Abstract
Quality of maternal care experienced during infancy is a key factor that can confer vulnerability or resilience to psychiatric disorders later in life. Research continues to indicate that early-life experiences can affect developmental trajectories through epigenetic alterations capable of affecting gene regulation and neural plasticity. Previously, our lab has shown that experiences within an adverse caregiving environment (i.e. maltreatment) produce aberrant DNA methylation patterns at various gene loci in the medial prefrontal cortex (mPFC) of developing and adult rats. This study aimed to determine whether caregiver maltreatment likewise affects expression levels of several genes important in regulating DNA methylation patterns (Dnmt1, Dnmt3a, MeCP2, Gadd45b, and Hdac1). While we observed minimal changes in gene expression within the mPFC of developing rats, we observed expression changes for all genes in adult animals. Specifically, exposure to maltreatment produced a significant decrease in mRNA levels of all epigenetic regulators in adult males and a significant decrease in Gadd45b in adult females. Our results here provide further empirical support for the long-term and sex-specific epigenetic consequences of caregiver maltreatment on the mPFC.
Keywords: epigenetics, early-life stress, maternal care, mPFC, Dnmt, Hdac, Gadd45b, MeCP2
1. Introduction
Infancy is a sensitive period of development in which alterations in the quality or quantity of maternal care are capable of producing dramatic effects on neurobiological and behavioral outcomes (Caldji et al., 2000; Cicchetti and Toth, 2005; Sanchez, 2006). Mechanisms underlying these environmentally-driven phenomena are not fully understood, but over the past decade the birth of epigenetics research has caused a revolution in our understanding of gene-environment interactions in this capacity in the developing and adult mammalian brain. Although epigenetic modifications were originally thought to only program patterns of gene expression during cellular development and differentiation, a significant body of research now indicates that such modifications occur in response to environmental signals from infancy to senescence, with significance for long-lasting changes in gene regulation and expression (Caldji et al., 2011; Roth, 2012) and neuronal plasticity (Day and Sweatt, 2011).
Various types of epigenetic modifications have been identified, such as DNA methylation and post-translational modifications to histone tails (i.e. acetylation and methylation), and together they help determine whether DNA is accessible for gene transcription (Strahl and Allis, 2000). Of these modifications, DNA methylation and histone acetylation have been the most extensively studied in the context of behavior, and are driven and maintained by various enzymes and proteins. DNA methylation, for example, is catalyzed by a family of DNA methyltransferases (DNMTs) that includes DNMT1 and DNMT3a (Moore et al., 2012). DNMT1 is a maintenance methyltransferase which is responsible for adding methyl groups to the complimentary strand of hemimethylated DNA, thereby maintaining DNA methylation patterns during the DNA replication process. DNMT3a is a de novo methyltransferase, capable of catalyzing new methylation patterns. While both Dnmt1 and Dnmt3a mRNA levels are highest in the mammalian cortex, hippocampus, striatum, and cerebellum during late prenatal and early postnatal development, both enzymes are present and functional throughout the lifetime (Feng et al., 2005; Goto et al., 1994; Inano et al., 2000; Matrisciano et al., 2012; Miller et al., 2012; Siegmund et al., 2007). Dnmt3a mRNA, for example, is at high levels within the rodent mPFC for the first three weeks of life but dramatically decreases to adult levels by postnatal day (PN) 21 (Miller et al., 2012). Dnmt1 mRNA levels within the rodent amygdala and subregions of the hippocampus are down-regulated within the first postnatal week (Simmons et al., 2012).
Another protein associated with DNA methylation is Methyl-CpG Binding Protein-2 (MeCP2), which binds to methylated cytosines and recruits either histone deacetylases (HDACs) and other corepressors to suppress gene transcription (Jones et al., 1998) or CREB1 and other coactivators to promote gene transcription (Chahrour et al., 2008; Uchida et al., 2011). A deficiency in MeCP2 has been widely associated with Rett syndrome, a neurodevelopmental disorder causing mental retardation with early onset in childhood, and MeCP2 mutant mice have been shown to exhibit many of the same cognitive deficits and neuroanatomical abnormalities associated with Rett syndrome (Chen et al., 2001; Jorgensen and Bird, 2002; Stearns et al., 2007). These studies have also emphasized the importance of MeCP2 in the developing and mature brain, with prefrontal cortical expression of this protein present during early fetal stages and increasing until late childhood when levels are maintained into adulthood (Akbarian et al., 2001; Balmer et al., 2003; Kaufmann et al., 2005; Shahbazian et al., 2002).
Recently, investigators have discovered that the Growth Arrest and DNA-Damage-Inducible beta (Gadd45b) protein plays an important role in DNA demethylation (i.e. the active removal of methyl groups). Instead of breaking the strong covalent bonds between methyl groups and cytosines, Gadd45b instead functions through a DNA-repair-like mechanism, in which an unmethylated cytosine replaces the methylated cytosine after a sequence of molecular events (Ma et al., 2009a; Ma et al., 2009b). The ontogenetic profile of Gadd45b has not been characterized, but recent work has demonstrated its role in regulation of memory formation (Leach et al., 2012; Sultan et al., 2012). Additional work in patients with psychosis has shown an increase in parietal cortical levels of Gadd45b mRNA and protein in comparison to healthy controls, demonstrating its possible contributory role to major psychosis (Gavin et al., 2012).
While the previously mentioned proteins and enzymes work to regulate DNA methylation patterns, the ability of histones to further regulate transcription adds another layer of complexity. The addition of an acetyl group to a lysine residue of the N-terminal histone tail is able to neutralize the positive charge and loosen the chromatin complex, allowing for increased binding of transcription factors and therefore increased gene expression. This process is catalyzed by histone acetyltransferases (HATs) and reversed by histone deacetylases (HDACs). HDACs are present throughout the lifetime, and the majority of these enzymes show changes in mRNA levels throughout development and adulthood (Levine et al., 2012). For example, while Hdac1 mRNA levels are relatively stable between PN21, PN28, and PN60 in the forebrain neocortex of mice, Hdac3 mRNA show a dramatic increase between PN28 and 60 (Levine et al., 2012) Alterations in mRNA levels of a number of HDACs have been implicated in psychiatric disorders (Sharma et al., 2008) and the lasting effects of early-life stress (Levine et al., 2012).
Previously, we found that exposing infant rats to an adverse caregiving environment produces DNA methylation alterations that are present in both the developing and adult whole (Roth et al., 2009) and medial (Blaze et al., 2013) prefrontal cortex. While our 2009 study measured methylation in the prefrontal cortex as a whole, the importance of the medial prefrontal cortex (mPFC) in developmental trajectories associated with early-life stress is beginning to be highlighted in the literature. For example, this region has long been recognized for its role in mediating the stress response (Diorio et al., 1993), and various groups have recently demonstrated changes in mPFC gene expression (Uchida et al., 2010) and structural abnormalities (Spinelli et al., 2009) following early-life stress. In our 2013 study, we found sex-specific alterations in methylation of the Brain-derived neurotrophic factor (Bdnf) gene in the mPFC of adolescent and adult rats that experienced early-life maltreatment. Additionally, differences in reelin methylation were only present in adulthood for maltreated males and both males and females exposed to nurturing care outside the home cage (Blaze et al., 2013). The aim of this study was to determine whether caregiver maltreatment likewise affects expression levels of several genes important in regulating epigenetic patterns. Here we examined whether caregiver maltreatment altered Dnmt1, Dnmt3a, MeCP2, Gadd45b, or Hdac1 mRNA levels in the developing and adult mPFC.
2. Materials and Methods
2.1 Subjects
All procedures were approved by the University of Delaware Animal Care and Use Committee. Brain tissue came from a total of twenty-two litters for all age cohorts combined, and this sample matches that previously used (Blaze et al., 2013). To generate subjects, male and female outbred Long-Evans rats from Harlan were housed in our temperature and light-controlled (12-hours light/dark cycle, lights on at 6:00am) colony room in standard polypropylene cages and given ample bedding material. Behavioral manipulations were all performed during the light cycle, and animals were provided with food and water ad libitum. Females were bred with males in our breeding colony and allowed to raise at least one litter of pups before the start of the experiment to eliminate confounds associated with first-time mothers. Dams and pups were left undisturbed on PN 0 (day of pup birth) and litters were culled to approximately 6 males and 6 females per litter on PN1.
2.2 Caregiving Manipulations
Our caregiving manipulations are based on a method previously reported (Blaze et al., 2013; Roth et al., 2009) and adapted from earlier studies (Ivy et al., 2008; Roth and Sullivan, 2005). On PN1, we used a within-litter design and split a litter into three equal groups for our three treatment conditions. For the first seven days of life (PN1-7), one group of pups (normally 2 females and 2 males) was exposed to a stressed, non-biological caregiver for thirty minutes each day (maltreatment condition). To produce maltreatment behaviors toward infants, the dam was provided insufficient nesting material (100 ml of wood shavings) while she was in a novel environment. Littermates were exposed to a nurturing caregiver outside of the home cage (cross-foster care condition), which consisted of another non-biological lactating dam who was allowed one hour to habituate to her new environment and given sufficient nesting material (approximately 2 cm layer of wood shavings). Both maltreatment and cross-foster care chambers were maintained between 24-29°C. Additional littermates were quickly marked for identification and weighed at the beginning of each session and immediately returned to the biological mother in the homecage (normal maternal care condition). Pups in all conditions were marked with the same non-toxic permanent marker for identification. After each day's thirty minute session, maltreatment and cross-foster care pups were returned to the homecage with the biological mother and all litters remained undisturbed except for weekly cage changes and treatment sessions. For our PN8 time-point, pups were sacrificed 24 hours after the last caregiving condition, but for older time-points pups were weaned at PN21 and housed in same-sex/treatment pairs until sacrifice in adolescence (PN30) or adulthood (PN90).
Maternal caregiving behaviors were scored via live observation or video recording by two trained observers. For each litter, maltreatment or nurturing behaviors were tallied in five-minute time bends and averaged across all seven exposure days. To measure pup response to maternal behaviors, we measured pup audible and 40 kHz ultrasonic vocalizations (Batbox III D, NHBS Ltd., UK) by marking the presence of a vocalization in each one-minute time bend and averaging across the seven exposure days. As previously reported for this cohort of rats (Blaze et al., 2013), we observed significant levels of aversive caregiving towards infants (stepped on, dropped during transport, dragged while nipple attached, actively avoided, and roughly handled infants) within the maltreatment condition, but similar high levels of nurturing care (infant licking, grooming, and nursing) within the cross-foster and normal care conditions. Furthermore, we observed high amounts of audible and ultrasonic distress vocalizations emitted within our maltreatment condition but not within the environments where pups experienced nurturing care (our normal and cross-foster groups).
2.3 Gene Expression Assays
Tissue was collected from animals at baseline conditions at PN8, PN30, or PN90. Brains were removed, sliced using a 1 mm brain matrix, flash frozen on untreated slides with 2-methylbutane, and placed in a -80 freezer until later processing. The mPFC (consisting of bilateral prelimbic and infralimbic tissue) was dissected on dry ice using stereotaxic coordinates and RNA was extracted (Qiagen Inc., Valencia, CA). Quantification and assessment of nucleic acid quality were determined using spectrophotometry (NanoDrop 2000). Reverse transcription was performed using a cDNA synthesis kit (Qiagen) on RNA, and cDNA was amplified by real-time PCR (Bio-Rad CFX96) with Taqman probes (Applied Biosystems) to target Dnmt1, Dnmt3a, MeCP2, Gadd45b, Hdac1, or tubulin (for a reference gene) mRNA. All reactions for each gene in the expression assays were run in triplicate. Product specificity was determined by gel electrophoresis.
2.4 Statistical Analyses
For gene expression assays, we used the comparative Ct method to obtain the relative fold change for experimental (maltreatment or cross-foster) vs. control (normal care) groups (Livak and Schmittgen, 2001). We used one-sample t-tests to compare fold change differences between experimental and control groups. Two-way ANOVAs were used to analyze differences across treatment groups and sex and were followed by Bonferroni post hoc tests if appropriate. Differences were considered to be statistically significant for p<0.05.
3. Results
3.1. Experiment 1: Infant Gene Expression
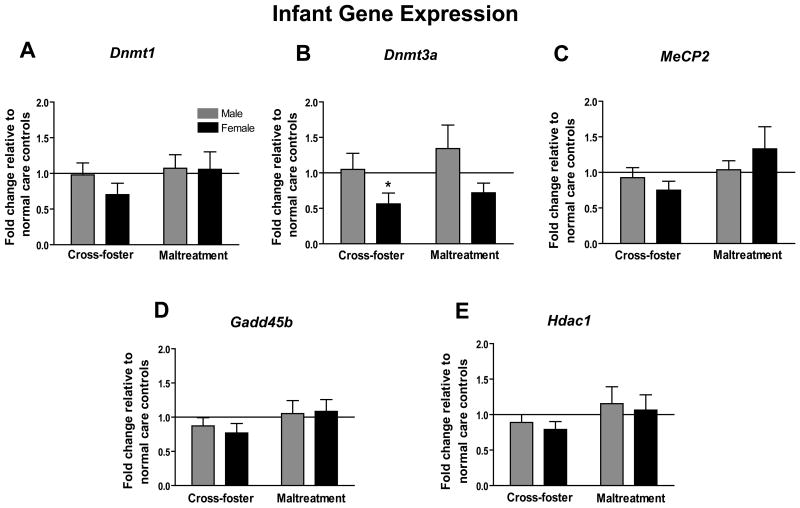
At PN8, brains were removed 24 hours after the last caregiving manipulation to assess whether mRNA levels of several epigenetic regulators within the mPFC had been affected by repeated exposures to our various caregiving conditions (Figure 1a-e). No significant differences were detected between our experimental and controls groups for Dnmt1, MeCP2, Gadd45b, or Hdac1 (one-sample t-tests, all p's>0.05). Likewise, no differences were detected between our experimental groups for these same gene loci (two-way ANOVAs, all p's>0.05). A one-sample t-test however revealed that females who experienced nurturing care outside the homecage had significantly less Dnmt3a mRNA compared to same sex normal care controls (t8=2.85, p<0.05) (Figure 1b). Likewise, maltreated-females had a trending decrease in Dnmt3a mRNA (t9=2.06, p=0.07). Additionally, a two-way ANOVA for this gene locus showed a significant main effect of sex (F (1,34)=5.85, p<0.05) but no main effect of infant condition or an infant condition by sex interaction.
Figure 1.
Infant (PN8) mPFC mRNA expression for epigenetic regulator genes (a) Dnmt1 (b) Dnmt3a (c) MeCP2 (d) Gadd45b (e) Hdac1. (for all genes- n=9-10 per group; *p<0.05 versus normal care controls; error bars represent SEM)
3.2 Experiment 2: Adolescent Gene Expression
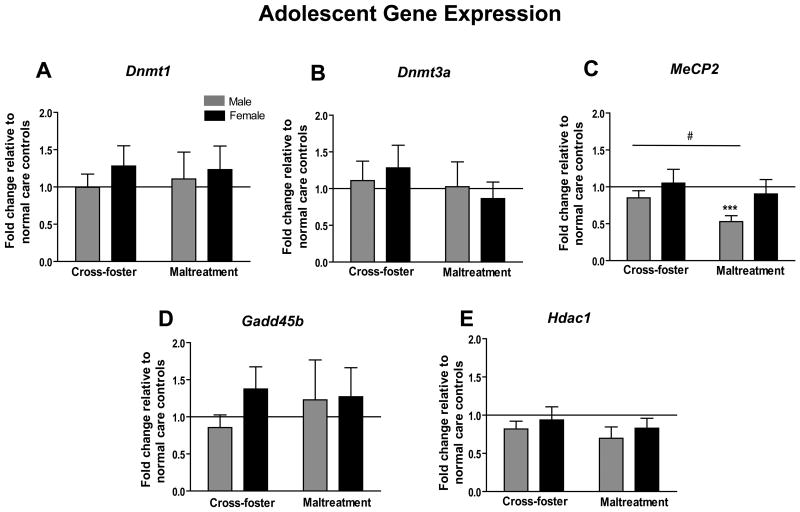
Another cohort of animals was sacrificed at PN30 to assess adolescent levels of epigenetic regulators within the mPFC (Figure 2a-e). At this time point, no significant differences were detected in mRNA levels for Dnmt1, Dnmt3a, Gadd45b, or Hdac1 (one-sample and two-way ANOVAs, all p's>0.05). For MeCP2 however, maltreated males had significantly less mRNA in comparison to normal care (t7=5.941, p<0.001) and cross-foster care controls (t15=2.54, p<0.05) (Figure 2c). A two-way ANOVA for this gene locus revealed a trending effect of sex (F(1,29)=3.73, p=0.06), and no main effect of infant condition or an interaction effect (p's>0.05).
Figure 2.
Adolescent (PN30) mPFC mRNA expression for epigenetic regulator genes (a) Dnmt1 (b) Dnmt3a (c) MeCP2 (d) Gadd45b (e) Hdac1. (n=7-9 per group; ***p<0.001 versus normal care controls; #p<0.05 between groups; error bars represent SEM)
3.3 Experiment 3: Adult Gene Expression
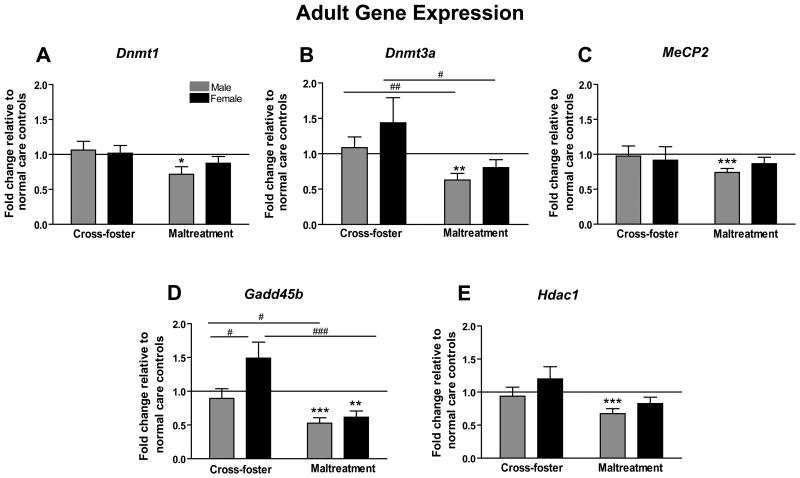
mPFC mRNA levels were likewise assessed on another cohort allowed to reach adulthood (PN90). Unlike our other two age groups, we observed significant group differences at all gene loci in our adult animals (Figure 3a-e). Males who experienced maltreatment had less Dnmt1 (t13=2.629, p<0.05) and MeCP2 (t13=4.689, p<0.001) mRNA compared to normal controls (Figure 3, panels a and c). Two-way ANOVAs for Dnmt1 and MeCP2 revealed no other significant differences (p's>0.05). Maltreated males also had less Dnmt3a mRNA in comparison to normal care controls (t13=3.934, p<0.01) (Figure 3b). In addition, a two-way ANOVA showed a main effect of infant condition (F(1,37)=12.41, p=0.001), with further analyses showing that both maltreated males (t22=2.703, p=0.01) and females (t15=2.269, p<0.05) had significantly less Dnmt3a mRNA compared to animals that experienced nurturing care outside the home cage (cross-foster care condition).
Figure 3.
Adult (PN90) mPFC mRNA expression for epigenetic regulator genes (a) Dnmt1 (b) Dnmt3a (c) MeCP2 (d) Gadd45b (e) Hdac1. (for all genes- n=5-6 cross-foster care females, n=10 cross-foster care males, n=12 maltreatment females, n=10 maltreatment males; *p<0.05, **p<0.01, ***p<0.001 versus normal care controls; #p<0.05 between sexes; ##p<0.01, ###p<0.001 between groups; error bars represent SEM)
For Gadd45b, maltreated males again had significantly less mRNA in comparison to normal care controls (t12=5.917, p<0.001), and maltreated females likewise had lower Gadd45b mRNA levels than controls (t10=4.202, p<0.01) (Figure 3d). A two-way ANOVA revealed a main effect of sex (F(1,35)=7.248, p=0.01), infant condition (F(1,35)=23.80, p<0.0001), and a sex by infant condition interaction (F(1,35)=3.965, p=0.05). Post-hoc analyses showed that maltreated males (t21=2.384, p<0.05) and females (t14=4.262, p<0.001) had less Gadd45b mRNA than sex-matched animals from the cross-foster care group. Additionally, females that experienced nurturing care outside of the home cage had significantly more Gadd45b mRNA than cross-foster males (t13=2.290, p<0.05). Finally, maltreated males had significantly lower Hdac1 mRNA levels than normal care controls (t12=4.354, p<0.001) (Figure 3e). A two-way ANOVA revealed a main effect of infant condition (F(1,37)=7.313, p=0.01) for Hdac1 mRNA, but no main effect of sex or a sex by condition interaction (p's>0.05).
4. Discussion
Using this rodent model, we have previously characterized the effects of repeated exposure to maltreatment or nurturing care during the first postnatal week on Bdnf and reelin DNA methylation patterns in the developing and adult mPFC (Blaze et al., 2013). Specifically, in adulthood, both males and females who experienced maltreatment in infancy displayed decreased methylation of DNA associated with Bdnf exon I. Maltreated-females also had increased methylation of Bdnf exon IV relative to normal and cross-foster care controls and maltreated-males. These group differences in methylation were not present in adolescence or infancy. Instead, a different pattern was present in adolescence, with maltreated males showing increased methylation at Bdnf exon I and females showing a decrease in methylation at exon IV relative to normal care controls. At the reelin promoter, we observed greater methylation in both adult males that were maltreated and nurtured outside of the homecage. Similar to our Bdnf effects, these group differences were not present earlier in development.
In the current study, we sought to determine the effects of these caregiving environments on genes important for establishing, maintaining, or reversing DNA methylation patterns. In the adult (PN90) mPFC, we found that maltreated-males exhibited less Dnmt1, Dnmt3a, MeCP2, Gadd45b, and Hdac1 mRNA than normal care controls. Various groups have demonstrated alterations in DNMT1 and DNMT3a mRNA or protein levels as the result of prenatal stress (Jensen Peña et al., 2012; Matrisciano et al., 2012). Though offspring of prenatally stressed mothers have been found to have increased Dnmt1 (cortex) and Dnmt3a (placenta) mRNA levels (Jensen Peña et al., 2012) and increased DNMT1 protein levels in adult brain tissue (Matrisciano et al., 2012), we found here decreased Dnmt1 and Dnmt3a mRNA levels in males who experienced maltreatment during the first seven postnatal days. This could demonstrate the differential sensitivity of these regulators to environmental factors during prenatal versus postnatal periods.
Maltreated-males also showed a change in MeCP2 expression, while female MeCP2 mRNA levels remained similar to that of controls. Transient sex differences in MeCP2 have previously been observed in the healthy amygdala and ventromedial hypothalamus, such that males had less MeCP2 mRNA and protein at PN1 but the difference was no longer present at PN10 (Kurian et al., 2007). It has also been shown that males with decreased MeCP2 protein (but not female) expression exhibit reductions in juvenile social play behavior (Kurian et al., 2008; McCarthy et al., 2009; Nagarajan et al., 2006). Our data here are consistent with other reports demonstrating the ability of repeated bouts of maternal separation to have lasting effects on MeCp2. For example, one report has shown reduced binding of MeCP2 in the hypothalamus of separated mice (Murgatroyd et al., 2009), and another more recent report has shown a reduction in MeCP2 cell counts in the adult nucleus accumbens of separated rats (Lewis et al., 2013). We also found changes at the Gadd45b gene locus, a recently identified candidate for active DNA demethylation (Ma et al., 2009a). To our knowledge, no studies to date have investigated the developmental profile of Gadd45b mRNA or protein levels, or the effects of early-life stress on this gene. In our study, both adult males and females with a history of maltreatment showed decreased levels of Gadd45b. While adult expression levels for all other epigenetic regulators that we examined were only lower in maltreated-males, this was the only locus reaching statistical significance in maltreated-females.
We also found a decrease in Hdac1 mRNA in males with a history of maltreatment. While the effects of early-life stress on DNA methylation have been widely studied by our lab and others, the effect of early-life stress on histone modifications is a less common area of research though these two mechanisms (histone modifications and DNA methylation) are not mutually exclusive. HDACs can participate in gene silencing by their binding to MeCP2 and working in parallel with other recruited corepressors. Adult offspring of low licking/grooming mothers have been shown to have decreased H3K9 acetylation in the hippocampus, which could be reversed with the HDAC inhibitor trichostatin-A (Weaver et al., 2004). Maternal separation during infancy has also been shown to produce changes in histone acetylation and methylation within the developing and adult hippocampus (Suri et al., 2013; Xie et al., 2013). A recent study revealed decreased Hdac1 mRNA in the forebrain neocortex of adult male mice that had experienced maternal separation in infancy (Levine et al., 2012), which parallels our findings. Their study also showed the capacity of maternal separation to alter mRNA levels of other classes of HDAC enzymes, suggesting it would be worthwhile to also characterize changes for additional HDACs in our model.
While the observed gene expression changes were robust in adulthood, several months post-manipulation, only few gene expression changes were present earlier in development. At PN30 (adolescence), maltreated-males showed a decrease in MeCP2 mRNA relative to normal and cross-foster controls. In infants, 24 hours after the last caregiving exposure, the only significant difference observed was a decrease in Dnmt3a mRNA levels in females who had experienced nurturing care outside of the homecage. Maltreated-females also showed a trending decrease in Dnmt3a levels. As previously mentioned, Dnmt3a is a de novo methyltransferase that establishes new methylation patterns, and this enzyme exhibits the highest mRNA levels during the first three weeks of life (Miller et al., 2012). There is also evidence that females have higher levels during the first two postnatal weeks than males (Jessen and Auger, 2011; Kolodkin and Auger, 2011). Our observation of a sex difference in Dnmt3a mRNA that is present at PN8 but no longer present at our adolescent time point may be in line with these sex and age effects. Furthermore, our data illustrate the sensitivity of Dnmt3a to changes in the caregiving environment.
A common theme among our observations from this study was aberrant gene expression specific to our maltreatment group, especially for males. While several of these genes are known to exhibit sexually-dimorphic expression patterns that could contribute to our group differences, it is possible that differences in maternal care toward male and female pups both within the homecage (before and after treatment) and during our manipulations contributed to gene differences. Though the design of the present experiment did not allow us to distinguish caregiving behaviors directed at males versus those at females, it is known that males are licked more by the dam within the nest (Hao et al., 2011; Moore and Morelli, 1979). Regardless of a mechanism of change, our data are consistent with the notion that there could be large-scale (global) methylation changes in our adult animals, especially males. Using the same paradigm, our previous work has shown sex-specific alterations in Bdnf DNA methylation levels in adulthood (Blaze et al., 2013), and other work is also in line with the idea of sexually-dimorphic DNA methylation changes throughout the lifespan (Schwarz et al., 2010). Future work will be necessary to identify whether other genes show similar experience-induced changes in DNA methylation, and to co-localize changes in DNMT, MeCP2, Gadd45b, and HDAC binding to specific gene loci.
A second common theme that continues to be present with our work is epigenetic differences that vary across or emerge later in development for our maltreatment group. This could be partially attributed to the late developing nature of the medial prefrontal cortex itself, with connections and circuits continuing to mature well past the postnatal period and into adolescence (Benes et al., 2000). It is also known that some of the enzymes/proteins measured as markers of epigenetic regulation are involved in neuronal plasticity (Ballas et al., 2005; Ma et al., 2009b), which may be potentiated at later developmental time points. Another factor that may be responsible for these observations is that epigenetic patterns (both DNA methylation and gene expression levels of the regulators we examined) are known to naturally change over the course of the lifespan in “normal” animals, thus our baseline (i.e. levels within the normal maternal care group) to which we compare our experimental groups is presumed to be changing too with age (Lister et al., 2013; Numata et al., 2012; Simmons et al., 2012). Finally, it is possible that social factors experienced later in life (i.e. experiences with cagemates after weaning) also contributed to the observed gene profiles in the maltreated-animals.
While our assays failed to detect significant gene expression changes for many of our regulators in infancy or adolescence, it is possible that our global approach missed changes that occurred at the level of specific genes or even within specific cell-types. For example, it has been observed that patterns of DNA methylation in the frontal cortex can depend on cell-type (Shulha et al., 2013), suggesting that expression levels of epigenetic regulators could differ between glia and neurons. While the techniques used here limit us from concluding anything about such changes in these tissue samples, it is a future avenue of research in our lab. Future research should also address the degree to which these differences in mRNA translate into differences in protein expression.
In conclusion, while it is clear that childhood maltreatment poses a significant risk for psychopathology, the biological basis of this phenomenon remains an important topic for investigation. Using our rodent model, we have shown that repeated exposure to caregiver maltreatment during the first postnatal week has the capacity to alter DNA methylation. Here we extend this work to show that postnatal maltreatment likewise alters expression levels of genes important for establishing, maintaining, or reversing DNA methylation. Additional studies are necessary to link these gene methylation and expression changes with histone alterations, protein expression, subsequent behavioral outcomes, and to explore their reversibility.
Highlights.
We assessed the impact of early adversity on expression of epigenetic regulators (fold changes in mRNA).
Minimal fold changes were detected within the infant and adolescent mPFC.
Adult males and females who experienced maltreatment displayed significant fold changes.
Data indicate long-term, sex-specific epigenetic changes after early adversity.
Acknowledgments
We thank Lisa Scheuing, Megan Warren, Hannah Evans, Hillary Porter, and Stephanie Matt for performing gel electrophoresis. This work was supported by the University of Delaware Research Foundation and The National Institute of General Medical Sciences (1P20GM103653).
Abbreviations
- mPFC
medial prefrontal cortex
- DNMT
DNA methyltransferase
- MeCP2
Methyl-CpG Binding Protein-2
- Gadd45b
Growth Arrest and DNA-Damage-Inducible beta
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- PN
postnatal day
- Bdnf
brain-derived neurotrophic factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akarian S, Chen RZ, Gribnau J, Rasmussen TP, Fong Hf, Jaenisch R, Jones EG. Expression pattern of the rett syndrome gene mecp2 in primate prefrontal cortex. Neurobiol Dis. 2001;8:784–791. doi: 10.1006/nbdi.2001.0420. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. Rest and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Balmer D, Goldstine J, Rao YM, LaSalle J. Elevated methyl-cpg-binding protein 2 expression is acquired during postnatal human brain development and is correlated with alternative polyadenylation. J Mol Med. 2003;81:61–68. doi: 10.1007/s00109-002-0396-5. [DOI] [PubMed] [Google Scholar]
- Benes FM, Taylor JB, Cunningham MC. Convergence and plasticity of monoaminergic systems in the medial prefrontal cortex during the postnatal period: Implications for the development of psychopathology. Cereb Cortex. 2000;10:1014–1027. doi: 10.1093/cercor/10.10.1014. [DOI] [PubMed] [Google Scholar]
- Blaze J, Scheuing L, Roth TL. Differential methylation of genes in the medial prefrontal cortex of developing and adult rats following exposure to maltreatment or nurturing care during infancy. Dev Neurosci. 2013;35:306–316. doi: 10.1159/000350716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldji C, Diorio J, Meaney MJ. Variations in maternal care in infancy regulate the development of stress reactivity. Biol Psychiatry. 2000;48:1164–1174. doi: 10.1016/s0006-3223(00)01084-2. [DOI] [PubMed] [Google Scholar]
- Caldji C, Hellstrom IC, Zhang TY, Diorio J, Meaney MJ. Environmental regulation of the neural epigenome. FEBS Lett. 2011;585:2049–2058. doi: 10.1016/j.febslet.2011.03.032. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. Mecp2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-cpg binding protein-2 in cns neurons results in a rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Cicchetti D, Toth SL. Child maltreatment. Ann Rev Clin Psych. 2005;1:409–438. doi: 10.1146/annurev.clinpsy.1.102803.144029. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic modifications in neurons are essential for formation and storage of behavioral memory. Neuropsychopharmacol. 2011;36:357–358. doi: 10.1038/npp.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diorio D, Viau V, Meaney M. The role of the medial prefrontal cortex (cingulated gyrus) in the regulation of hypothalamic-pituitary-adrenal responses to stress. The Journal of Neuroscience. 1993;13:3839–3847. doi: 10.1523/JNEUROSCI.13-09-03839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases dnmt3a and dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (gadd45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacol. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- Hao Y, Huang W, Nielsen DA, Kosten TA. Litter gender composition and sex affect maternal behavior and DNA methylation levels of the oprm1 gene in rat offspring. Front Psychiatry. 2011;2:21. doi: 10.3389/fpsyt.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, Tajima S. Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- Ivy AS, Brunson KL, Sandman C, Baram TZ. Dysfunctional nurturing behavior in rat dams with limited access to nesting material: A clinically relevant model for early-life stress. Neurosci. 2008;154:1132–1142. doi: 10.1016/j.neuroscience.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen Peña C, Monk C, Champagne FA. Epigenetic effects of prenatal stress on 11β-hydroxysteroid dehydrogenase-2 in the placenta and fetal brain. PLoS One. 2012;7:e39791. doi: 10.1371/journal.pone.0039791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen HM, Auger AP. Sex differences in epigenetic mechanisms may underlie risk and resilience for mental health disorders. Epigenetics. 2011;6:857–861. doi: 10.4161/epi.6.7.16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Jan Veenstra GC, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and mecp2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Jorgensen HF, Bird A. Mecp2 and other methyl-cpg binding proteins. Ment Retard Dev Disabil Res Rev. 2002;8:87–93. doi: 10.1002/mrdd.10021. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Johnston MV, Blue ME. Mecp2 expression and function during brain development: Implications for rett syndrome's pathogenesis and clinical evolution. Brain Dev. 2005;27:S77–S87. doi: 10.1016/j.braindev.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Kolodkin MH, Auger AP. Sex difference in the expression of DNA methyltransferase 3a in the rat amygdala during development. J Neuroendocrinol. 2011;23:577–583. doi: 10.1111/j.1365-2826.2011.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Bychowski ME, Forbes-Lorman RM, Auger CJ, Auger AP. Mecp2 organizes juvenile social behavior in a sex-specific manner. J Neurosci. 2008;28:7137–7142. doi: 10.1523/JNEUROSCI.1345-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Forbes-Lorman RM, Auger AP. Sex difference in mecp2 expression during a critical period of rat brain development. Epigenetics. 2007;2:173–178. doi: 10.4161/epi.2.3.4841. [DOI] [PubMed] [Google Scholar]
- Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, Gould TJ. Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Memory. 2012;19:319–324. doi: 10.1101/lm.024984.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Worrell TR, Zimnisky R, Schmauss C. Early life stress triggers sustained changes in histone deacetylase expression and histone h4 modifications that alter responsiveness to adolescent antidepressant treatment. Neurobiol Dis. 2012;45:488–498. doi: 10.1016/j.nbd.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CR, Staudinger K, Scheck L, Olive MF. The effects of maternal separation on adult methamphetamine self-administration, extinction, reinstatement, and mecp2 immunoreactivity in the nucleus accumbens. Front Psychiatry. 2013;4:55. doi: 10.3389/fpsyt.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013 doi: 10.1126/science.1237905. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2–δδct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009a;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009b;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Maccari S, Nicoletti F, Guidotti A. Pharmacological activation of group-ii metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacol. 2012;37:929–938. doi: 10.1038/npp.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. The epigenetics of sex differences in the brain. J Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. Microrna-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CL, Morelli GA. Mother rats interact differently with male amd female offspring. J Comp Physiol Psychol. 1979;93:677–684. doi: 10.1037/h0077599. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacol. 2012;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OFX, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nagarajan R, Hogart A, Gwye Y, Martin MR, LaSalle JM. Reduced mecp2 expression is frequent in autism frontal cortex and correlates with aberrant mecp2 promoter methylation. Epigenetics. 2006;1:172–182. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde Thomas M, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, Weinberger Daniel R, Kleinman Joel E, Lipska Barbara K. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL. Epigenetics of neurobiology and behavior during development and adulthood. Dev Psychobiol. 2012;54:590–597. doi: 10.1002/dev.20550. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the bdnf gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sullivan RM. Memory of early maltreatment: Neonatal behavioral and neural correlates of maternal maltreatment within the context of classical conditioning. Biol Psychiatry. 2005;57:823–831. doi: 10.1016/j.biopsych.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Sanchez MM. The impact of early adverse care on hpa axis development: Nonhuman primate models. Horm Behav. 2006;50:623–631. doi: 10.1016/j.yhbeh.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinol. 2010;151:4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into rett syndrome: Mecp2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: Analysis of the national brain databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type–specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9:e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RK, Howard JL, Simpson DN, Akil H, Clinton SM. DNA methylation in the developing hippocampus and amygdala of anxiety-prone versus risk-taking rats. Dev Neurosci. 2012;34:58–67. doi: 10.1159/000336641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli S, Chefer S, Suomi SJ, Higley J, Barr CS, Stein E. Early-life stress induces long-term morphologic changes in primate brain. Archives of General Psychiatry. 2009;66:658–665. doi: 10.1001/archgenpsychiatry.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns NA, Schaevitz LR, Bowling H, Nag N, Berger UV, Berger-Sweeney J. Behavioral and anatomical abnormalities in mecp2 mutant mice: A model for rett syndrome. Neurosci. 2007;146:907–921. doi: 10.1016/j.neuroscience.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri D, Veenit V, Sarkar A, Thiagarajan D, Kumar A, Nestler EJ, Galande S, Vaidya VA. Early stress evokes age-dependent biphasic changes in hippocampal neurogenesis, bdnf expression, and cognition. Biol Psychiatry. 2013;73:658–666. doi: 10.1016/j.biopsych.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Funato H, Hobara T, Otsuki K, Yamagata H, McEwen BS, Watanabe Y. Early life stress enhances behavioral vulnerability to stress through the activation of rest4-mediated gene transcription in the medial prefrontal cortex of rodents. The Journal of Neuroscience. 2010;30:15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Epigenetic status of gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Xie L, Korkmaz KS, Braun K, Bock J. Early life stress-induced histone acetylations correlate with activation of the synaptic plasticity genes arc and egr1 in the mouse hippocampus. J Neurochem. 2013;125:457–464. doi: 10.1111/jnc.12210. [DOI] [PubMed] [Google Scholar]