Abstract
MicroRNAs are predicted to regulate thousands of mammalian genes, but relatively few targets have been experimentally validated and few microRNA loss-of-function phenotypes have been assigned. As an alternative to chemically modified antisense oligonucleotides, we developed microRNA inhibitors that can be expressed in cells, as RNAs produced from transgenes. Termed ‘microRNA sponges’, these competitive inhibitors are transcripts expressed from strong promoters, containing multiple, tandem binding sites to a microRNA of interest. When vectors encoding these sponges are transiently transfected into cultured cells, sponges derepress microRNA targets at least as strongly as chemically modified antisense oligonucleotides. They specifically inhibit microRNAs with a complementary heptameric seed, such that a single sponge can be used to block an entire microRNA seed family. RNA polymerase II promoter (Pol II)-driven sponges contain a fluorescence reporter gene for identification and sorting of sponge-treated cells. We envision the use of stably expressed sponges in animal models of disease and development.
MicroRNAs are 20–24-nucleotide RNAs derived from hairpin precursors. Through pairing with partially complementary sites in 3′ untranslated regions (UTRs), they mediate post-transcriptional silencing of a predicted 30% of protein-coding genes in mammals1. MicroRNAs have been implicated in critical processes including differentiation, apoptosis, proliferation, and the maintenance of cell and tissue identity; furthermore, their misexpression has been linked to cancer and other diseases2–7. But relatively few micro-RNA-target interactions have been experimentally validated in cell culture or in mouse models, and the functions of most microRNAs remain to be discovered. Creating genetic knockouts to determine the function of microRNA families is difficult, as individual micro-RNAs expressed from multiple genomic loci may repress a common set of targets containing a complementary seed sequence. Thus, a method for inhibiting these functional classes of paralogous micro-RNAs in vivo is needed. Presently, loss-of-function phenotypes are induced by means of chemically modified antisense oligonucleotides—2′ O-methyl, locked nucleic acid (LNA) and others—which are presumed to pair with and block mature microRNAs through extensive sequence complementarity8–10. Typically, oligonucleotide inhibitors are transiently transfected into cells, providing a correspondingly transient derepression of microRNA targets. One type of inhibitor has been demonstrated to silence microRNAs in vivo: ‘antagomirs’, which are 2′ O-methyl, phosphorothioate, cholesterol-modified antisense oligonucleotides; their effect in an animal, however, is only achieved with a high dose11.
Antisense oligonucleotides work as competitive inhibitors of microRNAs, presumably by annealing to the mature microRNA guide strand after the RNA-induced silencing complex has removed the passenger strand12. Delivering a dose sufficient to saturate the cellular pool of microRNAs is critical to their function. We reasoned that a microRNA target expressed at a sufficiently high level could, analogously, function as a competitive inhibitor of cognate microRNA(s). To boost the affinity of a decoy target for its cognate microRNA, multiple binding sites could be inserted into its 3′ UTR. By designing the microRNA binding sites with a bulge at the position normally cleaved by Argonaute 2, these targets would be able to stably interact with, or ‘soak up’, microribonucleoprotein complexes (microRNPs) loaded with the corresponding micro-RNA. Such inhibitor RNAs could be expressed transiently from transfected plasmids or stably from chromosomal insertions. Because the interaction between microRNA and target is nucleated by and largely dependent on base-pairing in the seed region (positions 2–8 of the microRNA), a decoy target should interact with all members of a microRNA seed family. In so doing, it should better inhibit functional classes of microRNAs than do antisense oligonucleotides, which are thought to block single microRNA sequences.
We made decoy targets for several microRNA seed families, named them ‘microRNA sponges’, and tested their ability to derepress microRNA targets in mammalian cells. Here we present evidence that microRNA sponges are at least as effective as present antisense technology, that their activity is specific to microRNA seed families, and that they can be used to validate target predictions and assay microRNA loss-of-function phenotypes.
RESULTS
Construction of microRNA sponges
We constructed Pol II sponges by inserting tandemly arrayed microRNA binding sites into the 3′ UTR of a reporter gene encoding destabilized GFP driven by the CMV promoter (Fig. 1a). Binding sites for a particular microRNA seed family were perfectly complementary in the seed region with a bulge at positions 9–12 to prevent RNA interference–type cleavage and degradation of the sponge RNA (Fig. 1b). We also constructed perfectly complementary sponges for individual microRNAs. As a control, we constructed a sponge with repeated binding sites complementary to an artificial microRNA based on a sequence from the CXCR4 gene (but not complementary to any known microRNA). Binding site information for all sponge constructs is available in Supplementary Table 1 online.
Figure 1.
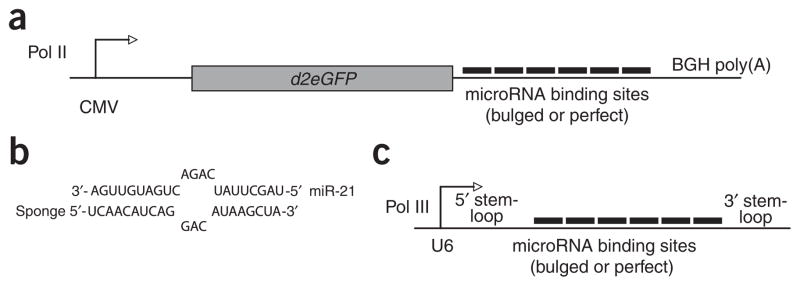
Design of microRNA sponges. (a) We constructed GFP sponges by inserting multiple microRNA binding sites into the 3′ UTR of a 2-h destabilized GFP reporter gene driven by the CMV promoter. (b) The imperfect pairing between a microRNA and a sponge with bulged binding sites is diagrammed for miR-21. We designed sponges with a bulge to protect against endonucleolytic cleavage by Argonaute 2. (c) We constructed U6 sponges by subcloning the microRNA binding site region into a vector containing a U6 snRNA promoter with 5′ and 3′ stem-loop elements.
We constructed a second class of microRNA sponges to take advantage of strong RNA polymerase III promoters (Pol III), which are known to drive expression of the most-abundant cellular RNAs (Fig. 1c). We subcloned tandemly arrayed microRNA binding sites from the GFP sponge constructs into a modified U6 small nuclear RNA promoter-terminator vector, which produces short (<300 nt) RNAs with structurally stabilized 5′ and 3′ ends13. As they lack an open reading frame, these U6 sponges are substrates for microRNA binding, but not for translation or translational repression.
Efficacy of microRNA sponges
We transfected HEK293T cells expressing abundant endogenous miR-20 with the CXCR4 control sponge plasmid (C-CX) or with sponge plasmids imperfectly (C-20b) or perfectly (C-20pf) complementary to miR-20. We cotransfected a sponge plasmid and a TK promoter-driven gene encoding Renilla reniformis luciferase (RLuc) regulated by 7 bulged miR-20 sites and an unregulated gene encoding firefly luciferase as a transfection control, at a ratio of 8:1 sponge plasmid to target plasmid. We assayed the expression of the RLuc target 24 h after transfection and observed that it was rescued by both Pol II– and Pol III–driven sponges with bulged or perfect miR-20 binding sites (Fig. 2a). At 48 h, we observed similar results (data not shown). We measured amounts of reporter mRNA by real-time PCR and found that derepression occurred mostly at the translational level (data not shown). For both sponge classes, sponges with 4–7 bulged binding sites produced stronger derepressive effects than sponges with two perfect binding sites. This difference may be due to the availability of more binding sites in the bulged sponges, and/or to the greater stability expected of bulged sponge RNAs compared to sponge RNAs that can be cleaved by miR-20–loaded Argonaute 2. Between the two sponge classes, the CMV sponges and U6 sponges derepressed the target reporter about equally well—nearly 50% rescue of a target with 7 miR-20 binding sites relative to an unrepressed control reporter—but the U6 sponges also produced a general inhibition of RLuc expression (Supplementary Fig. 1 online). Fluorescence in situ hybridization with a probe against the U6 sponge RNAs primarily labeled the nucleus, as in previous work13 (data not shown). How an inhibitor localized primarily to the nucleus can function against microRNA localized primarily in the cytoplasm is not clear. We speculate that a sufficient fraction of the U6 sponge RNA is present in the cytoplasm to inhibit mature microRNA.
Figure 2.
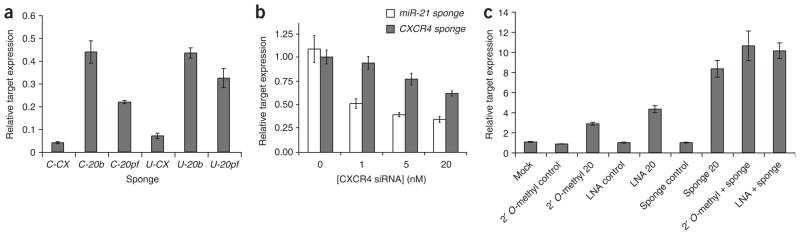
Efficacy of microRNA sponges. (a–c) RLuc activity relative to firefly luciferase activity was assayed in 293T cells 24 h after transfection with RLuc microRNA target reporters, firefly luciferase transfection control and microRNA sponge plasmids. An RLuc target regulated by 7 miR-20 sites was derepressed by GFP sponges and U6 sponges with bulged or perfect binding sites for miR-20 (a). C, CMV sponge; U, U6 sponge. CX, CXCR4 control; 20b, 7 bulged miR-20 sites; 20pf, two perfect miR-20 sites. Bars represent the expression of the miR-20 target relative to an untargeted control reporter. We measured an artificial CXCR4 target reporter with a single bulged binding site in the presence of a control GFP sponge against miR-21 (miR-21 sponge) or a GFP sponge containing seven CXCR4 binding sites (CXCR4 sponge; b). We transfected cells with 20 nM antisense oligonucleotide (2′ O-methyl 20 or LNA 20) or with the CMV bulged sponge against miR-20 (sponge 20; c). Negative controls; mock (no oligonucleotides or sponges), 2′ O-methyl against miR-30, LNA against miR-122, CXCR4 sponge. We performed each experiment at least three times and have shown a representative example. Error bars, s.d; n = 3.
We performed subsequent assays with the GFP bulged sponges, as they gave the highest activity on both microRNA target reporters tested (miR-16 and miR-20). Cells transfected with these sponge plasmids expressed large amounts of GFP, with only slight repression by endogenous microRNAs. Transfected at low doses, the sponge plasmids expressed GFP mRNA at a subsaturating level such that translation was visibly repressed by endogenous micro-RNAs relative to unregulated GFP control constructs (data not shown). Thus, the sponge mRNAs function by associating with active microRNPs.
To quantify the inhibition of cognate microRNAs by sponges, we used a target reporter with a single bulged binding site for an artificial microRNA based on the CXCR4 sequence. (This system, established in our laboratory, has been used to show that transfected small interfering RNA (siRNA) enters the same effector pathway as endogenous microRNA14.) The majority of predicted microRNA targets contain a single binding site in their 3′ UTR, so this target reporter probably mimics the response of a natural microRNA target. We cotransfected the CXCR4 siRNA at varying concentrations and included the CXCR4 sponge containing 7 bulged binding sites to the microRNA, or, as a negative control, a sponge containing 7 bulged binding sites to miR-21, a microRNA not expressed in 293T cells (Fig. 2b). At transfected siRNA concentrations of 1 and 5 nM, the luciferase target was repressed 2–2.5-fold, similar to the observed regulation by endogenous microRNAs of natural UTRs containing one binding site. Furthermore, flow cytometry analysis of GFP revealed that the CXCR4 sponge targeted by 5 nM CXCR4 siRNA was repressed to the same extent that a miR-21 sponge was repressed by endogenous miR-21 in T98G, a cell type that highly expresses that microRNA (data not shown). We infer that this range of transfected siRNA corresponds to the concentration range of natural endogenous microRNAs acting on typical target messages. In this range, the CXCR4 sponge rescued target gene expression 75–95% (1.8–1.9-fold derepression) and rescue was above 60% even at the highest siRNA concentration tested (20 nM). We conclude that the GFP sponge RNAs are being produced and accumulating to sufficiently high level to inhibit most endogenous microRNAs.
To compare the efficacy of inhibiting endogenous microRNAs by microRNA sponges to that of present antisense technology, we transfected 293T cells with target reporters and either a 2′ O-methyl antisense oligonucleotide, LNA antisense oligonucleotide, or a bulged GFP sponge, or with control inhibitors (Fig. 2c). The GFP sponge more strongly derepressed the target reporter than the 2′ O-methyl antisense oligonucleotide transfected at standard conditions (20 nM) for all microRNAs tested (miR-16, 18, 20, 21 and 30). This effect could be increased slightly by cotransfecting sponge and oligonucleotide. A miR-20 sponge outperformed, but a miR-16 sponge only performed about as well as, an LNA antisense oligonucleotide transfected at 20 nM (Fig. 2c and Supplementary Fig. 2 online). Perhaps the cross-reactivity of the sponges to seed family members, such as miR-17-5 in the case of the miR-20 sponge, allows them to rescue the effects of entire microRNA families more completely than specific antisense oligonucleotides.
We tested sponges with artificial target reporters and 3′ UTR reporters in two additional human cell lines and in mouse 3T3 cells and found them to be similarly active in all cell lines (data not shown).
To investigate the possibility of expressing sponges continuously from multicopy chromosomal insertions, we constructed polyclonal cell lines by cotransfecting 293T cells with linearized GFP sponge plasmids and a puromycin selection marker. After sorting the cell lines for a high-GFP fraction, we assayed the activity of endogenous microRNA in comparison to cells transiently transfected with sponge plasmids. The stable miR-16 sponge–expressing cell line allowed threefold higher expression of a miR-16 target (relative to an untargeted control reporter) than the stable CXCR4 sponge cell line or the parental 293T cells (Supplementary Fig. 3 online). This represents an activity approximately 40% as strong as that of the transiently transfected sponge. Thus, sponges expressed from transgenes have the potential to at least partially inhibit endogenous microRNAs.
Seed specificity of microRNA sponges
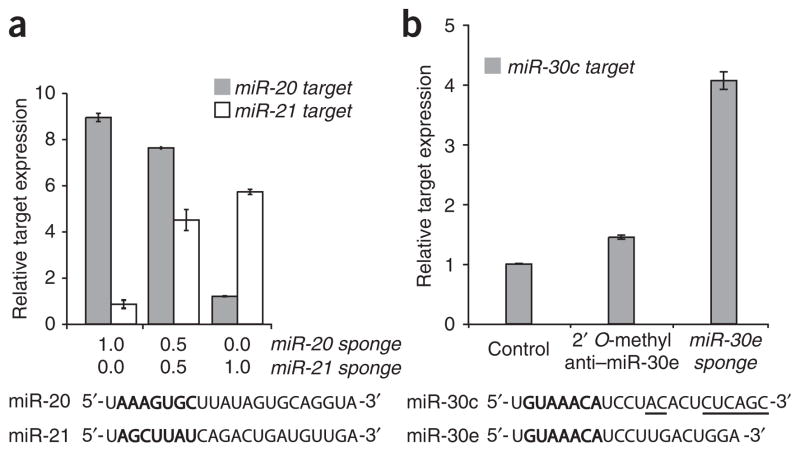
To assess the specificity of the Pol II–driven sponges, we transfected HeLa cells with target reporters and sponges against two micro-RNAs with different seeds: miR-20, miR-21 or a 50:50 combination of the two sponges (Fig. 3a). Dose-dependent derepression was apparent in samples treated with a 50 :50 mixture of the two plasmids. Each target was derepressed by its cognate microRNA sponge and unaffected by the other microRNA sponge relative to treatment with the CXCR4 sponge control. In contrast, we expected sponges based on the sequence of a given microRNA to be recognized as targets by multiple microRNAs that share the seed. In HeLa cells, microRNA expression profiling detects high levels of miR-30c and miR-30d, and a much lower level of miR-30e15. We reasoned that a sponge element based on the sequence of the low-abundance microRNA would recognize each family member through the common seed and thereby derepress a target of the high-abundance microRNA family member. Accordingly, we assayed a target reporter with perfect sites for miR-30c with either a 2′ O-methyl antisense oligonucleotide against miR-30e or a sponge with 6 bulged sites against miR-30e (Fig. 3b). As expected, the antisense oligonucleotide derepressed the miR-30c target to a very low degree, <1.5-fold, presumably by inhibiting only the low-abundance miR-30e. In contrast, the sponge designed to miR-30e derepressed the target by over fourfold, suggesting cross-reactivity with the more abundant miR-30 family members. Consistent with this, transfection of 20 nM 2′ O-methyl oligonucleotide against the more abundant miR-30c derepressed the miR-30c target to a slightly greater extent than the miR-30e sponge. Further supporting the generality of seed recognition by sponges, we observed derepression of perfect target reporters for miR-15a, miR-15b and miR-16, which share a common seed, by treatment with sponges based on the miR-16 sequence (data not shown).
Figure 3.
Specificity of microRNA sponges. (a) We assayed RLuc activity relative to firefly luciferase activity in HeLa cells 24 h after transfection with RLuc microRNA target reporters, firefly luciferase transfection control and microRNA sponge plasmids. Targets of miR-20 and miR-21 are specifically derepressed by the corresponding GFP sponge. Bars are normalized to the relative RLuc units of samples treated with the CXCR4 control sponge. (b) We assayed a perfect target reporter of miR-30c in HeLa cells transfected with oligonucleotide or sponge inhibitors of miR-30e. Controls: 2’ O-methyl anti–miR-181, CXCR4 sponge. MicroRNA sequences below show the heptameric seed sequence in bold, with nucleotide differences between the two family members underlined. We performed each experiment at least three times and have shown a representative example. Error bars, s.d; n = 3.
Validation of predicted microRNA targets
To test the ability of sponges to derepress natural microRNA targets, we assayed the E2F1 protein, a demonstrated target of the miR-20 seed family and a predicted target of miR-18 (ref. 16; Fig. 4a). The amount of the target protein increased by about 1.5-fold after treatment with the miR-18 GFP sponge and by about 2.5-fold after treatment with the miR-20 GFP sponge, as shown in relation to lanes loaded with 1 or 1.5 times the amount of lysate from the control CXCR4 sponge treatment. This difference likely results from the presence of two miR-20 binding sites and one miR-18 site in the E2F1 3′ UTR, plus the added inhibition of the coexpressed miR-20 family member miR-17-5. These effects were recapitulated in a luciferase assay wherein the RLuc reporter was fused to a fragment of the E2F1 UTR spanning the two miR-20 sites (Fig. 4b). Thus, sponges show direct effects on natural and endogenous targets, and can be used to validate target predictions. To test some predicted targets that had not yet been experimentally validated, we used a luciferase reporter regulated by a large fragment of the CD69 3′ UTR17 or by the E2F5 UTR. As predicted by the TargetScan 4.0 and miRanda algorithms, respectively, these UTRs are each regulated by a single miR-20 site18,19. Correspondingly, each reporter was derepressed upon treatment with a miR-20 sponge in 293T cells (Fig. 4c and Supplementary Fig. 4 online).
Figure 4.
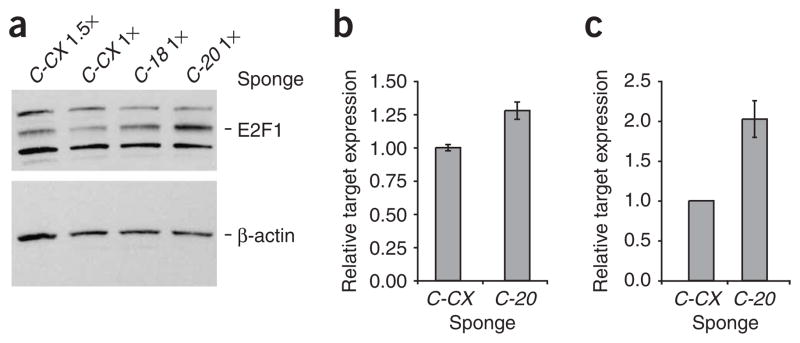
Validation of microRNA targets. (a) We assayed 293T cells transfected with GFP sponges against miR-18, miR-20 or the CXCR4 control by western blot 48 h after transfection. The increase in endogenous E2F1 upon inhibition of miR-18 or miR-20 is shown relative to the control samples loaded at indicated amounts; E2F1 is the ~60 kDa band indicated; the other bands are nonspecific (top). β-actin loading control (bottom). (b) We assayed 293T cells transfected with an RLuc reporter fused to a fragment of the E2F1 UTR spanning two miR-20 sites, firefly luciferase and GFP sponges. Bars represent RLuc units relative to firefly luciferase units. (c) We assayed RLuc activity relative to firefly luciferase activity in 293T cells transfected with an RLuc reporter fused to a fragment of the CD69 UTR containing a predicted miR-20 binding site, firefly luciferase and GFP sponges. We performed each experiment at least three times and have shown a representative example. Error bars, s.d; n = 3.
Effect of sponges on microRNA levels
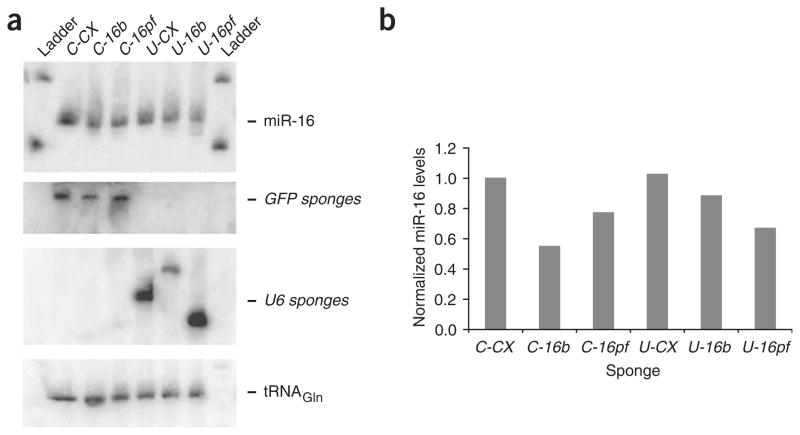
Antisense oligonucleotides have been shown to reduce the cellular concentration of their cognate microRNAs11,12. These results from northern blots are complicated by the possibility that the complementary RNA could compete with a labeled probe for base-pairing to the microRNA or prevent transfer of the short RNA to the hybridization matrix. We expected that the overexpression of a microRNA target, namely, expression of a microRNA sponge construct, would not alter the amount of endogenous microRNA. But northern blot analysis showed a modest (typically about twofold, ranging from 1.2–3-fold) specific decrease in free micro-RNA 24–48 h after transfection of the corresponding sponge (Fig. 5). We observed this effect for bulged and perfect sponges of both the Pol II and Pol III classes. The northern blots also showed microRNA signal near the location of the bands detected by probing against the GFP and U6 sponge RNAs, respectively. Thus, cellular microRNA concentration may be unchanged by sponge expression and the loss of a northern blot signal explained by microRNA retention at the top of the gel owing to interaction with the cognate sponge RNA. It is important to note that the signal of the GFP sponge RNAs is comparable to the signal of endogenous miR-16 detected with the same-length DNA probe and after the same exposure time, supporting the expected inhibition of micro-RNAs by excess binding sites in the form of Pol II–driven sponges. To evaluate the abundance of GFP sponge RNAs in transfected cells, we quantified GFP transcripts by real-time PCR in relation to GFP plasmid standards (data not shown). We estimated the copy number of bulged GFP mRNAs in transiently transfected 293T cells to be at least 1,000–2,000 per cell. If all seven binding sites in the sponge RNA’s UTR were used to bind microRNA, then this level of sponge expression should allow inhibition of approximately 104 microRNAs per cell, which would be sufficient to inhibit most microRNAs in most cell types.
Figure 5.
Effect of sponges on microRNA levels. (a) We transfected 293T cells with sponge plasmids and collected total RNA for northern blot analysis 48 h later. We probed the blot for miR-16 (top; 24-h exposure), then stripped the blot and reprobed it for GFP mRNA, U6 sponge RNA (both 24-h exposure) and a tRNA loading control (3-h exposure; bottom). (b) Quantitation of miR-16 relative to tRNA for each sponge-treated sample. We performed northern blots for miR-16 and miR-20 >10 times in 293T and HeLa cells, and show results from a representative blot.
DISCUSSION
Sponges designed as decoy targets for microRNAs were effective and specific inhibitors of microRNA seed families. Somewhat surprisingly, the sponges with perfectly complementary binding sites were not degraded so rapidly as to be ineffective at competing microRNA from targets. Although these sponge RNAs should be degraded by Argonaute 2–catalyzed cleavage, they probably also stably associate with microRNAs complexed to the cleavage-incompetent Argonautes 1, 3 and 4. They could also form stable interactions with other microRNAs that share the same seed but vary at nucleotides 10–11, producing a bulge that protects against endonucleolytic cleavage.
Inclusion of the GFP reporter in the sponge mRNA is useful for assessing transfection efficiency and for tracking those cells that express high levels of the inhibitor RNA. We envision multiple applications of the GFP sponges for target validation and phenotypic analysis. Cells with poor transfection rates can be subjected to fluorescence-activated cell sorting to isolate subpopulations expressing the sponge RNA and thus suppressing microRNA activity. This could be critical for detecting typically subtle (less than twofold) changes in the levels of proteins targeted by endogenous microRNA. Alternatively, transfected cells can be immunostained for predicted targets or phenotypic markers and two-color flow cytometry can be used to assess the correlation between GFP expression and target-protein level. In these applications GFP expression serves both as an indicator of sponge plasmid dose and as a sensor of cellular microRNA activity. By contrast, chemically modified antisense oligonucleotides, which lack a reporter function, limit the experimenter to pooled cell analyses and dilute the inhibitor’s effect often to unobservable levels in cell lines with low transfection rates. The properties of antisense oligonucleotides and sponges are summarized in Supplementary Table 2 online.
There might be several ways to improve the sponge technology described in this study. Addition of more microRNA binding sites to the sponge UTRs would increase the dose of antisense sequences and should therefore increase the potency of the sponges. Testing a microRNA sponge with 6, 10 or 18 sites showed a marginal increase in activity above 6 sites, with apparently saturating effect, but for sponges expressed at lower levels from chromosomal insertions, the additional sites may be beneficial (data not shown). Alternatively, the spacing between sites might be optimized to enhance the binding of miRNPs to every possible site, although previous results suggest that nearby sites are fully functional20. One could also construct sponges with combinations of seed binding sites for two or more microRNA families of interest. To express sponges at a high level transiently in vivo, one could use viral vectors as in a recent work using adenovirus delivered to cardiac tissue7. Finally, there may be Pol III elements other than U6 that would produce sponge RNAs at a high level that are transported to the cytoplasm where they would encounter mature microRNA. Just as sponges inhibit endogenous microRNAs, they could also be used to inhibit siRNAs. In a short hairpin RNA–expressing cell line, a siRNA sponge could provide another level of regulatory control.
An extension of the current technology would be to express sponges from stably integrated transgenes in vivo. Just as short hairpin RNAs, mRNA inhibitors expressed from transgenes, have expanded the experimental scope of siRNAs, transgenic sponges could expand the scope of antisense microRNA inhibitors. Beyond assaying long-term effects of microRNA loss of function in cell lines, we envision the use of drug-inducible sponges in xenograft models to investigate microRNA contributions to tumorigenesis; bone marrow reconstitution approaches to investigate microRNA roles in immune cell development; and, ultimately, germline transgenic sponge mice to ascertain the functions of microRNA families at cell, tissue, organ and organism levels. In principle, microRNA sponges expressed from appropriate promoters should be applicable in any transgenic model organism, including worm, fly and plants.
METHODS
Construction of sponge plasmids and reporters
We annealed, ligated, gel purified and cloned oligonucleotides for microRNA binding sites with 4-nt spacers for bulged sites, or with no spacers for perfect sites, into pcDNA5-CMV-d2eGFP vector (Invitrogen) digested with XhoI and ApaI. We constructed Pol III sponges by subcloning the UTR into pTZ-U6+27 vector (see Acknowledgments). We constructed luciferase reporters by the same oligonucleotide annealing method or by subcloning the UTR into pcDNA5-TK-RLuc vector. We PCR-amplified and ligated the E2F1 UTR fragment (nucleotides 393–978), the CD69 UTR fragment (nucleotides 25–899) and the E2F5 UTR (1–653) into the same vector.
Luciferase assays
We plated 293T cells or HeLa cells the day before transfection and transfected them in triplicate with Lipofectamine 2000 (Invitrogen) and 50 ng of pGL3 (Firefly luciferase plasmid), 90 ng of RLuc target reporter plasmid, and 700 ng of sponge plasmid. We transfected the E2F1 UTR reporter at 4.5 ng, the E2F5 UTR reporter at 0.9 ng. We cotransfected 2′ O-methyl antisense (Dharmacon) and LNA antisense (Exiqon, Dharmacon) oligonucleotides at 20 nM. We transfected the CXCR4 microRNA in the form of a siRNA mixed in varying ratios with negative control siRNA (Dharmacon) to maintain 20 nM total siRNA concentration. We performed all assays at 24 h after transfection with the dual luciferase assay (Promega) on an Optocomp I luminometer (MGM Instruments).
Additional methods
Primers used, western blot and northern blot analyses, construction of stable cell lines, and quantification of sponge RNAs are described in Supplementary Methods online.
Supplementary Material
Acknowledgments
This work was funded by US Public Health Service grants U19-AI056900 from the National Cancer Institute, by an Integrative Cancer Biology Program Grant U54 CA112967 from the National Institutes of Health to P.A.S. and partially by Cancer Center Support (core) P30-CA14051 from the National Cancer Institute. M.S.E. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship and a Paul and Cleo Schimmel Scholarship. J.R.N. is supported by the Cancer Research Institute. We thank A. Garfinkel and M. Kumar for luciferase reporter preparations, A. Leung for assistance with fluorescence in situ hybridization, D. Engelke (University of Michigan) for the U6 vector and members of the Sharp laboratory for helpful discussions.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
M.S.E. and J.R.N. conceived the experimental design and made the sponge constructs. M.S.E. performed the experiments and wrote the manuscript. P.A.S. supervised the work.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Cheng HY, et al. MicroRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:e98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 12.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul CP, et al. Localized expression of small RNA inhibitors in human cells. Mol Ther. 2003;7:237–247. doi: 10.1016/s1525-0016(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 14.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 17.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 19.John B, et al. Human microRNA targets. PLoS Biol. 2004;2:363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.