Summary
Noncoding RNAs control critical cellular processes, although their contribution to disease remains largely unexplored. Dyskerin associates with hundreds of H/ACA small RNAs to generate a multitude of functionally distinct ribonucleoproteins (RNPs). The DKC1 gene, encoding dyskerin, is mutated in the multisystem disorder X-linked Dyskeratosis Congenita (X-DC). A central question is whether DKC1 mutations affect the stability of H/ACA RNPs including those modifying ribosomal RNA (rRNA). We carried out comprehensive profiling of dyskerin-associated H/ACA RNPs, revealing remarkable heterogeneity in the expression and function of subsets of H/ACA small RNAs in X-DC patient cells. Using a novel mass spectrometry approach, we uncovered single-nucleotide perturbations in dyskerin-guided rRNA modifications, providing functional readouts of small RNA dysfunction in X-DC. Strikingly, we identified that the catalytic activity of dyskerin is required for accurate hematopoietic stem cell differentiation. Altogether, these findings reveal that small noncoding RNA dysfunctions may contribute to the pleiotropic manifestation of human disease.
Keywords: X-linked Dyskeratosis Congenita, H/ACA snoRNAs, dyskerin, pseudouridylation, rRNA modifications, bone marrow failure, hematopoietic stem cells
Introduction
H/ACA small RNAs are an evolutionarily conserved class of abundant noncoding RNAs (ncRNAs) involved in a diverse range of processes including post-transcriptional modifications of functional RNAs, pre-ribosomal RNA processing, and telomere maintenance (Baxter-Roshek et al., 2007; Lestrade and Weber, 2006; Matera et al., 2007; Reichow et al., 2007; Williams and Farzaneh, 2012). A central protein associated with all classes of H/ACA small RNAs is dyskerin, which is thereby at the nexus of controlling many diverse cellular processes (Lafontaine et al., 1998; Montanaro, 2010; Watkins et al., 1998). The largest subgroup of dyskerin-associated H/ACA small RNAs are small nucleolar RNAs (snoRNAs) that are responsible for modifying hundreds of specific nucleotides on the ribosome. The association of dyskerin with H/ACA snoRNAs is important for their stability and forms catalytically active ribonucleoprotein (RNP) complexes, which guide the site-specific conversion of uridine to pseudouridine (Ψ) residues on ribosomal RNA (rRNA)(Charette and Gray, 2000).
Importantly, the DKC1 gene encoding for dyskerin is mutated in a number of cancers and inherited human syndromes including X-linked Dyskeratosis Congenita (X-DC) and the clinically severe variant of Dyskeratosis Congenita (DC) known as Hoyeraal-Hreidarsson (HH) syndrome (Bellodi et al., 2010b; Cerami et al., 2012; Forbes et al., 2011; Heiss et al., 1998; Knight et al., 1999; Yip et al., 2012). These human syndromes are characterized by a wide range of defects including hematopoietic and cutaneous abnormalities (abnormal pigmentation, nail dysplasia, leukoplakia), increased risk of cancer, pulmonary fibrosis and liver disease, as well as severe congenital birth defects in brain development, growth, and the genitourinary system (Kirwan and Dokal, 2008). Among all H/ACA small RNAs, the telomerase RNA (TERC) is the most characterized and is widely employed as a prognostic molecular marker of DC (Alter et al., 2007; Mitchell et al., 1999). It remains largely unknown whether a multitude of small RNAs may be affected in human disease as a consequence of mutations in DKC1. In addition, a central unresolved question is whether the enzymatic activity of dyskerin is perturbed in and may contribute to some of the pathological features of diseases associated with mutations in DKC1. Interestingly, emerging evidence further suggests that specific subsets of dyskerin-associated H/ACA snoRNAs are deregulated in hematological disorders such as leukemia, lymphoma, and multiple myeloma (Ronchetti et al., 2012; Teittinen et al., 2012; Valleron et al., 2012a; Valleron et al., 2012b).
Here we sought to determine the landscape of H/ACA small RNA dysregulation in patients harboring DKC1 mutations that may underlie the wide range of pathological features observed. Surprisingly, by carrying out an extensive profiling screen for these small RNAs, we observed unexpected heterogeneity in their expression levels in X-DC patient cells harboring distinct DKC1 mutations. Thereby, these findings reveal unexpected complexity in the manifestation of impairments in small RNAs that may reflect an important genotype to phenotype relationship associated with human disease etiology. Strikingly, applying a novel and highly quantitative mass spectrometry approach, we identified reductions in site-specific rRNA Ψ modifications, which correspond to the decreased expression of H/ACA snoRNAs guiding the modification of these residues. Thereby, the catalytic activity of RNP complexes is also directly affected in X-DC human disease. Moreover, we uncovered a critical role for dyskerin's catalytic activity towards hematopoietic differentiation. Altogether, these findings provide compelling evidence that the deregulated expression and function of H/ACA snoRNPs may underlie specific pathological features of human disease. Moreover, complex deregulations in the patterns of H/ACA small RNA expression may serve as important molecular markers of human health and disease.
Results
Heterogeneous defects in all classes of H/ACA small RNAs are present in X-DC patients harboring distinct DKC1 mutations
To uncover whether DKC1 mutations affect functionally unique subsets of dyskerin-containing H/ACA small RNPs, we undertook a systematic expression analysis to examine a large panel of H/ACA small RNAs. First, we sought to examine the expression profiles of these small RNAs directly in highly purified CD34+ hematopoietic progenitor cells, as a common pathological feature and cause of lethality in X-DC patients is bone marrow failure (Dokal, 2011). To this end, we characterized patient cells harboring a mutation in the DKC1 promoter, a C to G substitution at position -141 hereafter referred to as DKC1(c.-141 C>G) (Figure 1A, top), which produces a hypomorphic allele that significantly reduces DKC1 transcript levels (Figure 1B). The same mutation in the putative Sp1 transcription binding sites within the DKC1 promoter has been previously reported (Knight et al., 2001; Salowsky et al., 2002). This patient presented at two years of age with several clinical features of DC including hematological defects, congenital anomalies involving the central nervous system and genitourinary system, and a family history of early onset colon and lung cancers (Figure 1A, bottom). Therefore, DKC1(c.-141 C>G) cells provide a very unique system to assess the impact of reducing the overall threshold of dyskerin activity.
Figure 1. Characterization of dyskerin-associated H/ACA small RNAs in X-DC patient cells.
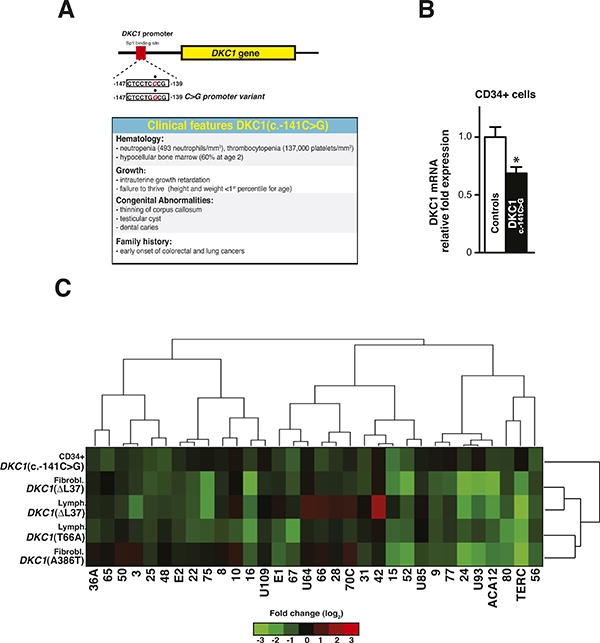
(A) Schematic of the DKC1(c.-141C>G) mutation illustrating the position of the base pair substitution on the DKC1 promoter (top). Clinical features of the DKC1(c.-141C>G) patient are shown (bottom). (B) Quantification of DKC1 mRNA levels in CD34+ cells from two healthy controls and DKC1(c.-141C>G) patient was measured by real-time qPCR. (C) Heatmap diagram displaying hierarchical clustering of 32 H/ACA small RNA relative expression levels in X-DC patient cells. The expression of H/ACA small RNAs was measured by real-time qPCR, relative to two type-matched control cells for each mutation, and normalized to the abundance of RN7SK small non-coding RNA from at least three independent experiments. Each row represents a different type of X-DC patient-derived cell. Each column illustrates the expression of individual H/ACA small RNAs relative to controls. The color bar indicates the color-coding of small RNA expression from +3 to -3 in log2 space (bottom). Only slight variation in the expression of H/ACA small RNAs was observed amongst controls either by real-time qPCR or northern blot analysis (Figure S1 and Tables S1-S2). Statistical information is included in Table S2.
We directly assessed the expression of functionally distinct H/ACA small RNAs in DKC1(c.-141 C>G) CD34+ cells, including 27 H/ACA snoRNAs, which are the most abundant class of dyskerin-associated small RNAs involved in site-specific rRNA pseudouridylation (Figure 1C, top row). In addition, the expression of small Cajal Body RNAs (scaRNAs), involved in pseudouridylation of spliceosomal small nuclear RNAs (snRNAs) (ACA12, U85, U93, and U109), as well as TERC, important for telomere function, was also examined. Unexpectedly, we observed differential reductions in the expression of distinct H/ACA small RNAs relative to matched healthy control CD34+ cells (Figure 1C, top row), which did not display significant variations in H/ACA small RNA levels (Figure S1A and Table S1). Notably, the expression of H/ACA snoRNA15 and snoRNA67, which guide site-specific modifications of two consecutive Ψ residues on 18S rRNA (i.e. Ψ1367 and Ψ 1445), displayed the greatest reduction, up to a 60% decrease (Figure 1C and Table S2). Interestingly, the levels of TERC remained unchanged in DKC1(c.-141 C>G) patient cells (Figure 1C and Table S2) and consistently no significant difference in telomere length was observed (Figure S2). These results suggest that missense mutations in DKC1 have a more profound effect on TERC levels (Batista et al., 2011; Walne et al., 2007) as compared to the DKC1 promoter mutation described in this study. For example, it is possible that missense mutations in dyskerin may disrupt specific protein-protein interactions within the telomerase complex that may be important for TERC stability. These results indicate that distinct classes of dyskerin-associated H/ACA small RNAs may be differentially affected by this specific DKC1 mutation.
In order to determine whether aberrant expression of H/ACA small RNAs is common among X-DC patients, we next extended our analysis to individuals harboring different DKC1 mutations. In this respect, we performed unsupervised hierarchical clustering of H/ACA small RNA relative expression in DKC1(c.-141 C>G), DKC1(T66A), DKC1(ΔL37), and DKC1(A386T) patient cells (Figure 1C). We observed remarkable heterogeneity in the expression of H/ACA small RNAs amongst X-DC patient cells harboring different mutations that have similar histologic origins, relative to matched controls, which did not display significant variations in H/ACA small RNA levels (Figures S1B–S1C and Table S2). Moreover, specific H/ACA small RNAs stratified according to their relative expression levels, cell type and nature of the DKC1 mutation (Figure 1C dendrograms). For example, the expression of a group of four H/ACA snoRNAs (snoRNA48, E2, 22, and 75) is perturbed in DKC1(ΔL37) fibroblasts, but remains unaltered in DKC1(A386T) fibroblasts (Figure 1C and Table S2). Similar variations in the expression of distinct H/ACA small RNAs were also observed amongst X-DC patient lymphoblasts harboring different DKC1 mutations (Figure 1C and Table S2). Furthermore, we also identified differences in H/ACA snoRNA expression in cells from different tissues (lymphoblasts versus fibroblasts) harboring the same DKC1(ΔL37) mutation (e.g. snoRNA3l, 42, 15 and U85) (Figure 1C), suggesting that tissue-specific defects in H/ACA small RNA expression may at least in part underlie the specific pathological features present in X-DC patients. Common nodes of dysregulation in H/ACA small RNA expression are also evident. For example, six H/ACA small RNAs (snoRNA16, 52, 24, U93, ACA12 and TERC) display the most dramatic reductions in the majority of X-DC patient cells analyzed (Figure 1C). Moreover, we confirmed these findings by northern blot analysis (Figure S1D). Importantly, not all the H/ACA small RNAs are reduced in X-DC patient cells. In particular, H/ACA snoRNAs such as 28 and U64 do not display differences in expression levels, in agreement with previous reports (Figure 1C and Table S2) (Batista et al., 2011; Mitchell et al., 1999). Interestingly, our observation that snoRNA42 expression is increased in DKC1(ΔL37) lymphocytes raises the possibility that certain snoRNAs may be selectively increased as a compensatory mechanism for perturbations in subsets of H/ACA small RNAs (Figure 1C). Strikingly, H/ACA sno/scaRNAs 24, U93, ACA12, 80, and 56 are decreased in the majority of X-DC patient cells, closely clustering with concomitant diminished TERC expression, and may therefore represent a central node of small RNA dysfunctions in human disease (Figure 1C). Importantly, the altered expression of H/ACA small RNAs in X-DC patients is highly specific, as a similar class of non-dyskerin associated small RNAs implicated in methyl modifications of rRNA, termed C/D snoRNAs (Bachellerie and Cavaille, 1997), are unaffected in X-DC patient cells (Figure 2 and Table S2).
Figure 2. Expression of dyskerin rescues H/ACA small RNA expression levels in primary X-DC fibroblasts.
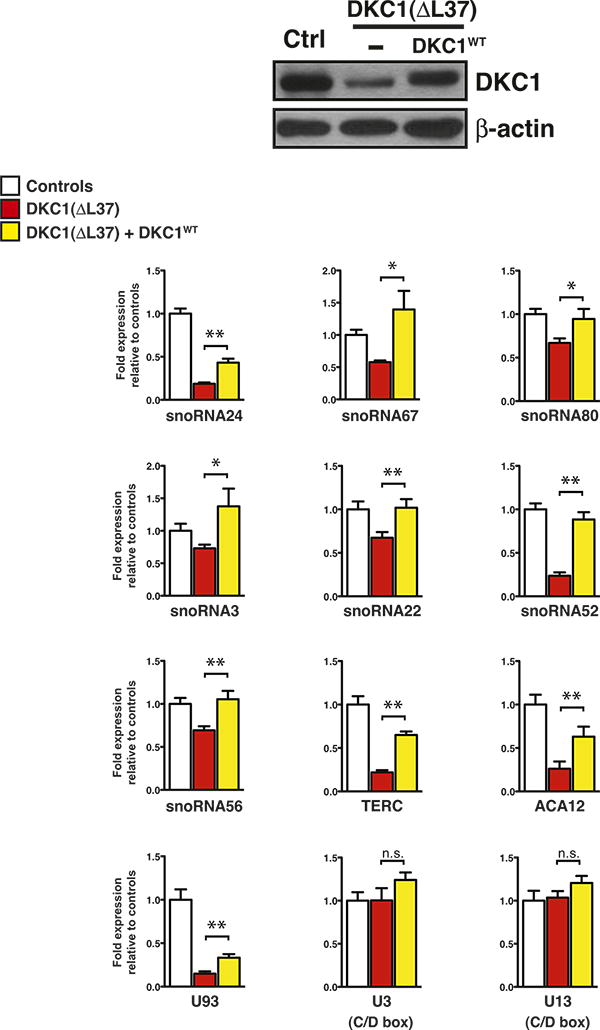
Expression levels of different classes of small RNAs in control, DKC1(ΔL37) primary fibroblasts, and DKC1(ΔL37) primary fibroblasts expressing exogenous wild type dyskerin (DKC1WT) measured by real-time qPCR. Graphs show mean fold expression ± SEM relative to control and normalized to the abundance of RN7SK small non-coding RNA from three independent experiments. Western blot analysis of dyskerin expression is shown in control and DKC1(ΔL37) primary fibroblasts in the absence or presence of DKC1WT (top). β-actin was used as loading control. Statistical analysis was performed using the unpaired Student's t-test, *P<0.05 and **P<0.01.
To establish whether the specific pattern of reductions in H/ACA small RNAs in X-DC cells is directly caused by dyskerin dysfunction, we restored dyskerin expression close to normal levels in primary X-DC fibroblasts (Figure 2). Importantly, reintroduction of dyskerin specifically rescued, to a large extent, the expression of dyskerin-associated H/ACA small RNAs in DKC1(ΔL37) primary fibroblasts compared to controls (Figure 2). Taken together, these results uncover aberrant expression of functionally diverse classes of dyskerin-associated H/ACA small RNAs in patients harboring distinct DKC1 mutations. Furthermore, these findings suggest that variations in the expression of specific H/ACA small RNAs may underlie, at least in part, the wide range and degree of clinical features observed amongst X-DC patients.
Defects in site-specific and global rRNA pseudouridylation are present in X-DC patients
Currently, it remains unknown whether dysregulations in H/ACA small RNA expression translate into defects in specific cellular processes orchestrated by these small RNAs. In particular, H/ACA snoRNAs are assembled into catalytically active RNP complexes through their association with dyskerin, which catalyzes the conversion of ∼100 uridine residues to Ψ in functionally important domains of rRNA (Ni et al., 1997). To ascertain the contribution of altered H/ACA snoRNA expression in X-DC patient cells on Ψ modifications at specific sites on the rRNA, we developed a novel highly sensitive mass spectrometry method for relative quantification of changes in Ψ residues on rRNA at nucleotide resolution (Figure 3A and experimental procedures for details). To our knowledge, this is the first technology that enables quantification of site-specific pseudouridine modifications on individual rRNA nucleotides. In this study we employed this novel technique to monitor the levels of site-specific Ψ modifications on 18S rRNA, as previously reported translational impairments upon dyskerin dysfunction are associated with defects involving the small ribosomal subunit (Bellodi et al., 2010a; Jack et al., 2011; Montanaro et al., 2006; Montanaro et al., 2010). Total 18S rRNA isolated from control and X-DC patient cells was cleaved to generate a mixture of Ψ-containing rRNA oligonucleotides that were subsequently resolved by liquid chromatography tandem mass spectrometry (LC/MS/MS). Briefly, Ψ modifications at specific sites on rRNA oligonucleotides (Figure S3A) were selected based on their m/z ratio and nucleotide sequence. The Ψ signal on each oligonucleotide was quantified and standardized to the amount of a non-Ψ containing oligonucleotide in the mixture (see experimental procedures for details). In particular, we employed two distinct X-DC patient cells including DKC1(ΔL37) primary fibroblasts and immortalized DKC1(T66A) cells. Utilizing this novel methodology, we detected reductions in the amount of Ψ residues at specific sites on rRNA in these X-DC patient cells (Figure 3B), which precisely correspond to reductions in H/ACA snoRNAs annotated to guide these modifications (Figure S3A) (Lestrade and Weber, 2006). For example, in DKC1(ΔL37) fibroblasts, pronounced reductions were observed in Ψ levels at Ψ119, Ψ1367, and Ψ1445 on 18S rRNA compared to control cells (Figure 3B and S3B). Conversely, no significant changes were detected at Ψ105 on 18S rRNA, in agreement with our findings that the expression of the corresponding guide H/ACA snoRNAs, 36A and 50, were not perturbed (Figures 1C and Table S2). In DKC1(T66A) cells, heterogeneous defects in site-specific pseudouridylation on 18S rRNA (Figures 3B and S3C) were also observed, including perturbations in sites that were either reduced or unaltered in DKC1(ΔL37) fibroblasts. For example, Ψ1445 on 18S rRNA is similarly reduced in both DKC1(ΔL37) and DKC1(T66A) cells and this is consistent with decreased expression of the corresponding guide H/ACA snoRNA (snoRNA67) in both cell types (Figures 1C and 3B). By contrast, the site-specific reductions of Ψ105, Ψ1692, or Ψ218 are exclusively observed in either DKC1(T66A) or DKC1(ΔL37) cells, respectively (Figures 3B and 3C). Based on these findings, we have uncovered a direct role for impaired H/ACA snoRNA expression toward defective patterns of rRNA pseudouridylation in X-DC patient cells. We further confirmed these findings by performing quantitative High Performance Liquid Chromatography (HPLC). Early passage DKC1(ΔL37) primary fibroblasts displayed significant reductions in total pseudouridine levels in both 18S and 28S rRNA (Figure 3D). Notably, no differences in the total amount of 18S or 28S rRNA were observed (data not shown). Moreover, significant reductions in the total pseudouridine levels from five different DKC1 point mutations including DKC1(T70I), DKC1(K314R) DKC1(K390Q), DKC1(A353V) and DKC1(T66A) were also observed (Figure 3E). The reductions in the total amount of rRNA pseudouridylation ranging from 10-25% may reflect unique differences in specific patterns of modifications on rRNA as revealed by mass spectrometry. Together, these findings provide the first evidence that rRNA modification defects are common amongst X-DC patients. Moreover, the patterns of Ψ modifications are distinct and directly correlate with specific decreases in subsets of H/ACA snoRNAs originating from distinct DKC1 point mutations.
Figure 3. Defective site-specific and global rRNA pseudouridylation manifest in X-DC patient cells.
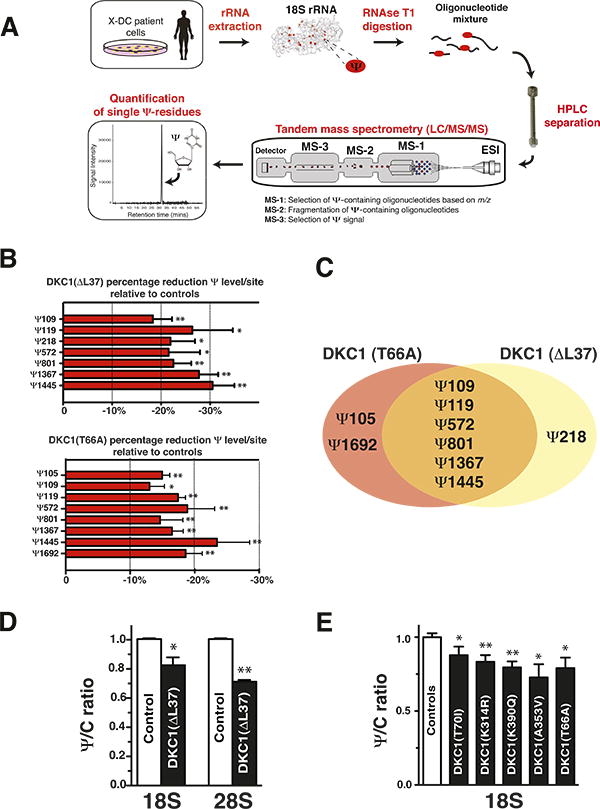
(A) Schematic representation of the mass spectrometry approach used to detect site-specific pseudouridine modifications on 18S rRNA from control and X-DC patient cells. m/z stands for mass-to-charge ratio. ESI is an abbreviation for electrospray ionization. (B) Site-specific quantification of Ψ levels in two controls and in DKC1(ΔL37) fibroblasts (top) and DKC1(T66A) lymphoblasts (bottom). Graph shows mean percentage Ψ reduction ± SEM relative to two controls at specific Ψ sites on 18S rRNA from three independent experiments. Quantification of all Ψ sites examined in this study is shown in Figures S3B-S3C. The accuracy of these measurements is well controlled for by performing calibration curves, demonstrating a linear response to different concentrations of synthetic Ψ-containing oligonucleotides within the range of Ψ values observed in patient samples (Figure S3D). The percentage reductions in Ψ measurements are relative and not absolute. (C) Venn diagram illustrates unique and commonly reduced Ψ sites in X-DC patient cells analyzed. (D) HPLC quantification of 18S and 28S rRNA Ψ levels in control and DKC1(ΔL37) primary fibroblasts. The graph shows mean Ψ to cytosine (Ψ/C) ratio ± SEM relative to controls from two independent experiments. (E) HPLC quantification of 18S rRNA Ψ levels in six independent controls and five X-DC lymphoblast cell lines harboring distinct DKC1 point mutations. The specific DKC1 mutation is presented on each column. The graph shows mean Ψ to cytosine (Ψ/C) ratio ± SEM for each X-DC lymphoblast cell line relative to controls from at least three independent experiments. Statistical analysis was performed using the unpaired Student's t-test, *P<0.05 and **P<0.01.
The pseudouridine synthase capacity of dyskerin is important for hematopoietic stem cell function
As individual H/ACA small RNAs from diverse classes, including those implicated in ribosome, splicing, and telomerase functions, are diminished in X-DC patient cells, an outstanding question is whether impaired dyskerin enzymatic activity is required for some of the pathological features of the disease. This is an important question as dyskerin is able to associate with diverse H/ACA small RNAs to maintain their stability. However, dyskerin acts as a pseudouridine synthase in the context of only a subset of these small RNAs. For example, dyskerin's association with TERC serves to stabilize the telomerase complex; however, its association with H/ACA scaRNAs and snoRNAs converts these RNP particles into catalytically active complexes guiding site-specific conversions of uridine to Ψ residues on snRNAs and rRNA, respectively (Meier, 2005). Given that bone marrow abnormalities are common and amongst the primary causes of early mortality in DC patients, we sought to determine the precise contribution of the pseudouridine synthase activity of dyskerin towards hematopoiesis. To this end, we employed DKC1(c.-141 C>G) primary CD34+ hematopoietic progenitor cells. The DKC1(c.-141 C>G) mutation is characterized by severe features of X-DC, including significant bone marrow hypoplasia and cytopenia (Figure 1A). We introduced expression constructs for wild type DKC1 (designated DKC1WT) or a catalytically inactive DKC1 mutant, D125A (designated DKC1D125A) so that they were both expressed at equivalent levels in DKC1(c.-141 C>G) CD34+ cells (Figures S4A-S4B). DKC1D125A harbors a mutation in a critical amino acid within the evolutionarily conserved TruB Ψ synthase domain that is essential for enzymatic activity (Figures 4A and S4C-S4D) (Hamma et al., 2005; Rashid et al., 2006; Zebarjadian et al., 1999). We next assessed hematopoietic progenitor colony formation on methylcellulose in the presence of a complete cocktail of cytokines that sustains the differentiation of progenitor cells into mature myeloid and erythroid cells (Figure 4B). DKC1(c.-141 C>G) CD34+ cells are greatly impaired in their capacity to differentiate and generate mature myeloid and erythroid colonies as compared to matched healthy human CD34+ cells (Figure 4B). Strikingly, the expression of DKC1WT completely restored the ability of DKC1(c.-141 C>G) CD34+ progenitors to differentiate into mature myeloid and erythroid cells (Figure 4B). In contrast, introduction of the DKC1D125A catalytic mutant failed to rescue the severe differentiation defect present in these cells (Figure 4B). These findings provide significant insight into the molecular basis that may account for specific hematopoietic defects in X-DC patients and strongly implicate dyskerin's pseudouridine synthase enzymatic activity as a requirement for efficient hematopoiesis.
Figure 4. Dyskerin pseudouridylation activity is important for HSC differentiation.
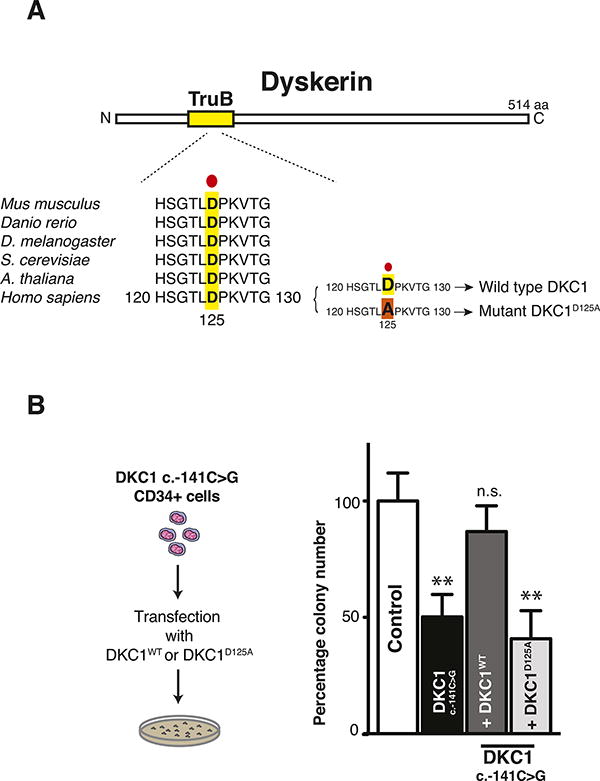
(A) Schematic presentation of dyskerin coding region highlighting evolutionary conservation of the TruB Ψ synthase domain across several species. A substitution of aspartic acid (D) with alanine (A) at position 125 abolishes dyskerin pseudouridylation activity and was employed as a catalytically inactive DKC1(D125A) mutant. (B) Hematopoietic colony forming assay was performed on CD34+ progenitor cells from a healthy control and DKC1(c.-141C>G) patient in the absence or presence of wild type DKC1WT or its catalytic mutant DKC1D125A using methylcellulose supplemented with complete cytokines cocktail sustaining the growth of all blood cell lineages. The total number of colonies formed was scored 10 days after plating. Graph shows the mean total percentage number of colonies ± SEM relative to controls from two independent experiments. Statistical analysis was performed using the unpaired Student's t-test, **P<0.01
Discussion
There is a growing realization that the majority of the human genome previously thought to be “junk DNA” encodes for both short and long ncRNAs, which may instead exert important RNA-based cellular functions. However, the relative contribution of these RNAs to human disease is poorly understood. Our studies reveal that a large class of H/ACA small RNAs are found deregulated in human disease, displaying variable expression patterns that may dictate the number and the severity of disease features. With respect to H/ACA snoRNAs, deregulations in their expression patterns in X-DC directly produce site-specific defects in RNA modifications present on the ribosome, as revealed by state-of-the-art mass spectrometry. These findings provide a functional link between variations in expression patterns of H/ACA snoRNAs in disease and specific alterations in the array of nucleotide modifications present on the ribosome. Moreover, these studies suggest that heterogeneous pools of ribosomes harboring unique differences in modification patterns are present in X-DC patient cells. This heterogeneity in rRNA modifications may have important functional implications for expression of the X-DC patient genome at the post-transcription level and is consistent with previous studies revealing molecular impairments in accurate translational control in X-DC patient cells (Bellodi et al., 2010a; Yoon et al., 2006). Moreover, a specific subgroup of H/ACA snoRNAs is commonly reduced among all DKC1 patient mutations. Intriguingly, these H/ACA snoRNAs guide modifications that cluster primarily within two defined regions of the ribosome (Piekna-Przybylska et al., 2008), including expansion segment 6 (ES6) on 18S rRNA and domain II of 28S rRNA. These findings suggest that additional levels of specificity may coordinate the assembly of specific subsets of H/ACA snoRNPs required for modifications that cluster within functional regions of rRNA and are commonly deregulated in X-DC. Moreover, the mass spectrometry approach we have developed will now make it possible to test the intriguing possibility that different patterns of rRNA pseudouridylation may serve as a mechanism to modulate ribosome function in a cell-type and tissue specific manner.
Our study further reveals an important contribution of the pseudouridine synthase activity of dyskerin in hematopoiesis, which is critically impaired in X-DC patients (Dokal, 2000). This suggests a critical requirement for modifications of specific RNA species in accurate stem cell activity. This is further supported by the diminished expression of H/ACA snoRNAs, which require the catalytic activity of dyskerin. Thus, in addition to impaired telomere maintenance through reductions in TERC expression (Mitchell et al., 1999), heterogeneity in the expression and function of several additional H/ACA small RNAs may cause molecular defects that contribute to X-DC. The analysis of specific subsets of H/ACA small RNAs, such as those identified in our study, may be of great medical importance as novel prognostic markers, where the genetic basis of approximately 50% of DC patients remains unknown (Dokal, 2011). Further evidence for small RNAs in the pathogenesis of DC is highlighted by recent findings that the gene product of C16orf57, mutated in autosomal recessive DC and additional inherited diseases including poikiloderma with neutropenia, is required for the 3′ end processing of spliceosomal U6 small nuclear RNA (Hilcenko et al., 2012; Mroczek et al., 2012; Walne et al., 2010). Intriguingly, hypermethylation of several H/ACA snoRNA loci including snoRNA70C, which our findings reveal is deregulated in X-DC cells, have been reported in solid tumors (Ferreira et al., 2012). Thereby, small ncRNAs such as H/ACA snoRNAs may serve as important molecular markers for human health and should therefore be widely examined at the genomic and expression level in disease pathogenesis.
Our findings suggest that a common feature of human disease arising from perturbations in ncRNAs may be the selective pressure for specific core components of RNP complexes to invariably associate with a multitude of small RNAs, providing greater coordination between RNA-based cellular processes. For example, dyskerin's interactions with small H/ACA RNAs link the activity of a single protein to hundreds of RNAs involved in splicing, telomere activity, and ribosome function (Meier, 2006). Therefore, DKC1 mutations simultaneously affect the expression and function of multiple classes of dyskerin-associated H/ACA small RNAs (Figure 1C), at the nexus of diverse RNA-based cellular processes. Thereby, mutations in genes encoding proteins such as dyskerin may reflect a particular vulnerability to the underlying biology of small RNAs, contributing to human disease. An outstanding question is the molecular nature of the extreme variability in the phenotypic spectrum of DKC1 point mutations that range from severe birth defects, bone marrow failure, to cancer. Impaired modifications of distinct functional RNAs may represent an important, novel molecular mechanism underlying these specific pathological features.
Experimental Procedures
NCI participant lymphocytes
The patients with T70I, K314R, A353V, and K390Q mutations in DKC1 are participants in the Institutional Review Boards (IRBs)-approved longitudinal cohort study at the National Cancer Institute (NCI) entitled “Etiologic Investigation of Cancer Susceptibility in Inherited Bone Marrow Failure Syndromes” (www.marrowfailure.cancer.gov, NCI 02-C-0052, ClinicalTrials.gov Identifier: NCT00027274 (Alter et al., 2010). See Extended Experimental Procedures for detailed methods.
Primary bone marrow progenitor cells
Informed consent was obtained from the DKC1(c.-141 C>G) patient in accordance with a human subjects study protocol approved by the IRBs of the Seattle Children's Hospital and Fred Hutchinson Cancer Research Center. Anonymous healthy control bone marrow cells were obtained from discarded bone marrow harvest screens as approved by the Fred Hutchinson Cancer Research Center IRB.
HPLC and Mass Spectrometry quantification of human rRNA pseudouridylation
Global analysis of rRNA pseudouridine levels was performed by HPLC analysis on a C18 250×4.6 mm (particle size 5 μm) Reverse phase HPLC column (Agilent) as previously described (Jack et al., 2011). For quantifications of site-specific Ψ modifications all LC/MS/MS analyses were performed using an in-house packed capillary column (320 μm ID × 150 mm length, packed with Jupiter 4μm Phenomenex Proteo 90 A material). The solvents used were as described with minor modifications (Apffel et al., 1997). A stock solvent solution was made consisting of 460 ml of HPLC grade water (Fisher Scientific), 42 ml of 1,1,1,3,3,3-hexafluoro-2-propanol (Fluka) and 1.2 ml of Triethylamine (Pierce). Solvent A was prepared by diluting the stock 1:1 V:V with HPLC grade water. Solvent B was prepared by diluting the stock 1:1 V:V with HPLC grade methanol (Fisher). The HPLC system consisted of an Eldex Micropro LC and a Dionex LC-Packings FAMOS auto sampler. The flow rate was 5 to 7 μl/min on a 45 mins linear gradient from 0-70 % B. All samples and standards were dissolved in Solvent A prior to injection. Selected Ψ sites on human 18S rRNA (Figure S3A) were determined by LC/MS/MS following RNase T1 (Roche) digestion using 10U/μg of gel purified 18S rRNA (0.2-1.0 μg) in 10 mM Tris-HCl, 1 mM EDTA, pH 7.4 on a Thermo LTQ Orbitrap Velos mass spectrometer using 90 mins gradients and operated in negative ion detection mode. The IS voltage was set at 4.0 to 5.0 kV, with source temperature at 225°C and a sheath gas flow of 10. The instrument was run in a data dependent mode, using a survey scan from 400-1600, resolution 30,000, and with a 2E6 AGC setting for full scan with one microscan of 250 millisecond (ms). After every survey scan, the top three most intense ions were selected for HCD fragmentation with 2E5 AGC setting, and 3 × 200 ms microscans. The HCD mass range was set at 100-2000. Ions selected for HCD were added to an exclusion list for the next 60 secs. The data files were processed using Xcalibur software suite (Thermo Scientific). Extracted ion profiles for previously characterized Ψ ions at 165.0302 (when the Ψ is at the 5′ end of an oligo) and 207.0400 (when elsewhere in the oligo) (Pomerantz and McCloskey, 2005) were created for all data files and were used to locate and map distinct Ψ-containing oligonucleotides from purified human 18S rRNA. The HCD spectrum was manually interpreted with the aid of Ariadne(Nakayama et al., 2009) and the Mongo Oligo Mass Calculator program (http://medlib.med.utah.edu//massspec/mongo.htm). The identifications of Ψ-containing oligonucleotides were facilitated by the accurate masses (+/- 5 ppm) obtained from the Velos instrument in both full scan and HCD scan mode. See Extended Experimental Procedures for detailed methods.
Telomere length measurements
Telomere length was measured by multicolor flow FISH (Repeat Diagnostics) as described (Alter et al., 2007; Baerlocher et al., 2006).
Supplementary Material
Highlights.
Heterogeneous defects in H/ACA small RNAs manifest in X-DC patients
DKC1 mutations lead to impaired H/ACA small RNA-guided rRNA pseudouridylation
Dyskerin's pseudouridine synthase activity affects hematopoietic stem cell differentiation
Acknowledgments
We thank M. Barna for critical discussion and reading of this manuscript; K. Tong for editing the manuscript; the Fujimori laboratory for technical support and equipment; J. Johl for technical assistance and Lizette Caballero and the UCSF BMT Laboratory for their generous assistance. We also thank Drs. B. P. Alter, and N. Giri at the National Cancer Institute, for clinical characterization of patients and biospecimen collection. We are grateful to the patients for their valuable contributions to this study. C.B. is a fellow of the Leukemia & Lymphoma Society and Aplastic Anemia & MDS International Foundation. D.R. is a Leukemia & Lymphoma Society Scholar. This work was supported, in part, by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health (S.S.). This work is supported by NIH R01DK098057-06A1 (D.R.), NIH R01HL085572 (D.R.), NIH 3R01HL085572-05S1 (D.R.), NIH NIGMS 8P41GM103481 (A.B.) and NIH NIGMS 8P41GM103481 (A.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, Peters JA, Giri N, Lansdorp PM. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, Carr AG, Greene MH, Rosenberg PS. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. British journal of haematology. 2010;150:179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of Oligonucleotides by HPLC-Electrospray Ionization Mass Spectrometry. Analytical chemistry. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaille J. Guiding ribose methylation of rRNA. Trends in biochemical sciences. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat Protoc. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS One. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010a;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010b;70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- Dokal I. Dyskeratosis congenita. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- Ferreira HJ, Heyn H, Moutinho C, Esteller M. CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer. RNA biology. 2012;9:881–890. doi: 10.4161/rna.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Reichow SL, Varani G, Ferre-D'Amare AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol. 2005;12:1101–1107. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Hilcenko C, Simpson PJ, Finch AJ, Bowler FR, Churcher MJ, Jin L, Packman LC, Shlien A, Campbell P, Kirwan M, et al. Aberrant 3′ oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood. 2012 doi: 10.1182/blood-2012-10-461491. [DOI] [PubMed] [Google Scholar]
- Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. rRNA Pseudouridylation Defects Affect Ribosomal Ligand Binding and Translational Fidelity from Yeast to Human Cells. Molecular cell. 2011;44:660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- Knight SW, Heiss NS, Vulliamy TJ, Aalfs CM, McMahon C, Richmond P, Jones A, Hennekam RC, Poustka A, Mason PJ, et al. Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene, DKC1. Br J Haematol. 1999;107:335–339. doi: 10.1046/j.1365-2141.1999.01690.x. [DOI] [PubMed] [Google Scholar]
- Knight SW, Vulliamy TJ, Morgan B, Devriendt K, Mason PJ, Dokal I. Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum Genet. 2001;108:299–303. doi: 10.1007/s004390100494. [DOI] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & development. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic acids research. 2006;34:D158–162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nature reviews Molecular cell biology. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT. How a single protein complex accommodates many different H/ACA RNAs. Trends Biochem Sci. 2006;31:311–315. doi: 10.1016/j.tibs.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Montanaro L. Dyskerin and cancer: more than telomerase. The defect in mRNA translation helps in explaining how a proliferative defect leads to cancer. The Journal of pathology. 2010;222:345–349. doi: 10.1002/path.2777. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Brigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, Taffurelli M, Calienni M, Teruya-Feldstein J, Trere D, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–18. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Calienni M, Bertoni S, Rocchi L, Sansone P, Storci G, Santini D, Ceccarelli C, Taffurelli M, Carnicelli D, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer research. 2010;70:4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kucinski I, Ginalski K, Dziembowski A. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes & development. 2012;26:1911–1925. doi: 10.1101/gad.193169.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T. Ariadne: a database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data. Nucleic Acids Research. 2009;37 doi: 10.1093/nar/gkp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic acids research. 2008;36:D178–183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz SC, McCloskey JA. Detection of the common RNA nucleoside pseudouridine in mixtures of oligonucleotides by mass spectrometry. Anal Chem. 2005;77:4687–4697. doi: 10.1021/ac058023p. [DOI] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Molecular cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic acids research. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchetti D, Todoerti K, Tuana G, Agnelli L, Mosca L, Lionetti M, Fabris S, Colapietro P, Miozzo M, Ferrarini M, et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012;2:e96. doi: 10.1038/bcj.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salowsky R, Heiss NS, Benner A, Wittig R, Poustka A. Basal transcription activity of the dyskeratosis congenita gene is mediated by Sp1 and Sp3 and a patient mutation in a Sp1 binding site is associated with decreased promoter activity. Gene. 2002;293:9–19. doi: 10.1016/s0378-1119(02)00725-4. [DOI] [PubMed] [Google Scholar]
- Teittinen KJ, Laiho A, Uusimaki A, Pursiheimo JP, Gyenesei A, Lohi O. Expression of small nucleolar RNAs in leukemic cells. Cell Oncol (Dordr) 2012 doi: 10.1007/s13402-012-0113-5. [DOI] [PubMed] [Google Scholar]
- Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, Agirre X, Prosper F, Kiss T, Brousset P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012a;26:2052–2060. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- Valleron W, Ysebaert L, Berquet L, Fataccioli V, Quelen C, Martin A, Parrens M, Lamant L, de Leval L, Gisselbrecht C, et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood. 2012b;120:3997–4005. doi: 10.1182/blood-2012-06-438135. [DOI] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. Mutations in C16orf57 and normal-length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Human molecular genetics. 2010;19:4453–4461. doi: 10.1093/hmg/ddq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, Masunari Y, Al-Qurashi FH, Aljurf M, Dokal I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Human Molecular Genetics. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Garlp in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nature reviews Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- Yip S, Butterfield YS, Morozova O, Chittaranjan S, Blough MD, An J, Birol I, Chesnelong C, Chiu R, Chuah E, et al. Concurrent CIC mutations, IDH mutations, and lp/19q loss distinguish oligodendrogliomas from other cancers. The Journal of pathology. 2012;226:7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Molecular and cellular biology. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


