatives when there are many more variables than observations. Bernoulli. 2004;10(6): 989–1010 [Google Scholar]
- 18.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97 (457): 77–87 [Google Scholar]
- 19.Kuehn HS, Caminha I, Niemela JE, Rao VK, Davis J, Fleisher TA, et al. FAS haploinsufficiency is a common disease mechanism in the human autoimmune lymphoproliferative syndrome. J Immunol. 2011;186(10): 6035–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magerus-Chatinet A, Neven B, Stolzenberg MC, Daussy C, Arkwright PD, Lanzarotti N, et al. Onset of autoimmune lymphoproliferative syndrome (ALPS) in humans as a consequence of genetic defect accumulation. J Clin Invest. 2011;121(1): 106–12 [DOI] [PMC free article] [PubMed] [Google Scholar]


