Abstract
Altered expression of oncogenic and tumor-suppressing microRNAs (miRNAs) is widely associated with tumorigenesis. However, the regulatory mechanisms underlying these alterations are poorly understood. We sought to shed light on the deregulation of miRNA biogenesis promoting the aberrant miRNA expression profiles identified in these tumors. Using sequencing technology to perform both whole-transcriptome and small RNA sequencing of glioma patient samples, we examined precursor and mature miRNAs to directly evaluate the miRNA maturation process, and interrogated expression profiles for genes involved in the major steps of miRNA biogenesis. We found that ratios of mature to precursor forms of a large number of miRNAs increased with the progression from normal brain to low-grade and then to high-grade gliomas. The expression levels of genes involved in each of the three major steps of miRNA biogenesis (nuclear processing, nucleo-cytoplasmic transport, and cytoplasmic processing) were systematically altered in glioma tissues. Survival analysis of an independent data set demonstrated that the alteration of genes involved in miRNA maturation correlates with survival in glioma patients. Direct quantification of miRNA maturation with deep sequencing demonstrated that deregulation of the miRNA biogenesis pathway is a hallmark for glioma genesis and progression.
Keywords: microRNA, biogenesis, glioma
INTRODUCTION
MicroRNAs (miRNAs) are a class of conserved, small, noncoding RNAs that control gene expression by binding to complementary sequences at the 3′ untranslated regions (UTRs) of target messenger RNAs (mRNAs) resulting in translational repression or mRNA degradation [1]. MiRNAs have been shown to play important roles in mammalian systems by influencing genes involved in processes like cell proliferation, apoptosis, and tumorigenesis [2]. Many miRNAs have been designated as oncogenes (“oncomiRs”) or tumor suppressors based on the effects of the miRNAs on cells and the functions of the mRNA target genes [3–6].
Similar to mRNAs, miRNAs can be regulated at the transcriptional level by DNA-binding transcription factors or epigenetic mechanisms [4, 7] and posttranscriptionally by a multistep processing pathway [8]. The lack of correlation between primary miRNA (pri-miRNA) transcripts and mature miRNAs in tumors and the association of miRNA processing factors with tumorigenesis in cell culture and mouse model studies indicate that deregulation of the biogenesis pathway is likely to be a key player in the aberrant miRNA expression profiles observed in cancer [9].
MiRNA biogenesis is controlled by the multistep miRNA processing pathway [10]. The pri-miRNA transcript is transcribed by RNA polymerase II (in some cases polymerase III) in the nucleus. For intergenic miRNA, the pri-miRNA is cleaved into a short, 60 to 70 nucleotide (nt), hairpin precursor miRNA (pre-miRNA) by a microprocessor unit containing the nuclease, RNASEN (also known as DROSHA), and other factors [11]. miRNA located in intronic regions or within exons of protein-coding genes can be cleaved by splicing machinery to generate the pre-miRNA [12]. This nuclear processing is followed by the transport of pre-miRNA from the nucleus into the cytoplasm via exportin-5 (XPO5) [13]. In the cytoplasm, the pre-miRNA is cleaved into an approximately 19–25-nt long mature form of the miRNA by the ribonuclease Dicer (DICER1), [14] or by a Dicer-independent maturation process that is beginning to be revealed [15]. The mature miRNA is loaded into the RNA-induced silencing complex (RISC), where it initiates translational repression or degradation of target mRNAs [16, 17]. Thus, miRNA biogenesis is tightly controlled by a set of protein-coding genes that eventually lead to production of functionally mature miRNAs in the cytoplasm.
The deregulation of the miRNA biogenesis pathway has been associated with various cancers. This is supported by the large number of differentially expressed mature miRNA that have been identified in multiple cancers [18]. Cell culture studies and animal models have demonstrated that deregulation of miRNA expression by knockdown of key miRNA biogenesis factors enhances tumorigenesis [19]. Expression levels of these factors have also been associated with prognosis. For example, elevated expression of RNASEN accelerates the proliferation of esophageal squamous cell carcinoma cells and is a negative prognostic marker in esophageal cancer patients [20], and reduced expression of DICER1 is associated with poor prognosis in lung and in ovarian cancer patients [21, 22] whereas overexpression has been linked to poor prognosis in colon cancer [23]. Mutations have also been identified in biogenesis components, including XPO5 and the Dicer cofactor TARBP2 in colon cancers [24, 25]. It should be noted that the role of these processing genes in tumorigenesis remains complex. For example, while some studies indicate that for DICER1, loss of one copy is advantageous to tumor growth while complete loss is disadvantageous; others have demonstrated that in some instances DICER1 null cells can maintain tumorigenic capacity [26, 27].
A number of studies have reported changes in the steady-state levels of mature miRNAs in glioma [28–30]. One of the best characterized events is the elevation of oncomiRs miR-21 and miR-221 [30–35]. However, the mechanisms underlying deregulation of miRNA biogenesis in glioma are unknown. As mutations in biogenesis components such as XPO5 and TARBP2 have not been identified in glioma, the clinical significance of deregulated biogenesis genes remains uncertain.
We sought to thoroughly examine the miRNA processing pathway by taking advantage of deep sequencing technology to directly measure the maturation process of all the miRNAs and to identify key changes in biogenesis gene expression. This analysis provides a comprehensive view of the deregulation of miRNA biogenesis in glioma and the subsequent miRNA expression-profile alterations. This analysis shows that miRNA maturation is tightly regulated, and that the multilevel regulatory process is altered at each step during glioma genesis and progression. Correlation of miRNA biogenesis gene expression with survival information from a major glioma database further demonstrates the significance of these findings.
MATERIALS AND METHODS
Patient Samples
Human glioma samples were obtained from The University of Texas MD Anderson Cancer Center Brain Tumor Center tissue bank and were collected under an institutional review board-approved protocol. Commercially available normal brain RNA pooled from multiple donors was used as reference (Ambion, Carlsbad, CA).
Library Preparation and RNA Sequencing
Library preparation for both whole-transcriptome sequencing and small RNA sequencing was performed using Applied Biosystems Incorporated’s (ABI, Carlsbad, CA) whole-transcriptome and small RNA sequencing protocols. Sequencing runs were performed using ABI’s SOLiD System version 3.5 for both whole-transcriptome sequencing and small RNA sequencing. From the whole transcriptome and small RNA sequencing over 610 million and 230 million 50nt long sequencing reads were obtained, respectively. Detailed description is provided in Supplementary Methods.
Sequencing Data Analysis
Sequencing reads were aligned against transcript sequences from the National Center for Biotechnology Information (NCBI) reference sequence build version 38 using Bowtie version 0.12.5 [36]. Gene expression levels were quantified as reads per kilobase per million (RPKM)-normalized expression values and quantile normalized across sample pools.
Small RNA sequencing reads were aligned against 721 pre-miRNA and 904 mature miRNA sequences from miRBase build 14 [37]. We excluded from further analysis miRNAs with a median number of reads less than 20 in mature miRNAs or pre-miRNAs across all sample pools, which left us with 505 miRNAs for subsequent analysis. Expression levels were normalized by the total number of mappable reads per sequencing experiment.
Prior to sequencing, we used Agilent miRNA microarray to characterize each individual sample. For each sequencing pool, the mean Pearson correlation between the samples was above 0.91 indicating good consistency of pooled samples (data described in Supplemental Methods).
Microarray Data Analysis
Raw gene expression array data were downloaded from the Repository of Molecular Brain Neoplasia Data (https://caintegrator.nci.nih.gov/rembrandt/) and summarized into gene expression values using the robust multichip average (RMA) algorithm [38]. Differential expession between high- and low-grade was tested with t-test under means are equal null hypothesis. Bonferroni corrected p-values < 0.05 were considered significant. Enrichment was tested with hypergeometric distribution, using all the genes on the array as a background.
Clustering Analysis
Gene expression profiles of biogenesis genes were scaled to an interval of zero-to-one. A K-means clustering algorithm with squared Euclidean distance was applied to identify gene clusters. Based on a silhouette plot, three clusters provided a good separation for the data.
Survival Analysis
Kaplan-Meier analysis was used to determine the survival effect based on gene expression data. Two groups of samples were defined by applying K-means clustering to the expression profiles of a group of genes. Statistical significance of Kaplan-Meier curves was evaluated with the log-rank test.
Cell Culture
SNB19 human glioblastoma cells were cultured in Dulbecco’s modified Eagle’s medium nutrient mixture F-12 (DMEM:F12) (Life Technologies, Carlsbad, CA) with 10% fetal bovine serum (FBS). Small interfering RNA (siRNA) SMART pools (Dharmacon, Lafayette, CO) targeting DICER1, EIF2C1, and EIF2C2 were transfected using RNAiMax (Life Technologies, Carlsbad, CA), according to manufacturer’s instructions. Cells were harvested after 48 hours, and total RNA was isolated using the mirVana kit (Ambion, Carlsbad, CA).
Quantitative RT-PCR
Total RNA from individual patient samples represented in the pooled samples for the sequencing analysis was used for these experiments. The precursor forms of miRNAs were detected using SYBR green and normalized to GAPDH. The mature forms of miRNAs were assayed using TaqMan MicroRNA assay kit (ABI, Carlsbad, CA) according to manufacturer’s protocol, and normalized to RUN6B.
Primer sequences:
Human pre-miR-21: 5′ TGTCGGGTAGCTTATCAGAC and 5′
TGTCAGACAGCCCATCGACT; Human pre-miR-1912: 5′
CTCTAGGATGTGCTCATTGC and 5′ AATCTCTATTATGTTCACAC;
Human GAPDH: 5′ ACCACAGTCCATGCCATCAC and 5′
TCCACCACCCTGTTGCTGTA
miRNA Northern Blot Analysis
miRNA Northern blot analysis was performed as previously described [39]. Briefly, 5 to 20 μg of total RNA was resolved on a 12% acrylamide/urea gel and was transferred to nylon membrane. Blots were hybridized overnight with 10 pmol of 32P-labeled locked nucleic acid (LNA) miRNA probes (Exiqon, Woburn, MA).
RESULTS
Alteration of miRNA Maturation and miRNA Biogenesis Genes in Gliomas
We performed deep sequencing analysis of both pre-miRNAs and mature miRNAs from 40 glioma patient samples and normal brain reference RNA. The glioma samples included patients diagnosed with both low-grade gliomas (oligodendroglioma [Oligo]) and high-grade gliomas (anaplastic oligodendroglioma [AO], anaplastic astrocytoma [AA], and glioblastoma [GBM]) glioma. In the interest of cost, we pooled 20 GBM samples into 4 groups and pooled each of the other glioma subtypes into individual pools (1 AA, 1 AO, and 2 Oligo pools; Table 1).
Table 1. Glioma patient sample information.
Samples were grouped into sequencing pools based on available clinical information. The total number of sequencing reads for each pool is also shown.
| Sample Number | Grade | Survival (mo) | Sequencing Pool | Average Survival | Whole Transcriptome Reads | Small RNA reads |
|---|---|---|---|---|---|---|
| 1 | GBM | 6 | A | < 6 month | 75321717 | 20724499 |
| 2 | 3 | |||||
| 3 | 4 | |||||
| 4 | 1 | |||||
| 5 | 6 | |||||
|
| ||||||
| 6 | GBM | 11 | B | 10–15 months | 75295418 | 21275202 |
| 7 | 10 | |||||
| 8 | 11 | |||||
| 9 | 13 | |||||
| 10 | 15 | |||||
|
| ||||||
| 11 | GBM | 15 | C | Intermediate > 1 year | 52438870 | 18597970 |
| 12 | 18 | |||||
| 13 | >12 | |||||
| 14 | 20 | |||||
| 15 | >12 | |||||
|
| ||||||
| 16 | GBM | >12 | D | > 1 to 2 years | 67066205 | 22944568 |
| 17 | >24 | |||||
| 18 | >48 | |||||
| 19 | >84 | |||||
| 20 | 29 | |||||
|
| ||||||
| 21 | Oligo | 48 | E | NA | 58344119 | 19075166 |
| 22 | >48 | |||||
| 23 | >48 | |||||
| 24 | >72 | |||||
|
| ||||||
| 25 | Oligo | >72 | F | NA | 66169704 | 18095113 |
| 26 | >36 | |||||
| 27 | 36 | |||||
| 28 | >36 | |||||
|
| ||||||
| 29 | AO | >36 | G | NA | 87724772 | 34399996 |
| 30 | 2 | |||||
| 31 | 24 | |||||
| 32 | >60 | |||||
| 33 | 24 | |||||
| 34 | 60 | |||||
| 35 | NA | |||||
|
| ||||||
| 36 | AA | >36 | H | NA | 70196537 | 36178145 |
| 37 | 35 | |||||
| 38 | NA | |||||
| 39 | >36 | |||||
| 40 | >36 | |||||
|
| ||||||
| 41 | Adult Normal | NA | I | NA | 57701862 | 42319647 |
Abbreviations: GBM, glioblastoma; Oligo, oligodendroglioma; AO, anaplastic oligodendroglioma; AA, anaplastic astrocytoma; NA, no survival data available or not applicable.
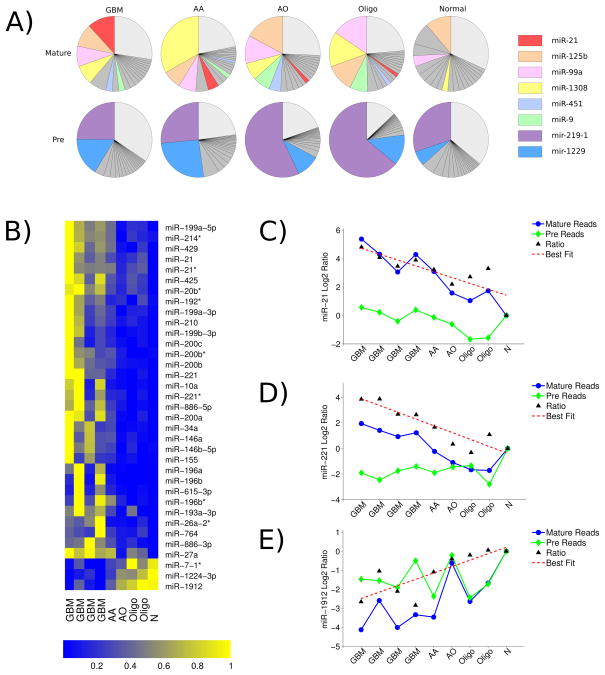
Analysis of both pre-miRNA and mature miRNA expression levels revealed widespread alterations in the abundance of many miRNAs. The twenty miRNAs with the highest number of sequencing reads are shown (Figure 1A). While the abundance of many mature miRNAs, including miR-21, change with glioma grade (Figure 1A, top), the abundance of corresponding precursor forms remains unchanged, suggesting that the alterations in mature miRNA levels may be due to deregulated miRNA processing (Figure 1A, bottom). Similar discrepancies between mature and pre-miRNA levels have also been previously observed [40]. For the most abundant pre-miRNA (Figure 1A, bottom) we do not observe increased processing and thus, they do not appear among the most abundant mature miRNA. We calculated the ratio of mature miRNA to pre-miRNA (M/P) for each miRNA as a measure of miRNA maturation (processing from precursor to mature form) (Supplementary Figure 1) and identified 35 miRNAs whose change in M/P ratio correlated with tumor grade (Spearman rank correlation; P < 0.01) (Figure 1B). The expression levels of mature and pre-miRNA forms of these 35 miRNAs are indicated (Supplementary Figure 2). The M/P ratios of three of the miRNAs were decreased in gliomas and were inversely associated with tumor grade. M/P ratios of the other 32 miRNAs were increased in gliomas and were positively associated with increasing glioma grade. Among the identified differentially matured miRNAs, several have been previously associated with tumor development and prognosis in glioma [24], including miR-21 and miR-221 (Figure 1C–E). We subsequently validated these findings by measuring the precursor and mature forms of miR-21 and miR-1912 by quantitative RT-PCR in a subset of patient samples which were represented in the sequencing pools (Supplementary Figure 3). These results indicate that the changes in mature miRNA expression are likely a function of the deregulated miRNA processing.
Figure 1. Discovery of miRNAs with increased maturation in glioma.
(A) Abundance of mature miRNA and pre-miRNA in different tumor grades revealed by small RNA sequencing. Twenty miRNAs with the highest number of sequencing reads are shown as individual sectors with the sector size representing the number of reads for a given miRNA. The remaining miRNAs are contained in a single sector (light gray). Some of the miRNAs with large changes in their abundance are highlighted in color. (B) M/P miRNA ratio across all samples. Shown are miRNAs with M/P ratios that are significantly correlated with glioma grade (correlation, P < 0.01) and display differential maturation (M/P ratio > 2-fold) between gliomas and normal brain. For visualization, the M/P ratio of each miRNA is scaled to a zero (lowest expression ratio) to one (highest expression ratio) interval, as indicated by the color bar. (C–E) Log2 fold change of pre-miRNA (green) and mature miRNA (blue) read counts relative to read counts of normal brain, M/P expression ratio relative to the M/P expression ratio of normal brain, and linear fit to the M/P expression ratios are shown for typical examples (C) miR-21, (D) miR-221, and (E) miR-1912.
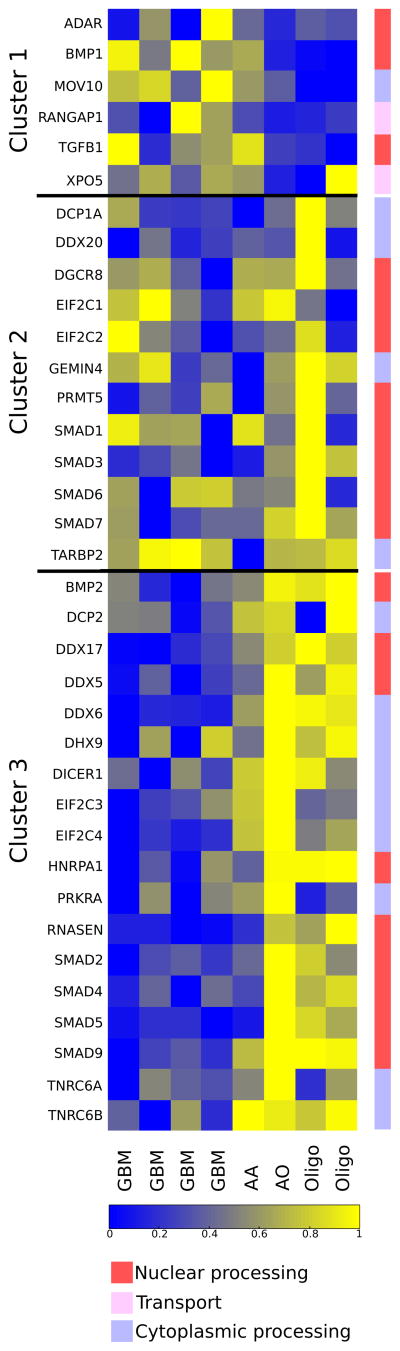
We reasoned that the extensive changes in miRNA maturation revealed by our deep sequencing analysis were a result of altered expression of the genes that regulate the miRNA biogenesis pathway. We therefore compiled a list of genes with known roles in the miRNA biogenesis pathway and examined their gene expression patterns by whole transcriptome sequencing in the same glioma patient samples (Table 2). Gene expression analysis revealed that 14 genes examined were deregulated in the glioma progression, demonstrating deregulation at each of the three major steps of miRNA biogenesis (Figure 2, Table 2). Statistical analysis with independent cohort of samples from the Rembrandt glioma database validated 8 of these 14 genes to be deregulated (Table 2). Enrichment analysis indicated that more biogenesis pathway genes are aberrated in glioma than expected by random (p=2.0711e-04, hypergeometric test). Clustering analysis of miRNA biogenesis pathway genes identified three clusters: one that positively correlated with increasing tumor grade (Cluster 1), one that negatively correlated with tumor grade (Cluster 3), and one that was without a distinctive pattern among the glioma samples (Cluster 2).
Table 2. miRNA biogenesis pathway genes and statistical analysis.
The list is compiled by literature review. Roles of the genes in the pathway are annotated in the table. Bonferroni corrected p-values from Students t-test for miRNA biogenesis genes using Rembrandt glioma data is shown. NaN: probeset for gene was not included in microarray.
| Gene Name | Entrez ID | Role in Pathway | Whole Transcriptome Sequencing Dataset P-value (U-test H0: medians are equal) | Rembrandt Glioma Dataset P-value (t-test H0: means are equal) |
|---|---|---|---|---|
| ADAR | 103 | Nuclear Processing | 0.785714 | 1 |
| BMP1 | 649 | Nuclear Processing | 0.035714 | 0.000137 |
| BMP2 | 650 | Nuclear Processing | 0.035714 | 0.007533 |
| DCP1A | 55802 | Cytoplasmic Processing | 0.142857 | 1 |
| DCP2 | 167227 | Cytoplasmic Processing | 0.571429 | 1 |
| DDX17 | 10521 | Nuclear Processing | 0.035714 | 1 |
| DDX20 | 11218 | Cytoplasmic Processing | 0.785714 | 0.00959 |
| DDX5 | 1655 | Nuclear Processing | 0.035714 | 1 |
| DDX6 | 1656 | Cytoplasmic Processing | 0.035714 | 0.000055 |
| DGCR8 | 54487 | Nuclear Processing | 0.571429 | 1 |
| DHX9 | 1660 | Cytoplasmic Processing | 0.071429 | 1 |
| DICER1 | 23405 | Cytoplasmic Processing | 0.142857 | 1 |
| EIF2C1 | 26523 | Cytoplasmic Processing | 0.571429 | 1 |
| EIF2C2 | 27161 | Cytoplasmic Processing | 1 | 1 |
| EIF2C3 | 192669 | Cytoplasmic Processing | 0.392857 | 1 |
| EIF2C4 | 192670 | Cytoplasmic Processing | 0.142857 | 1 |
| GEMIN4 | 50628 | Cytoplasmic Processing | 0.25 | 1 |
| HNRPA1 | 3178 | Nuclear Processing | 0.035714 | 0.002531 |
| MOV10 | 4343 | Cytoplasmic Processing | 0.035714 | 0.000003 |
| PRKRA | 8575 | Cytoplasmic Processing | 0.785714 | NaN |
| PRMT5 | 10419 | Cytoplasmic Processing | 0.142857 | 0.128026 |
| RANGAP1 | 5905 | Transport | 0.25 | 1 |
| RNASEN | 29102 | Nuclear Processing | 0.035714 | 0.000953 |
| SMAD1 | 4086 | Nuclear Processing | 1 | 1 |
| SMAD2 | 4087 | Nuclear Processing | 0.035714 | 1 |
| SMAD3 | 4088 | Nuclear Processing | 0.035714 | 1 |
| SMAD4 | 4089 | Nuclear Processing | 0.035714 | 0.242406 |
| SMAD5 | 4090 | Nuclear Processing | 0.035714 | 1 |
| SMAD6 | 4091 | Nuclear Processing | 1 | 1 |
| SMAD7 | 4092 | Nuclear Processing | 0.035714 | 0.046877 |
| SMAD9 | 4093 | Nuclear Processing | 0.035714 | 0.006042 |
| TARBP2 | 6895 | Cytoplasmic Processing | 1 | 1 |
| TGFB1 | 7040 | Nuclear Processing | 0.142857 | 3.04E-008 |
| TNRC6A | 27327 | Cytoplasmic Processing | 0.321429 | 1 |
| TNRC6B | 23112 | Cytoplasmic Processing | 0.25 | 0.000297 |
| XPO5 | 57510 | Transport | 0.571429 | 1 |
Figure 2. Altered gene expression in the miRNA biogenesis pathway.
Expression levels of key miRNA biogenesis genes revealed by whole transcriptome sequencing are shown in the heat map. Data are shown as log 2 expression ratios, relative to normal brain. Data of each gene is scaled such that minimum value is shown with blue and maximum with yellow. Differential expression among different grades of glioma is indicated by the color intensity, represented by the accompanied color bar. The best known association of each gene with the specific miRNA biogenesis step is also indicated. Clustering analysis identifies three groups. Genes within cluster 1 have a positive correlation with tumor grade, and genes within cluster 3 have negative correlation with tumor grade. Cluster 2 does not demonstrate a distinct pattern relative to glioma grade.
Glioma miRNA Maturation Network Signatures Are Predictive of Survival
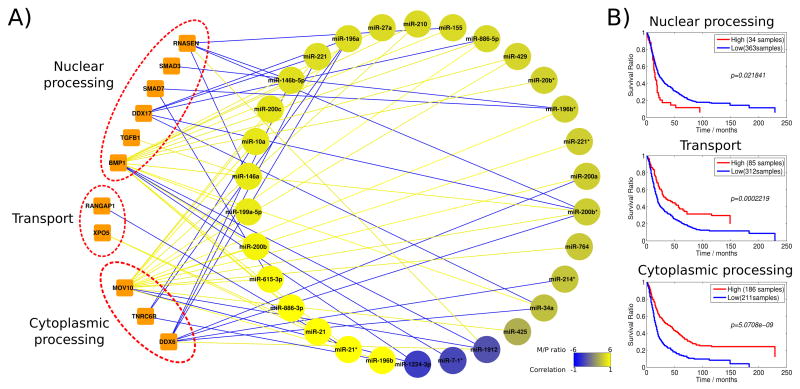
To take advantage of both mRNA and miRNA expression from the same samples, we constructed a network representation of miRNA maturation by correlating the M/P ratio of each of the 35 miRNAs with the gene expression of miRNA biogenesis genes. In Figure 3A, statistically significant correlations (Pearson correlation; P < 0.01) between expression levels of the biogenesis genes and the M/P ratios of miRNAs are shown. Biogenesis genes were grouped based on the biogenesis pathway steps with which they are primarily associated. This network indicates that several genes are correlated with the maturation of each miRNA. Furthermore, we found that the M/P ratio of each miRNA is correlated with genes from different steps of the biogenesis process, indicating the regulation of maturation at multiple steps of the miRNA processing pathway. For example, the transforming growth factor β (TGFβ)/ bone morphogenetic protein (BMP)/SMAD signaling pathway genes, which are part of the nuclear processing pathway, are connected to 20 out of the 35 differentially matured miRNAs. Overall, the network analysis uncovers complex relationships between miRNA maturation and genes involved in biogenesis process. This suggests that the processing of pre-miRNA to mature miRNA can be affected by changes in the expression of genes in all steps of the biogenesis pathway.
Figure 3. Network analysis identifies miRNA biogenesis gene signatures predictive of survival.
(A) Statistically significant (P < 0.01) correlations between biogenesis genes and miRNA maturation (M/P ratio) are shown. The network is highly connected and includes 30 out of 35 miRNA. Genes have been grouped and labeled in three groups based on their role in the miRNA biogenesis pathway. Link colors indicate positive (yellow) or negative (blue) correlation between the gene and miRNA. The same color code is used for coloring the miRNA nodes in the networks and to show M/P ratio differences between glioma and normal. Network is visualized using Cytoscape version 2.8.1 [41]. (B) Survival analysis of the network-based gene signatures on the independent data set from Rembrandt. Shown from top to bottom are nuclear processing, transport from nucleus, and cytoplasmic processing. High expression of genes in cytoplasmic processing and nucleo-cytoplasmic transport signatures is related to poor survival. Nuclear processing genes indicate an opposite pattern.
In order to evaluate the clinical relevance of the deregulated miRNA biogenesis pathway, we used the genes in the network representation to define a gene expression signature for each of the three major biogenesis steps (nuclear processing, nucleo-cytoplasmic transport, and cytoplasmic processing). Using the Rembrandt glioma data as an independent data set, we examined the relationship between these signatures and patient survival. All three signatures were associated with survival (Figure 3B). The strongest correlation was observed for cytoplasmic processing, where overall low expression indicates poor survival (p=5*10e-9). Nuclear processing shows the opposite trend, but the result is only marginally significant (p=0.02). This survival correlation is well in agreement with the clusters identified in Figure 2. Cluster 1 includes primarily genes involved in the nuclear processing of pri-miRNA, and cluster 3 includes genes involved in the cytoplasmic processing.
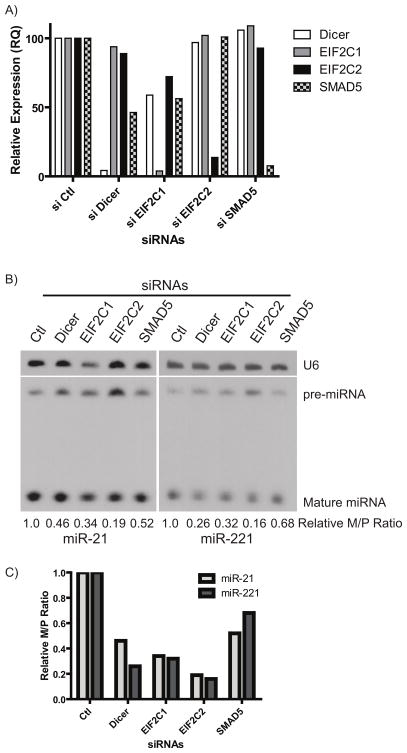
Knockdown of miRNA Biogenesis Genes Inhibits miRNA Maturation in Glioma Cell Lines
Our analysis suggested that changes in biogenesis gene expression resulted in changes in the maturation of a set of miRNAs. To confirm this regulatory relationship in glioma, we examined the effect of altered biogenesis gene expression on miRNA M/P ratio in the SNB19 glioma cell line. Focusing on biogenesis factors involved in cytoplasmic and nuclear miRNA processing, we performed knockdown studies of DICER1, EIF2C1, EIF2C2, and SMAD5 genes to demonstrate a direct relationship between gene expression and miRNA maturation. Knockdown of DICER1, EIF2C1, EIF2C2, and SMAD5 expression by siRNA in the SNB19 GBM cells was confirmed using real-time PCR (Figure 4A). We then examined the expression of both pre-miRNAs and mature miRNAs by Northern blot analysis using probes specific for miR-21 and miR-221 (Figure 4B). Inhibition of each of these miRNA processing genes promoted a change in the M/P ratio (Figure 4C). Specifically, decreasing the levels of miRNA processing genes that were found to be deregulated in glioma (DICER1, EIF2C1, EIF2C2, and SMAD5) directly resulted in decreased M/P ratios, validating our observations of M/P ratio changes in the sequencing data. Knockdown of the cytoplasmic processing genes (DICER1, EIF2C1, and EIF2C2) had a greater impact on the M/P ratio than knockdown of a nuclear processing gene (SMAD5), which is indicative of the more direct effects of the cytoplasmic processing genes in this validation experiment. However, this study confirms that deregulation of either the cytoplasmic or the more upstream nuclear processing steps can ultimately affect the M/P ratio. These studies confirmed that the miRNA M/P ratios are modulated by changes in expression of the processing genes.
Figure 4. Loss of several biogenesis genes decreases the miRNA maturation.
(A) Quantitative, real-time PCR demonstrates siRNA-mediated knockdown of miRNA processing genes. Graph represents an average of two independent experiments. (B) Northern blot analysis of pre-miRNA and mature miRNAs. Knockdown of these genes inhibits the maturation of miR-21 and miR-221. (C) Quantitation of Northern blots in (B).
DISCUSSION
MiRNAs represent key nodes in a regulatory network that modulates diverse biological processes. Thus, it is not surprising that deregulation of miRNAs markedly contributes to tumorigenesis [42]. Published literature has focused on changes in the steady-state levels of oncogenic and tumor-suppressing miRNAs and their protein-coding targets. Recent studies have begun to reveal upstream alterations of genes that regulate miRNA production [43].
The studies presented here demonstrate that the altered miRNA-expression profiles of glioma are at least in part due to widespread gene expression changes that affect miRNA processing. Using deep sequencing technologies, we dissected part of the miRNA biogenesis process by directly measuring the miRNAs at the pre- and mature steps in a set of 40 glioma samples representing different grades. We calculated the ratios of mature to pre-miRNAs to obtain the M/P ratio, which represents the biogenesis, or maturation process, of each miRNA. The M/P ratios revealed that the miRNA maturation process is markedly activated for a number of miRNAs in glioma genesis, including miR-21 and miR-221, in such a way that mature forms are actively produced. We also identified a small number of miRNAs whose maturation process is suppressed in glioma compared with normal brain, suggesting a role in glioma suppression.
By integrated analysis of both whole transcriptome and small RNA sequencing results from the same set of samples, we gained insight into specific defects in the miRNA regulatory network in glioma and revealed signatures of genes that correlate with survival in a large and independent data set. This study implicates the miRNA biogenesis pathway as a key pathway targeted for deregulation in glioma cells.
One limitation of our analysis is that the M/P ratio is an indicator of pre-miRNA-to-mature miRNA processing only. Pri-miRNAs are problematic for the analysis as they have not been systematically annotated. In addition, it has been shown that pri-miRNAs are processed co-transcriptionally, which makes the expression quantification from RNA sequencing data problematic [44]. We were unable to examine the miRNA transport process where pre-miRNAs are exported into the cytoplasm. Dissection of this step requires isolation of nuclear and cytoplasmic small RNA for library construction and deep sequencing. Whereas this can be achieved using cultured cells, it was not feasible with the small amounts of frozen tumor tissue available for our study. However, these limitations can be partially ameliorated by quantifying the expression of the genes that are known to regulate the nuclear processing and transport steps of miRNA biogenesis. In this study, we actually performed integrated analysis of miRNAs and genes that regulate the three major steps in miRNA biogenesis—nuclear processing, transport from nucleus, and cytoplasmic processing. Interestingly, the network-based gene signatures for each of these steps are associated with grade and survival in glioma.
Some of the most striking gene expression changes identified in our study can be seen in of the TGFβ/BMP/SMAD signaling pathway. The BMP-specific receptor SMADs (R-SMADS: SMAD1 and SMAD5) were first implicated in miRNA biogenesis by the observation that they interacted with the RNA helicase p68 (DDX5) to aid in the processing of pri-miR-21 to pre-miR-21 [45]. Later, the TGFβ/BMP pathway was found to regulate a larger subset of miRNAs via interaction with pri-miRNAs containing a R-SMAD consensus sequence (R-SBE) and enhancing Drosha-mediated processing [46]. Our network analysis identified this mechanism as part of the gene signature whose expression is correlated with most of the 35 miRNAs. Within the nuclear processing network, the BMP1 and TGFB1 genes were highly connected and positively correlated with the maturation of a large number of miRNAs. Other members of the SMAD family were included in the network, albeit with a smaller and more distinct set of correlated miRNAs. This indicates that the above described Drosha-mediated processing, where individual co-factors such as the members of the TGFβ/BMP/SMAD signaling pathway control the processing in miRNA specific manner, is also likely to occur in glioma. Further investigation will be necessary to fully understand the specificity that occurs with SMAD-mediated processing.
The TGFβ pathway has also been shown to enhance epidermal growth factor (EGF)-mediated effects in gliomas and heighten expression of SMAD2 and SMAD4 in glioma cell lines [47]. Furthermore, TGFβ induces leukemia inhibitory factor (LIF) through the SMAD-dependent pathway, which increases the self-renewing capacity of glioma-initiating cells [48]. In gliomas with an unmethylated PDGFB gene, the TGFβ/SMAD pathway increases the expression of PDGFB, which can induce the proliferation of glioma cells [49]. High TGFβ/SMAD activity is associated with poor prognosis in patients with glioma, [49] which is confirmed in our current study. The emerging picture suggests that this pathway may be a key regulator of glioma development and progression through miRNA-mediated effects.
In summary, by utilizing the comprehensive data provided by deep sequencing, we have uncovered a complicated network of gene expression changes within the miRNA biogenesis pathway that impact miRNA maturation and correlate with glioma progression. This study not only provides a newly characterized mechanism of miRNA alterations in glioma but also leads to a number of potentially new oncogenic and tumor-suppressing miRNAs that are important for glioma. Further studies on these miRNAs will likely yield important insights as well as opportunities for prognosis and therapeutics for glioma.
Supplementary Material
292 miRNAs were identified as differentially maturated (M/P ratio > 2-fold) between gliomas and normal brain. For visualization, the M/P ratio of each miRNA is scaled to interval of zero (lowest expression ratio)-to-one (highest expression ratio), as indicated by the color bar.
Levels are shown across all samples for the 35 differentially matured miRNA which correlate with glioma grade. The expression of each miRNA is scaled to interval of zero-to-one.
Individual patient samples from different sequencing pools were selected at random for validation by qRT-PCR. Fold change of pre-miRNA (green) and mature miRNA (blue) relative quantity normalized to normal brain control, M/P expression ratio relative to the M/P expression ratio of normal brain.
The bar height at each base represents the number of reads overlapping that position. Locations of the pre-miRNA loop and mature sequences are labeled. While mature sequences are substantially more abundant, there are a sufficient number of reads also for the pre-miRNA allowing the quantification of the expression level.
Acknowledgments
Supported in part by the Paul and Joann Oreffice Fund for Brain Tumor Research (R.S., G.N.F., and W.Z.), U24 CA143835 from the National Institute of Health (I.S. and W.Z.), the Academy of Finland project no. 132877 (M.N.), the Finnish Funding Agency for Technology and Innovation Finland Distinguished Professor Programme (M.N. and O.Y.-H.), Sigrid Juselius Foundation (M.N.) and The University of Texas MD Anderson Cancer Center core grant CA016672 from the National Institutes of Health. We would like to thank Sue Moreau from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing this manuscript.
Footnotes
Conflict of interest statement: None of the authors has a conflict of interest regarding this study.
Deep sequencing data has been deposited in short read archives under accession number SRA057775.
AUTHORS’ CONTRIBUTIONS
LMM, VK, YL, MA, DG, MN, WZ analyzed data. LMM, DC, XL, CGL performed experiments. MA implemented the sequence analysis. LMM, MN, WZ, IS conceived of the study, OYH, RS, GNF participated in its design and coordination, LMM, VK, MN, WZ drafted the manuscript. All authors read and approved the final manuscript.
Detailed information on library preparation, sequencing, and data analysis.
References
- 1.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 4.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling H, Zhang W, Calin GA. Principles of microRNA involvement in human cancers. Chin J Cancer. 2011;11:739–48. doi: 10.5732/cjc.011.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 8.Thomson JM, Newman M, Parker JS, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 12.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Cifuentes D, Xue H, Taylor DW, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevignani C, Calin GA, Siracusa LD, et al. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 18.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 19.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 20.Sugito N, Ishiguro H, Kuwabara Y, et al. RNASEN Regulates Cell Proliferation and Affects Survival in Esophageal Cancer Patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 21.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer science. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and Outcomes in Patients with Ovarian Cancer. New Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;9:1414–9. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Melo SA, Moutinho C, Ropero S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Melo SA, Ropero S, Moutinho C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Kumar MS, Pester RE, Chen CY, Lane K, et al. Dicer1 functions as a haploinsufficient tumor suppressor. 2009;23:2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravi A, Gurtan AM, Kumar MS, Bhutkar A, et al. Proliferation and tumorigenesis of a murine sarcoma cell line in the absence of DICER1. Cancer Cell. 2012;21:848–55. doi: 10.1016/j.ccr.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 29.Kim TM, Huang W, Park R, et al. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sana J, Hajduch M, Michalek J, et al. MicroRNAs and glioblastoma: roles in core signalling pathways and potential clinical implications. J Cell Mol Med. 2011;15:1636–1644. doi: 10.1111/j.1582-4934.2011.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 32.Conti A, Aguennouz M, La Torre D, et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol. 2009;93:325–332. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- 33.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 Targets a Network of Key Tumor-Suppressive Pathways in Glioblastoma Cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Han L, Ge Y, et al. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–920. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- 35.Moore LM, Zhang W. Targeting miR-21 in glioma: a small RNA with big potential. Expert Opin Ther Targets. 2010;11:1247–57. doi: 10.1517/14728222.2010.527334. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protocols. 2008;3:190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 40.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblstoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 41.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2. 8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 43.Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morlando M, Ballarino M, Gromak N, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis BN, Hilyard AC, Lagna G, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis BN, Hilyard AC, Nguyen PH, et al. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Held-Feindt J, Lütjohann B, Ungefroren H, et al. Interaction of transforming growth factor-beta (TGF-beta) and epidermal growth factor (EGF) in human glioma cells. J neurooncol. 2003;63:117–127. doi: 10.1023/a:1023943405292. [DOI] [PubMed] [Google Scholar]
- 48.Peñuelas S, Anido J, Prieto-Sánchez RM, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 49.Bruna A, Darken RS, Rojo F, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
292 miRNAs were identified as differentially maturated (M/P ratio > 2-fold) between gliomas and normal brain. For visualization, the M/P ratio of each miRNA is scaled to interval of zero (lowest expression ratio)-to-one (highest expression ratio), as indicated by the color bar.
Levels are shown across all samples for the 35 differentially matured miRNA which correlate with glioma grade. The expression of each miRNA is scaled to interval of zero-to-one.
Individual patient samples from different sequencing pools were selected at random for validation by qRT-PCR. Fold change of pre-miRNA (green) and mature miRNA (blue) relative quantity normalized to normal brain control, M/P expression ratio relative to the M/P expression ratio of normal brain.
The bar height at each base represents the number of reads overlapping that position. Locations of the pre-miRNA loop and mature sequences are labeled. While mature sequences are substantially more abundant, there are a sufficient number of reads also for the pre-miRNA allowing the quantification of the expression level.