Abstract
Transient receptor potential (TRP) channels are a large family of cation channels. The 28 TRP channel subtypes in rodent are divided into 6 subfamilies: TRPC1-7, TRPV1-6, TRPM1-8, TRPP2/3/5, TRPML1-3 and TRPA1. TRP channels are involved in peripheral olfactory transduction. Several TRPC channels are expressed in unidentified neurons in the main olfactory bulb (OB), but the expression of most TRP channels in the OB has not been investigated. The present study employed RT-PCR as an initial survey of the expression of channel mRNAs in the mouse OB and in 3 cell types: external tufted, mitral and granule cells. All TRP channel mRNAs except TRPV5 were detected in OB tissue. Single cell RT-PCR revealed that external tufted, mitral and granule cell populations expressed in aggregate 14 TRP channel mRNAs encompassing members of all 6 subfamilies. These different OB neuron populations expressed 7 to 12 channel mRNAs. Common channel expression was more similar among external tufted and mitral cells than among these cells and granule cells. These results indicate that a large number of TRP channel subtypes are expressed in OB neurons, providing the molecular bases for these channels to regulate OB neuron activity and central olfactory processing.
Keywords: transient receptor potential (TRP) channel, olfactory bulb, external tufted cell, mitral cell, granule cell, RT-PCR
INTRODUCTION
Transient receptor potential (TRP) channels are a large family of cation channels with wide distribution in the CNS and periphery. The 6 TRP channel subfamilies are present in diverse neuronal and non-neuronal cell types and are involved in numerous functions such as intracellular signaling and cellular responses to changes in the local environment, neuronal excitability, cellular development and death [10, 19, 27]. TRP channels participate in transduction of external stimuli in several sensory systems with a notable TRPV channel role in nociception [4,34].
TRP channels are involved in chemosensory transduction. TRPC2 channels expressed by rodent vomeronasal organ receptor neurons are essential for transduction of pheromones [14,38]. TRPM5 was recently identified in a subset of olfactory receptor neurons involved in responses to urine [15]. In contrast to peripheral olfactory sensory neurons, far less is known about TRP channels in the rodent OB. mRNAs for several members of the TRPC family have been detected in OB tissue and unidentified OB neurons [16,17,20,22,35]. Cation currents with TRP-like electrophysiological and pharmacological properties have also been reported in several OB neuron types [7,24,25]. Beyond these studies however, there is a paucity of information on TRP channel expression in identified OB neuronal populations. The goal of this study, therefore, was to survey the expression of the 28 TRP channel mRNAs in the mouse OB and individual cell types (mitral, external tufted and granule cells) using reverse transcriptase-polymerase chain reaction (RT-PCR) analysis to provide the molecular bases for these channels to regulate OB neuron activity and olfactory processing.
MATERIAL and METHODS
OB tissue RNA extraction
Total tissue RNA was extracted from the OB of 6 male C57BL/6J mice (28-30 days old, Jackson Laboratory). Mice were deeply anesthetized and decapitated in accordance with Institutional Animal Care and Use and National Institutes of Health Guidelines. The brain was removed and the middle one-third of the OB was manually isolated for total RNA extraction with an AM 1560 MirVana miRNA isolation kit (Ambion, Austin, TX). OB tissue (~30 mg) was homogenized with a 10X volume (300 μl) of lysis buffer, then mixed by vortexing with a 1/10 volume of RNA homogenate additive for organic extraction. After 10 min incubation on ice a volume of Acid-Phenol Chloroform equal to the initial lysate volume was added and centrifuged (10,000 x g, 5 min) for aqueous and organic phase separation. A 1.25 volume of 100% ethanol was added to the aqueous phase, the mixture was transferred into filter cartridge and RNA was isolated by centrifugation (10,000 × g, 15 s). Filters were washed twice with 100 μl nuclease-free water and centrifuged (10,000 × g, 30 s) to recover RNA. RNA concentration and purity was detected by UV spectrometry. Only RNA with an A260/280 and A260/230 ratio >1.8 was used for RT-PCR. Ten ng of total RNA from each OB was used for RT.
Slice electrophysiology
Male and female C57BL/6J mice (n=38, 18-30 days old) were deeply anesthetized, decapitated and 400 μm-thick horizontal OB slices were prepared as previously described [7]. A standard artificial cerebrospinal fluid [7] was made with autoclaved ddH2O. Brief visually-guided whole-cell current clamp recordings (<2 min) at room temperature identified external tufted, mitral and granule cells by soma location, input resistance and spontaneous discharge as previously described [6,7,11]; granule cells in the granule cell layer were sampled. Recording pipettes (2-4 μm tip diam., 3-5 MΩ) were heated overnight to 180°C to inactivate RNase and filled with DNase-RNAse free water solution containing (in mM): 124 potassium gluconate, 1 NaCl, 10 phosphocreatine ditris salt, 3 MgATP, 0.3 Na2GTP, 0.5 EGTA, and 10 HEPES; pH 7.3, 290 mOsm. Gentle suction aspirated cytoplasm into the recording pipette; seal resistance was monitored and only cells maintained in whole cell configuration during suction were used. The pipette content was expelled into a 0.2 ml PCR tube. Due to the small size and limited cytoplasmic volume of granule cells, aspirates of two or three cells were pooled. To rule out extracellular mRNA contamination, in 3 experiments patch pipettes were lowered into the tissue without aspirating cytoplasm. Pipette contents from these experiments yielded no positive bands after RT-PCR with GAPDH primers.
RT-PCR
Identical RT-PCR protocols were used for OB tissue and single cell samples. Genomic DNA was digested by DNAse I (5 min at 25°C). cDNA was synthesized using SuperScript III Reverse Transcriptase-based Cells-Direct cDNA Synthesis kit (Invitrogen, Eugene, OR). RT was performed at 50°C for 90 min and the reaction was terminated by heating to 85°C for 5 min. A total of 30 μl cDNA was obtained for immediate PCR reaction or storage at −20°C for later use. Nested PCR amplification was performed as described previously [36]. In the first stage, 5 μl cDNA was amplified using the outer-primer pairs listed in Supplemental Table I; 20 pmol of forward and reverse primer at 0.5 μl was added to the PCR tube with 45 μl high specificity and high-yield Platinum PCR SuperMix (Invitrogen). Thermal cycling was 2 min at 94°C for the initial denaturation, then 35 cycles of 30 s at 94°C to denature, 30 s at 50°C to anneal, and 1 min at 72°C to extend, followed by a 10 min final extension at 72°C. In the second stage, a 5 μl sample from the first stage amplification was used as template for an inner nested primer pair (Supplemental Table I). Forty cycles were performed and annealing temperature was increased to 55°C and the extension was shortened to 50 s. Then 20 μl of individual PCR products was run on a 1.5% agarose gel with 50-650 bp sequence DNA fragments as molecular weight markers. The size of PCR-generated amplicons was as predicted by the mRNA sequences (Supplemental Table I). PCR products from the second stage amplification were separated by 2.5% agarose gel electrophoresis for sequencing study. cDNA bands were cut under UV light. DNA fragments were extracted using a QIAquick gel extraction kit (QIAGEN, MD) and sequenced at the University of Tennessee Health Science Center Molecular Resource Center. Primer pairs for TRP channels, GAPDH and GAD1 were designed based on mouse GenBank sequences (Supplemental Table I) using web-based Primer3 software (http://fokker.wi.mit.edu/primer3/input.htm)(Massachusetts). Except for TRPML2, intron-spanning primer pairs that help detect genomic DNA contamination were used. Primers were synthesized by Integrated DNA Technologies (Coralville, IA). Primer pairs were previously found to positively detect the 28 TRP mRNAs in mouse brain tissue [37]. GAPDH and GAD1 amplicons were not obtained in control experiments performed without RT (n=6 OB samples), indicating that genomic DNA was completely digested by DNase. Amplicons of expected sizes (Supplemental Table I) were obtained from OB tissue and served as positive controls for the single cell results. These positive controls indicate that negative detection in single cells was due to lack or undetectable level of expression. GAPDH mRNA expression for single cells confirmed the success of cytoplasmic aspiration and cDNA synthesis. Samples that lacked GAPDH mRNA expression were discarded. Granule cell samples were further characterized by expression of GAD1 mRNA (GAD67 GABA biosynthetic enzyme).
RESULTS
TRP Channel mRNA in OB Tissue
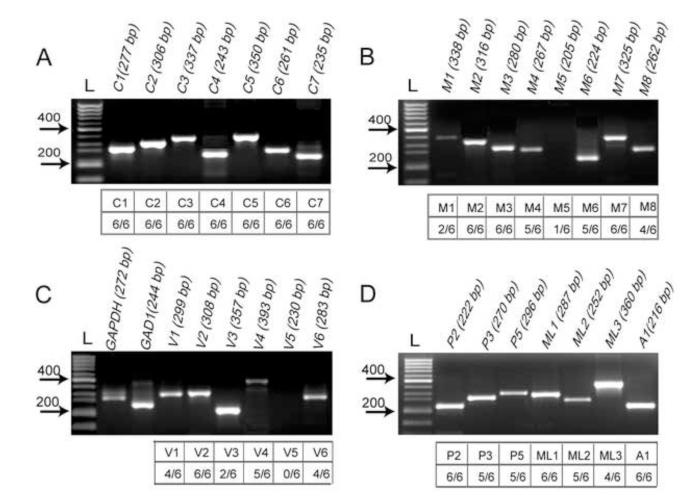
RT-PCR of total RNA from 6 OBs detected the expression of 27 of the 28 known mouse TRP channel mRNAs; TRPV5 was the only TRP channel mRNA not detected (Fig. 1). All PCR amplicons were sequenced and positively identified with TRP channel, GAPDH, and GAD1 mouse gene bank mRNA sequences. In the 6 OB samples, 14 TRP mRNAs were consistently expressed: TRPC1-7, TRPM2/3/7, TRPV2, TRPP2, TRPML1, and TRPA1. mRNA for 10 TRP channels was expressed at a relatively high frequency (i.e., 4-5 of 6 OBs): TRPM4/6/8, TRPV1/4/6, TRPP3/5 and TRPML2/3. By contrast, TRPM1/5 and TRPV3 mRNA were detected in only one or two samples, perhaps due to very low expression level or variability in the portion of the OB sampled.
Fig.1.
RT-PCR analysis of TRP channel mRNA in mouse OB tissue. (A) Representative gel electrophoresis images illustrating mRNA expression of TRPC1-7 amplicons. (B) TRPM1-8 amplicons. (C) GAPDH, GAD1 and TRPV1-6, amplicons. (D) TRPP2/3/5, TRPML1-3 and TRPA1 amplicons. Lanes marked L show the 650bp molecular weight marker used for the determination of amplicons of the expected size (indicated at top in parentheses). The lower tables show the number of positive detections of each TRP channel mRNA in OBs from 6 mice.
TRP Channel mRNA in External Tufted, Mitral and Granule Cells
Single cell RT-PCR was performed on cytoplasmic contents of cells recorded in OB slices from 38 mice. Electrophysiological properties of recorded and aspirated external tufted, mitral and granule cells are shown in Figure 2. All rodent TRP channel mRNAs were examined except TRPV5 which was not detected in OB tissue. Each TRP mRNA was examined in 4-11 individual external tufted or mitral cells, or 8-24 granule cells. OB neurons expressed a more restricted subset of TRP channels – 14 of 27 – than detected in the tissue.
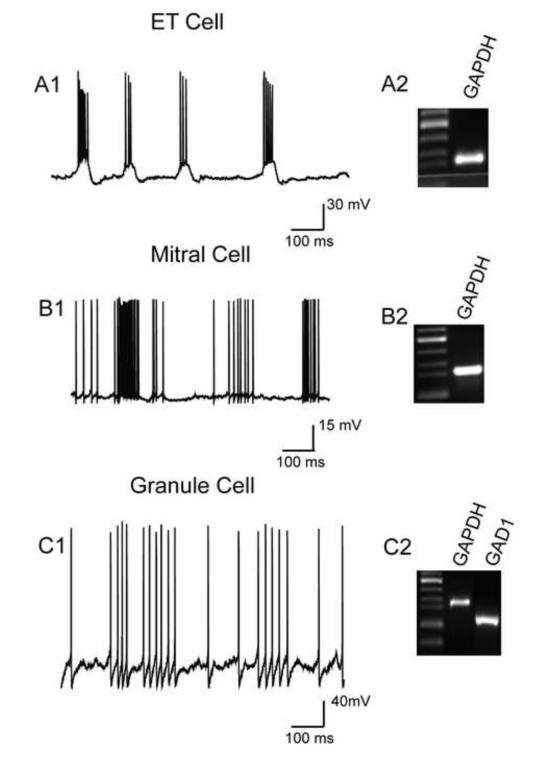
Fig.2.
Identification of external tufted, mitral and granule cells for single cell RT-PCR. (A1) External tufted cells were identified by soma in the glomerular layer and in current clamp recordings by spontaneous rhythmic spike bursts; their input resistance was 186±8.6 MΩ. (B1) Mitral cells were spontaneously active, tended to exhibit non-rhythmic spike clusters, and their input resistance was 170±12.6 MΩ. (C1) Granule cells in the internal granule cell layer had high input resistances (905±34.2 MΩ) and exhibited low levels of spontaneous spikes (n=37) or did not spike (n=21). This example was silent at rest and was induced to spike by 5.7 pA current injection. (A2-C2) Gel electrophoresis of GAPDH amplicons detected in aspirates from single external tufted or mitral cells, and GAPDH and GAD1 amplicons in pooled aspirates from 2-3 granule cells.
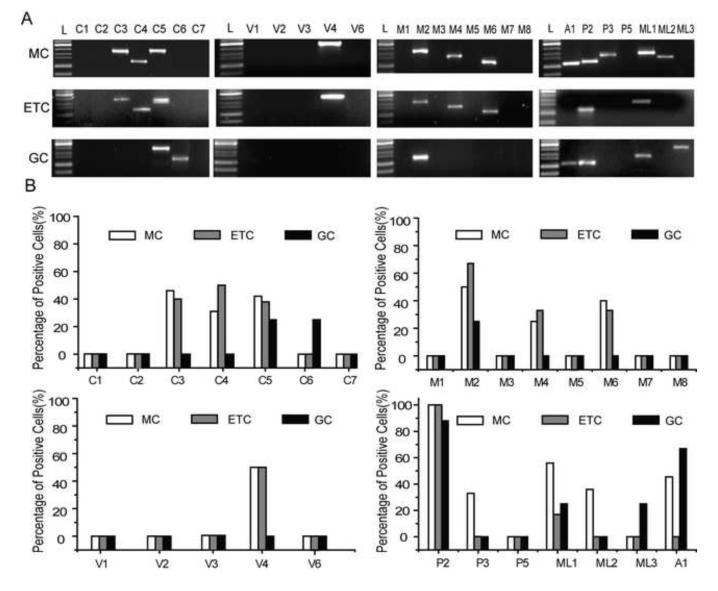
First, we examined the expression of TRP channel mRNAs in mitral cells. Sixty three electrophysiologically identified mitral cells with positive GAPDH mRNA detection were included in our analysis. As shown in Figure 3, in these mitral cells, we detected 12 TRP channel mRNAs: TRPC3-5, TRPM2/4/6, TRPV4, TRPP2-3, TRPML1-2 and TRPA1. TRPP2 mRNA was detected in all mitral cells, while the detection of the other TRP mRNAs was variable (Fig. 3B).
Fig.3.
Single cell RT-PCR analysis of TRP channel mRNA expression in OB neurons. (A) Representative gel images illustrating mRNA expression of TRP mRNA from mitral (MC), external tufted (ETC) and granule cell (GC) aspirates. (B) The percentage of neurons positive for detection of different TRP channel mRNAs. Pooled GC aspirates were considered as single samples.
We next surveyed TRP channel mRNAs in 51 external tufted cells with positive GAPDH mRNA detection. In this population, we detected 9 TRP channel mRNAs: TRPC3-5, TRPM2/4/6, TRPV4, TRPP2 and TRPML1 (Fig. 3A). Like mitral cells, TRPP2 was detected in all external tufted cells and the other channel mRNAs were detected less frequently (Fig. 3B). The detection profile of TRP mRNAs in mitral and external tufted cells was similar. Specifically, these two cell types expressed 9 common TRP channel mRNAs. Expression of members of the TRPC, TRPM and TRPV channel subfamilies was identical among mitral and external tufted cell populations. Unlike mitral cells, TRPP3, TRPML2 and TRPA1 mRNA were not detected in external tufted cells. Thus, mitral cells expressed all TRP channels detected in external tufted cells, whereas external tufted cells expressed the majority (75%) of the TRP channels expressed in mitral cells.
Finally, we investigated TRP channel mRNA expression in granule cells. Thirty pooled granule cell samples (from 60 granule cell aspirates) with positive detection for both GAPDH and GAD1 mRNA were included in our analysis. We detected 7 TRPC channel mRNAs: TRPC5-6, TRPM2, TRPP2, TRPML1/3 and TRPA1 (Fig. 3A-B). Like mitral and external tufted cells, TRPP2 was detected at a high frequency (~88%). Unlike mitral and external tufted cells, in these granule cells, we detected TRPC6 and TRPML3, whereas TRPV subfamily members were not detected. Furthermore, in contrast to the high similarity of TRP channel expression among mitral and external tufted cells, granule cells expressed 42.5% and 44% of the channels detected in mitral and external tufted cells. However, it should be noted that TRP channel expression in granule cells detected here is likely to be an underestimate. We used GAD1 mRNA, which codes for the GAD67 isoform, as a positive marker for GABAergic granule cells. Approximately 50% of the deep granule cell population expresses GAD65, but not the GAD67, isoform [21].
DISCUSSION
This study provides the first comprehensive survey of the TRP channel family expression in mouse OB tissue and in several OB neuronal subtypes. Our results show that OB tissue expresses mRNA for a large number of TRP channels including members of all 6 TRP channel subfamilies. mRNAs for 27 of the 28 known TRP channels were detected in OB tissue with TRPV5 the one exception. TRP channel mRNA expression in OB neurons was also diverse and included members of all 6 TRP channel subfamilies. In aggregate, mitral, external tufted and granule cells expressed 14 TRP channel mRNAs, ranging from 7 to 12 channels per neuronal subtype. These results suggest that TRP channels participate in diverse functions in OB in general and in OB neuronal function specifically.
We are unaware of any comparable comprehensive surveys of the TRP channel family mRNA in other brain regions or neurophysiologically identified neurons. Certainly, these results are tempered by the caveat mRNA detection may not equate to protein translation or that mRNA expression may not linearly correspond to protein expression. Also, TRP channel mRNAs detected in OB tissue include potential contributions from OB neurons and non-neuronal components including blood cells, endothelial cells and neurovasculature pericytes, the meninges, and glia. TRP channels are widely expressed by non-neuronal cell types elsewhere [2], with scant data on their expression in nonneuronal OB cells. TRPV1 protein is expressed in hippocampal astrocytes and pericytes and TRPV1 protein is present in OB tissue [31]. TRPV1 mRNA was detected in OB tissue here, but not in the 3 neuronal types examined. Thus, TRPV1 and other channels may be present in non-neuronal OB cell types. It is noteworthy that olfactory ensheathing cells in the OB exhibit TRPC-mediated calcium fluxes [5]. These cells and astrocytes in the glomeruli, express metabotropic glutamate receptors which have been linked to TRP channel activation [23,29]. OB tissue may also contain mRNA derived from olfactory nerve terminals as they are known to express olfactory receptor mRNA and protein [28, 33]. Olfactory receptor neurons have been reported to express TRPC2, TRPM5/8, TRPV1-4, TRPML3 and TRPA1 [1,14,15,18]. Our results indicate that mRNA for most of the preceding channels was found in OB tissue, but not in the 3 neuron types examined. It is not known, however, if TRP channel mRNA expressed by olfactory receptor neurons is transported to their axon terminals in the OB.
Our results indicate that mitral, external tufted and granule cell populations express multiple TRP channel mRNAs spanning several subfamilies. Expression of the entire TRP family in specific neuronal subtypes has not been studied elsewhere in the brain. Several studies, however, indicate that individual neurons express multiple channels within a specific TRP family. For example, dopaminergic substantia nigra neurons as a population were found to express mRNA for 5 TRPC channels [32]. Similarly, populations of hypothalamic neurons containing proopiomelanocortin, gonadotropin-releasing hormone or hypocretin/orexin express 5-6 members of the TRPC family [3,26,36]. Expression of other TRP family channels was not examined in these studies, but it is reasonable to speculate that these and other neuronal types may express channels for other TRP families. In comparison to these studies, our results indicate that OB neurons express a more restricted subset (2-3) of TRPC channel mRNAs.
With the exception of the TRPC family, there is little information available on the expression of TRP channels in OB neurons. Unidentified cells in the mouse glomerular layer have been reported to express TRPC1/3/4/5/6 as determined by in situ hybridization [20,22]. Our results indicate that external tufted cells express TRPC3-5, suggesting that the other members of the TRPC family expressed in the glomerular layer (i.e., TRPC1/6) may be localized to periglomerular or short axon interneurons. Cells in the mitral cell layer were reported to express mRNA for TRPC3/4/5 [20,22], but since the mitral cell layer contains more granule cells than mitral cells, the cell type was unclear in these studies. However, our results show that mitral cells specifically express TRPC3/4/5, in agreement with the preceding studies. Neurons in the granule cell layer express TRPC1/4 [20,22,30]. We did not detect TRPC1/4 expression in granule cells. The reason for this discrepancy is unclear, but it is possible that these TRPC channels are restricted to granule cells that express the GAD65 isoform.
Among the TRP superfamily, TRPC channels are engaged by G-protein coupled receptors such as metabotropic glutamate receptors [2,8,9,13,32]. In this regard, it is interesting to note that activation of the metabotropic glutamate receptor mGluR1 subtype elicits a cation current in external tufted cells [7]. This current is blocked by flufenamic acid and SKF96365, compounds that have been reported to block two of the TRPC channels expressed by external tufted cells, TRPC3/5 [2,27]. mGluR1 activation also elicits a cation current in mitral cells [12]. Mitral cells, like external tufted cells, were found to express TRPC3/5 channels in the present study, suggesting that these channels may mediate the mGluR1-activated current in mitral cells. Serotonin recently was found to elicit a cation current in external tufted cells that was blocked by 2-aminoethyl diphenylborinate (2-ABP). TRPC3/5 channels are blocked by 2-ABP and are thus potential channels for the serotonin current in external tufted cells [2,27].
Granule cells were found to express TRPC5 mRNA, in common with external tufted and mitral cells, and also TRPC6 mRNA. Granule cells exhibit an afterdepolarization mediated by a nonselective cation current that is enhanced by muscarinic receptor activation and attenuated by flufenamic acid [25]. Our results indicate that granule cells express two channels sensitive to flufenamic acid, TRPC5 and TPRM2 [2,27]. Taken together with these findings, our results suggest that TRP channels may mediate G-protein coupled receptor responses in several classes of OB neurons and play important roles in the processing of olfactory signals.
Supplementary Material
Highlight.
TRP channel mRNAs were detected in OB tissue and its neurons using RT-PCR.
All 28 TRP channel mRNAs except TRPV5 were detected in OB tissue.
Total 14 TRP channel mRNAs over 6 subfamilies were screened in three cell types.
Common channel expression was similar between external tufted and mitral cells.
Acknowledgements
This work was supported by NIH grants DC009049, DC003195, NS058850 and University of Tennessee William Webster Endowment Fund Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Castiglioni AJ, Remis NN, Flores EN, Garcia-Anoveros J. Expression and vesicular localization of mouse Trpml3 in stria vascularis, hair cells, and vomeronasal and olfactory receptor neurons. J. Comp. Neurol. 2011;19:1095–1114. doi: 10.1002/cne.22554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Clapham DE, Julius D, Montell C, Schultz G. International Union of Pharmacology, XLIX, nomenclature and structure-function relationships of transient receptor potential channels. Pharmacol. Rev. 2005;57:427–450. doi: 10.1124/pr.57.4.6. [DOI] [PubMed] [Google Scholar]
- [3].Cvetkovic-Lopes V, Eggermann E, Uschakov A, Grivel J, Bayer L, Jones BE, Serafin M, Muhlethaler M. Rat hypocretin/orexin neurons are maintained in a depolarized state by TRPC channels. Plos One. 2010;5:e15673. doi: 10.1371/journal.pone.0015673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Damann N, Voets T, Nilius B. TRPs in our sense. Curr Biol. 2008;18:R880–R889. doi: 10.1016/j.cub.2008.07.063. [DOI] [PubMed] [Google Scholar]
- [5].Davies R, Hayat S, Wigley CB, Robbins J. The calcium influx pathway in rat olfactory ensheathing cells shows TRPC channel pharmacology. Brain Res. 2004;1023:154–156. doi: 10.1016/j.brainres.2004.07.032. [DOI] [PubMed] [Google Scholar]
- [6].Dong HW, Hayar AM, Ennis M. Activation of group I metabotropic glutamate receptors on main olfactory bulb granule cells and periglomerular cells enhances synaptic inhibition of mitral cells. J. Neurosci. 2007;27:5654–5663. doi: 10.1523/JNEUROSCI.5495-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dong HW, Hayar AM, Callaway J, Yang XH, Nai Q, Ennis M. Group I mGluR activation enhances Ca2+ dependent nonselective cation currents and rhythmic bursting in main olfactory bulb external tufted cells. J. Neurosci. 2009;29:11943–11953. doi: 10.1523/JNEUROSCI.0206-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Faber ES, Sedlak P, Vidovic M, Sah P. Synaptic activation of transient receptor potential channels by metabotropic glutamate receptors in the lateral amygdale. Neurosci. 2006;137:781–794. doi: 10.1016/j.neuroscience.2005.09.027. [DOI] [PubMed] [Google Scholar]
- [9].Gee CE, Benquet P, Gerber U. Group I metabotropic glutamate receptors activate a calcium-sensitive transient receptor potential-like conductance in rat hippocampus. J. Physiol. 2003;546:655–664. doi: 10.1113/jphysiol.2002.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hardie RC. TRP channels and lipids: from drosophila to mammalian physiology. J. Physiol. (Lond) 2007;578(Pt 1):9–24. doi: 10.1113/jphysiol.2006.118372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Heinbockel T, Hamilton KA, Ennis M. Metabotropic glutamate receptors in the main olfactory bulb drive granule cell-mediated inhibition. J. Neurophysiol. 2007;97:858–870. doi: 10.1152/jn.00884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heinbockel T, Heyward P, Conquet F, Ennis M. Regulation of main olfactory bulb mitral cell excitability by metabotropic glutamate receptor mGluR1. J. Neurophysiol. 2004;92:3085–3096. doi: 10.1152/jn.00349.2004. [DOI] [PubMed] [Google Scholar]
- [13].Kim SJ, Kim YS, Yuan JP, Petralia RS, Worley PF, Linden DJ. Activation of the TRPC1 cation channel by metabotropic glutamate receptor mGluR1. Nature. 2003;426:285–291. doi: 10.1038/nature02162. [DOI] [PubMed] [Google Scholar]
- [14].Liman ER, Corey DP, Dulac C. TRP2: a candidate transduction channel for mammalian pheromone sensory signaling. Proc. Natl. Acad. Sci. USA. 1999;96:5791–5796. doi: 10.1073/pnas.96.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lin WH, Margolskee R, Donnert G, Hell SW, Restrepo D. Olfactory neurons expressing transient receptor potential channel M5 (TRPM5) are involved in sensing semiochemicals. Proc. Natl. Acad. Sci. USA. 2007;104:2471–2476. doi: 10.1073/pnas.0610201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mizuno N, Kitayama S, Saishin Y, Shimada S, Morita K, Mitsuhata C, Kurihara H, Dohi T. Molecular cloning and characterization of rat trp homologues from brain. Mol. Brain Res. 1999;64:41–51. doi: 10.1016/s0169-328x(98)00296-4. [DOI] [PubMed] [Google Scholar]
- [17].Mori Y, Takada N, Okada T, Wakamori M, Imoto K, Wanifuchi H, Oka H, Oba A, Ikenaka K, Kurosaki T. Differential distribution of TRP Ca2+ channel isoforms in mouse brain. Neuroreport. 1998;9:507–515. [PubMed] [Google Scholar]
- [18].Nakashimo Y, Takumida M, Fukuiri T, Anniko M, Hirakawa K. Expression of transient receptor potential channel vanilloid (TRPV) 1ߝ4, melastin (TRPM) 5 and 8 and ankyrin (TRPA1) in the normal and methimazole-treated mouse olfactory epithelium. Acta Otolaryngol. 2010;130:1278–1286. doi: 10.3109/00016489.2010.489573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol. Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- [20].Otsuka Y, Sakagami H, Owada Y, Kondo H. Differential localization of mRNAs for mammalian trps, presumptive capacitative calcium entry channels, in the adult mouse brain. Tohoku J. Exp. Med. 1998;185:139–146. doi: 10.1620/tjem.185.139. [DOI] [PubMed] [Google Scholar]
- [21].Parrish-Aungst S, Shipley MT, Erdelyi F, Szabo G, Puche AC. Quantitative analysis of neuronal diversity in the mouse olfactory bulb. J. Comp. Neurol. 2007;50:825–836. doi: 10.1002/cne.21205. [DOI] [PubMed] [Google Scholar]
- [22].Philipp S, Hambrecht J, Braslavski L, Schroth G, Freichel M, Murakami M, Cavalie A, Flockerzi V. A novel capacitative calcium entry channel expressed in excitable cells. EMBO J. 1998;17:4274–4282. doi: 10.1093/emboj/17.15.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].van den Pol AN, Santarelli JG. Olfactory ensheathing cells: time lapse imaging of cellular interactions, asonal support, rapid morphologic shifts, and mitosis. J. Comp. Neurol. 2003;458:175–794. doi: 10.1002/cne.10577. [DOI] [PubMed] [Google Scholar]
- [24].Pressier RT, Strowbridge BW. Blanes cells mediate persistent feedforward inhibition onto granule cells in the olfactory bulb. Neuron. 2006;49:889–904. doi: 10.1016/j.neuron.2006.02.019. [DOI] [PubMed] [Google Scholar]
- [25].Pressler RT, Inoue T, Strowbridge BW. Muscarinic receptor activation modulates granule cell excitability and potentiates inhibition onto mitral cells in the rat olfactory bulb. J. Neurosci. 2007;27:10969–10981. doi: 10.1523/JNEUROSCI.2961-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Qiu J, Fang Y, Rønnekleiv OK, Kelly MJ. Leptin excites proopiomelanocortin neurons via activation of TRPC channels. J. Neurosci. 2010;30:1560–1565. doi: 10.1523/JNEUROSCI.4816-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu. Rev. Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- [28].Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- [29].Rieger A, Deitmer JW, Lohr C. Axon-glia communication evokes calcium signaling in olfactory ensheathing cells of the developing olfactory bulb. Glia. 2007;55:352–359. doi: 10.1002/glia.20460. [DOI] [PubMed] [Google Scholar]
- [30].Stroh O, Freichel M, Kretz O, Birnbaumer L, Hartmann J, Egger V. NMDA receptor-dependent synaptic activation of TRPC channels in olfactory bulb granule cells. Neurosci. 2012;32:5737–5746. doi: 10.1523/JNEUROSCI.3753-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Toth A, Boczan J, Kedel N, Lizanecz E, Bagi Z, Papp Z, Edes I, CSIBA L, Blumberg PM. Expression and distribution of vaqnilloid receptor 1(TRPV1) in the adult rat brain. Brain Res. Mol. Brain Res. 2005;135:162–168. doi: 10.1016/j.molbrainres.2004.12.003. [DOI] [PubMed] [Google Scholar]
- [32].Tozzi A, Bengtson CP, Longone P, Carignani C, Fusco FR, Bernardi G, Mercuri NB. Involvement of transient receptor potential-like channels in responses to mGluR-1 activation in midbrain dopamine neurons. Eur. J. Neurosci. 2003;18:2133–2145. doi: 10.1046/j.1460-9568.2003.02936.x. [DOI] [PubMed] [Google Scholar]
- [33].Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- [34].Venkatachalam K, Montell C. TRP Channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zechel S, Werner S, von Bohlen, Halbach O. Distribution of TRPC4 in developing and adult murine brain. Cell Tissue Res. 2007;321:651–656. doi: 10.1007/s00441-007-0388-4. [DOI] [PubMed] [Google Scholar]
- [36].Zhang C, Roepke TA, Kelly MJ, Rønnekleiv OK. Kisspeptin depolarizes gonadotropin-releasing hormone neurons through activation of TRPC-like cationic channels. J. Neurosci. 2008;28:4423–4434. doi: 10.1523/JNEUROSCI.5352-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou FW, Matta SG, Zhou FM. Constitutively active TRPC3 channels regulated basal ganglia output neurons. J. Neurosci. 2008;28:473–482. doi: 10.1523/JNEUROSCI.3978-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zufall F. TRPC2 ion channel and pheromone sensing in the accessory olfactory system, Naunyn-Schmiedeberg′s Arch. Pharmacol. 2005;371:245–250. doi: 10.1007/s00210-005-1028-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.