Abstract
Plant non-symbiotic hemoglobins possess hexa-coordinate heme geometry similar to the heme protein neuroglobin. We recently discovered that deoxygenated neuroglobin converts nitrite to nitric oxide (NO), an important signaling molecule involved in many processes in plants. We sought to determine whether Arabidopsis thaliana non-symbiotic hemoglobins class 1 and 2 (AHb1 and AHb2) might function as nitrite reductases. We found that the reaction of nitrite with deoxygenated AHb1 and AHb2 generates NO gas and iron-nitrosyl-hemoglobin species. The bimolecular rate constants for nitrite reduction to NO are 19.8 ± 3.2 and 4.9 ± 0.2 M−1s−1, at pH = 7.4 and 25°C, respectively. We determined the pH dependence of these bimolecular rate constants and found a linear correlation with the concentration of protons, indicating the requirement for one proton in the reaction. Release of free NO gas during reaction in anoxic and hypoxic (2% oxygen) conditions was confirmed by chemiluminescence detection. These results demonstrate that deoxygenated AHb1 and AHb2 reduce nitrite to form NO via a mechanism analogous to that observed for hemoglobin, myoglobin and neuroglobin. Our findings suggest that during severe hypoxia and in the anaerobic plant roots, especially in water submerged species, non-symbiotic hemoglobins provide a viable pathway for NO generation via nitrite reduction.
Nitric oxide (NO) is a diffusible and short-lived free radical gas with a wide range of functions in both eukaryotes and prokaryotes (1). NO is produced in plants in response to bacterial or viral exposure and plays a role in multiple processes (2, 3). Currently, two major pathways for NO formation in plants are accepted: one route involves enzymatic and non-enzymatic nitrate and nitrite reduction (4–6) and the other arginine oxidation (7, 8). However, the mechanisms of NO production in plants are not determined and controversial (9, 10). Although the metabolic source of NO in plants remains uncertain, accumulating evidence suggests that nitrite can be a source of NO in mammals under hypoxic/ischemic conditions (11–13). Several hemeproteins, iron–sulfur cluster containing proteins, and molybdenum-based reductases, have been recently proposed as nitrite reductases (14). In addition, the pentacoordinate respiratory hemeproteins hemoglobin (Hb) and myoglobin (Mb) and hexacoordinate neuronal protein neuroglobin (Ngb) have shown the ability to reduce nitrite to NO under both physiological and pathological hypoxia via reactions in equation 1–2 (15–17).
| (equation 1) |
| (equation 2) |
Hemoglobins are an ancient class of molecules ubiquitous in eukaryotes and uniting almost all forms of life (18). Plant hemoglobins were first found in root nodules of plants capable of symbiotic nitrogen fixation where they function to regulate oxygen delivery and were then termed symbiotic Hb. The subsequent discovery of non-symbiotic plant hemoglobins (nsHbs) renewed research interest (19, 20). Plant nsHbs have been divided into two molecular species, class 1 and class 2, which have a sequence identity of approximately 60% but differ from each other in phylogenetic characteristics, gene expression pattern and oxygen binding properties (21, 22). Both classes possess at least partial hexacoordinate heme geometry with proximal and distal histidines directly bound to the heme iron like mammalian neuroglobin and cytoglobin. However, despite competing with the distal histidine for the same iron binding site, both nsHbs bind oxygen with very high affinity at physiological pH (Kd ~ 2 nM and ~100 nM respectively for Arabidopsis thaliana nsHb class 1 and class 2, AHb1 and AHb2) (23). For this reason it has been suggested that they do not function for oxygen transport or storage but are involved in metabolic reactions (24), although their physiological function remains unclear. Oxygen bound AHb1 has been shown to have NO-dioxygenase activity using NADPH as electron donor and producing nitrate and ferric hemoglobin (25). This reaction has been proposed in NO detoxification by acting as an NO scavenger (26). AHb2, is induced by low temperatures (cold-stress) and ubiquitously expressed at low levels: transgenic over-expression leads to a significant increase (40%) in the plant metabolic performance (27) but its role in plant physiology remains elusive.
The recent finding that deoxygenated Hb, Mb and Ngb can reduce nitrite to form NO under hypoxic and anoxic conditions (16, 17, 28), suggests that this activity is inherent to the heme moiety of the globins. This hypothesis is further supported by a recent report by Sturms and Hargrove (29) that extends the ability of nitrite reduction to ferrous cyanobacterial hemoglobin from Synechocystis and rice non-symbiotic hemoglobin 1. However, their direct attempts to measure NO release using an NO electrode were unsuccessful and whether these hemoglobins can produce authentic NO from nitrite has not been evaluated to date.
Here we tested whether Arabidopsis thaliana nsHb class 1 and 2 can function as nitrite reductases and pursued kinetic characterization of the reaction with nitrite during anoxia and 2% oxygen hypoxia. We found that AHb1 and AHb2 are capable of reducing nitrite to NO and the reaction is proton dependent in the physiological range.
MATERIALS AND METHODS
All reagents were purchased from Sigma-Aldrich unless otherwise specified. UV-visible spectra and kinetic data were recorded on a HP8453 UV-Vis spectrophotometer (Agilent, Santa Clara, CA) using 1 cm pathlength quartz or special optical glass cuvettes. Solutions of sodium dithionite and nitrite were prepared and kept at 25°C with argon degassed 0.1 M phosphate buffer (pH 7.5) under inert gas.
Cloning, expression and purification of recombinant AHb1 and AHb2
Recombinant AHb1 and AHb2 were expressed in E. coli BL21(DE3). Restriction digestions, ligation, transformation, cloning, bacterial growth and isolation of DNA fragments were performed using standard techniques. Purification was carried out as described with minor modification (30). To increase purification yield, AHb1 and AHb2 cDNA was fused with a 6xHis tag in the N-termini and cloned into pET28a. Purification of His tagged AHb1 and AHb2 was performed using Ni-NTA-agarose (Qiagen) affinity column according to the manufacturer’s manual, and the additional amino acids at the N-terminus were removed using a thrombin cleavage capture kit (Novagen). The eluted protein was dialyzed against PBS at 4°C, concentrated with a 10-kD cutoff filter and stored in aliquots at −80°C. The purity of each recombinant batch prepared was assessed by SDS-PAGE and UV-visible spectroscopy.
Sample Preparation
Proteins were fully oxidized with excess potassium ferricyanide or fully reduced by incubation with 100 mM sodium dithionite; excess reagents were removed by passing the mixture through two sequential Sephadex G-25 desalting columns. Concentrations were estimated by measuring the Soret peak absorbance at 425 nm of the deoxy ferrous form using ε = 92 mM−1cm−1 (26).
Reference spectra of individual species
Standard reference species of recombinant AHb1 and AHb2 were prepared following procedures previously described for other hemoglobins (15, 17). Deconvolution of spectra into individual species was accomplished with multilinear regression analysis, using a set of pure spectra of deoxy-, iron-nitrosyl-, met-, and oxy- AHb1 and AHb2 species as a reference basis (Supplemental Figure 1).
Anaerobic reactions of globins with nitrite
Anaerobic reduced samples were prepared in a glovebox under a 2–4% H2 atmosphere of catalyst-deoxygenated nitrogen, collected directly in cuvettes and sealed with rubber septa inside the glovebox before use. Reaction kinetics of known amounts of AHbs with nitrite were monitored by absorption spectroscopy for the indicated time in a cuvette in the absence or in the presence of 3 mM sodium dithionite. All reactions were run at 25°C in 0.1 M phosphate buffer at controlled pH. Previously deoxygenated nitrite was added, using an airtight syringe, to a sealed anaerobic cuvette to initiate the reaction. Oxygen contamination was prevented by application of positive argon pressure. The time-dependent changes of single species of AHb1 and AHb2 were calculated by least squares deconvolution of the reaction spectra. To vary pH, deoxy species and nitrite were prepared in phosphate buffer adjusted to the target pH values. Determined values are the mean ± S.D. from triplicate experiments representative of two independent enzyme preparations.
Measurement of NO emission in the gas phase
NO gas liberated from a reaction mixture was measured by ozone-based chemiluminescence NO analyzer (CLD88Y; Eco Physics Inc., Ann Arbor, MI and Sievers, GE Analytical Instruments, Boulder, CO) in real-time. The reactions of deoxy- AHb1 and AHb2 (prepared by titrating stock proteins with a stoichiometric amount of dithionite in an anaerobic cuvette and then diluted to a final concentration of 25 μM) were carried out in 100 mM phosphate buffer, pH = 7.4 at 25 °C, either in a vessel purged with helium gas in studies under anaerobic conditions or in a flat flasks (surface area = 25 cm2) purged with 2% O2/98% N2 gas mixture without bubbling and strictly regulating the flow rate (50 ml/min) for hypoxic conditions. Once a stable baseline was established the indicated amount of nitrite was injected in the mixture as previously described (15). In order to test whether the release of NO into the gas phase from the solutions could be used as a continuous measurement for the NO production, we build a calibration curve using amounts of NO validated by injection of sodium nitrite standards into tri-iodide solution.
RESULTS
Nitrite is reduced to NO via reaction with deoxygenated AHb1 and AHb2
To examine the reaction of nitrite with plant Arabidopsis thaliana nsHbs we used recombinant AHb1 and AHb2. Spectrophotometric analysis of the purified proteins confirmed the presence of hexacoordinated heme in both the ferrous and ferric states with visible peaks at 529 nm (α band) and 558 nm (β band), typically observed for hexacoordinate systems (supplemental Figure 1). However, a substantial fraction of the AHb1 molecules displayed a pentacoordinate heme geometry as previously reported (estimated about 40% by spectral deconvolution) (31).
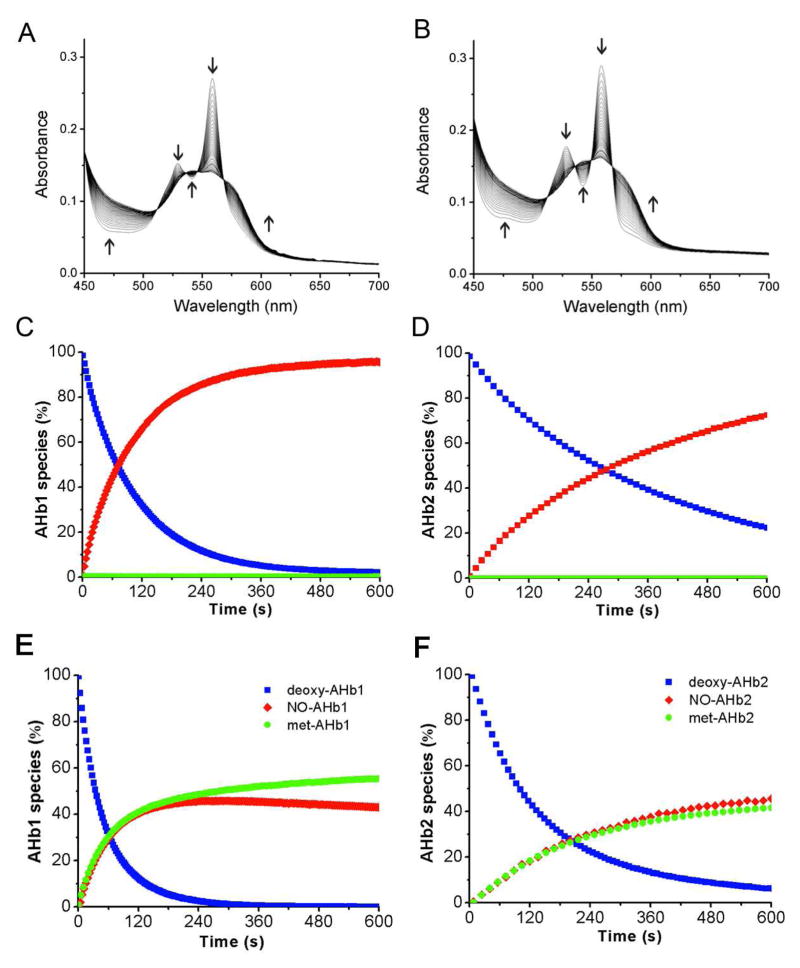
We prepared ferrous deoxygenated AHb1 and AHb2 in an anaerobic glove box as detailed in the Methods and performed anaerobic experiments both in the presence and in the absence of 3 mM sodium dithionite. We recorded the visible spectra of the reaction between the proteins and deoxygenated nitrite at 25°C in a spectrophotometer at constant intervals in a sealed air tight cuvette as exemplified in Figure 1A, B with 0.25 mM nitrite at pH = 7.4 and in the presence of excess dithionite. Upon addition of nitrite the spectrum shifts from ferrous heme species to ferrous-nitrosyl species with clear isosbestic points. The traces extracted at 558 and 576 nm were fitted to a single exponential equation and the concentrations of single species of AHb1 and AHb2 as a function of time were calculated by least squares deconvolution of the reaction spectra. The reaction in the presence of dithionite resulted in the full conversion of deoxygenated AHb1 and AHb2 to the respective iron-nitrosyl-heme species (Figure 1C, D) while in the absence of dithionite we observed that two molecules of deoxy protein form approximately one iron-nitrosyl- and one ferric species both for AHb1 and AHb2 (Figure 1E, F). This stoichiometry is consistent with previous results obtained for the anaerobic reaction of Hb, Mb and Ngb with nitrite. The presence of dithionite in the reaction mixture helps to avoid the formation of oxygenated nsHbs species and, at the same time, quickly reduce the resulting ferric AHb1 and AHb2 (equation 1) to the deoxy species. We performed reactions at different dithionite concentrations (1 to 5 mM) to verify that the dithionite reduction of ferric to ferrous heme is not the driving force of the reaction. We also had previously verified that under these conditions dithionite does not effectively reduce nitrite to NO (32) thus the observed formation of iron-nitrosyl-Hbs results from the deoxy-Hbs mediated reduction of nitrite.
Fig 1. Anaerobic reaction of deoxygenated AHb1 and AHb2 with nitrite in the presence and absence of dithionite.
(A, B) Visible spectra of the reaction between 20 μM deoxy- AHb1 or AHb2 and 250 μM nitrite in the presence of 3 mM dithionite recorded respectively at 5 and 20 s intervals. (C, D) Concentrations changes (in percentage) of deoxy- (blue), met- (green) and iron-nitrosyl- (red) AHb1 and AHb2 species versus time. (E, F) Anaerobic reaction as in panels C, D in the absence of dithionite. All measurements were made in 100 mM phosphate buffer and 25 °C as described in “Materials and Methods”.
We then performed the reactions with nitrite in the concentration range 0.05 – 2.5 mM, at pH = 7.5 and T = 25°, in the presence of excess dithionite and found that the observed rate constants depended linearly with the nitrite concentration increase (Figure 2A). In the pseudo-first order conditions of the assay the observed rate constant corresponds to kobs = kBRC × [NO2−]; being kBRC the bimolecular rate constant of the reaction between the ferrous protein and nitrite. The kBRC obtained from the linear fit for reactions at pH = 7.5 and T = 25°, are 18.6 ± 1.1 and 3.9 ± 0.1 M−1s−1 respectively for AHb1 and AHb2.
Fig 2. Kinetics of nitrite reaction with Arabidopsis thaliana nsHbs.
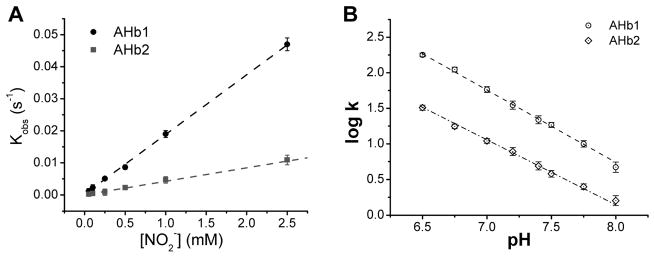
(A) Plot of observed rate constants (kobs) obtained at pH = 7.5 and 25 °C versus nitrite concentration, the second-order bimolecular rate constant obtained from the linear fit of the data are 18.6 ± 1.1 and 3.9 ± 0.1 M− 1s−1. (B) Effect of pH on the nitrite reductase reaction rates. The bimolecular rate constant (k) is linear with the proton concentration (lines show linear regression analysis of the data).
Proton dependence of the nitrite reductase reaction with plant hemoglobins
The reduction of nitrite by AHb1 and AHb2 requires a proton to form the reactive nitrous acid species according to equation 1, and therefore increasing concentration of protons will accelerate the nitrite reductase rate by 10-fold for each pH unit decrease. We determined the pH dependence of the observed bimolecular rate constants of the nitrite reductase reaction for AHb1 and AHb2 in the range 6.5 – 8.0. (Figure 2B). The slopes of the linear fittings of the log of the observed bimolecular rate constants k versus pH are 1.04 ± 0.07 and 0.93 ± 0.08 for AHb1 and AHb2, respectively. These values are in agreement with the value of 1 as expected according to the reaction in equation 1. We conclude that deoxygenated plant nsHbs class 1 and 2 reduce nitrite via an electron and proton transfer to form NO analogous to the mechanism proposed for neuroglobin and other mammalian hemoglobin (17, 33).
Nitrite reduction by plant hemoglobins generates NO in the gas phase under anaerobic and hypoxic conditions
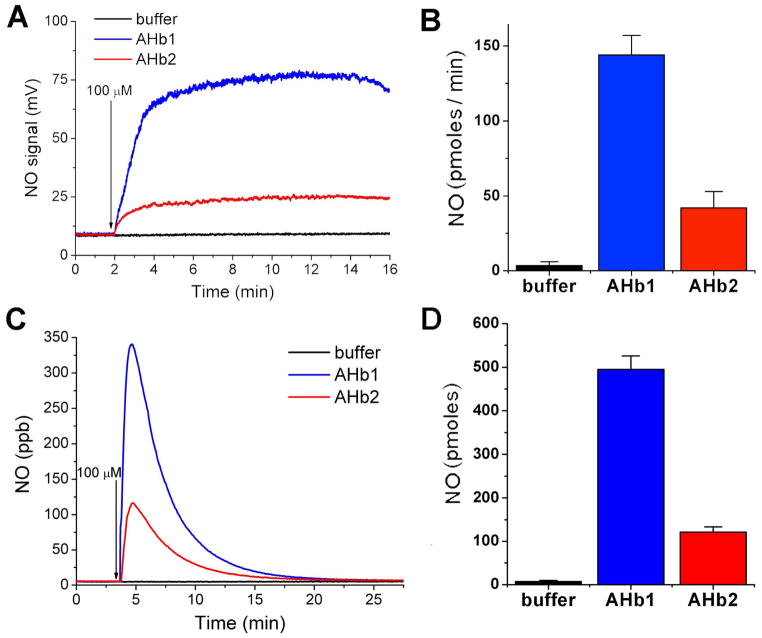
Although in our in vitro conditions deoxygenated AHbs can recapture NO, we explored whether NO gas can escape ferrous heme binding at measurable rates. We used chemiluminescence to monitor NO release during the reaction of deoxy- AHb1 and AHb2 (25 μM) and nitrite (up to 100 μM) both under anoxic and hypoxic conditions. In Figure 3A we report the detection of NO gas liberated during the anaerobic reaction in a vessel purged with helium: after injection of nitrite in the mixture, the NO level increased till it reached a plateau and then it was stable for several minutes before decreasing (not shown). Figure 3B shows that the average rate of NO generation calculated during the plateau segment (in pmoles/min) for AHb1 is about 3.5 fold higher than the value obtained for AHb2. This result is comparable to the ratio of the relative rates of nitrite reduction obtained by spectroscopy however, as expected, only a small but significant amount of the NO generated during the reaction was detected as gas phase NO.
Fig 3. Nitrite reduction by deoxy- AHb1 and AHb2 generates NO gas during anoxia and hypoxia (2% oxygen).
Chemiluminescence detection of NO gas emission during the anaerobic (Panel A) and hypoxic (2% O2) (Panel C) reaction of nitrite with buffer alone (black), 25 μM deoxy-AHb1 (blue) or 25 μM deoxy-AHb2 (red). Arrows indicate the addition of nitrite in the specified amounts. Traces are representative or 3 or more separate experiments. (B, D) Quantification of the amount of NO detected for the reaction with 100 μM nitrite.
Next we measured the NO emission in a more physiological hypoxic state: analogous reactions were performed in a flask vented with a mixture of 2% O2/98% N2 (PaO2 of approximately 14 mm Hg) without bubbling and in the absence of dithionite. This method allows for the detection of the NO gas released in the headspace, which is a small fraction of the NO produced. In these conditions the addition of deoxy- AHb1 and AHb2 to a buffered solution of 100 μM nitrite (or vice-versa) generates in both cases an initial large increase of NO that slowly decreases until it returns to the baseline (Figure 3C). Similarly to the ratio obtained in anoxic conditions, the total amount of NO measured from the nitrite reduction of AHb1 was about 4 fold larger than the amount for AHb2 (Figure 3D).
DISCUSSION
The primary finding of our study is that non-symbiotic Hbs class 1 and 2 from Arabidopsis thaliana are capable of reducing nitrite to NO and the reaction rates increase linearly as [H+] increases. This reactivity is similar to the reaction of nitrite with Hb, Mb and Ngb (17, 33) and the bacterial nitrite reductase (34) in which a coupled electron and proton transfer to nitrite to generate NO. Recent report by Sturms et al. (29) showed that hemoglobin from cyanobacterium Synechocystis and class 1 rice non-symbiotic Hb are also able to convert nitrite to NO under anoxic conditions and our work extends their findings. In an anaerobic environment deoxygenated nsHbs reduce nitrite to NO according to equation 1 as first proposed for hemoglobin by Doyle et al. (35). Although the results we have obtained do not directly reveal the chemical nature of the electron transfer process, the formation of either N-nitro- or O-nitrito heme iron bound nitrite or alternatively the direct binding of nitrous acid to five-coordinate heme iron, as previously proposed for Ngb (17), offers an attractive pathway for the formation of end products ferric heme and NO. The NO generated has very high affinity (kon = 108 M−1s−1) for the ferrous heme, thus yielding iron-nitrosyl-heme (Fe(II)-NO) as a final reaction product (equation 2). The overall stoichiometry is two molecules of deoxygenated protein form one iron-nitrosyl- and one ferric species. This ratio is confirmed respectively for AHb1 and AHb2 by the spectra deconvolution results shown in Figure 1E and 1F (despite AHb1 showing a late conversion of iron-nitrosyl into the ferric species). In the presence of dithionite however the ferric heme species formed by oxidation of the deoxygenated Hbs is reduced back to the ferrous form that reacts again according to equation 1. Thus, the stoichiometry is now one molecule of deoxy- protein forming one molecule of Fe(II)-NO Hb and the latter nitrosyl species is produced at the same rate of the deoxy species consumption rate. In Table 1 we compare the rate constants we obtained for AHb1 and AHb2 to those of other mammalian globins at pH = 7.4 and the values reported for rice nsHb1 and cyanobacterium Synechocystis Hb (synHb) at pH = 7.0. AHb1 has a rate constant approximately 4- to 5-fold higher than AHb2, and comparable to the values reported by Sturms et al. (36) for rice Hb1 and synHb when compared at the same proton concentration. However both AHb1 and AHb2 rate constants are an order of magnitude higher than the values reported for wild-type Ngb, which also presents a hexa-coordinate heme. We have shown in human Ngb that mutation of the distal histidine with either a Leu (H64L) or Gln (H64Q) residue locks the heme iron center in the penta-coordination state and results in about a 2000-fold increase of the nitrite reductase reaction rate. The distal HisE7 side chain position of AHb1 and AHb2 has been reported to modulate the equilibrium between penta- and hexa-coordinate species and the differences in external ligands binding affinity to their heme iron (31, 37). Therefore, the faster nitrite reductase rate of AHb1 versus AHb2 could be associated with the substantial fraction of AHb1 molecules in penta-coordinate heme geometry (approximately 40%), and the distal HisE7 residue position might play a major role in regulating the nitrite reductase activity of AHb1 and AHb2.
TABLE 1.
Nitrite reductase rates of non symbiotic hemoglobins from Arabidopsis Thaliana and related heme-globin proteins.
| Protein | Nitrite reductase rate (M−1s−1) pH = 7.4 | Nitrite reductase rate (M−1s−1) pH = 7.0 |
|---|---|---|
| AHb1 | 19.8 ± 3.2 a | ~ 58 a |
| AHb2 | 4.9 ± 0.2 a | ~ 11 a |
| rice nsHb1 | ND | ~ 83 d |
| SynHb | ND | ~ 68 d |
| Human Hb (T-state) | ~ 0.12 b | ND |
| Human Hb (R-state) | ~ 6 b | ND |
| Sperm whale Mb | 5.6 ± 0.6 c | ~ 11 d |
| Human Ngb | 0.12 ± 0.02 c | ~ 0.25 c |
| Human Ngb, H64L | 259 ± 8 c | ~ 956 c |
The apparent kcat values were determined at 25 °C, in sodium phosphate buffer 100 mM, pH 7.4 in the presence or in the absence of dithionite as described in “Materials and Methods”;
values from Huang et al. JCI 2005 in Sodium Phosphate buffer 100 mM, pH 7.4 at 37 °C;
values from Tiso et al. 2011;
values from Sturms et al. 2011.
ND, not determined.
Functional and Biological Implications
NO signaling in plants involves various messenger molecules such as cGMP, cADP ribose, and Ca2+ (38) and modulates several physiological functions during the entire life of the plant. NO can have a protective or a toxic effect on cells, depending on its concentration. It can alter the expression of specific genes and plays a key role in metabolism, defense, root development and cell elongation (39). Perazzolli et al. (26) showed that oxygen bound AHb1 has a role in NO detoxification by catalyzing the NADH-dependent oxidation of NO back to nitrate via the NO-dioxygenase reaction. However, AHb1 expression is strongly induced in roots under conditions of hypoxia (40) and is required for survival of plants after a severe hypoxic challenge (41) suggesting that oxygenated AHb1 might not be the species exerting a function related to hypoxic stress. Nitrite levels in plants vary according to the kind of plant, tissue localization and the nitrogen content of the soil (nitrate/nitrite levels may change enormously by agricultural fertilization) however nitrite concentrations are kept in the high micromolar range by nitrate reductase enzyme activity. Of particular notice exposure to nitrate and nitrite induce gene expression of AHb1 (42, 43). Our results shown that during hypoxic and anoxic conditions AHb1 and AHb2 are able to produce NO by nitrite reduction (Figure 3). This indicates that in conditions of poor soil oxygenation and especially for water submerged species the pH-dependent reduction of nitrite by deoxygenated nsHbs is a viable pathway for hypoxic/anoxic NO generation. In our in vitro conditions the resulting formation of ferric (equation 1) and nitrosyl-bound Hbs (equation 2) limits the NO turnover to the initial concentration of deoxygenated nsHbs however in vivo ferric nsHbs can be directly reduced by NADPH (26), or by a mixture of NADH and FAD, as for alfalfa nsHb (44), or by a met-hemoglobin reductase, as for barley nsHb (45). In mammalian cells, deoxy-hemeproteins related NO production was observed to inhibit mitochondrial respiration (at the complex I and complex IV level) and prevent electron leakage with subsequent formation of superoxide and peroxynitrite at the site of complex I and III (16, 46). An analogous mechanism might serve to prevent the formation of reacting oxygen and nitrogen species during plant hypoxia. As suggested for other hemoglobins, it is feasible that via the nitrite reductase and NO dioxygenase reactions nsHbs control NO levels serving both as NO scavenger and generator, and indirectly regulating the multiple effect of NO on plant physiology based on ambient oxygen tension. The few in vivo studies available on AHb1 and AHb2 cellular interactions and regulation of plant metabolism are insufficient to draw a clear picture of their physiological role; further studies on transgenic plants will be helpful to elucidate the cellular function of these proteins.
Hypothesis on Primordial Hemoglobins Function
Phylogenic analyses suggest that plant nsHbs precede evolution of more specialized symbiotic leghemoglobin (47, 48) and that a common ancient evolutionary origin of all hemoglobins can be traced to a prokariotic era (49). In addition, the discovery of ”protoglobins” in strictly anaerobic Archaea (50) suggests that the last common globin ancestor might have arisen before oxygenic photosynthesis. In the Earth’s pre-aerobic atmosphere (during the first 2,000 million years of its existence), nitrates and nitrites were successfully employed as electron acceptors in bacterial and plant respiration (51), and oxygen was indeed a toxic compound for cells. The evolution of denitrification processes and nitrite reductase enzymes may have constituted the earliest pathway for NO formation and signaling predating the evolution of NOS enzymes. Based on these considerations, we hypothesize that the nitrite reductase reaction of heme containing globins, such as Hb, Mb, Ngb and nsHbs, represent conserved biochemical processes from a pre-aerobic Earth and could represent a primordial form of “NO synthases” retained by modern hemoglobins during hypoxia/anoxia.
Supplementary Material
The abbreviations used are
- AHb1
type 1 non-symbiotic hemoglobin
- AHb2
type 2 non-symbiotic hemoglobin
- Hb
hemoglobin
- Mb
myoglobin
- Ngb
neuroglobin
- wt
wild-type
Footnotes
Dr. Gladwin receives research support from NIH grants R01HL098032, RO1HL096973, and PO1HL103455, the Institute for Transfusion Medicine and the Hemophilia Center of Western Pennsylvania.
The authors declare no competing financial interest.
Visible standard reference spectra of AHb1 and AHb2 proteins are available in supplemental figure 1. This material is available via the Internet at http://pubs.acs.org.
References
- 1.Ignarro LJ. Nitric oxide: biology and pathobiology. 1. Academic Press; San Diego: 2000. [Google Scholar]
- 2.Delledonne M, Xia Y, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature. 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 3.Besson-Bard A, Pugin A, Wendehenne D. New insights into nitric oxide signaling in plants. Annu Rev Plant Biol. 2008;59:21–39. doi: 10.1146/annurev.arplant.59.032607.092830. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki H. Nitrite-dependent nitric oxide production pathway: implications for involvement of active nitrogen species in photoinhibition in vivo. Philosophical Transactions of the Royal Society B: Biological Sciences. 2000;355:1477–1488. doi: 10.1098/rstb.2000.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot. 2002;53:103–110. [PubMed] [Google Scholar]
- 6.Meyer C, Lea US, Provan F, Kaiser WM, Lillo C. Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth Res. 2005;83:181–189. doi: 10.1007/s11120-004-3548-3. [DOI] [PubMed] [Google Scholar]
- 7.Besson-Bard A, Courtois C, Gauthier A, Dahan J, Dobrowolska G, Jeandroz S, Pugin A, Wendehenne D. Nitric oxide in plants: production and cross-talk with Ca2+ signaling. Mol Plant. 2008;1:218–228. doi: 10.1093/mp/ssm016. [DOI] [PubMed] [Google Scholar]
- 8.Wimalasekera R, Tebartz F, Scherer GF. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 2011;181:593–603. doi: 10.1016/j.plantsci.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Moreau M, Lindermayr C, Durner J, Klessig DF. NO synthesis and signaling in plants--where do we stand? Physiol Plant. 2010;138:372–383. doi: 10.1111/j.1399-3054.2009.01308.x. [DOI] [PubMed] [Google Scholar]
- 10.Frohlich A, Durner J. The hunt for plant nitric oxide synthase (NOS): is one really needed? Plant Sci. 2011;181:401–404. doi: 10.1016/j.plantsci.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Lundberg JO, Weitzberg E, Gladwin MT. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 12.van Faassen EE, Bahrami S, Feelisch M, Hogg N, Kelm M, Kim-Shapiro DB, Kozlov AV, Li H, Lundberg JO, Mason R, Nohl H, Rassaf T, Samouilov A, Slama-Schwok A, Shiva S, Vanin AF, Weitzberg E, Zweier J, Gladwin MT. Nitrite as regulator of hypoxic signaling in mammalian physiology. Med Res Rev. 2009;29:683–741. doi: 10.1002/med.20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machha A, Schechter AN. Dietary nitrite and nitrate: a review of potential mechanisms of cardiovascular benefits. Eur J Nutr. 2011;50:293–303. doi: 10.1007/s00394-011-0192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutruzzola F, Rinaldo S, Castiglione N, Giardina G, Pecht I, Brunori M. Nitrite reduction: a ubiquitous function from a pre-aerobic past. Bioessays. 2009;31:885–891. doi: 10.1002/bies.200800235. [DOI] [PubMed] [Google Scholar]
- 15.Huang Z, Shiva S, Kim-Shapiro DB, Patel RP, Ringwood LA, Irby CE, Huang KT, Ho C, Hogg N, Schechter AN, Gladwin MT. Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J Clin Invest. 2005;115:2099–2107. doi: 10.1172/JCI24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiva S, Huang Z, Grubina R, Sun J, Ringwood LA, MacArthur PH, Xu X, Murphy E, Darley-Usmar VM, Gladwin MT. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ Res. 2007;100:654–661. doi: 10.1161/01.RES.0000260171.52224.6b. [DOI] [PubMed] [Google Scholar]
- 17.Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, Frizzell S, Jayaraman T, Geary L, Shapiro C, Ho C, Shiva S, Kim-Shapiro DB, Gladwin MT. Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem. 2011;286:18277–18289. doi: 10.1074/jbc.M110.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 19.Trevaskis B, Watts RA, Andersson CR, Llewellyn DJ, Hargrove MS, Olson JS, Dennis ES, Peacock WJ. Two hemoglobin genes in Arabidopsis thaliana: the evolutionary origins of leghemoglobins. Proc Natl Acad Sci U S A. 1997;94:12230–12234. doi: 10.1073/pnas.94.22.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. Structure and reactivity of hexacoordinate hemoglobins. Biophys Chem. 2010;152:1–14. doi: 10.1016/j.bpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kundu S, Trent JT, 3rd, Hargrove MS. Plants, humans and hemoglobins. Trends Plant Sci. 2003;8:387–393. doi: 10.1016/S1360-1385(03)00163-8. [DOI] [PubMed] [Google Scholar]
- 22.Igamberdiev AU, Bykova NV, Hill RD. Structural and functional properties of class 1 plant hemoglobins. IUBMB Life. 2011;63:146–152. doi: 10.1002/iub.439. [DOI] [PubMed] [Google Scholar]
- 23.Kundu S, Premer SA, Hoy JA, Trent JT, 3rd, Hargrove MS. Direct measurement of equilibrium constants for high-affinity hemoglobins. Biophys J. 2003;84:3931–3940. doi: 10.1016/S0006-3495(03)75121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smagghe BJ, Hoy JA, Percifield R, Kundu S, Hargrove MS, Sarath G, Hilbert JL, Watts RA, Dennis ES, Peacock WJ, Dewilde S, Moens L, Blouin GC, Olson JS, Appleby CA. Review: correlations between oxygen affinity and sequence classifications of plant hemoglobins. Biopolymers. 2009;91:1083–1096. doi: 10.1002/bip.21256. [DOI] [PubMed] [Google Scholar]
- 25.Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell. 2004;16:2785–2794. doi: 10.1105/tpc.104.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perazzolli M, Romero-Puertas MC, Delledonne M. Modulation of nitric oxide bioactivity by plant haemoglobins. J Exp Bot. 2006;57:479–488. doi: 10.1093/jxb/erj051. [DOI] [PubMed] [Google Scholar]
- 27.Vigeolas H, Hühn D, Geigenberger P. Nonsymbiotic hemoglobin-2 leads to an elevated energy state and to a combined increase in polyunsaturated fatty acids and total oil content when overexpressed in developing seeds of transgenic Arabidopsis plants. PLANT PHYSIOLOGY. 2011;155:1435–1444. doi: 10.1104/pp.110.166462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang KT, Keszler A, Patel N, Patel RP, Gladwin MT, Kim-Shapiro DB, Hogg N. The reaction between nitrite and deoxyhemoglobin. Reassessment of reaction kinetics and stoichiometry. J Biol Chem. 2005;280:31126–31131. doi: 10.1074/jbc.M501496200. [DOI] [PubMed] [Google Scholar]
- 29.Sturms R, DiSpirito AA, Hargrove MS. Plant and cyanobacterial hemoglobins reduce nitrite to nitric oxide under anoxic conditions. Biochemistry. 2011;50:3873–3878. doi: 10.1021/bi2004312. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto A, Sakurao SH, Fukunaga K, Matsubara T, Ueda-Hashimoto M, Tsukamoto S, Takahashi M, Morikawa H. Three distinct Arabidopsis hemoglobins exhibit peroxidase-like activity and differentially mediate nitrite-dependent protein nitration. FEBS Lett. 2004;572:27–32. doi: 10.1016/j.febslet.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Bruno S, Faggiano S, Spyrakis F, Mozzarelli A, Abbruzzetti S, Grandi E, Viappiani C, Feis A, Mackowiak S, Smulevich G, Cacciatori E, Dominici P. The reactivity with CO of AHb1 and AHb2 from Arabidopsis thaliana is controlled by the distal HisE7 and internal hydrophobic cavities. J Am Chem Soc. 2007;129:2880–2889. doi: 10.1021/ja066638d. [DOI] [PubMed] [Google Scholar]
- 32.Grubina R, Huang Z, Shiva S, Joshi MS, Azarov I, Basu S, Ringwood LA, Jiang A, Hogg N, Kim-Shapiro DB, Gladwin MT. Concerted nitric oxide formation and release from the simultaneous reactions of nitrite with deoxy- and oxyhemoglobin. J Biol Chem. 2007;282:12916–12927. doi: 10.1074/jbc.M700546200. [DOI] [PubMed] [Google Scholar]
- 33.Gladwin MT, Grubina R, Doyle MP. The new chemical biology of nitrite reactions with hemoglobin: R-state catalysis, oxidative denitrosylation, and nitrite reductase/anhydrase. Acc Chem Res. 2009;42:157–167. doi: 10.1021/ar800089j. [DOI] [PubMed] [Google Scholar]
- 34.Rinaldo S, Giardina G, Castiglione N, Stelitano V, Cutruzzola F. The catalytic mechanism of Pseudomonas aeruginosa cd1 nitrite reductase. Biochem Soc Trans. 2011;39:195–200. doi: 10.1042/BST0390195. [DOI] [PubMed] [Google Scholar]
- 35.Doyle MP, Pickering RA, DeWeert TM, Hoekstra JW, Pater D. Kinetics and mechanism of the oxidation of human deoxyhemoglobin by nitrites. J Biol Chem. 1981;256:12393–12398. [PubMed] [Google Scholar]
- 36.Sturms R, Dispirito AA, Hargrove MS. Plant and Cyanobacterial Hemoglobins Reduce Nitrite to Nitric Oxide under Anoxic Conditions. Biochemistry. 2011 doi: 10.1021/bi2004312. [DOI] [PubMed] [Google Scholar]
- 37.Nienhaus K, Dominici P, Astegno A, Abbruzzetti S, Viappiani C, Nienhaus GU. Ligand migration and binding in nonsymbiotic hemoglobins of Arabidopsis thaliana. Biochemistry. 2010;49:7448–7458. doi: 10.1021/bi100768g. [DOI] [PubMed] [Google Scholar]
- 38.Wendehenne D, Durner J, Klessig DF. Nitric oxide: a new player in plant signalling and defence responses. Curr Opin Plant Biol. 2004;7:449–455. doi: 10.1016/j.pbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Stohr C, Stremlau S. Formation and possible roles of nitric oxide in plant roots. J Exp Bot. 2006;57:463–470. doi: 10.1093/jxb/erj058. [DOI] [PubMed] [Google Scholar]
- 40.Morard P, Silvestre J, Lacoste L, Caumes E, Lamaze T. Nitrate uptake and nitrite release by tomato roots in response to anoxia. J Plant Physiol. 2004;161:855–865. doi: 10.1016/j.jplph.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Dordas C. Nonsymbiotic hemoglobins and stress tolerance in plants. Plant Science. 2009;176:433–440. doi: 10.1016/j.plantsci.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Ohwaki Y, Kawagishi-Kobayashi M, Wakasa K, Fujihara S, Yoneyama T. Induction of class-1 non-symbiotic hemoglobin genes by nitrate, nitrite and nitric oxide in cultured rice cells. Plant Cell Physiol. 2005;46:324–331. doi: 10.1093/pcp/pci030. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seregelyes C, Igamberdiev AU, Maassen A, Hennig J, Dudits D, Hill RD. NO-degradation by alfalfa class 1 hemoglobin (Mhb1): a possible link to PR-1a gene expression in Mhb1-overproducing tobacco plants. FEBS Lett. 2004;571:61–66. doi: 10.1016/j.febslet.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 45.Igamberdiev AU, Seregelyes C, Manac’h N, Hill RD. NADH-dependent metabolism of nitric oxide in alfalfa root cultures expressing barley hemoglobin. Planta. 2004;219:95–102. doi: 10.1007/s00425-003-1192-3. [DOI] [PubMed] [Google Scholar]
- 46.Shiva S, Sack MN, Greer JJ, Duranski M, Ringwood LA, Burwell L, Wang X, MacArthur PH, Shoja A, Raghavachari N, Calvert JW, Brookes PS, Lefer DJ, Gladwin MT. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J Exp Med. 2007;204:2089–2102. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrocho-Villegas V, Gopalasubramaniam SK, Arredondo-Peter R. Plant hemoglobins: what we know six decades after their discovery. Gene. 2007;398:78–85. doi: 10.1016/j.gene.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 48.Hoy JA, Robinson H, Trent JT, 3rd, Kakar S, Smagghe BJ, Hargrove MS. Plant hemoglobins: a molecular fossil record for the evolution of oxygen transport. J Mol Biol. 2007;371:168–179. doi: 10.1016/j.jmb.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 49.Freitas TA, Saito JA, Hou S, Alam M. Globin-coupled sensors, protoglobins, and the last universal common ancestor. J Inorg Biochem. 2005;99:23–33. doi: 10.1016/j.jinorgbio.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 50.Freitas TA, Hou S, Dioum EM, Saito JA, Newhouse J, Gonzalez G, Gilles-Gonzalez MA, Alam M. Ancestral hemoglobins in Archaea. Proc Natl Acad Sci U S A. 2004;101:6675–6680. doi: 10.1073/pnas.0308657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reutov VP. Nitric oxide cycle in mammals and the cyclicity principle. Biochemistry (Mosc) 2002;67:293–311. doi: 10.1023/a:1014832416073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.