Abstract
Studies aimed towards glycan biomarker discovery have focused on glycan characterization by the global profiling of released glycans. Site-specific glycosylation analysis is less developed but may provide new types of biomarkers with higher sensitivity and specificity. Quantitation of peptide-conjugated glycans directly facilitates the differential analysis of distinct glycoforms associated with specific proteins at distinct sites. We have developed a method using MRM to monitor protein glycosylation normalized to absolute protein concentrations to examine quantitative changes in glycosylation at a site-specific level. This new approach provides information regarding both the absolute amount of protein and the site-specific glycosylation profile and will thus be useful to determine if altered glycosylation profiles in serum/plasma are due to a change in protein glycosylation or a change in protein concentration. The remarkable sensitivity and selectivity of MRM enable the detection of low abundant IgG glycopeptides, even when IgG was digested directly in serum with no clean-up prior to the liquid chromatography. Our results show a low limit of detection of 60 attomoles, and a wide dynamic range of 3 orders magnitude for IgG protein quantitation. The results show that IgG glycopeptides can be analyzed directly from serum (without enrichment) and yield more accurate abundances when normalized to the protein content. This report represents the most comprehensive study so far of the use of multiple reaction monitoring for the quantitation of glycoproteins and their glycosylation patterns in biofluids.
Keywords: Immunoglobulin G, absolute quantitation, Glycopeptide, Site specific Glycan analysis
INTRODUCTION
Multiple reaction monitoring (MRM) is a technology that is valued for its potential towards the reliable quantitation of analytes of low abundance in complex mixtures.1 MRM is often performed on triple quadrupole (QqQ) instruments whereby a predetermined precursor ion is selected in the first quadrupole (Q) and fragmented in the collision quadrupole (q); a predetermined fragment ion is selected in the third quadrupole (Q) for detection. A high selectivity is obtained due to the two selection steps that filter out co-eluting background ions. Moreover, the non-scanning nature of the method allows for high sensitivity and a linear response over a wide dynamic range. These features facilitate the robust quantitative analysis of lower abundant compounds in complicated mixtures. While MRM has often been applied for quantitation in metabolomics2–3 and proteomics4–5 settings, its use for glycan and glycoconjugate analyses has been limited. A possible reason being that MRM is mostly performed on QqQ instruments that are often optimized for small molecules with mass ranges and ion optics often not suited for large glycopeptides.
Glycosylation is one of the most common protein post-translation modifications. Nearly 70% of human proteins are glycosylated,6 and glycans play an important role in cell-cell and cell-matrix interactions. Many diseases are associated with aberrant protein glycosylation.7–11 As a result, there has been an increasing interest in glycans as biomarkers for diseases.12–16
Approaches towards glycan biomarkers have focused on released glycans, where generally no information is obtained about the originating protein or the site. Often, glycans are released from the total pool of serum glycoproteins and each component is monitored relative to the glycan pool.17–19 This strategy has identified altered glycosylation patterns in numerous physiological states, such as aging, pregnancy and several different types of cancer.14–15, 20–22 However due to the release of the glycans from the proteins, the original site and protein of attachment is lost. A technique that distinguishes glycosylation patterns on individual proteins as well as site-specific glycoforms would provide greater utility to glycan markers.23–24 To this end, recent biomarker studies have started to focus on the glycosylation of individual proteins, mostly immunoglobulin G (IgG). These studies involve some form of protein enrichment, often immunoprecipitation, prior to analysis. Analysis is performed either on the released glycan25–28 or on the glycopeptide from the Fc region.29–34 These methods have relied on quantitation of the specific glycoforms relative to the total profile with little or no regard to protein expression.18, 31, 35
Methods employing MRM have been used to examine glycoproteins, with a focus on peptide quantitation. Sialylated or fucosylated glycopeptides were examined using enrichment, followed by de-glycosylation prior to peptide analysis.36–37 This approach quantitates glycosylation on peptides but yields no information regarding individual glycoforms. MRM of glycopeptides was developed using precursor ion scans to monitor oxonium ions and identify intact glycoproteins in depleted serum. MRM transitions were then developed for glycopeptides, but no quantitation of the glycopeptides or their corresponding proteins was obtained.38 A similar method was employed for haptoglobin, whereby the glycopeptide signal was normalized to the protein peptide signal, thus taking both the protein concentration as well as the glycan heterogeneity into account.39 However, protein quantitation was performed using a second method, and the accuracy of the MRM method was not determined.
In this report, we employ the power of MRM to observe and quantify glycoforms of glycoproteins directly from serum without protein enrichment and with glycoform quantitation relative to absolute protein abundances. We performed studies on tuning the MS conditions for glycopeptides and systematically studied the MRM behavior of peptides and glycopeptides from the serum glycoprotein IgG for the absolute quantification of IgG in biofluids. Furthermore, the site-specific glycosylation profile was obtained and normalized to the absolute protein concentration. The optimized method yields both the absolute amount of the glycoprotein and the site-specific quantitation of individual glycoforms. This strategy is applicable to all glycoproteins in biological samples but can be performed directly in the fluid for relatively abundant proteins, without protein enrichment. It is anticipated that this method will have considerable value for use in disease biomarker discovery.
EXPERIMENTAL PROCEDURES
A brief description of the experimental procedures is given here. A more detailed version of the materials and methods used can be found in the supplementary information.
Sample preparation
100 μg of IgG standard and 2 μL of serum sample were reduced and alkylated prior to trypsin digestion in a 37°C water bath for 18 h. The resulting peptide samples were used directly for mass spectrometric (MS) analysis without further sample cleanup or dilution.
UPLC-ESI-QqQ analysis
The peptide samples were analyzed using an Agilent 1290 infinity UPLC system coupled to an Agilent 6490 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA). An Agilent Eclipse plus C18 column (RRHD 1.8 μm, 2.1×100 mm) was used for UPLC separation.
Upon injection of 1 μL of sample, peptides and glycopeptides were separated using a 10-minute binary gradient consisting of solvent A of 3% acetonitrile, 0.1% formic acid; solvent B of 90% acetonitrile, 0.1% formic acid in nano-pure water (v/v) at a flow rate of 0.5 mL/min. To reduce the cycle time, the instrument was used in the dynamic MRM mode at unit resolution and peptide and glycopeptides ionization was performed in the positive mode. The MRM results were analyzed using Agilent MassHunter Quantitative Analysis B.4.0 software.
Nano-LC-Chip-QTOF analysis
For profiling, 3 μL of sample was injected into an Agilent 1200 series HPLC-Chip system coupled to an Agilent 6520 QTOF (Agilent Technologies, Santa Clara, CA). The microfluidic chip was consisted of two C18 columns (300 Å, 5 μm): one for enrichment (4 mm, 40 nL) and one for separation (43 mm × 75 μm). A 60-minute LC separation was performed using a binary gradient at 0.3 μL/min flow rate: solvent A consisted of 3% acetonitrile, 0.1% formic acid in nano-pure water (v/v); solvent B consisted of 90% acetonitrile, 0.1% formic acid in nano-pure water (v/v). The mass spectrometer was operated in the positive mode. The collision energy used for the tandem experiment was calculated on the basis of the m/z value using the relationship:
| eq (1) |
RESULTS AND DISCUSSION
We developed a method for the absolute quantitation of human immunoglobulin G and its glycoforms directly from serum using multiple reaction monitoring on a QqQ-MS. A human IgG standard was used to systematically study the behavior of IgG peptides and glycopeptides in MRM experiments and develop a method for the quantitative analysis of IgG and its glycoforms; the method was then applied to a pooled serum sample and subsequently sera from ten healthy individuals. Because MRM is based on the detection and quantitation of specific fragment ions, the collision induced dissociation (CID) behavior of the tryptic peptides and glycopeptides of the four different subclasses was first studied using QTOF-MS. Based on these results, MRM transitions were developed on the QqQ-MS, and the instrument parameters were optimized to get the best sensitivity for glycopeptides. Both peptides and glycopeptides were quantified using MRM in the same run. The absolute amount of the IgG protein was determined using a peptide calibration curve, while the degree of glycosylation was normalized to the total protein content. Using this strategy the abundance of the protein and the degree of protein glycosylation was monitored simultaneously and at the site-specific level.
Fragmentation of IgG peptides and glycopeptides
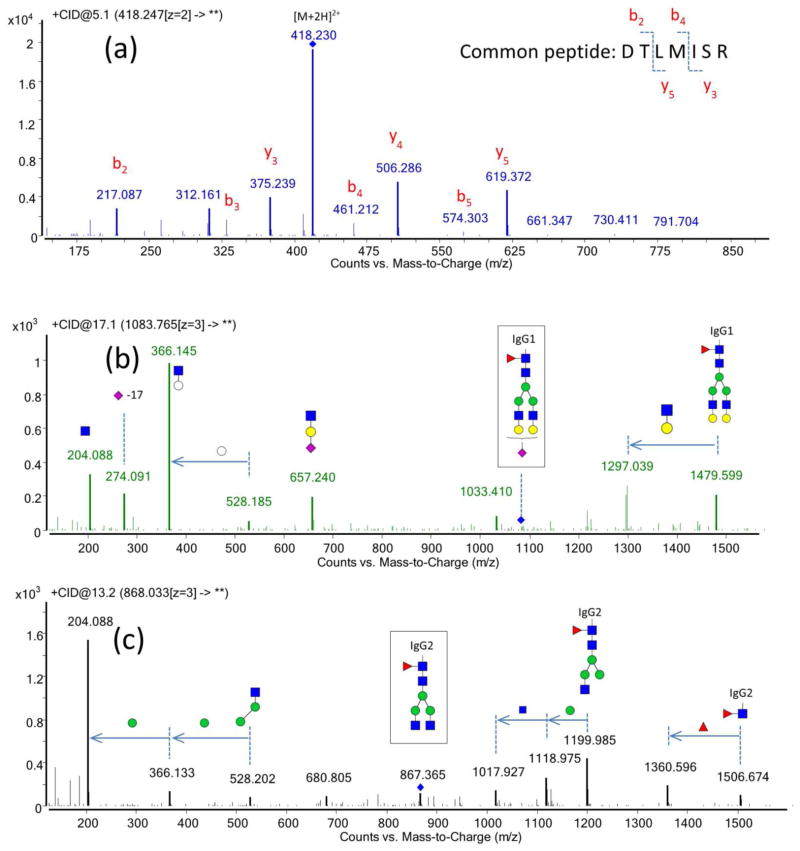
An IgG protein standard was subjected to trypsin digestion, and the resulting peptides and glycopeptides were analyzed using LC-Q-TOF MS/MS to evaluate the fragmentation behavior of peptides and glycopeptides. The IgG standard was observed to contain all four IgG subclasses: IgG1 (UniProtKB ID: P01857), IgG2 (P01859), IgG3 (P01860), and IgG4 (P01861). The tryptic peptide DTLMISR is common to the Fc region of all four subclasses of IgG and may therefore be used for the absolute quantitation of IgG. In Figure 1a, the tandem mass spectrum of this peptide is shown to consist of mostly b- and y- ions. The CID fragmentation patterns for peptides that are unique to each subclass of IgG were also examined including FNWYVDGVEVHNAK for IgG1, CCVECPPCPAPPVAGPSVFLFPPKPK for IgG2, WYVDGVEVHNAK for IgG3 and TTPPVLDSDGSFFLYSR for IgG4. As shown in Figure SI-1 in the supplementary information, mostly b- and y- ions were observed from the tandem MS of these peptides.
Figure 1.
Representative Q-TOF tandem mass spectra of peptides and glycopeptides. (a) MS/MS spectrum of the peptide DTLMISR. (b) MS/MS spectrum of glycopeptide Hex5HexNAc4Fuc1Neu5Ac1-EEQYNSTYR from IgG1 (c) MS/MS spectrum of Hex3HexNAc4Fuc1- EEQFNSTFR from IgG2. (
 ) N-acetylglucosamine (
) N-acetylglucosamine (
 ) mannose (
) mannose (
 ) galactose (○) hexose (
) galactose (○) hexose (
 ) fucose (
) fucose (
 ) N-acetyl neuraminic acid
) N-acetyl neuraminic acid
Nano-flow LC-MS analysis with the Q-TOF instrument allowed identification of the glycopeptides based on the accurate mass and the tandem MS. The fragmentation spectra contained the typical glycan fragment ions, the so-called “oxonium ions” corresponding to the small glycan fragments, that have been observed previously by this group40–41 and others,33, 42–43 including m/z 204.08 (HexNAc), 366.14 (Hex1HexNAc1), 292.09 (Neu5Ac) and 657.24 (Hex1HexNAc1Neu5Ac1). The tandem mass spectra of two of the glycopeptides (Hex5HexNAc4Fuc1NeuAc1-IgG1 and Hex3HexNAc4Fuc1-IgG2) are shown in Figure 1(b, c), and the predicted glycan fragments are clearly visible with high intensities. As listed in Table 1, 26 glycopeptides compositions were readily identified from the IgG standard. These results are in good correspondence with literature.24, 31, 33, 44 Other studies have reported six sialylated glycopeptides for the IgG Fc region,33, 44 of which four have very low abundances.
Table 1.
MRM transitions used to monitor glycopeptides.
| Compound Namea | Precursor Ion (m/z) | Product Ion (m/z) | Collision Energy (eV) | Ret. Time (min) | Delta Ret. Timeb (min) |
|---|---|---|---|---|---|
| H3N5F1-IgG1 | 946.5 | 204.1 | 15 | 2.2 | 0.6 |
| H4N4-IgG1 | 884.1 | 204.1 | 14 | 2.2 | 0.6 |
| H4N4F1-IgG1 | 932.8 | 204.1 | 15 | 2.2 | 0.6 |
| H3N4F1-IgG1 | 878.8 | 204.1 | 13 | 2.25 | 0.6 |
| H5N4-IgG1 | 938.1 | 366.1 | 14 | 2.25 | 0.8 |
| H5N4F1-IgG1 | 986.8 | 366.1 | 15 | 2.25 | 0.8 |
| H4N5-IgG1 | 951.7 | 204.1 | 15 | 2.3 | 0.8 |
| H4N5F1-IgG1 | 1000.5 | 204.1 | 16 | 2.3 | 0.8 |
| H5N5F1-IgG1 | 1054.5 | 366.1 | 17 | 2.3 | 0.6 |
| H4N4F1S1-IgG1 | 1029.8 | 204.1 | 16 | 2.6 | 0.6 |
| H5N4F1S1-IgG1 | 1083.8 | 366.1 | 17 | 2.6 | 0.6 |
| H3N4F1-IgG3/4 | 873.4 | 204.1 | 13 | 3.65 | 0.6 |
| H4N4F1-IgG3/4 | 927.4 | 204.1 | 14 | 3.65 | 0.6 |
| H3N5F1-IgG3/4 | 941.1 | 204.1 | 15 | 3.68 | 0.6 |
| H4N5F1-IgG3/4 | 995.1 | 204.1 | 15 | 3.7 | 0.6 |
| H5N4F1S1-IgG3/4 | 1078.4 | 366.1 | 17 | 3.75 | 0.6 |
| H3N5F1-IgG2 | 935.8 | 204.1 | 14 | 4.1 | 0.6 |
| H4N4F1-IgG2 | 922.1 | 204.1 | 25 | 4.1 | 0.6 |
| H4N5F1-IgG2 | 989.9 | 204.1 | 15 | 4.1 | 0.6 |
| H5N4F1-IgG2 | 976.1 | 366.1 | 15 | 4.1 | 0.6 |
| H3N4F1-IgG2 | 868.1 | 204.1 | 13 | 4.15 | 0.6 |
| H4N5_IgG2 | 941.1 | 204.1 | 15 | 4.15 | 0.6 |
| H4N4-IgG2 | 873.4 | 204.1 | 13 | 4.15 | 0.6 |
| H5N4F1S1-IgG2 | 1073.1 | 366.1 | 25 | 4.2 | 0.6 |
| H5N5F1-IgG2 | 1043.8 | 366.1 | 17 | 4.2 | 1 |
| H4N4F1S1-IgG2 | 1019.1 | 204.1 | 25 | 4.25 | 0.6 |
| Peptide_IgG1234 (DTLMISR) | 418.2 | 506.3 619.4 |
9 9 |
4.5 4.5 |
0.5 0.5 |
| Peptide IgG3 (WYVDGVEVHNAK) | 472.9 | 697.4 534.3 |
6 6 |
4.7 4.7 |
0.5 0.5 |
| Peptide IgG1 (FNWYVDGVEVHNAK) | 839.4 | 968.5 1067.6 |
30 30 |
5.35 5.35 |
0.6 0.6 |
| Peptide IgG2 CCVECPPCPAPPVAGPSVFLFPPKPK |
970.1 | 1100.6 839.5 |
28 28 |
6.06 6.06 |
0.8 0.8 |
| Peptide IgG4 (TTPPVLDSDGSFFLYSR) | 635.0 | 1217.6 425.2 |
9 9 |
6.2 6.2 |
0.6 0.6 |
H: hexose; N: HexNAc; F: fucose; S: N-acetyl neuraminic acid.
Dynamic MRM was used. Delta retention time is the retention time window for the target transition.
IgG quantitation using MRM
Quantitation of proteins
It should be noted that the choice of the peptides for quantitation has several requirements. First, the peptides should be unique, i.e. they should not be present in other serum proteins. Second, peptides that contain post-translational modifications (PTMs), such as phosphorylation, methylation, etc., should be avoided to limit variation across samples. Third, peptides should not contain amino acids that are found to undergo deamination or oxidation because these modifications are normally incomplete and can vary across experiments. Some amino acids are found to undergo modifications more easily than others. Examples include glutamine and tryptophan, which deaminates45 and oxidizes,46 respectively. Peptides containing these amino acids must be carefully examined by performing extensive repeatability studies to ensure that they are not modified biologically or during the processing. In this study, only peptides with high repeatability characteristics were used for quantitation.
From the fragmentation patterns obtained with the LC-Q-TOF-MS/MS, MRM transitions were developed for the peptides. The IgG peptide common to all four types and selected for quantitation is DTLMISR. The MRM transition from the quasimolecular ion ([M+2H]2+ m/z 418.3) to fragment ion m/z 506.3 was selected as the quantifier, while a second transition to fragment ion m/z 619.4 was used as qualifier. The optimized fragmentation voltage was determined to be 9 eV for the quantifier. For the subclass-specific peptides the following transitions were determined to be optimal: ([M+2H]2+ 839.4 → m/z 968.5 and m/z 1067.6) for IgG1, ([M+3H]3+ 970.1 → m/z 1100.6 and m/z 839.5) for IgG2, ([M+3H]3+ 472.9 → m/z 697.4 and m/z 534.3) for IgG3 and ([M+3H]3+ 635.0 → m/z 1217.6 and m/z 425.2) for IgG4. The MRM transitions for all peptides, together with their respective fragmentation voltages are listed in Table 1.
Quantitation of glycopeptides
Tandem MS of glycopeptides yielded both m/z 204.8 and m/z 366.14. MRM transitions were developed from the quasimolecular ions to either fragments depending on which was more abundant. The MRM transitions for all the glycopeptides monitored are shown in Table 1. Glycopeptides from IgG3 and IgG4 could not be distinguished, as their peptide moieties (IgG3: EEQYNSTFR, IgG4: EEQFNSTRY) share the same amino acid composition and therefore identical masses. However, IgG1 and IgG2 yielded distinct glycopeptides and could thus be monitored individually.
MRM is a non-scanning technique, where each transition is detected individually, and the detection of multiple transitions occurs concurrently in duty cycles. Important parameters in the MRM method are therefore the cycle time, which is the time spent monitoring all transitions in one duty cycle, and the dwell time, which is the time spent acquiring a specific transition during each duty cycle. Increasing the cycle time will result in limited sampling and thus poor data quality, while shorter dwell times would results in a poorer signal-to-noise ratio, especially for lower abundant analytes. Retention of glycopeptides on C18 stationary phases relies mainly on the peptide moiety of the glycoconjugates. Therefore, glycopeptides that originate from the same site and thus share the same peptide generally elute closely together. As a result, a large number of concurrent transitions will result in either longer cycle time and thus lower frequency of data points or shorter dwell time. To reduce the number of concurrent transitions in our experiments, only one transition was monitored for each glycopeptide. Monitoring just one transition for the glycopeptides provides enough information, as the transition to the oxonium ion both identifies the compound as a glycopeptide, and provides good quantitation. Moreover, a dynamic MRM method was applied to further reduce the number of concurrent transitions. In dynamic MRM, a specific analyte is only monitored at the time it elutes, which greatly reduces the number of concurrent transitions and saves the duty cycle only for the co-eluting compounds. In our experiment, the cycle time was fixed at 500 ms to ensure enough data points per compound and obtain similar sampling across analytes, but the dwell time was variable (32ms – 499 ms) based on the number of concurrent transitions.
Reverse phase-ultra high pressure liquid chromatography (RP-UPLC)
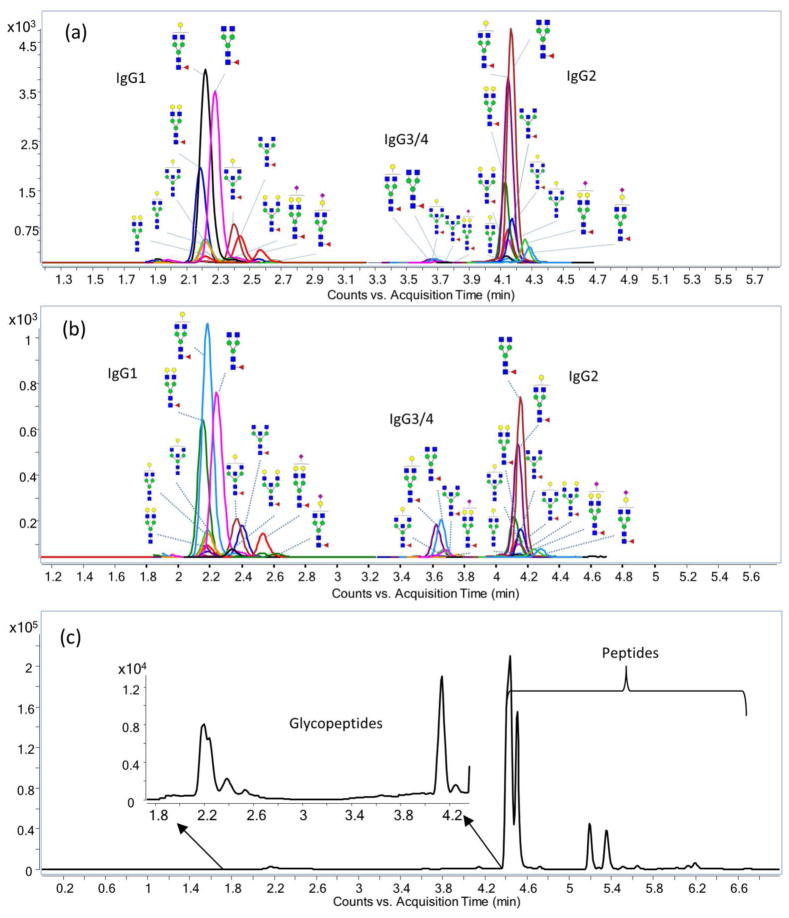
UPLC separation is significantly faster than standard HPLC. An example chromatogram obtained from the MRM transitions is shown in Figure 2. Good separation was obtained with total run times as low as 10 minutes. IgG glycopeptides elute earlier than most of the peptides (Figure 2), which is advantageous, as it reduces the charge competition during electrospray ionization resulting in higher glycopeptide sensitivity. Peptides ionize more easily than glycopeptides47–48 and will suppress their signal when they elute simultaneously.
Figure 2.
Total MRM chromatogram for IgG using UPLC-C18 chromatography. MRM chromatograms for 26 glycopeptides from (a) tryptic digested IgG standard and (b) tryptic digested pooled serum and (c) total MRM chromatogram for tryptic peptides from pooled serum. The MRM transitions are shown in Table 1. One MRM transition was monitored for each glycopeptide; two MRM transitions were monitored for each peptide.
IgG1 glycopeptides were all eluted at 2.2 min, followed by IgG3/4 glycopeptides at 3.6 min and then IgG2 glycopeptides at 4.2 min. Generally, the sialylated glycopeptides show slightly greater retention on the C18, resulting in somewhat later elution, however the retention was influenced mostly by the peptide moiety. This elution pattern is in accordance with previous literature.33 Generally, no glycan isomer separation was observed using a 10- minute gradient.
Quantitation of protein glycosylation
Normalized glycoform abundances
Quantitation of protein glycosylation is currently limited to relative quantitation due to the lack of glycan/glycopeptide standards. Absolute ion abundances are used to obtain relative quantitative information.47, 49–50 Absolute ion abundances, however, are greatly affected by protein concentration. Comparing glycoforms between samples yields changes in both glycan and protein abundances. To determine changes in glycosylation at the protein specific level, it is necessary to monitor protein concentration. We developed a method to monitor the absolute protein abundances of each IgG subclass as well as glycopeptides for each subclass. Here, the glycopeptide signals were normalized to the protein abundances to separate out the contribution of protein concentration (Equation shown below).
| eq (2) |
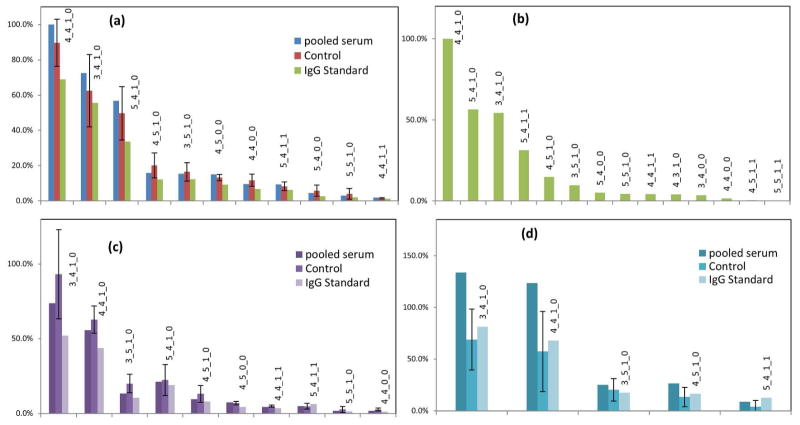
This method allows normalization to IgG1 and IgG2. Glycopeptides from IgG3 and IgG4 could not be distinguished, thus the signals were normalized to the sum of the two peptides from IgG3 and IgG4. This strategy allows both the determination of the glycoprotein concentration, as well as the absolute and relative abundances of the different glycoforms in a given sample. The normalized glycan intensities for IgG 1, 2 and 3/4 are shown in Figure 3a, 3c and 3d, respectively.
Figure 3.
Normalized abundance of IgG glycopeptides. (a) IgG1 glycopeptide MRM signal normalized to IgG1 peptide (FNWYVDGVEVHNAK) and ranked by the order of relative abundance. (b) IgG1 glycopeptide profiling using nano-chip-ESI-TOF-MS for the IgG standard. (c) IgG2 glycopeptide normalized to IgG2 peptides (CCVECPPCPAPPVAGPSVFLFPPKPK). (d) IgG3/4 glycopeptide normalized to IgG3 (WYVDGVEVHNAK) and IgG4 (TTPPVLDSDGSFFLYSR) peptides. The glycoform compositions are annotated above the histogram. For example, 4_4_1_1 corresponds to Hex4HexNAc4Fuc1Neu5Ac1.
Comparison of abundances across glycoforms using MRM
The lack of glycopeptide standards hampers relative comparison between glycoforms as their response factors may differ due to the differences in ionization efficiency and fragmentation efficiency. Recently, it was reported that the ionization efficiencies of different glycoforms with the same peptide moiety are very similar in ESI.51 Thus, the major concern for relative comparisons using MRM is the differences in fragmentation efficiencies. Direct mass spectrometry using, for example, LCESI- TOF instrument is expected to provide more accurate relative quantitation compared to a MRM when comparing different ionic species, because it is not dependent on fragmentation. Indeed the technology is widely used to perform relative quantitation of different compounds.25, 44, 52 We therefore compared the IgG glycopeptide abundances obtained from MRM using a UPLC-QqQ-MS with the abundances obtained from direct MS scans using a nano- LC-Chip-Q-TOF to assess the utility of MRM for quantitation of different glycoforms. Figure 3(a, b) shows that both QTOF and QqQ yield similar relative abundances that very closely resembles the distribution observed by MALDI-TOF-MS.31 That is, glycoforms that are observed with high abundances in the ESI-QTOF and the MALDI-TOF also produce strong MRM signals in the QqQ. Furthermore, the relative distribution appears very similar suggesting that it may be possible to compare abundances between different IgG glycoforms through their MRM signals.
Repeatability of the method
To evaluate the intra-day repeatability of the MRM quantitation, 1.0 μL of a tryptic digest of the IgG standard was injected three times. The results are depicted in Figure SI-2(a, b) of the supplementary information. Clearly, the peak areas of both the peptides as well as the glycopeptides were highly repeatable, with a RSD lower than 10%.
The inter-day repeatability of the instrument response was evaluated by analyzing the same sample (stored at −80°C) once per week for a total period of three weeks. The results depicted in Figure SI-2(c, d) of the supplementary information show high inter-day instrument repeatability for the glycopeptide (RSD=8.1%). However, for the peptide NQVSLTCLVK, a higher standard deviation was observed (RSD=25.5%). The larger variation in this peptide was found to be due to the deamination of the glutamine residue. As a result, this peptide was not chosen for protein quantitation. Studies of the peptides used for quantitation showed better inter-day instrument repeatability (RSD 3%–14%, data not shown).
Tryptic digestion is based on the enzymatic activity of the trypsin, and it is widely known that different degrees of amino acid modification and missed cleavages during enzyme digestion may generate different tryptic peptide profiles. Such variations in peptide sequence will decrease the repeatability and accuracy of the MRM quantification. Therefore, the interday and intra-day repeatability of the trypsin digestion were assessed using the IgG standard. Results for the analysis of three samples that were digested on the same day are depicted in Figure SI-2(e, f) of the supplementary information. A high intra-day repeatability of less than 6% was observed, even for peptides which were found to have modifications, such as NQVSLTCLVK. However, as depicted in Table SI-1, tryptic digestions performed on different days are somewhat less consistent; while most peptides show inter-day RSDs of less than 6%, some peptides have clearly higher RSDs. The larger variation is likely due to incomplete trypsin digestion and different degrees of modifications such as deamination and oxidation. Since only one peptide is needed for quantitation, it was chosen to monitor peptides with high intra- and inter-day repeatability. Interestingly, the glycopeptide responses were generally very repeatable in both intra-day and inter-day experiments.
Application
Direct analysis of IgG and its glycosylation pattern from human serum
While the method development was performed on an IgG standard sample, glycosylation analyses of IgG are necessarily performed with biofluids such as serum or plasma. Currently, capturing methods using protein A or protein G are used to enrich IgG prior to analysis;29, 31 however, with the targeted MRM strategy these methods may not be necessary. Indeed, when a tryptic digest of serum was analyzed directly using our optimized method, all 5 peptides and 26 glycopeptides were readily observed as shown in Figure 2(b, c).
To determine the absolute amount of IgG in the pooled serum sample, the peak area of the peptide from the Fc region and common to all four IgG subclasses was used. A serial dilution of the tryptic digest of the IgG standard was made to concentrations of 9.00 × 10−3 μg/mL, 9.00 × 10−2 μg/mL, 9.00 × 10−1 μg/mL, 9.00 μg/mL, 18.0 μg/mL 90.0 μg/mL, 1.80 × 102 μg/mL, 4.50 × 102 μg/mL, and 9.00 × 102 μg/mL, and 1.0 μL of each of the concentrations was injected. The resulting calibration curve is shown in Figure SI-3 and shows a dynamic range over 1000, a limit of detection (LOD) of 0.060 femtomoles, and a limit of quantitation (LOQ) of 0.60 femtomoles. The calibration was fitted linearly with an R2 of 1.000 for concentrations from 0.901 μg/mL to 4.50 × 102 μg/mL and the absolute amount of IgG in the pooled serum was determined to be 10.1±0.2 mg/mL. The glycosylation profile of the pooled serum sample was also determined, and could be shown to be similar to the glycosylation profile of the IgG standard in Figure 2(a, b).
To assess the effects of the tryptic digestion on the IgG quantitation, different digestion times and enzyme amounts were evaluated, and it was observed that 1 μg of trypsin is sufficient to digest 2 μL of pooled serum in 18 hours (data not shown). Furthermore, no difference was observed by increasing the alkylation time from 30min to 60min.
To study the matrix effects of the serum on the absolute IgG quantitation, a standard addition experiment was performed. Equal portions of tryptic digest of pooled serum and IgG standard solutions (18.0 μg/mL, 90.0 μg/mL, and 1.80 × 102 μg/mL, respectively) were combined and analyzed using LC-QqQ-MS. Figure SI-4 of the supplemental information shows the linear regression of the experiment. From the standard addition experiment, the IgG concentration of the pooled serum was determined to be 11.5±0.5 mg/mL, while the original result was 10.1±0.2 mg/mL. The result shows a high recovery rate of 87.8%, indicating that the matrix effects from serum are limited. Better recoveries may still be obtained using a longer HPLC gradient to reduce ion suppression due to the co-eluting interfering ions; however this will substantially increase the analysis time. We decided therefore to apply the shorter analysis time to the serum samples.
Application to samples from ten healthy individuals
To evaluate the variation in absolute IgG concentration as well as the relative glycosylation profile in the general population, the optimized method was applied to ten serum samples from healthy individuals. First, the absolute concentrations of IgG in the sera were determined, and the results are shown in Table SI-2. The average concentration of IgG in serum was 10.6 mg/mL with a standard deviation of 3.1 mg/mL. The observed concentrations are in the same range as those previously reported.53–54 There is a relatively large variation between individuals (RSD=29%), which further supports the need for considering protein concentrations in the glycosylation analyses.
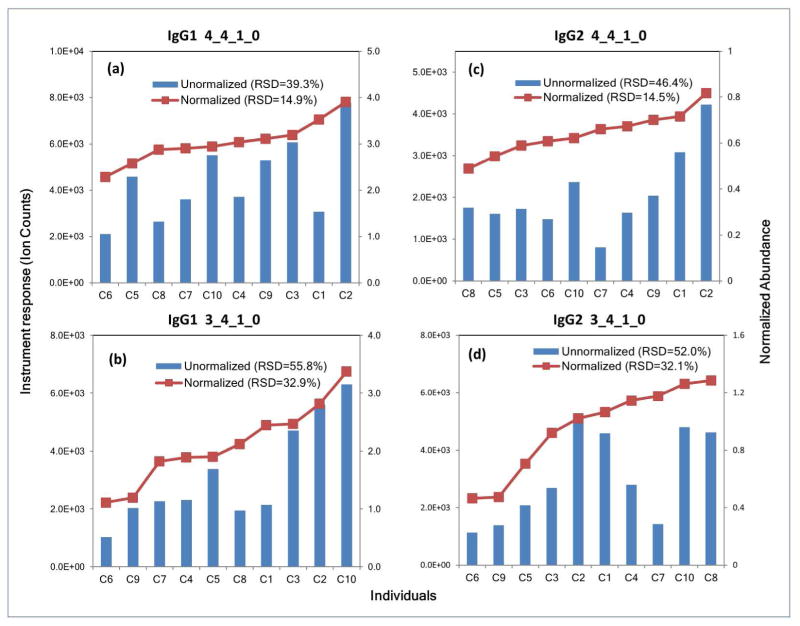
We then measured the glycopeptides of the ten individuals. The results of two specific glycoforms (Hex4HexNAc4Fuc1 and Hex3HexNAc4Fuc1) on two subtypes (IgG1 and IgG2) are shown in Figure 4, while the result for all other glycoforms monitored are depicted in Figure SI-5 in the supplementary information. For most glycans, the variation in the glycan abundances, here representing the inter-individual differences, is significant when using the absolute glycopeptide ion abundance. However, the variation often becomes smaller when the glycopeptides are normalized to the protein content suggesting that protein concentration contributes considerably to the variation in glycopeptide abundance. It also has to be noted that the normalized and unnormalized abundance may show completely different trends for some of the samples, suggesting that the true effects of the changes in glycosylation may be attenuated by the absolute glycopeptide (and hence protein) abundances.
Figure 4.
Normalized (red squares) and unnormalized (blue bars) glycopeptide abundances of 4 IgG glycopeptides monitored in the sera of individuals. Normalization was performed using equation 2. (a) IgG1 glycopeptide, Hex4HexNAc4Fuc1-EEQYNSTYR (b) IgG1 glycopeptide, Hex3HexNAc4Fuc1-EEQYNSTYR (c) IgG2 glycopeptide, Hex4HexNAc4Fuc1-EEQFNSTFR (d) IgG2 glycopeptide, Hex3HexNAc4Fuc1-EEQFNSTFR. For each glycoform, the normalized trends in abundances do not directly match the unnormalized ones.
Conclusion
We conducted a systematic evaluation of multiple reaction monitoring for the quantitation and glycoprofiling of immunoglobulin G, one of the most abundant proteins in serum. It was observed that MRM is a powerful technique for the absolute quantitation of immunoglobulin G and the site-specific glycosylation quantitation relative to the protein content. The selectivity of the technique allows for the direct analysis of IgG in serum, without purification. The sensitivity of the MRM method allows detection of IgG at the attomole level. The method facilitates simultaneous determination of IgG protein content as well as glycan profiles, which may be normalized to the protein content. It is anticipated that this method will facilitate further studies towards protein- and site- specific glycosylation patterns, whether for protein characterization or biomarker discovery studies.
Supplementary Material
Acknowledgments
The authors are thankful for the funding provided by the National Institutes of Health (RO1 CA136647, R01HD061923, R01GM049077)
References
- 1.Lange V, Picotti P, Domon B, Aebersold R. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li AC, Alton D, Bryant MS, Shou WZ. Rapid Commun Mass Spectrom. 2005;19:1943–1950. doi: 10.1002/rcm.2008. [DOI] [PubMed] [Google Scholar]
- 3.Xiao JF, Zhou B, Ressom HW. TrAC Trends in Analytical Chemistry. 2012;32:1–14. doi: 10.1016/j.trac.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. J Chromatogr, B. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Gallien S, Duriez E, Domon B. J Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 6.Apweiler R, Hermjakob H, Sharon N. Biochimica et Biophysica Acta (BBA) - General Subjects. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 7.Kellokumpu S, Sormunen R, Kellokumpu I. FEBS Lett. 2002;516:217–224. doi: 10.1016/s0014-5793(02)02535-8. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko Y, Nimmerjahn F, Ravetch JV. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 9.Londrigan SL, Turville SG, Tate MD, Deng YM, Brooks AG, Reading PC. J Virol. 2011;85:2990–3000. doi: 10.1128/JVI.01705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y, Aglipay JA, Bernstein JD, Goswami S, Stanley P. Cancer Res. 2010;70:3361–3371. doi: 10.1158/0008-5472.CAN-09-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakomori S-i, Cummings R. Glycoconjugate J. 2012;29:565–566. doi: 10.1007/s10719-012-9448-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Qiu W, Simeone DM, Lubman DM. J Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 13.Hashii N, Kawasaki N, Itoh S, Nakajima Y, Kawanishi T, Yamaguchi T. Immunology. 2009;126:336–345. doi: 10.1111/j.1365-2567.2008.02898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. Mol Cell Proteomics. 2011;10:M110 002717. doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillard M, Morava E, van Delft FL, Hague R, Köner C, Adamowicz M, Wevers RA, Lefeber DJ. Clinical Chemistry. 2011;57:593–602. doi: 10.1373/clinchem.2010.153635. [DOI] [PubMed] [Google Scholar]
- 16.Ueda K, Takami S, Saichi N, Daigo Y, Ishikawa N, Kohno N, Katsumata M, Yamane A, Ota M, Sato TA, Nakamura Y, Nakagawa H. Mol Cell Proteomics. 2010;9:1819–1828. doi: 10.1074/mcp.2010/000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papac DI, Briggs JB, Chin ET, Jones AJS. Glycobiology. 1998;8:445–454. doi: 10.1093/glycob/8.5.445. [DOI] [PubMed] [Google Scholar]
- 18.Thobhani S, Yuen CT, Bailey MJA, Jones C. Glycobiology. 2009;19:201–211. doi: 10.1093/glycob/cwn099. [DOI] [PubMed] [Google Scholar]
- 19.Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Anal Bioanal Chem. 2010;397:3457–3481. doi: 10.1007/s00216-010-3532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pucic M, Knezevic A, Vidic J, Adamczyk B, Novokmet M, Polasek O, Gornik O, Supraha-Goreta S, Wormald MR, Redzic I, Campbell H, Wright A, Hastie ND, Wilson JF, Rudan I, Wuhrer M, Rudd PM, Josic D, Lauc G. Mol Cell Proteomics. 2011;10:M111 010090. doi: 10.1074/mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Axford JS. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1999;1455:219–229. doi: 10.1016/s0925-4439(99)00057-5. [DOI] [PubMed] [Google Scholar]
- 22.Ruhaak LR, Uh HW, Beekman M, Hokke CH, Westendorp RGJ, Houwing-Duistermaat J, Wuhrer M, Deelder AM, Slagboom PE. J Proteome Res. 2010;10:1667–1674. doi: 10.1021/pr1009959. [DOI] [PubMed] [Google Scholar]
- 23.Kolarich D, Altmann F. Anal Biochem. 2000;285:64–75. doi: 10.1006/abio.2000.4737. [DOI] [PubMed] [Google Scholar]
- 24.Harazono A, Kawasaki N, Itoh S, Hashii N, Matsuishi-Nakajima Y, Kawanishi T, Yamaguchi T. J Chromatogr B. 2008;869:20–30. doi: 10.1016/j.jchromb.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Mimura Y, Ashton PR, Takahashi N, Harvey DJ, Jefferis R. J Immunol Methods. 2007;326:116–126. doi: 10.1016/j.jim.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Balog CI, Stavenhagen K, Koeleman CA, Scherer HU, Selman MH, Deelder AM, Huizinga TW, Toes RE, Wuhrer M. Mol Cell Proteomics. 2011;10:M110 004655. doi: 10.1074/mcp.M110.004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauko L, Nordborg A, Hutchinson JP, Lacher NA, Hilder EF, Haddad PR. Anal Biochem. 2011;408:235–241. doi: 10.1016/j.ab.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 28.Wacker C, Berger CN, Girard P, Meier R. European Journal of Pharmaceutics and Biopharmaceutics. 2011;79:503–507. doi: 10.1016/j.ejpb.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Bakovic MP, Selman MH, Hoffmann M, Rudan I, Campbell H, Deelder AM, Lauc G, Wuhrer M. J Proteome Res. 2013;12:821–831. doi: 10.1021/pr300887z. [DOI] [PubMed] [Google Scholar]
- 30.Scherer HU, Wang J, Toes REM, van der Woude D, Koeleman CAM, de Boer AR, Huizinga TWJ, Deelder AM, Wuhrer M. PROTEOMICS – Clinical Applications. 2009;3:106–115. doi: 10.1002/prca.200800098. [DOI] [PubMed] [Google Scholar]
- 31.Selman MHJ, McDonnell LA, Palmblad M, Ruhaak LR, Deelder AM, Wuhrer M. Anal Chem. 2010;82:1073–1081. doi: 10.1021/ac9024413. [DOI] [PubMed] [Google Scholar]
- 32.Wuhrer M, Hokke CH, Deelder AM. Rapid Commun Mass Spectrom. 2004;18:1741–1748. doi: 10.1002/rcm.1546. [DOI] [PubMed] [Google Scholar]
- 33.Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, Verrips CT, Dolhain RJ, Hokke CH, Deelder AM. Proteomics. 2007;7:4070–4081. doi: 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- 34.Ruhaak LR, Uh HW, Beekman M, Koeleman CAM, Hokke CH, Westendorp RGJ, Wuhrer M, Houwing-Duistermaat JJ, Slagboom PE, Deelder AM. PLoS One. 2010;5:e12566. doi: 10.1371/journal.pone.0012566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kam RKT, Poon TCW, Chan HLY, Wong N, Hui AY, Sung JJY. Clinical Chemistry. 2007;53:1254–1263. doi: 10.1373/clinchem.2007.085563. [DOI] [PubMed] [Google Scholar]
- 36.Kurogochi M, Matsushista T, Amano M, Furukawa J-i, Shinohara Y, Aoshima M, Nishimura S-I. Mol Cell Proteomics. 2010;9:2354–2368. doi: 10.1074/mcp.M110.000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Jia W, Wang J, Ying W, Zhang Y, Qian X. Anal Chem. 2011;83:8802–8809. doi: 10.1021/ac201676a. [DOI] [PubMed] [Google Scholar]
- 38.Song E, Pyreddy S, Mechref Y. Rapid Commun Mass Spectrom. 2012;26:1941–1954. doi: 10.1002/rcm.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, German JB, Lebrilla CB. J Proteome Res. 2012;11:2912–2924. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German BJ, Lebrilla CB. J Proteome Res. 2011;10:2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conboy JJ, Henion JD. J Am Soc Mass Spectrom. 1992;3:804–814. doi: 10.1016/1044-0305(92)80003-4. [DOI] [PubMed] [Google Scholar]
- 43.Wuhrer M, Deelder AM, van der Burgt YEM. Mass Spectrom Rev. 2011;30:664–680. doi: 10.1002/mas.20337. [DOI] [PubMed] [Google Scholar]
- 44.Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. Proteomics. 2008;8:2858–2871. doi: 10.1002/pmic.200700968. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Go EP, Desaire H. Anal Chem. 2008;80:3144–3158. doi: 10.1021/ac702081a. [DOI] [PubMed] [Google Scholar]
- 46.Thiede B, Lamer S, Mattow J, Siejak F, Dimmler C, Rudel T, Jungblut PR. Rapid Commun Mass Spectrom. 2000;14:496–502. doi: 10.1002/(SICI)1097-0231(20000331)14:6<496::AID-RCM899>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 47.Roth Z, Yehezkel G, Khalaila I. International Journal of Carbohydrate Chemistry. 2012;2012:1–10. [Google Scholar]
- 48.Rebecchi KR, Wenke JL, Go EP, Desaire H. J Am Soc Mass Spectrom. 2009;20:1048–1059. doi: 10.1016/j.jasms.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Kronewitter SR, de Leoz MLA, Peacock KS, McBride KR, An HJ, Miyamoto S, Leiserowitz GS, Lebrilla CB. J Proteome Res. 2010;9:4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harvey DJ, Royle L, Radcliffe CM, Rudd PM, Dwek RA. Anal Biochem. 2008;376:44–60. doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 51.Sinha S, Pipes G, Topp EM, Bondarenko PV, Treuheit MJ, Gadgil HS. J Am Soc Mass Spectrom. 2008;19:1643–1654. doi: 10.1016/j.jasms.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Selman MHJ, Derks RJE, Bondt A, Palmblad M, Schoenmaker B, Koeleman CAM, van de Geijn FE, Dolhain RJEM, Deelder AM, Wuhrer M. Journal of Proteomics. 2012;75:1318–1329. doi: 10.1016/j.jprot.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Fahey JL, McKelvey EM. The Journal of Immunology. 1965;94:84–90. [PubMed] [Google Scholar]
- 54.Stoop JW, Zegers BJ, Sander PC, Ballieux RE. Clin Exp Immunol. 1969;4:101–112. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.