In the April 2013 issue of Haematologica, Lee et al. have described the TET1 genomic breakpoints and clinical features of MLL-TET1 rearranged cases of acute leukemia.1 So far, 13 cases have been reported in the literature, 11 in acute myeloid leukemia (AML) patients1–8 and 2 in B-cell precursor acute lymphoblastic leukemia (ALL).9 It was also recently reported that MLL is fused to TET1 in only 5 out of 1,590 MLL rearranged AML cases (0.3%).10 Although those cases are very uncommon, their study can improve our current understanding of leukemogenesis. We report here the first t(10;11) MLL-TET1 positive case of T-cell lymphoblastic lymphoma occurring in a 31-year old male patient, with a subsequent transformation to AML.
The patient was referred for a large mediastinal mass and right pleural effusion. Blood cell count showed no abnormalities. Mediastinal and bronchus biopsies led to the diagnosis of a precursor-T-cell lymphoblastic lymphoma (pre-T LBL), expressing CD3, CD5, CD4, CD8 and CD10 antigens, together with a high expression of Ki67 (90%). No expression of CD34 or CD79a was observed. The same cells were observed in pleural fluid (Figure 1A and B) that expressed CD3, CD4, CD8, CD2, CD7, CD10 antigens but neither CD34 nor myeloperoxidase. Bone marrow examination and central nervous system imaging did not show any other specific localization. The patient was treated following the Groupe d’Etudes des Lymphomes de l’Adulte (GELA) LL03 protocol, and was considered in complete remission after induction and consolidation phases. A 32×22×48 mm residual mediastinal mass remained after treatment, without hypermetabolic abnormality on the FDG-PET scan and was considered to be fibronecrotic scar tissue. Fourteen months after the diagnosis, during the maintenance therapy, a bone marrow examination was performed for thrombopenia (6 g/L) that revealed a myelomonocytic acute leukemia with trilineage dysplasia (Figure 1C). The mediastinal mass remained unchanged on the imaging scan. The patient achieved complete remission after intensive chemotherapy based on cytarabine and daunorubicin, followed by a consolidation course with high-dose cytarabine. A non-familial donor allogeneic bone marrow transplant (10/10 match) was performed four months after the diagnosis of the acute myeloid leukemia that was complicated by a Grade IV acute graft-versus-host disease involving digestive tract, liver and skin. The patient died 54 days after the transplant of bacterial sepsis leading to multi-organ failure.
Figure 1.
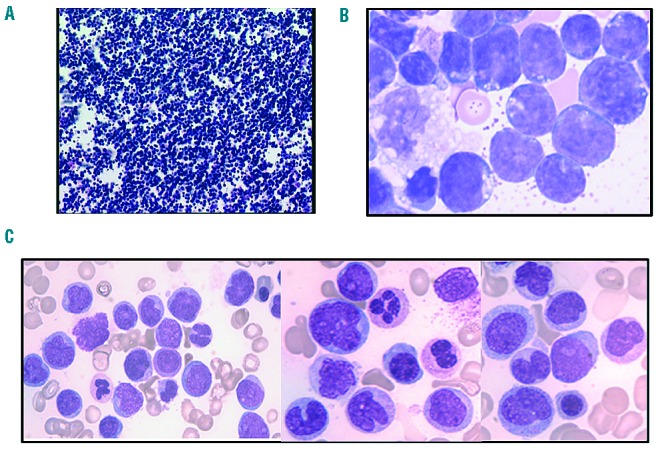
Morphology of the malignant cells observed at the two phases of the evolution (A and B) T-cell lymphoblastic lymphoma; (C) myelomonocytic acute leukemia). (A) Section of the embedded pleural cell pellet used for pathology showing a dense infiltration of malignant cells. (B) Cytology of the leukemic cells observed in the pleural fluid. (C) Bone marrow infiltration by monocytic cells, hypogranular myeloid precursors and myeloid blast cells (May-Grünwald-Giemsa staining).
Molecular analyses of the malignant T cells showed a clonal T-cell receptor gamma-chain gene and a minor IgG kappa gene rearrangement (Biomed 2 protocol) together with HOXA10 overexpression. The presence of MLL rearrangement was detected using fluorescence in situ hybridization (FISH) analysis (XL MLL Break Apart Probe, Metasystems, Altlussheim, Germany). The partner gene, TET1, was identified using a RP11-9E13 and RP11-314J18 BACs (BlueGnome, Cambridge, UK (Figure 2), corresponding to the recurrent t(10;11)(q22;q23) translocation. This translocation was found as the sole chromosomal abnormality in blast cells (Figure 2), as frequently observed (4 of 10 cases with available karyotype reported by Lee et al.).1MLL-TET1 and its reciprocal fusion transcript were amplified using published primers,6 and direct sequencing of PCR products showed a fusion of the exon 11 of MLL (Ensembl ENSG00000118058) to exon 9 of TET1 (Ensembl ENSG00000138336), within the main breakpoint cluster of TET1 localized from exon 8 to 12.1 In the case reported here, 87 nucleotides of the intron 8 of TET1 were inserted in the fusion transcript, which contained cryptic splice donor and acceptor sites.
Figure 2.
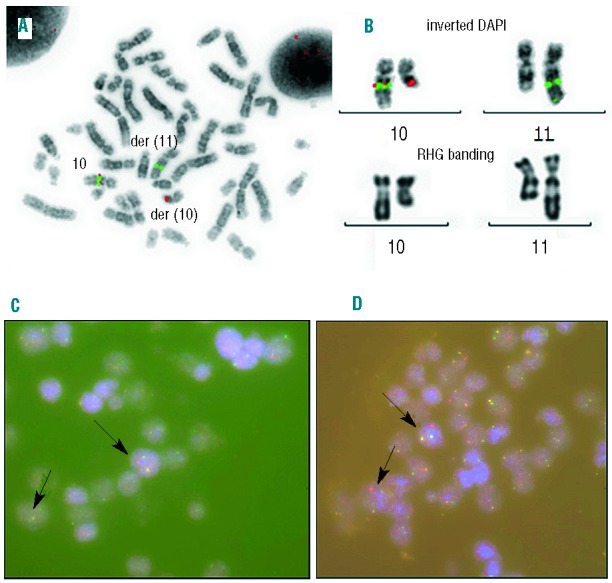
Molecular and conventional cytogenetics of bone marrow in the acute myelomonocytic leukemia stage (A and B) and paraffin embedded pleural fluid pellet in the T-cell lymphoblastic lymphoma (C and D). (A) Fluorescent in situ hybridization (FISH) analysis using BACs RP11-9E13 (spectrum orange) and RP11-314J18 (spectrum green, BlueGnome®) flanking TET1gene that are separated in the t(10;11)(q22;q23) translocation. (B) Magnification of the chromosomes 10 and 11 in inverted DAPI and XL MLL break apart probe (Metasystems®) FISH and RHG banding, demonstrating the rearrangement of MLL. (C) FISH study using an XL MLL break apart probe in pleural effusion. A split of the MLL probe had been observed within the malignant cell population (arrows). (D) Interphasic FISH study of TET1 using BACs probes RP11-9E13 (spectrum orange) and RP11-314J18 (spectrum green). A separation of the signals for one allele had been observed within the malignant cell population.
The MLL-TET1 fusion transcript, as well as HOXA10 overexpression, was still present at the myelomonocytic acute leukemia phase, but neither TCR nor IGH rearrangements were detected in the myeloid blasts. Also, neither NPM mutation nor FLT3 internal tandem duplication were observed.
MLL-TET1 fusions have been described in 11 AML cases (mostly AML M4 or M5) and in 2 B-ALL cases.1 The case reported here presents two interesting features. Firstly, this patient harbors the first MLL-TET1 fusion reported to date in T-ALL, and, secondly, a lymphoid to myeloid phenotype switch occurred during the course of the disease. Indeed, MLL-TET1 fusion was detected at both steps, demonstrating a common origin of the two malignancies. This strongly suggests that the translocation occurred very early during hematopoietic differentiation, prior to the lymphoid/myeloid commitment. As described for the Ph1 chromosome rearrangement, the t(10;11) translocation may arise in hematopoietic stem cell rather than in committed progenitors.
Recent studies on TET1 properties provide a possible explanation as to how such a switch may have occurred. TET family enzymes convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and play a key role in active DNA demethylation. TET1 and TET2 are also the key enzymes responsible for the presence of 5hmC in mouse embryonic stem cells (ESCs), and TET1 functions to regulate the lineage differentiation potential of ESCs.11 In addition to its role in DNA demethylation, TET1 interacts physically with NANOG, synergistically enhancing the efficiency of NANOG in somatic cell reprogramming in a manner that is dependent on its catalytic activity.12 NANOG/TET1 co-occupy genomic loci of genes associated with both maintenance of pluripotency and lineage commitment in embryonic stem cells, and may deposit 5hmC to target genes before the establishment of pluripotency.12 Taken together, these observations suggest a possible mechanism for the lineage switch observed in this patient, and provide a rationale for using demethylating agents in MLL-TET1 neoplasms.
On the other hand, MLL is a histone lysine specific methyltransferase frequently rearranged in lineage switching in acute leukemia,13 and may lead to a stem cell/progenitor phenotype.14 As both direct and reciprocal transcripts were detected, the respective roles of TET1 and MLL may be additive for lineage switching in this patient.
This case, together with those previously published, indicates that MLL-TET1 may induce myeloid leukemia, B and T lymphoma, possibly arising from the hematopoietic stem cell compartment. These features are also described in the 8p11 stem cell syndrome that involves FGFR1, which arise in myeloid or lymphoid neoplasms with possible subsequent transformation to acute myeloid leukemia. As proposed by Lee et al., we confirm the need to collect and report more cases and to test the leukemogenic activity of MLL-TET1 associated to demethylating agents in a murine bone marrow transduction/transplantation model system.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Lee S-G, Cho SY, Kim MJ, Oh SH, Cho EH, Lee S, et al. Genomic breakpoints and clinical features of MLL-TET1 rearrangement in acute leukemias. Haematologica. 2013;98(4): e55–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thirman MJ, Gill HJ, Burnett RC, Mbangkollo D, McCabe NR, Kobayashi H, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329(13): 909–14 [DOI] [PubMed] [Google Scholar]
- 3.Harrison CJ, Cuneo A, Clark R, Johansson B, Lafage-Pochitaloff M, Mugneret F, et al. Ten novel 11q23 chromosomal partner sites. European 11q23 Workshop participants. Leukemia. 1998;12(5): 811–22 [DOI] [PubMed] [Google Scholar]
- 4.Aventín A, La Starza R, Martínez C, Wlodarska I, Boogaerts M, Van den Berghe H, et al. Involvement of MLL gene in a t(10;11)(q22;q23) and a t(8;11)(q24;q23) identified by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1999;108(1): 48–52 [DOI] [PubMed] [Google Scholar]
- 5.Kim HJ, Cho HI, Kim EC, Ko EK, See CJ, Park SY, et al. A study on 289 consecutive Korean patients with acute leukaemias revealed fluorescence in situ hybridization detects the MLL translocation without cytogenetic evidence both initially and during follow-up. Br J Haematol. 2002;119(4): 930–9 [DOI] [PubMed] [Google Scholar]
- 6.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002;62(14): 4075–80 [PubMed] [Google Scholar]
- 7.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17(3): 637–41 [DOI] [PubMed] [Google Scholar]
- 8.Shih L-Y, Liang DC, Fu JF, Wu JH, Wang PN, Lin TL, et al. Characterization of fusion partner genes in 114 patients with de novo acute myeloid leukemia and MLL rearrangement. Leukemia. 2006;20(2): 218–23 [DOI] [PubMed] [Google Scholar]
- 9.Burmeister T, Meyer C, Schwartz S, Hofmann J, Molkentin M, Kowarz E, et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: results from the GMALL study group. Blood. 2009;113(17): 4011–5 [DOI] [PubMed] [Google Scholar]
- 10.Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013. April 30. 10.1038/leu.2013.135 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2): 200–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495(7441): 370–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorantes-Acosta E, Pelayo R. Lineage switching in acute leukemias: a consequence of stem cell plasticity? Bone Marrow Res. 2012; 2012:406796. 10.1155/2012/406796 Epub 2012 Jul 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;(11):823–33 [DOI] [PubMed] [Google Scholar]


