Synopsis
High-density lipoproteins (HDL) remove cell cholesterol and protect from atherosclerosis. The major HDL protein is apolipoprotein A-I (apoA-I). Most plasma apoA-I circulates in lipoproteins, yet ~5% forms monomeric lipid-poor/free species. This metabolically active species is a primary cholesterol acceptor and is central to HDL biogenesis. Structural properties of lipid-poor apoA-I are unclear due to difficulties in isolating this transient species. We used thermal denaturation of human HDL to produce lipid-poor apoA-I. Analysis of the isolated lipid-poor fraction showed protein:lipid weight ratio 3:1, with apoA-I, phosphatidylcholine and cholesterol ester at approximate molar ratios of 1:8:1. Compared to lipid-free apoA-I, lipid-poor apoA-I showed slightly altered secondary structure and aromatic packing, reduced thermodynamic stability, lower self-associating propensity, increased adsorption to phospholipid surface, and comparable ability to remodel phospholipids and form reconstituted HDL. Lipid-poor apoA-I can be formed by heating of either plasma or reconstituted HDL. We propose the first structural model of lipid-poor apoA-I which corroborates its distinct biophysical properties and postulates the lipid-induced ordering of the labile C-terminal region. In summary, HDL heating produces folded functional monomolecular lipid-poor apoA-I that is distinct from lipid-free apoA-I. Increased adsorption to phospholipid surface and reduced C-terminal disorder may help direct lipid-poor apoA-I towards HDL biogenesis.
Keywords: HDL remodeling, protein adsorption to phospholipid surface, apolipoprotein structure and stability, protein self-association, reverse cholesterol transport
High-density lipoproteins (HDL) are nanoparticles containing two or more protein molecules and from about 160 to 300 lipids per particle (ref. [1] and references therein). HDL surface is comprised of amphipathic proteins and polar lipids, mainly phosphatidylcholine (PC) and unesterified cholesterol (Ch). These lipids are thought to form a bilayer in nascent discoidal HDL, with the protein molecules wrapped around the disk perimeter [1, 2]. Mature spherical HDL contain a core of apolar lipids, mainly cholesterol esters (CE), that are sequestered between the protein-containing PC monolayers [2–4]. HDL are the primary vehicles of cholesterol removal from peripheral cells via the reverse cholesterol transport pathway, and possess other cardioprotective properties [5–8]. It is well-established that the risk of developing atherosclerosis correlates inversely with the plasma levels of HDL and the main HDL protein, apolipoprotein A-I (apoA-I, 243 a. a.) [2, 5–8]. The current consensus is that both quantity and quality of HDL are important for cardioprotection [9–12]. This notion stems from the observation that plasma HDL form an heterogeneous population of particles differing in shape (nascent discoidal or mature spherical), size (d=7.7–12 nm), charge, protein and lipid composition, and function [2]. Understanding the distinct properties of these subspecies is part of the on-going efforts to design new HDL-based biomarkers and therapies for atherosclerosis [13, 14]. One potentially important but elusive target of these studies is lipid-poor apoA-I.
Although over 90% of plasma apoA-I is found in lipoproteins, nearly 5% is present as transient lipid-poor or lipid-free species that is generated either de novo by the liver or intestine or via the metabolic remodeling of mature HDL by lipases and lipid transfer proteins [15–18]. Lipid-poor/free apoA-I is particularly metabolically active and is rapidly incorporated into lipoproteins, either by binding to the existing particles or by forming new ones [18, 19]. Importantly, lipid-poor/free apoA-I has been proposed to act as the primary acceptor of cell cholesterol and phospholipids via the interaction with the plasma membrane mediated by the ATP-binding cassette transporter ABCA1 [20–22]. This interaction promotes the efflux of cholesterol from peripheral cells to lipid-poor/free apoA-I at an essential early step of HDL biogenesis. This step is central to the reverse cholesterol transport from cholesterol-overloaded macrophages in the arterial wall, which significantly contributes to the cardioprotective action of apoA-I and HDL. Therefore, the presence of adequate amounts of biologically active lipid-poor/free apoA-I in the arterial wall is essential for the cardioprotection [22]. However, excess of lipid-poor/free apoA-I is not necessarily beneficial, since this labile species is susceptible to proteolysis and is rapidly catabolized and/or cleared by the kidney [18, 23]; in contrast, clearance of the larger lipoprotein particles is hampered by the glomerular filtration barrier [23]. Hence, optimal removal of cell cholesterol probably requires a balance between the production of lipid-poor/free apoA-I, its recruitment in HDL biogenesis, and its catabolism [18]. Our goal is to provide the molecular basis underlying this delicate balance. To this end, we have determined the biophysical and structural properties of a lipid-poor form of apoA-I that dissociates from HDL upon heating. We propose that this form provides a useful model for studying the lipid-poor apoA-I that is central to HDL biogenesis in vivo.
Despite its functional importance as a primary acceptor of cell cholesterol, the overall conformation and stability of lipid-poor apoA-I are not well defined, in part, because of the difficulty in obtaining this transient species and its apparent heterogeneity. The methods used to isolate and characterize lipid-poor apoA-I from plasma include preparative gel electrophoresis [24–26], size exclusion chromatography [27] and isotachophoresis [28]. Analytical techniques using 2D electrophoresis [24–26, 29, 30] and isotachophoresis [31] have been developed to quantify lipid-poor apoA-I in plasma. These methods produce minimal amounts of lipid-poor apoA-I, and the apparent size and electrophoretic mobility of this labile species can vary depending on the sample concentration [25] and the method of preparation. This leads to variability in the reported lipid content that ranges from 1 to 30 lipid molecules (mostly phospholipid and some cholesterol) per apoA-I molecule. Nonetheless, the consensus is that lipid-poor apoA-I is monomeric (ref. [32, 33] and references therein), as opposed to HDL that contain 2–5 molecules of apoA-I per particle [1–4]. Lipid-poor apoA-I can also be obtained via the interaction of lipid-free apoA-I with cells overexpressing ABCA1 [32, 33]. The resulting lipid-poor species contains apoA-I monomer associated with several PC molecules, with the overall conformation and lipid binding properties similar to those of free apoA-I [33].
Here, we used for the first time heat denaturation of HDL to produce milligrams of human lipid-poor apoA-I and carry out its physicochemical analysis. Earlier we postulated that thermal denaturation mimics important aspects of metabolic remodeling of HDL during reverse cholesterol transport, including lipoprotein fusion, rupture and apolipoprotein dissociation [34–36]. In the current work, we show that the apolipoprotein dissociated upon HDL fusion and rupture comprises folded functional lipid-poor apoA-I whose structural, stability, self-association and lipid binding properties are significantly different from those of the lipid-free protein monomer. We propose that these distinct properties help protect lipid-poor apoA-I from catabolism and direct it towards HDL biogenesis.
MATERIALS AND METHODS
Isolation of human lipoproteins
Lipoproteins from single-donor plasma of three healthy volunteers were used. Fresh ethylenediaminetetraacetic acid-treated plasma was obtained from the blood bank according to the rules of the institutional review board and with written consent from the donors. Lipoproteins were isolated by density gradient ultracentrifugation in the density range 1.063–1.21 g/mL for HDL [37]. HDL migrated as a continuous band on the agarose and non-denaturing gels. Lipoprotein stock solutions were dialyzed against 10 mM Na phosphate, pH 7.5 (which is the standard buffer used in this work), degassed, stored at 4 °C, and used during 2–3 weeks. To ensure reproducibility, all experiments in this study were repeated 3–5 times by using plasma from three different donors. The key results of this work were fully reproducible for all batches explored.
Reconstitution of model HDL
Single-donor apoA-I, which was obtained by HDL delipidation using 6M Gdn HCl following established protocols [38], was purified to 95% purity and refolded as described (ref. [39] and references therein). The protein obtained by this method contains less than one PC molecule per apoA-I, as indicated by mass spectrometry [32], and is essentially lipid-free. Lipids, including dimyristoyl PC (DMPC), palmitoyl PC (POPC), and cholesterol (Ch), were 95+ % pure from Avanti Polar Lipids. Discoidal rHDL containing human apoA-I, POPC and Ch were prepared by cholate dialysis [40] using apoA-I:POPC:Ch molar ratio of 1:80:4, and were isolated by density gradient centrifugation to remove uncomplexed protein. Spherical reconstituted HDL (rHDL) were obtained from these discoidal apoA-I: POPC:Ch complexes via the lecithin-cholesterol acyltransferase reaction [40] as described in the supplement. Biochemical composition of the rHDL was determined by using a modified Lowry assay for protein, Bartlett assay for PC, and colorimetric analysis for Ch. [ref. [41] and references therein].
Characterization of HDL and the products of their thermal remodeling
Molecular size of intact lipoproteins and the products of their heat denaturation were assessed by non-denaturing gel electrophoresis (NDGE) using 4–20% gradient gels. Samples containing 10 µg protein were run for 2 h at 120 V. The gels were stained with Denville protein stain. Negative stain EM was performed to visualize intact and heated lipoproteins and SUV by using a CM12 transmission electron microscope (Philips Electron Optics) as described [34]. To establish the absence of lipid peroxidation upon heating, intact and heated HDL were analyzed by UV absorbance at 234 nm for conjugated diene formation, and by TLC as described [41] (Fig. S1).
Size-exclusion chromatography (SEC) was used to separate and characterize the protein-containing fractions. The samples were run on a Superose 6 10/300 column (GE Biosciences) at a flow rate of 0.5 mL/min in standard buffer containing 150 mM NaCl. The protein size was assessed from the calibration plot (R2=0.95) by using molecular-size markers (Biorad) [42].
The lipid-poor protein peak fraction obtained from the heat-denatured lipoproteins was purified by SEC. The net charge of the purified species, which was assessed by agarose gel electrophoresis as described [41], showed pre-β electrophoretic mobility (supplemental Fig. S2). The absence of lipoprotein particles was confirmed by NDGE and EM. Lipid composition in this fraction and in intact HDL was determined by TLC as described [41]. Protein composition was determined by SDS PAGE using 10–20 % gradient. In addition, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry was used as described in the supplement to compare protein composition in HDL and in the lipid-poor protein (Fig. S3).
Circular dichroism (CD) and fluorescence spectroscopy
CD data were recorded by using an AVIV 400 spectropolarimeter with thermoelectric temperature controller, to monitor apolipoprotein secondary and tertiary structure and thermal unfolding. Far-UV CD spectra (190–250 nm) were recorded from samples of 0.02 mg/mL protein concentration. Heat-induced changes were monitored at 222 nm for α-helical unfolding during sample heating and cooling at a rate of 10 °C/h. Far-UV CD data were normalized to protein concentration and expressed as molar residue ellipticity, [Θ]. Protein α-helical content was assessed from the value of [Θ] at 222 nm, [Θ]222 [41]. Near-UV CD spectra were recorded from 250–350 nm using solutions of 0.5 mg/mL protein concentration placed in a 5 mm cell, and were reported as molar ellipticity.
Trp emission spectra were recorded at 22 °C using a Fluoromax-2 spectrofluorometer. The samples contained 0.1 mg/mL protein in standard buffer. The excitation wavelength was 295 nm, and the emission was recorded from 310–450 nm with 3 nm excitation and emission slit widths. The wavelength of maximum fluorescence, λmax, was determined from the peak position in the uncorrected spectra.
Differential scanning and pressure perturbation calorimetry (DSC and PPC)
DSC and PPC data were recorded by using a VP-DSC microcalorimeter (MicroCal) as described [42, 43]. In DSC, the samples (1 mg/mL protein in standard buffer) were heated from 5–115 °C at a rate of 90 °C/h. Differential heat capacity, Cp(T), was recorded in mid-gain mode; the buffer–buffer baselines were subtracted. In PPC, the heat effects associated with pressure jumps by ±5 bar were used to determine volume expansion coefficient of the solute on an absolute scale, αv(T)=1/V·(∂V/∂T)P. The PPC data were recorded in mid-gain, low-noise mode as described [42, 43]. Partial specific volumes used for the data processing were 0.735 cm3 g−1 for free protein and 0.805 cm3 g−1 for lipid-poor apoA-I; the latter is based on the value of 1.02 cm3 g−1 for 100% lipid and the protein:lipid weight ratio of 3:1.
Lipid binding and clearance
The ability of apoA-I to bind to the surface of POPC small unilamellar vesicles (SUV) was assessed by SEC. Homogenous SUV (d~22 nm) of POPC (16:0, 16:1) were prepared as described [42, 44]. Apolipoprotein binding to the POPC surface was rapid, as evidenced by the SEC data recorded immediately after mixing apoA-I with POPC SUV. Such rapid adsorption is distinct from the apolipoprotein insertion into the bilayers of shorter-chain phospholipids such as DMPC (14:0, 14:0) and their remodeling to form reconstituted HDL. The latter was assessed by clearance of DMPC multilamellar vesicles (MLV) at 24 °C at which the protein-DMPC reconstitution is fastest [45]. The time course of lipid clearance was monitored by absorbance at 325 nm using Varian Cary Biomelt spectrometer as described [46]. The protein:lipid molar ratio was 1:80, which is typical of discoidal rHDL [40]. To eliminate the effects of protein self-association on the clearance kinetics, we used 20 µg/mL protein concentration at which apoA-I is fully monomeric.
RESULTS
Heating of human HDL leads to apolipoprotein dissociation
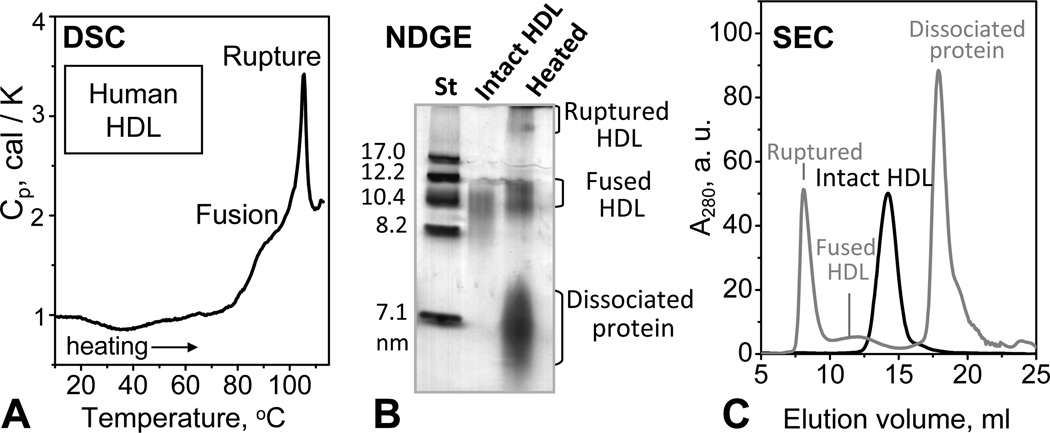
Human plasma HDL were heated from 10 °C to 115 °C in differential scanning calorimetry (DSC) experiments, and the products of thermal denaturation were analyzed by NDGE and SEC (Fig. 1). HDL heating produced no changes in UV absorbance at 234 nm or in the lipid composition assessed by TLC (Fig. S1), indicating the absence of heat-induced lipid peroxidation. This and earlier studies of HDL showed two DSC peaks corresponding to two irreversible transitions (Fig. 1A). Circular dichroism (CD) spectroscopy, negative stain electron microscopy (EM) and NDGE showed that the first broad transition reflects partial apolipoprotein unfolding, dissociation, and HDL fusion; the following sharp transition involves additional apolipoprotein unfolding, dissociation and lipoprotein rupture and release of apolar core lipids that coalesce into droplets [35]. Such protein-containing lipid droplets, along with fused HDL and dissociated protein, were clearly observed by NDGE and/or SEC of HDL samples that had been heated to 115 °C (Fig. 1B, C). This work focuses on the dissociated protein fraction that eluted in SEC near 18 mL (Fig. 1C). EM does not detect any lipoprotein-like particles in this fraction (data not shown).
Figure 1.
Thermal denaturation products of plasma spherical HDL. Human plasma HDL solutions of 4 mg/mL protein concentration in 10 mM Na phosphate, pH 7.5 (which is the standard buffer used throughout this work) were heated from 10 to 115 °C at a rate of 90 °C/h. Heat capacity, Cp(T), was recorded by differential scanning calorimetry (DSC) (A). The DSC peaks centered near 85 and 105 °C reflect HDL fusion and rupture, respectively. (B) Non-denaturing gel electrophoresis (NDGE) of HDL that were intact or heated to 115 °C and cooled to 22 °C in a DSC experiment. (C) Size exclusion chromatography (SEC) of HDL that were intact (black) or heated to 115 °C and cooled to 22 °C (grey). Protein-containing fractions in panels B and C are indicated.
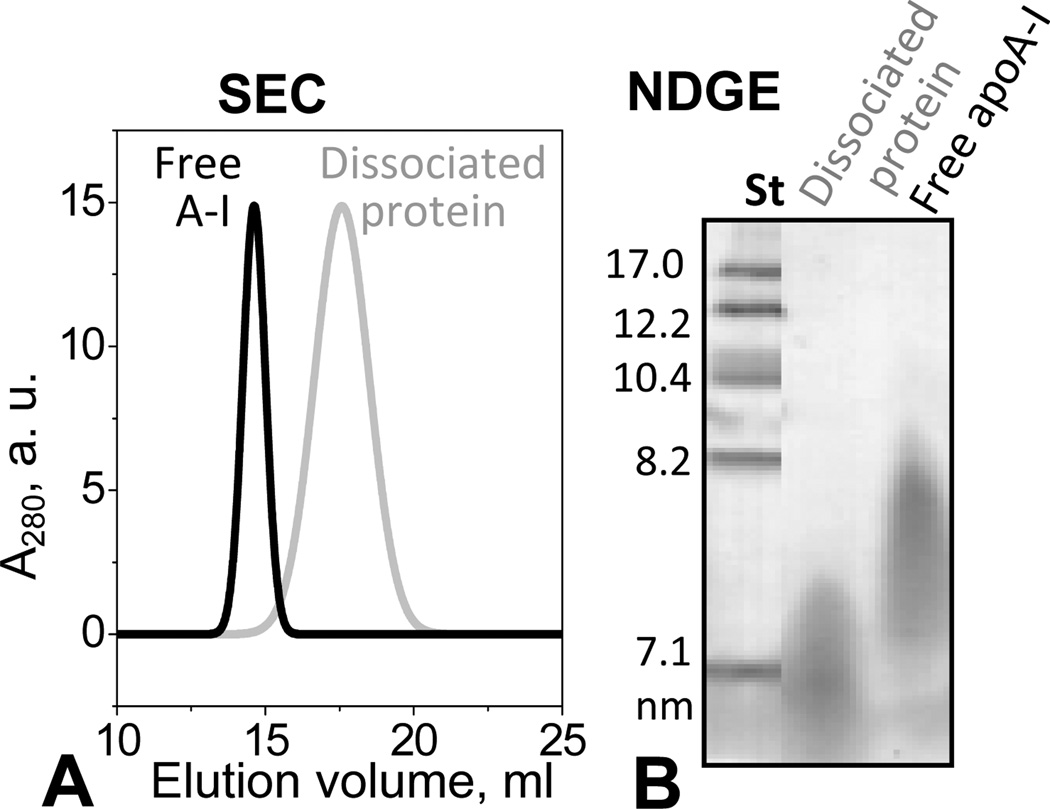
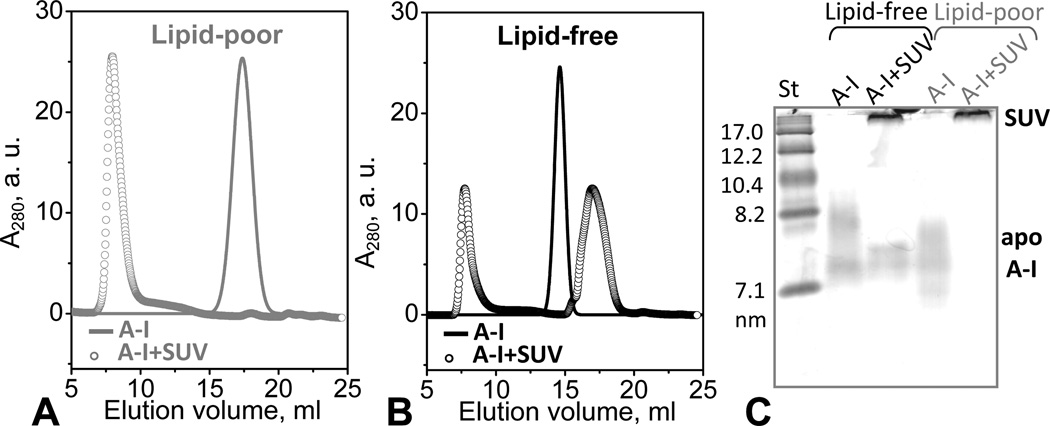
The dissociated protein was purified by SEC and characterized by NDGE and SEC. The protein fraction that peaked at 18 mL was collected and subjected to a second SEC run, collected, and concentrated to 1 mg/mL. At this concentration, lipid-free apoA-I isolated from human plasma is largely self-associated [47] and shows batch-to-batch variations in the degree of self-association, from dimers to higher-order oligomers. For example, in the SEC profile in Fig. 2A, lipid-free apoA-I elutes as an apparent hexamer, when injected at a concentration of 1 mg/mL. In contrast, SEC of the protein dissociated from heated HDL invariably showed a single peak whose position was consistent with the elution volume of monomeric apoA-I (~18 mL) (Fig. S4B). Because of the relatively broad nature of the peak, the presence of dimers could not be excluded (Fig. 2A). By NDGE this dissociated protein was similar to lipid-free apoA-I but was less self-associated (Fig. 2B), as confirmed by chemical crosslinking (data not shown). Its pre-β mobility on the agarose gel (Fig. S2) was also similar to that of lipid-free apoA-I. In summary, our gel electrophoresis and SEC data show that HDL heating leads to dissociation of a protein-rich fraction that has similar electrophoretic mobility to lipid-free apoA-I but is less prone to self-association.
Figure 2.
Isolation of the protein-rich fraction from the thermally denatured plasma HDL. Human HDL were heated to 115 °C as described in Figure 1. The protein-rich peak fraction marked “dissociated protein” in the SEC chromatogram in Fig. 1C was isolated and subjected to a second run of SEC (A) and NDGE (B). The protein concentration was 1 mg/mL in standard buffer and in 10 mM PBS, pH 7.5 for NDGE and SEC, respectively. At these concentrations, lipid-free apoA-I forms high-order oligomers (apparent hexamers in panel A, black line). The lipid-poor protein obtained upon HDL heating is less self-associated (predominantly monomers) as it appears by SEC analysis (panel A, grey line).
Biochemical analysis of the protein-rich fraction dissociated from heated human HDL
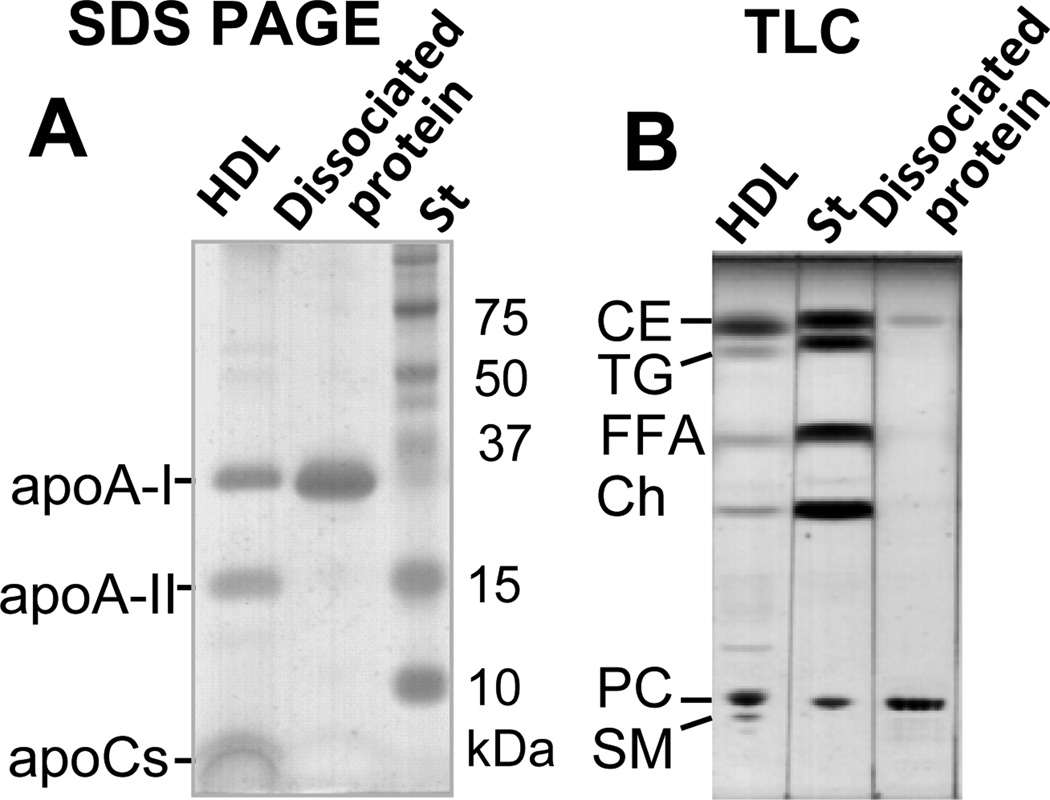
The SEC-purified protein-rich fraction was analyzed by SDS PAGE and mass spectrometry for protein composition. The results clearly showed the major protein, apoA-I (28 kDa), but no minor HDL proteins such as apoA-II (17 kDa) or apoCs (6–9 kDa) (Fig. 3A and supplemental Fig. S3). For lipid analysis of the protein-rich fraction, the lipids were extracted from samples containing 0.50–0.85 mg protein. Based on the initial protein content and the dry weight of the extracted lipids, the protein-rich fraction contained 73–74% protein and 26–27% lipid by weight. Thin-layer chromatography (TLC) analysis of the lipids extracted from this fraction showed the presence of PC and CE but no significant amounts of any other lipids (Fig. 3B). Comparison of the TLC band intensities suggested that 8–10% of the total lipid weight was CE and 90–92% was PC. In summary, the protein fraction dissociated from heated human HDL contained apoA-I, PC, and a small amount of CE. Considering the average molecular weights of PC (780 Da) and CE (670 Da), our results suggest that each apoA-I molecule in the dissociated protein binds, on average, 8–9 molecules of PC and 1 molecule of CE. Notably, these average values do not necessarily represent the true stoichiometric ratios because of the possible heterogeneity of the lipid-poor species which may contribute to the broad peak width in SEC (Fig. 2A, grey line).
Figure 3.
Biochemical composition of the protein-rich fraction isolated from heated HDL. The protein-rich fraction was isolated from thermally denatured human HDL as described in Fig. 2. Protein and lipid compositions of this fraction were assessed by 10–20 % gradient SDS PAGE (A) and thin-layer chromatography (TLC) (B), respectively. Intact HDL are shown for comparison. The main lipid and protein constituents of HDL are indicated.
Conformation of the lipid-poor apoA-I dissociated from human HDL
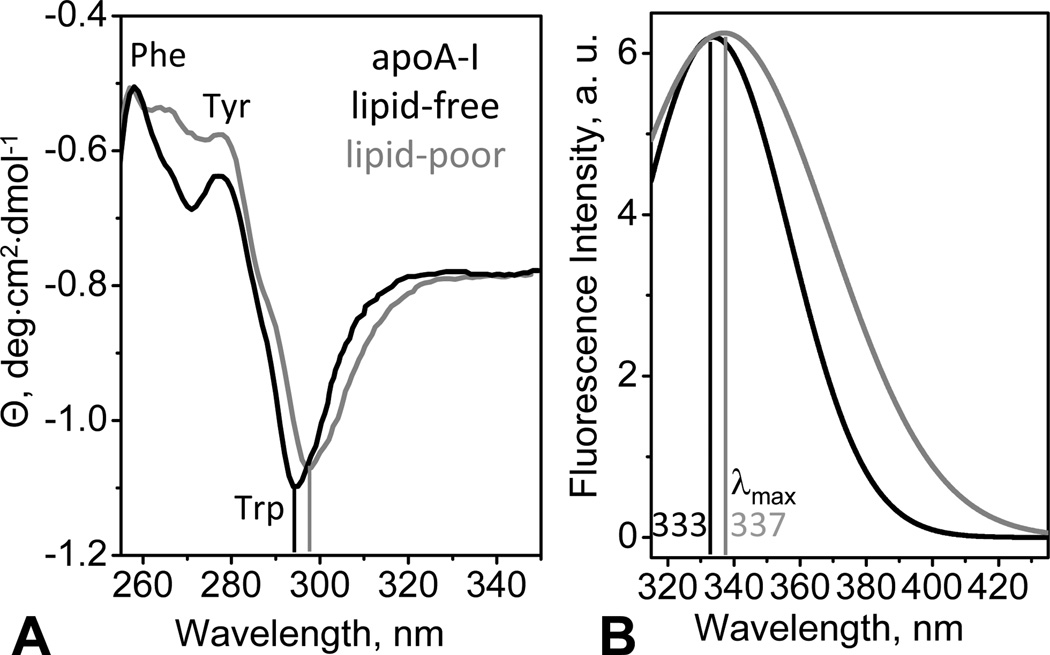
Protein conformation of the lipid-poor apoA-I was assessed by CD and fluorescence spectroscopy. Far-UV CD spectra of lipid-poor apoA-I were similar to those of the free protein but showed a slight reduction in intensity corresponding to about 7% reduction in the α α-helical content (Fig. S4A). Since under conditions of our far-UV CD experiments (0.02 mg/mL protein) both lipid-free and lipid-poor apoA-I are largely monomeric, this spectral difference reflects small lipids-induced secondary structural changes in the monomolecular apoA-I. This contrasts with the increase in the α-helical content upon conversion from free protein to lipoprotein particles containing multiple protein copies.
Near-UV CD spectra of lipid-poor and free apoA-I show a large negative peak near 295 nm corresponding to Trp (Fig. 4A). In contrast, plasma and model HDL show a positive CD peak at these wavelengths [42]. Therefore, the overall aromatic packing in lipid-poor apoA-I resembles that of lipid-free protein but is very different from that in HDL. Compared to lipid-free apoA-I, lipid-poor apoA-I shows a small red shift in the Trp peak, along with the spectral changes at 260–280 nm reflecting mainly Tyr contribution (Fig. 4A), which indicates distinctly different environment of the aromatic groups. Furthermore, tertiary structure of lipid-poor apoA-I was probed by Trp emission recorded under conditions where both lipid-free and lipid-poor apoA-I is largely monomeric. Compared to lipid-free protein, lipid-poor protein showed a red shift in the wavelength of maximal fluorescence (Fig. 4B) which is consistent with the red shift in the near-UV CD peak (Fig. 4A) and suggests more polar Trp environment. Taken together, our spectroscopic results show that, despite overall structural similarity, there are small but significant differences between the monomolecular lipid-poor and lipid-free apoA-I: compared to free protein, lipid-poor apoA-I has slightly lower α-helical content and slightly altered aromatic packing, with increased polarity of the Trp environment.
Figure 4.
Tertiary structure of lipid-poor (grey) and free apoA-I (black) in standard buffer monitored by near-UV circular dichroism (CD) and fluorescence spectroscopy at 25 °C. (A) Near-UV CD spectra recorded from solutions of 0.5 mg/mL protein concentration. Aromatic groups dominating specific spectral regions are indicated. Vertical lines mark the main Trp peak. (B) Trp emission spectra recorded from samples of 0.1 mg/mL protein concentration (excitation at 295 nm). The wavelength of maximal fluorescence, λmax, is indicated.
Thermal denaturation of lipid-poor apoA-I dissociated from human HDL
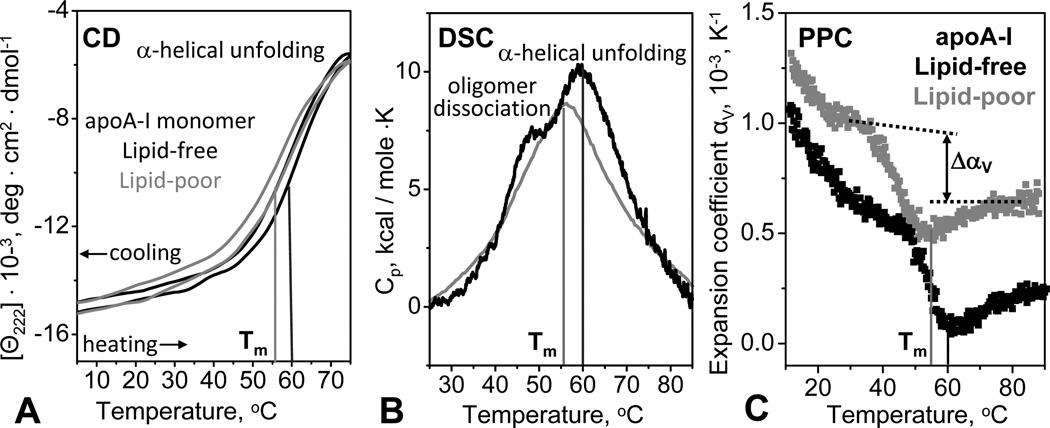
Thermal stability of the lipid-poor apoA-I was assessed by far-UV CD and calorimetry. In CD experiments, lipid-poor or lipid-free apoA-I (0.02 mg/mL protein in standard buffer) was heated and cooled from 10 to 75 °C at a constant rate of 10 or 80 °C/h, and α-helical unfolding and refolding was monitored at 222 nm, Θ222(T). Similar to lipid-free protein, lipid-poor apoA-I showed thermodynamically reversible unfolding evident from the close superimposition of the heating and cooling CD data (Fig. 5A) and from the lack of the heating rate effects on these data (not shown). This is in stark contrast with the thermodynamically irreversible unfolding of lipoproteins characterized by the hysteresis and large scan rate effects in the melting data [35, 48]. Consequently, similar to lipid-free protein but in contrast to HDL or rHDL, lipid-poor apoA-I is stabilized by a thermodynamic mechanism. Furthermore, compared to lipid-free apoA-I, lipid-poor apoA-I showed a decrease in the melting temperature Tm by 5 °C, suggesting lower thermodynamic stability (Fig. 5A, B, C).
Figure 5.
Comparison of thermal stability of lipid-poor (grey) and lipid-free (black) apoA-I in standard buffer. (A) Heat-induced unfolding of the α-helical protein structure was monitored by CD at 222 nm. The samples (0.02 mg/mL protein) were heated and cooled from 5–98 °C at a rate of 80 °C/h. The melting temperature Tm corresponding to the inflexion point in the heating curves is indicated. (B) Differential heat capacity, Cp(T), recorded by DSC during heating of 1 mg/mL protein solution at a rate of 90 °C/h. In lipid-free apoA-I, which is highly self-associated at this concentration (Fig. 2B), the shoulder near 45 °C represents oligomer dissociation and the main peak at Tm=60 °C represents monomer unfolding (black line). Lipid-poor apoA-I, which is less prone to self-association (Fig. 2), shows only one main peak at Tm=55 °C (grey line). (C) Thermal expansion coefficient, αv(T), measured by pressure perturbation calorimetry (PPC) during heating. Protein concentration was 0.5 mg/mL in standard buffer. Similar to lipid-free apoA-I, lipid-poor protein showed: i) a negative peak in Δαv(T) corresponding to a relatively large volume decrease upon unfolding, ΔV/V~−0.3 %, and ii) a reduction in thermal expansion coefficient upon unfolding, Δαv<0, which is unique to apoA-I [43]. Compared to the lipid-free protein, lipid-poor apoA-I shows lower peak temperature, Tm=55 °C, and higher absolute value of αv(T) that is indicative of the presence of lipids that have higher αv than the protein [42, 43].
In DSC experiments, heat capacity Cp(T) was recorded from 1 mg/mL protein solutions during heating from 10–90 °C. Lipid-free apoA-I showed a major peak centered at Tm=60 °C that corresponds to α-helical unfolding; in addition, a shoulder near 45 °C was observed (black line in Fig. 5B). A similar but less well resolved shoulder was observed by using less sensitive calorimeter and higher apoA-I concentrations, and was attributed to tertiary structural unfolding [39]. To test for possible heat-induced changes in the quaternary protein structure, we used SEC to analyze 0.5–1.0 mg/mL protein solutions at 22 °C prior to and immediately after heating to final temperatures ranging from 40 to 90 °C. The results showed that lipid-free apoA-I, which was oligomeric prior to heating, dissociated into monomers upon heating to 45 °C and beyond (Fig. S4B), but gradually self-associated upon prolonged incubation at ambient temperatures. This suggests that the shoulder near 45 °C in the DSC peak reflects oligomer dissociation rather than tertiary structure unfolding in lipid-free apoA-I. The absence of this shoulder from the consecutive DSC scans of lipid-free apoA-I strongly supports this notion (supplemental Fig. S4C).
The DSC data of lipid-poor apoA-I show only the main peak corresponding to α-helical unfolding (Fig 5B, grey line). The absence of the low-temperature shoulder in this peak is consistent with the monomolecular state of lipid-poor apoA-I observed by SEC (Fig. 2A). Hence, DSC and SEC data consistently show that lipid-poor apoA-I is less prone to self-association as compared to lipid-free apoA-I. Furthermore, DSC and CD data consistently show that lipid-poor apoA-I has Tm about 5 °C lower than the free protein.
The latter result is supported by pressure perturbation calorimetry (PPC) data showing a negative peak in the volume expansion coefficient αV(T) corresponding to protein unfolding. Consistent with the DSC results, this transition is centered at Tm= 55 °C in lipid-poor apoA-I and at 60 °C in lipid-free apoA-I (Fig. 5C). Furthermore, in lipid-poor apoA-I the value of αV(T) at any temperature is slightly higher than that in lipid-free protein (Fig. 5C), reflecting higher expansivity of lipid as compared to protein [ref. [42] and references therein]. Integration of the αV(T) peak indicates that, similar to lipid-free apoA-I [43], lipid-poor apoA-I shows a relatively large volume contraction upon unfolding, ΔV/V≅−0.3%. Also, similar to free apoA-I, lipid-poor apoA-I shows a reduction in thermal expansivity upon unfolding, ΔαV(T)<0, which has not been detected in any other proteins [43]. Since large negative ΔV and ΔαV(T) are observed not only in free apoA-I that is self-associated but also in lipid-poor apoA-I that is largely monomeric under conditions of our calorimetric experiments (Fig. 5C), these values reflect unique hydration properties of the apoA-I molecule. We propose that these values reflect increased charged residue hydration resulting from disruption of multiple salt bridges upon α-helical unfolding in apoA-I [43]. This notion is supported by the rapid decline in αV(T) of lipid-free and lipid-poor apoA-I observed upon heating from 5 to 40 °C, which indicates dominant effects of the charged residue hydration [43].
In summary, our CD, DSC and PPC melting data indicate that lipid-poor apoA-I undergoes thermal unfolding that is reversible, endothermic, and involves negative volume changes. Such unfolding behavior is similar to lipid-free apoA-I but is in stark contrast with the apoA-I-containing lipoproteins whose heat denaturation occurs at higher temperatures, is thermodynamically irreversible, and shows no detectable volume changes by PPC [42]. Furthermore, compared to lipid-free apoA-I, lipid-poor apoA-I has lower Tm suggesting lower thermodynamic stability (Fig. 5). Also in contrast to lipid-free apoA-I that self-associates in near-neutral low-salt solutions above 0.1 mg/mL protein concentrations, lipid-poor apoA-I remains monomeric at 0.5–1.0 mg/mL protein (Fig. 2). This is supported by our limited cross-linking studies with bis(sulfosuccinimidyl)suberate which detected monomeric lipid-poor protein under conditions when the free apoA-I forms oligomers (data not shown).
Lipid binding and lipid clearance by lipid-poor apoA-I
To compare the lipid binding ability of lipid-poor and lipid-free apoA-I, we first assessed the protein adsorption to the phospholipid surface by using SUV of POPC as a model membrane. Apolipoproteins such as apoA-I rapidly adsorb to the surface of POPC vesicles but do not spontaneously remodel them into smaller particles [49, 50]. To compare such adsorption, lipid-poor and lipid-free apoA-I at 0.5 mg/mL protein concentration were incubated for 12 h at 22 °C in the presence of POPC SUV at 1:100 protein:lipid molar ratio, followed by SEC and NDGE analyses (Fig. 6). The results in Fig. 6 A–C support the notion that, in contrast to lipid-free apoA-I, lipid-poor apoA-I is less prone to self-association and does not form high-order oligomers at 0.5 mg/mL protein in the absence of SUV. Upon addition of SUV, all lipid-poor apoA-I rapidly adsorbed to the lipid surface (Fig. 6A, C). In contrast, addition of POPC SUV promptly disrupted self-association of lipid-free apoA-I, but only a fraction of the monomeric protein was able to bind the SUV surface (Fig. 6B, C); the distribution between monomeric lipid-free and SUV-bound apoA-I did not change upon 24 h incubation, suggesting an equilibrium between the two species. Furthermore, our SEC, NDGE (Fig. 6) and EM data (not shown) demonstrate that incubation of POPC SUV with lipid-poor or lipid-free apoA-I does not lead to vesicle remodeling into smaller particles. In summary, our results show that lipid-poor apoA-I adsorbs to the POPC surface more readily than the lipid-free protein.
Figure 6.
Apolipoprotein adsorption to phospholipid surface. Protein-containing fractions formed upon adsorption of lipid-poor or lipid-free apoA-I to POPC SUV were detected by SEC (A, B) and NDGE (C). Protein concentration was 0.5 mg/mL in 10 mM PBS, pH 7.5; at this concentration, lipid-free apoA-I is significantly self-associated. ApoA-I:POPC molar ratio was 1:100 (open circles in A, B). Solid lines show protein in the absence of POPC SUV. (C) For NDGE (4–20 % gradient), 10 µg of protein from a 0.8 mg/mL solution were loaded per lane. Lipid-free apoA-I at this concentration is largely self-associated. Bands corresponding to apoA-I in solution or bound to SUV (d>22 nm) are indicated.
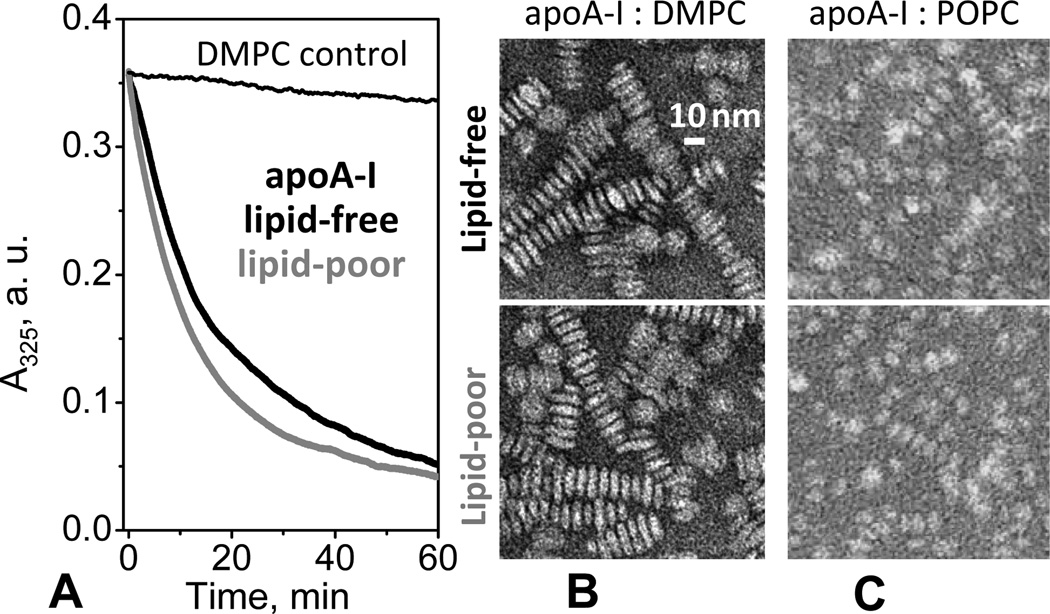
In contrast to POPC, vesicles of shorter-chain PCs such as DMPC (14:0, 14:0) can be spontaneously remodeled by apolipoproteins to form discoidal rHDL that resemble nascent plasma HDL. To test for such remodeling, we used absorption spectroscopy to monitor the time course of turbidity changes upon addition of apoA-I to the suspension of MLV of DMPC. DMPC clearance by lipid-poor and lipid-free apoA-I showed very similar time course, including similar initial rate and similar overall amplitude of turbidity changes (Fig. 7A). EM data showed that DMPC incubation with lipid-poor or free apoA-I at 1:80 protein:lipid molar ratio led to formation of very similar discoidal rHDL (Fig. 7B). NDGE results confirmed this observation and showed that the particle size distribution in these rHDL was very similar and that all protein was incorporated into rHDL (lanes 3, 4 in Fig. S5). To test whether lipid-poor and lipid-free apoA-I form similar rHDL with physiologic lipids such as POPC, we reconstituted apoA-I: POPC complexes by cholate dialysis using lipid-free or lipid-poor apoA-I. The resulting rHDL had similar size, morphology (Fig. 7C), secondary structure, and stability (CD and DSC data not shown). Taken together, our results show that lipid-poor and lipid-free apoA-I have comparable ability to remodel phospholipids and form rHDL.
Figure 7.
Clearance of lipid vesicles and formation of discoidal rHDL by lipid-poor or lipid-free apoA-I. (A) Time course of spontaneous clearance of multilamellar vesicles (MLV) of dimyristoyl phosphatidylcholine (DMPC) by lipid-poor (grey) or lipid-free (black) apoA-I monitored by turbidity at 24 °C. The final sample concentrations are 80 µg/mL DMPC and 20 µg/mL apoA-I in standard buffer; at this concentration, lipid-free apoA-I is fully monomeric. Data for DMPC MLV in the absence of the protein are shown as a control (thin line). (B) Electron micrographs of the negatively stained discoidal complexes formed upon DMPC clearance by lipid-poor (bottom) or lipid-free (top) apoA-I at 24 °C. Discoidal rHDL particles are stacked in rouleaux formation. NDGE of these particles is shown in supplemental Fig. S5. (C) Discoidal apoA-I:POPC complexes reconstituted by cholate dialysis using lipid-poor (bottom) or lipid-free (top) apoA-I and visualized by negative-stain EM.
In summary, compared to lipid-free apoA-I, lipid-poor apoA-I adsorbs more readily to the POPC surface (Fig. 6) but has similar ability to insert into the DMPC surface and gradually remodel it to form rHDL (Fig. 7). This illustrates the fact that protein adsorption and penetration into the phospholipid surface are distinct events.
Lipid-poor apoA-I dissociated from model rHDL
To test whether the composition of the parent particles affects the properties of the lipid-poor apoA-I that dissociates upon heating, we used rHDL of controlled composition. Discoidal apoA-I:POPC:Ch complexes (POPC:A-I:Ch molar ratio 80:4:1, <d>=9.6 nm) were reconstituted by cholate dialysis and were converted into spherical CE-containing rHDL (<d>=9.3 nm) via the lecithin-cholesterol acyltransferase reaction [51] (see supplement for detail). These purified spherical rHDL were heated to 110 °C at a rate of 90 °C/h in DSC experiments (Fig. S6A). The EM, NDGE and SEC data in Fig. S6 show that, similar to spherical plasma HDL, heating of spherical rHDL led to formation of fused and ruptured particles and dissociation of monomolecular apoA-I (fraction 3, Fig. S6D). TLC analysis of this purified fraction showed that it contained PC and CE (Fig. S6D, lane 3). NDGE and SEC showed that this fraction was indistinguishable from the lipid-poor apoA-I dissociated from plasma HDL (Fig. 3). Hence, similar to plasma HDL, thermal remodeling of spherical rHDL also leads to dissociation of monomolecular lipid-poor apoA-I that contains PC and CE.
The lipid-poor apoA-I that dissociated from spherical rHDL or from plasma HDL had very similar secondary structure and stability (CD data not shown). Furthermore, all these lipid-poor species cleared DMPC MLV and formed very similar discoidal complexes with DMPC. Consequently, lipid-poor apoA-I that dissociated from plasma HDL or from spherical rHDL upon heating beyond 100 °C fully retained its lipid binding function.
DISCUSSION
Physicochemical properties of lipid-poor apoA-I
We show that thermal denaturation of HDL generates milligrams of lipid-poor apoA-I that provides a useful model for studying the transient lipid-poor species formed in vivo. The yield of the lipid-poor protein obtained by this method is 10–15% of the initial protein content in HDL. The resulting monomeric lipid-poor protein is folded and functional, as evident from the far- and near-UV CD spectra and the ability to clear lipid and form rHDL (Figs. 4, 5, 7, S2A).
The lipid-poor protein formed upon heating of plasma HDL has apparent Stokes diameter d=7.1±0.4 nm (Fig. 2B), molecular weight Mw~37 kDa (based on Mw(apoA-I)=28 kDa and 3:1 w/w protein:lipid ratio), and contains one apoA-I, eight-to-nine PC, and one CE. The composition, conformation, self-association, and lipid binding properties of this species resemble those of the lipid-poor apoA-I formed upon interaction of free apoA-I with ABCA1-expressing cells [32, 33, 52]. For example, Fielding’s team reported that ABCA1 facilitates extracellular conversion of free apoA-I into a monomeric lipid-poor species that promotes reverse cholesterol transport [52]; this species has d~7.2 nm and Mw~34 kDa, comparable to those observed in our work. Phillips’ group reported transient formation of monomolecular lipid-poor apoA-I upon interaction of free apoA-I with ABCA1-overexpressing cells, followed by conversion of this lipid-poor into lipid-rich HDL-like form [33]. The size (d=7.5±0.4 nm), composition (one apoA-I, three-to-four PC, and one-to-two Ch) and the α-helical content of this transient lipid-poor species are comparable to those observed in our work. Parks’ team reported formation of various-size products upon apoA-I incubation with ABCA1-expressing cells [32]. The smallest product (d~7.1 nm, one apoA-I and five-to-nine PC) was similar to the lipid-poor protein of our study, while the larger products were discoidal HDL. Our results reveal that heating of core-containing spherical HDL (plasma or reconstituted) generates lipid-poor apoA-I that contains PC and a small amount of CE (Fig. 3B, S4E). This suggests that apoA-I can interact with the sterols in the parent HDL, either directly or via the PC.
We propose that the relative flexibility of the secondary structural elements, which is characteristic of the molten globule [39], helps to accommodate several lipid molecules varying in their number with minimal perturbations to the structure of the apoA-I monomer. Importantly, our CD and calorimetric melting data reveal that lipid-poor apoA-I has low thermodynamic stability (Fig. 5A–C) that is slightly lower than that of free protein, ΔG(25 °C)~2.5 kcal/mol [39]. This contrasts with high kinetic stability of HDL and other lipoproteins [34–36, 42]. Consequently, high kinetic stability does not necessarily result from the apolipoprotein-lipid binding, but is characteristic of large macromolecular complexes such as HDL that contain at least 2 copies of apoA-I and over 150 lipid molecules. We propose that low thermodynamic stability facilitates conformational changes in the lipid-poor apoA-I monomer, such as domain swapping, which are required for its dimerization and HDL formation.
The general drawback of low thermodynamic stability is that it facilitates protein degradation. In fact, compared to HDL, lipid-poor/free apoA-I is rapidly catabolized in normal and in pathologic conditions [18]. The latter include Tangier disease, an HDL deficiency disorder resulting from loss of function of ABCA1 [54], as well as acute phase response in which lipid-poor apoA-I is displaced from HDL by serum amyloid protein [55]. We envision two mechanisms that help to direct lipid-poor apoA-I towards HDL biogenesis as opposed to degradation. One is suggested by our SEC data showing that apoA-I, particularly in its lipid-poor form, readily adsorbs to the phospholipid surface (Fig. 6). Such adsorption must be rapid since it involves no high-energy barriers associated with lipid penetration and remodeling. We hypothesize that this rapid adsorption, which in lipid-poor apoA-I may be mediated by its lipid moiety, helps recruit the protein to the plasma membrane for HDL biogenesis. Furthermore, apoA-I adsorption to the PC surface involves the hydrophobic C-terminal segment 190–243 [ref. [33] and references therein]. In lipid-free apoA-I, this flexible segment is highly susceptible to proteolysis [49]. Adsorption of this segment to the PC surface, along with its reduced flexibility in lipid-poor apoA-I (described below), is expected to reduce its proteolytic degradation.
Structural model of lipid-poor apoA-I
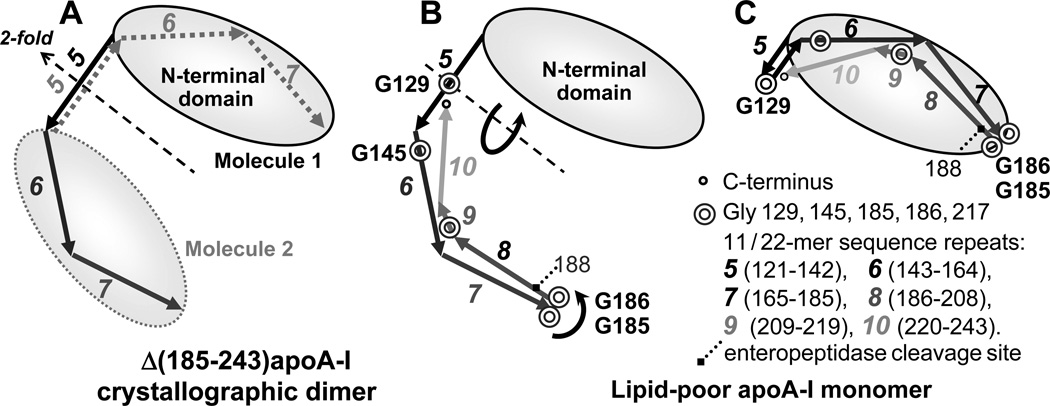
We propose a structural model for the lipid-poor apoA-I monomer which is based on previous low-resolution structural studies of apoA-I [2, 56, 57] and the high-resolution x-ray crystal structure of the C-terminal truncated lipid-free protein, Δ(185–234)apoA-I, recently solved by Mei and Atkinson [58]. Our model is consistent with the physicochemical properties determined in this and earlier studies [32, 33, 59] and takes advantage of the modular structure of apoA-I comprised of ten 11/22-mer tandem repeats with high propensity to form amphipathic α-helices [60, 61]. These repeats are punctuated by Pro that induce helical kinks, and/or Gly that confer flexibility to the polypeptide chain. In the crystal, Δ(185–243)apoA-I forms an antiparallel dimer that adopts a highly α-helical semi-circular conformation with central repeats 5 (residues 121–142) in registry [58], in agreement with the “double belt” model postulated for apoA-I on HDL [1–4, 60]. The dimer stability is conferred by two bundles at its termini. Each bundle is comprised of the N-terminal segments from one apoA-I molecule (residues 1–100), while an additional helical segment (repeats 6–7) comes from the second molecule (Fig. 8A). Since the highly hydrophobic C-terminal repeats 8–10 form the primary lipid binding site in apoA-I, and deletion of repeat 10 prevents formation of lipid-poor species [33], the C-terminal repeats probably interact with the lipids in lipid-poor apoA-I. To sequester the apolar lipid moieties in the lipid-poor apoA-I, this C-terminal segment must fold against the protein molecule [33]. We posit that such folding can be accomplished by pivoting of the polypeptide chain around two flexible Gly-containing hinges (Fig. 8B). The first hinge is formed by G185 and G186, a pair of highly conserved Gly that confer flexibility to the polypeptide chain of apoA-I which is necessary for its adaptation to various environments [61]. The second hinge likely encompasses G129 and/or G145 in the central part of apoA-I that has increased flexibility both on HDL and in the crystal structure of lipid-free Δ(185–243)apoA-I [58]. We hypothesize that pivoting around these hinges can pack repeats 8–10 and 6–7 against the N-terminal bundle in lipid-poor apoA-I (Fig. 8C). In fact, packing of repeats 6–7 against the N-terminal bundle was observed in the crystal structure of lipid-free Δ(185–243)apoA-I, in which repeats 6–7 from one molecule interact with the N-domain from the second molecule within the dimer (Fig. 8A). In monomeric apoA-I, all repeats must belong to the same molecule (Fig. 8C), suggesting “domain swapping” of repeats 6–7 upon apoA-I monomer-to-dimer conversion [58].
Figure 8.
Cartoon illustrating molecular dimer observed in the crystal structure of the C-terminally truncated apoA-I by Mei and Atkinson (A), and the proposed folding of the lipid-poor apoA-I monomer in a compact conformation (B). In panel A, molecule 1 (black solid lines) and molecule 2 (grey dotted lines) within the dimer are related by 2-fold symmetry axis passing through the middle of sequence repeat 5. Amino acid sequence of apoA-I is comprised of the N-terminal 43 residues (encoded by exon 3) followed by residues 44–243 (encoded by exon 4) that form 11- or 22-mer sequence repeats punctuated by Pro or Gly. These repeats have high propensity to form amphipathic class-A α-helices that have high affinity for lipid surface [60, 61]. Repeats 5–10 encompassing residues 121–243 are shown by line arrows and numbered. Gly positions in the C-terminal half of the molecule are indicated. The N-terminal part of apoA-I forms a loosely folded “buckle” (shown by an oval). We posit that in monomeric lipid-poor apoA-I repeats 6, 7 and 8–10 from the same molecule pack against this N-terminal “buckle” by pivoting around two Gly-containing flexible hinges, G129 and G185, G186 (circular arrows, panel B). The resulting conformation of the lipid-poor apoA-I monomer is shown in panel C. In lipid-free apoA-I, the C-terminal repeats 8–10 (residues 186–243) are highly flexible and susceptible to cleavage by enteropeptidase at R118 [59]. In lipid-poor apoA-I (panel C), the lipids (not shown) are packed between the N- and the C-terminal parts of the molecule.
The crystal structure of Δ(185–243)apoA-I solved by Mei and Atkinson demonstrates the propensity of apoA-I to form dimers that resemble the intermediate species during formation of nascent HDL from the monomeric protein. Our current data provide novel information on the conformation of monomeric lipid-poor protein and the similarities and differences between it and the lipid-free apoA-I monomer. We propose that, despite the overall structural similarity of lipid-free and lipid-poor apoA-I monomers, the distinct differences in their behavior result, in part, from the packing of the C-terminal region. In the lipid-free protein, the C-terminal segment 185–243 is largely disordered and flexible [ref. [33] and references therein]. We speculate that in lipid-poor protein, the lipid sequestration between the C-terminal and the N-terminal segments [33] increases the ordering of the C-terminal segment and packs it against the three N-terminal segments in the bundle. This explains the reduced self-associating propensity of the lipid-poor protein observed in this (Fig. 2) and earlier studies [32, 33], and its reduced susceptibility to proteolysis at R188 [59]. Furthermore, in the lipid-poor protein, the interactions of the N-terminal bundle with the C-terminal segment and/or the bound lipids induce subtle changes in the Trp packing in the N-domain (W8, W50, W72, and W108) detected by near-UV CD and fluorescence (Fig. 4). These interdomain interactions also slightly reduce the thermodynamic stability of the N-domain, as observed by CD, DSC and PPC (Fig. 5). Such reduced stability is consistent with the increased Trp polarity in lipid-poor apoA-I observed by CD and fluorescence (Fig. 4), and with a slightly reduced α-helical content observed by far-UV CD. We speculate that this reduced stability primes the N-domain for the conformational rearrangement that is necessary for HDL formation [62].
Taken together, our CD, fluorescence and calorimetric data (Figs. 4, 5, S2) along with the biophysical data from Phillips’ group [33] indicate that lipid-poor and lipid-free apoA-I monomers adopt similar overall conformation (Fig. 8C), particularly in their N-terminal domain, but have distinct dynamic properties, particularly in their C-terminal segment 186–243. In the lipid-free protein, this hydrophobic segment is largely unordered and forms the primary self-association and lipid binding site [ref. [33] and references therein]. We posit that in the lipid-poor protein this segment is less flexible due to its direct involvement in lipid sequestration.
Potential functional implications
We speculate that the subtle structural and dynamic differences between the lipid-poor and the lipid-free apoA-I monomer help direct the lipid-poor species towards HDL biogenesis as opposed to degradation. Compared to the lipid-free protein, lipid-poor apoA-I: i) is better protected from the post-translational modifications and proteolytic degradation of its most labile C-terminal part [33, 59], ii) adsorbs more readily to the phospholipid surface (Fig. 6), which helps to recruit the protein from plasma to the cell membrane for HDL biogenesis, and iii) has reduced thermodynamic stability (Fig. 5), which facilitates conformational re-arrangement of the apoA-I monomer necessary for its dimerization and HDL formation [62]. Taken together, these properties could help to shift the balance, from protein degradation to HDL biogenesis, and thereby contribute to the removal of excess cell cholesterol by lipid-poor apoA-I.
Supplementary Material
ACKNOWLEDGEMENT
We thank Cheryl England and Michael Gigliotti for help with isolation of human plasma HDL and with HDL delipidation to obtain lipid-free apoA-I, and Donald L. Gantz for expert help with electron microscopy. We are grateful to Drs. John Parks and Kerry-Ann Rye for their kind gift of lecithin-cholesterol acyltransferase. We are indebted to Xiaohu Mei and Dr. David Atkinson for sharing the privileged information on the crystal structure of the C-terminally truncated lipid-free apoA-I prior to publication.
Funding: This work was supported by the National Institutes of Health grants HL026335 and RO1 GM067260 to OG and by a New Investigator Award from the Tobacco-Related Disease Research Program of California (#18KT-0021) to GC.
Abbreviations
- HDL
high-density lipoprotein
- rHDL
reconstituted HDL
- apo
apolipoprotein
- PC
phosphatidylcholine
- DMPC
dimyristoyl PC (14:0,14:0)
- POPC
palmitoyloleoyl PC (16:0,16:1)
- Ch
unesterified cholesterol
- CE
cholesterol ester
- MLV
multilamellar vesicles
- SUV
small unilamellar vesicles
- DSC
differential scanning calorimetry
- PPC
pressure perturbation calorimetry
- CD
circular dichroism
- EM
electron microscopy
- NDGE
non-denaturing gel electrophoresis
- SEC
size-exclusion chromatography
- TLC
thin-layer chromatography
- PBS
phosphate buffer saline
- ABCA1
ATP-binding cassette transporter A1
REFERENCES
- 1.Gu F, Jones MK, Chen J, Patterson JC, Catte A, Jerome WG, Li L, Segrest JP. Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 2010;285(7):4652–4665. doi: 10.1074/jbc.M109.069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lund-Katz S, Phillips MC. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 2010;51:183–227. doi: 10.1007/978-90-481-8622-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catte A, Patterson JC, Bashtovyy D, Jones MK, Gu F, Li L, Rampioni A, Sengupta D, Vuorela T, Niemelä P, Karttunen M, Marrink SJ, Vattulainen I, Segrest JP. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys J. 2008;94(6):2306–2319. doi: 10.1529/biophysj.107.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang R, Silva RA, Jerome WG, Kontush A, Chapman MJ, Curtiss LK, Hodges TJ, Davidson WS. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 2011;18(4):416–422. doi: 10.1038/nsmb.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fielding CJ, Fielding PE. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 6.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005;96(12):1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 7.Rye KA, Bursill CA, Lambert G, Tabet F, Barter PJ. The metabolism and anti-atherogenic properties of HDL. J. Lipid Res. 2009;50:S195–S200. doi: 10.1194/jlr.R800034-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 2010;30:139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy D, Rader DJ. Update on strategies to increase HDL quantity and function. Nat. Rev. Cardiol. 2009;6(7):455–463. doi: 10.1038/nrcardio.2009.94. [DOI] [PubMed] [Google Scholar]
- 10.Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, Thompson A, Wood AM, Lewington S, Sattar N, Packard CJ, Collins R, Thompson SG, Danesh J. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302(18):1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kontush A, Chapman MJ. Antiatherogenic function of HDL particle subpopulations: focus on antioxidative activities. Curr. Opin. Lipidol. 2010;21:312–318. doi: 10.1097/MOL.0b013e32833bcdc1. [DOI] [PubMed] [Google Scholar]
- 12.Asztalos BF, Tani M, Schaefer EJ. Metabolic and functional relevance of HDL subspecies. Curr. Opin. Lipidol. 2011;22(3):176–185. doi: 10.1097/MOL.0b013e3283468061. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls SJ, Nissen SE. New targets of high-density lipoprotein therapy. Curr. Opin. Lipidol. 2007;18(4):421–426. doi: 10.1097/MOL.0b013e32821f603b. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, Yuan S. High density lipoproteins-based therapies for cardiovascular disease. J. Cardiovasc. Dis. Res. 2010;1(3):99–103. doi: 10.4103/0975-3583.70898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rye KA, Clay MA, Barter PJ. Remodelling of high density lipoproteins by plasma factors. Atherosclerosis. 1999;145:227–238. doi: 10.1016/s0021-9150(99)00150-1. [DOI] [PubMed] [Google Scholar]
- 16.Young IS, Nicholls DP. Lipid metabolism. Curr. Opin. Lipidol. 2010;21(6):550–551. doi: 10.1097/MOL.0b013e328340b4d9. [DOI] [PubMed] [Google Scholar]
- 17.Liang H-Q, Rye K-A, Barter PJ. Remodeling of reconstituted high density lipoproteins by lecithin: cholesterol acyltransferase. J. Lipid Res. 1996;37:1962–1970. [PubMed] [Google Scholar]
- 18.Lee JY, Lanningham-Foster L, Boudyguina EY, Smith TL, Young ER, Colvin PL, Thomas MJ, Parks JS. Prebeta high density lipoprotein has two metabolic fates in human apolipoprotein A-I transgenic mice. J. Lipid Res. 2004;45(4):716–728. doi: 10.1194/jlr.M300422-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Cavigiolio G, Geier EG, Shao B, Heinecke JW, Oda MN. Exchange of apolipoprotein A-I between lipid-associated and lipid-free states: a potential target for oxidative generation of dysfunctional high density lipoproteins. J. Biol. Chem. 2010;285(24):18847–18857. doi: 10.1074/jbc.M109.098434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanotti I, Favari E, Bernini F. Cellular cholesterol efflux pathways: Impact on intracellular lipid trafficking and methodological considerations. Curr. Pharm. Biotechnol. 2011 doi: 10.2174/138920112799095383. (in press) [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S. Assembly of high-density lipoprotein. Arterioscler. Thromb. Vasc. Biol. 2006;26:20–27. doi: 10.1161/01.ATV.0000195789.39418.e8. [DOI] [PubMed] [Google Scholar]
- 22.Oram JF, Heinecke JW. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 2005;85(4):1343–1372. doi: 10.1152/physrev.00005.2005. [DOI] [PubMed] [Google Scholar]
- 23.Moestrup SK, Nielsen LB. The role of the kidney in lipid metabolism. Curr. Opin. Lipidol. 2005;16(3):301–306. doi: 10.1097/01.mol.0000169350.45944.d4. [DOI] [PubMed] [Google Scholar]
- 24.Castro GR, Fielding CJ. Early incorporation of cell-derived cholesterol into pre-beta-migrating high-density lipoprotein. Biochemistry. 1988;27:25–29. doi: 10.1021/bi00401a005. [DOI] [PubMed] [Google Scholar]
- 25.Asztalos BF, Roheim PS. Presence and formation of ‘free apolipoprotein A-I-like’ particles in human plasma. Arterioscler. Thromb. Vasc. Biol. 1995;15:1419–1423. doi: 10.1161/01.atv.15.9.1419. [DOI] [PubMed] [Google Scholar]
- 26.Jaspard B, Collet X, Barbaras R, Manent J, Vieu C, Parinaud J, Chap H, Perret B. Biochemical characterization of pre-beta 1 high-density lipoprotein from human ovarian follicular fluid: Evidence for the presence of a lipid core. Biochemistry. 1996;35:1352–1357. doi: 10.1021/bi950938i. [DOI] [PubMed] [Google Scholar]
- 27.Nanjee MN, Brinton EA. Very small apolipoprotein A-I-containing particles from human plasma: isolation and quantification by high-performance size-exclusion chromatography. Clin. Chem. 2000;46:207–223. [PubMed] [Google Scholar]
- 28.Bottcher A, Schlosser J, Kronenberg F, Dieplinger H, Knipping G, Lackner KJ, Schmitz G. Preparative free-solution isotachophoresis for separation of human plasma lipoproteins: apolipoprotein and lipid composition of HDL subfractions. J. Lipid Res. 2000;41:905–915. [PubMed] [Google Scholar]
- 29.Kunitake ST, La Sala KJ, Kane JP. Apolipoprotein A-I-containing lipoproteins with pre-beta electrophoretic mobility. J. Lipid Res. 1985;26:549–555. [PubMed] [Google Scholar]
- 30.O'Kane MJ, Wisdom GB, McEneny J, McFerran NV, Trimble ER. Pre-beta high-density lipoprotein determined by immunoblotting with chemiluminescent detection. Clin. Chem. 1992;38:2273–2277. [PubMed] [Google Scholar]
- 31.Schmitz G, Mollers C, Richter V. Analytical capillary isotachophoresis of human serum lipoproteins. Electrophoresis. 1997;18:1807–1813. doi: 10.1002/elps.1150181015. [DOI] [PubMed] [Google Scholar]
- 32.Mulya A, Lee JY, Gebre AK, Thomas MJ, Colvin PL, Parks JS. Minimal lipidation of pre-beta HDL by ABCA1 results in reduced ability to interact with ABCA1. Arterioscler. Thromb. Vasc Biol. 2007;27(8):1828–1836. doi: 10.1161/ATVBAHA.107.142455. [DOI] [PubMed] [Google Scholar]
- 33.Duong PT, Weibel GL, Lund-Katz S, Rothblat GH, Phillips MC. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 2008;49(5):1006–1014. doi: 10.1194/jlr.M700506-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta R, Gantz DL, Gursky O. Human plasma high-density lipoproteins are stabilized by kinetic factors. J. Mol. Biol. 2003;328(1):183–192. doi: 10.1016/s0022-2836(03)00155-4. [DOI] [PubMed] [Google Scholar]
- 35.Jayaraman S, Gantz DL, Gursky O. Effects of salt on thermal stability of human plasma high-density lipoproteins. Biochemistry. 2006;45:4620–4628. doi: 10.1021/bi0524565. [DOI] [PubMed] [Google Scholar]
- 36.Guha M, Gao X, Jayaraman S, Gursky O. Structural stability and functional remodeling of high-density lipoproteins: The importance of being disordered. Biochemistry. 2008;47(44):11393–11397. doi: 10.1021/bi8014746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumaker VN, Puppione DL. Sequential flotation ultracentrifugation. Methods Enzymol. 1986;128:155–170. doi: 10.1016/0076-6879(86)28066-0. [DOI] [PubMed] [Google Scholar]
- 38.Nichols AV, Gong EL, Blanche PJ, Forte TM, Anderson DW. Effects of guanidine hydrochloride on human plasma high density lipoproteins. Biochim. Biophys. Acta. 1976;446:226–239. doi: 10.1016/0005-2795(76)90113-6. [DOI] [PubMed] [Google Scholar]
- 39.Gursky O, Atkinson D. Thermal unfolding of human high-density apolipoprotein A-1: implications for a lipid-free molten globular state. Proc. Natl. Acad. Sci. USA. 1996;93(7):2991–2995. doi: 10.1073/pnas.93.7.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonas A. Reconstitution of high-density lipoproteins. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 41.Jayaraman S, Gantz DL, Gursky O. Effects of phospholipase A(2) and its products on structural stability of human LDL: relevance to formation of LDL-derived lipid droplets. J. Lipid Res. 2011;52(3):549–557. doi: 10.1194/jlr.M012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayaraman S, Jasuja R, Zakharov MN, Gursky O. Pressure perturbation calorimetry of lipoproteins reveals an endothermic transition without detectable volume changes. implications for adsorption of apolipoprotein to a phospholipid surface. Biochemistry. 2011;50(19):3919–3927. doi: 10.1021/bi200090y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjwal S, Gursky O. Pressure perturbation calorimetry of apolipoproteins in solution and in model lipoproteins. Proteins. 2010;78:1175–1185. doi: 10.1002/prot.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean LR, Phillips MC. Cholesterol transfer from small and large unilamellar vesicles. Biochim. Biophys. Acta. 1984;776(1):21–26. doi: 10.1016/0005-2736(84)90246-3. [DOI] [PubMed] [Google Scholar]
- 45.Pownall H, Pao Q, Hickson D, Sparrow JT, Kusserow SK, Massey JB. Kinetics and mechanism of association of human plasma apolipoproteins with dimyristoyl phosphatidyl choline: Effect of protein structure and lipid clusters on reaction rates. Biochemistry. 1981;20:6630–6635. doi: 10.1021/bi00526a017. [DOI] [PubMed] [Google Scholar]
- 46.Benjwal S, Jayaraman S, Gursky O. Role of secondary structure in protein-phospholipid surface interactions: reconstitution and denaturation of apolipoprotein C-I:DMPC complexes. Biochemistry. 2007;46(13):4184–4194. doi: 10.1021/bi062175c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitello LB, Scanu AM. Studies on human serum high density lipoproteins. Self-association of apolipoprotein A-I in aqueous solutions. J. Biol. Chem. 1976;251:1131–1136. [PubMed] [Google Scholar]
- 48.Gursky O, Ranjana and Gantz DL. Complex of human apolipoprotein C-1 with phospholipid: Thermodynamic or kinetic stability? Biochemistry. 2002;41:7373–7384. doi: 10.1021/bi025588w. [DOI] [PubMed] [Google Scholar]
- 49.Saito H, Dhanasekaran P, Nguyen D, Deridder E, Holvoet P, Lund-Katz S, Phillips MC. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 2004;279:20974–20981. doi: 10.1074/jbc.M402043200. [DOI] [PubMed] [Google Scholar]
- 50.Arnulphi C, Sánchez SA, Tricerri MA, Gratton E, Jonas A. Interaction of human apolipoprotein A-I with model membranes exhibiting lipid domains. Biophys. J. 2005;89:285–295. doi: 10.1529/biophysj.104.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonas A, Wald JH, Toohill KL, Krul ES, Kezdy KE. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J. Biol. Chem. 1990;265:22123–22129. [PubMed] [Google Scholar]
- 52.Chau P, Nakamura Y, Fielding CJ, Fielding PE. Mechanism of prebeta-HDL formation and activation. Biochemistry. 2006;45(12):3981–3987. doi: 10.1021/bi052535g. [DOI] [PubMed] [Google Scholar]
- 53.Vedhachalam C, Chetty PS, Nickel M, Dhanasekaran P, Lund-Katz S, Rothblat GH, Phillips MC. Influence of apolipoprotein (Apo) A-I structure on nascent high density lipoprotein (HDL) particle size distribution. J. Biol. Chem. 2010;285(42):31965–33173. doi: 10.1074/jbc.M110.126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang C, Oram JF. The cell cholesterol exporter ABCA1 as a protector from cardiovascular disease and diabetes. Biochim. Biophys. Acta. 2009;1791(7):563–572. doi: 10.1016/j.bbalip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 55.Jahangiri A, de Beer MC, Noffsinger V, Tannock LR, Ramaiah C, Webb NR, van der Westhuyzen DR, de Beer FC. HDL remodeling during the acute phase response. Arterioscler. Thromb. Vasc. Biol. 2009;29(2):261–267. doi: 10.1161/ATVBAHA.108.178681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci USA. 1997;94(23):12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chetty PS, Mayne L, Lund-Katz S, Stranz D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106(45):19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mei X, Atkinson D. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of HDL by dimerization. J. Biol. Chem. 2011;286(44):38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Safi W, Maiorano JN, Davidson WS. A proteolytic method for distinguishing between lipid-free and lipid-bound apolipoprotein A-I. J. Lipid Res. 2001;42(5):864–872. [PubMed] [Google Scholar]
- 60.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: A review of secondary structure and function. J. Lipid Res. 1992;33(2):141–166. [PubMed] [Google Scholar]
- 61.Bashtovyy D, Jones MK, Anantharamaiah GM, Segrest JP. Sequence conservation of apolipoprotein A-I affords novel insights into HDL structure-function. J. Lipid Res. 2011;2(3):435–450. doi: 10.1194/jlr.R012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka M, Dhanasekaran P, Nguyen D, Nickel M, Takechi Y, Lund-Katz S, Phillips MC, Saito H. Influence of N-terminal helix bundle stability on the lipid-binding properties of human apolipoprotein A-I. Biochim. Biophys. Acta. 2011;1811(1):25–30. doi: 10.1016/j.bbalip.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.