Abstract
The present study investigated the induction and possible role of activating transcription factor 3 (ATF3) in the corpus luteum. Postpubertal cattle were treated at midcycle with prostaglandin F2α(PGF) for 0–4 hours. Luteal tissue was processed for immunohistochemistry, in situ hybridization, and isolation of protein and RNA. Ovaries were also collected from midluteal phase and first-trimester pregnant cows. Luteal cells were prepared and sorted by centrifugal elutriation to obtain purified small (SLCs) and large luteal cells (LLCs). Real-time PCR and in situ hybridization showed that ATF3 mRNA increased within 1 hour of PGF treatment in vivo. Western blot and immunohistochemistry demonstrated that ATF3 protein was expressed in the nuclei of LLC within 1 hour and was maintained for at least 4 hours. PGF treatment in vitro increased ATF3 expression only in LLC, whereas TNF induced ATF3 in both SLCs and LLCs. PGF stimulated concentration- and time-dependent increases in ATF3 and phosphorylation of MAPKs in LLCs. Combinations of MAPK inhibitors suppressed ATF3 expression in LLCs. Adenoviral-mediated expression of ATF3 inhibited LH-stimulated cAMP response element reporter luciferase activity and progesterone production in LLCs and SLCs but did not alter cell viability or change the expression or activity of key regulators of progesterone synthesis. In conclusion, the action of PGF in LLCs is associated with the rapid activation of stress-activated protein kinases and the induction of ATF3, which may contribute to the reduction in steroid synthesis during luteal regression. ATF3 appears to affect gonadotropin-stimulated progesterone secretion at a step or steps downstream of PKA signaling and before cholesterol conversion to progesterone.
The corpus luteum, a transient endocrine gland formed from the ovarian follicle after ovulation, plays an important role in the regulation of normal reproductive cycles and in the maintenance of pregnancy. In the absence of pregnancy, the corpus luteum undergoes the degenerative process of regression or luteolysis. As a result of the luteolytic process, a new reproductive cycle ensues. Luteolysis is characterized by the rapid cessation of progesterone production (functional regression) followed by involution of the corpus luteum glandular structure via apoptotic death (structural regression) (1, 2). Prostaglandin F2α (PGF), a major luteolytic factor, was identified more than 40 years ago in mammals (3), but the cellular mechanisms by which PGF initiates the luteolytic cascade remain poorly understood.
Luteinized follicular cells acquire the capacity to respond to PGF through increased expression of the Gq-coupled prostaglandin F receptor (PTGFR). It is well known that activation of the PTGFR rapidly induces calcium mobilization and activation of protein kinase C (PKC) (4). These initial events lead to activation of MAPK signaling cascades including ERK1/2 (5, 6), p38, and c-Jun N-terminal kinase (JNK) (7, 8) in vivo and in vitro, with subsequent activation of multiple transcription factors. These signaling pathways, particularly activated MAPK signaling, are involved in inducing a rapid and dramatic increase in the immediate early genes such as FOS and JUN (9, 10), NR4A1 (11, 12), and EGR1 (11, 13) in the corpus luteum. Recent studies demonstrate that PGF treatment induces global changes in gene expression in corpus luteum, including immediate early genes, steroidogenic genes, prostaglandin-related genes, immune-related genes, and angiogenesis-related genes (11, 14–18).
Activating transcription factor 3 (ATF3) is a member of the activating transcription factor/cAMP responsive element binding (CREB) protein family of transcription factors. The basal level of ATF3 mRNA is typically low or undetectable in most tissues and cell lines, but greatly increases under stress conditions, and is considered to be an adaptive-response gene (19, 20). Extracellular signals including cytokines, chemokines, growth factors/hormones, or DNA damage can induce ATF3 expression (21). This protein binds the cAMP response element (CRE) and may repress transcription by stabilizing the binding of inhibitory cofactors at the promoter (19, 22). Target genes of ATF3 have been reported to associate with metabolism (23–25), differentiation (26), inflammation (27, 28), survival, and apoptosis (29, 30) in multiple cell types. ATF3 is one of the immediate-early response genes in vascular endothelial cells in response to atherogenic stimuli and may play a role in the endothelial cell death (31, 32). ATF3 has been implicated as a regulator in host defense against invading pathogens and as a tumor suppressor in various forms of cancer (22, 33).
Mondal et al (34) found that PGF treatment increased ATF3 protein expression in the bovine corpus luteum, rendering the hypothesis that ATF3 may induce changes in luteal function associated with luteolysis. Many signaling pathways have been demonstrated to be involved in the induction of ATF3 by stress signals, including the JNK, p38, nuclear factor-κB (NFκB), PKC, and calcium signal pathways (21). However, the mechanism of induction and the function of ATF3 in the corpus luteum are unknown. In this report, we found that PGF induced ATF3 mRNA and protein expression in corpora lutea from midcycle cows in vivo and in luteal cells isolated from both midcycle and pregnant corpora lutea in vitro. We found that PGF selectively induced a concentration- and time-dependent increase in ATF3 expression in large luteal cells (LLCs), whereas TNF-α stimulated ATF3 expression in small luteal cells (SLCs) and LLCs. To begin to understand the role of ATF3 in luteal cells, we expressed ATF3 using an adenoviral construct. We found that expression of ATF3 reduced LH-stimulated CRE-dependent transcription of a luciferase promoter reporter and progesterone synthesis in both SLCs and LLCs, but did not alter the viability of LLCs or SLCs. The induction of ATF3, therefore, may serve to modulate gonadotropin responsiveness during luteolysis.
Materials and Methods
Reagents
PGF, protease and phosphatase inhibitor cocktails, 22R-hydroxycholesterol (22R-OH-CH), 4′,6-diamidino-2-phenylindole, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), hydrogen peroxide, and hematoxylin were from Sigma. Human TNF-α and Fas ligand were from R&D. Bovine IGF-1 was from GroPep. The MEK1/2 inhibitor (U0126) was from Enzo. The p38 MAPK inhibitor (SB203580) and JNK inhibitor (SP600125) were from EMD Millipore. Bovine LH was purchased from Tucker Endocrine Research Institute. M199 was from Lonza. Fetal bovine serum (FBS) was obtained from Valley Biomedical. Penicillin-streptomycin and Gentamycin were from Gibco, and Amphotericin B was from MP Biomedical, LLC. Type II collagenase was obtained from Atlantic Biologicals. The unmasking solution, antirabbit VECTASTAIN ABC kit, 3,3′diaminobenzidine detection kit, and VectaMount were obtained from Vector Laboratories. All antibodies were from Cell Signaling Technology except for ATF3 (Santa Cruz Biotechnology), SR-B1 (Novus Biological; β-actin and antimouse horseradish peroxidase (HRP)-conjugated IgG (Sigma); steroidogenic acute regulatory protein (StAR) (abcam); CYP11A1 (Millipore); 3 β-hydroxysteroid dehydrogenase (HSD3B) (a gift from Dr. Ian Mason); vimentin (Millipore); and antirabbit HRP-conjugated IgG (Jackson ImmunoResearch Laboratories, Inc.). The bovine luteal endothelial cell line was prepared in our laboratory as previously described (35).
Bovine corpus luteum tissue
Postpubertal cattle of composite breeding at midluteal phase (days 9–10) were injected with saline (n = 3) or a luteolytic dose (25 mg) of the PGF analog Lutalyse (n = 9) (Upjohn Co). Ovaries were surgically removed from saline-injected controls (0 hours) or at 1, 2, and 4 hours after PGF injection. Luteal tissues were rapidly dissected from the ovary, sliced, and either snap-frozen in liquid nitrogen and stored at −80°C for protein and RNA, or fixed for immunohistochemistry and in situ hybridization. All procedures were approved by the University of Nebraska-Lincoln and University of Nebraska Medical Center Institutional Animal Care and Use Committees.
Isolation of steroidogenic luteal cells
Bovine ovaries were collected from first-trimester pregnant cows (fetal crown rump length < 10 cm) and midcycle cows as described by Ireland et al (36) at a local slaughterhouse (XL Four Star Beef, Inc.). Enriched steroidogenic cells were prepared from the luteal slices by enzymatic digestion with type II collagenase as described previously (37, 38).
SLCs and LLCs were separated using a Beckman Coulter Avanti J-20 XP centrifuge equipped with a Beckman JE-5.0 elutriator rotor. The mixed luteal cells from early pregnant corpus luteum were resuspended and subjected to centrifugal elutriation using elutriation medium (calcium-free DMEM (US Biological; catalog no. D9800–10), 25 mM HEPES, 0.1% BSA, 0.02 mg/mL deoxyribonuclease, 3.89 g/L sodium bicarbonate, 3 mg/mL glucose, and antibiotics). The eluates were collected with continuous flow. The viability, concentration, and size of cells in each fraction were determined using a hemocytomer and the trypan blue exclusion test. Cells with a diameter of 15–25 μm were classified as SLCs, and cells with a diameter greater than 30 μm were classified as LLCs. A 100 mL fraction (F1) containing predominantly erythrocytes and endothelial cells (ϕ< 10 μm) and a variable degree of SLCs was harvested using a flow rate of 16 mL/min at 1800 rpm. The following 100 mL fraction (F2) containing predominantly SLCs (ϕ10∼20 μm) was harvested using a flow rate of 16 mL/min at 1400 rpm. A third fraction (F3) containing small-cell clumps mixed with large cells was collected using a flow rate of 24 mL/min at 1200 rpm. The remaining fraction (F4), a highly enriched LLC fraction (ϕ> 30 μm) containing less than 5% enucleated cells, was collected using a flow rate of 30 mL/min at 680 rpm. The cells were then pelleted and resuspended in M199. Cells in F2 and F4 were used in experiments as reported in the figure legends. The average purity of SLCs in F2 and LLCs in F4 were 81.6 ± 2.78% and 76.9% ± 3.77% (n = 15).
Cell culture
The cells were seeded at a density of 1 × 105 cells/cm2 for mixed cells and SLCs, and a density of 4 × 104 cells/cm2 for LLCs. Cells were allowed to attach in a 5% CO2 incubator at 37°C in M199 medium, containing 5% FBS, 0.1% BSA, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 μg/mL Gentamycin, and 2.5 μg/mL Amphotericin B. The next day, media was removed and cells washed with PBS, after which cells were incubated in serum-free medium overnight. On the day of the experiment, the medium was replaced with fresh preequilibrated media, and cells were equilibrated for 2–3 hours before applying various treatments as described in the legends to the figures.
Real-time PCR
Total RNA was extracted from corpora lutea using the Absolutely RNA Miniprep Kit (Agilent Technologies) according to the manufacturer's directions. Quantification, purity, and integrity of the RNA were verified using a Nanodrop instrument (Nanodrop Technologies). cDNA was synthesized from 1 μg of total RNA using iScript Reverse Transcription Supermix (Bio-Rad Laboratories) with a final reaction volume of 20 μL following the manufacturer's recommendations. Primers were designed based on bovine ATF3 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA sequences using online primer 3.0 (www.SimGene.com). Quantitative real-time PCRs were carried out on the CFX96 Real-Time PCR Detection System (Bio-Rad) using 1 μL of prepared cDNA, 250 nM primers, and 10 μL of FAST SYBR Green Master Mix with a final reaction volume of 20 μL. Cycling conditions were as follows: 30 seconds at 95°C, and 40 cycles of 95°C for 5 seconds and 60°C for 5 seconds. The amount of specific mRNA was calculated from the linear portion of the amplification signal. Samples containing no RNA or RNA without reverse transcriptase were used as negative controls. The sequences for forward and reverse primers were as follows: ATF3 (NM_001046193.2): forward, 5′-AGCACCTCTGCCACCGGATGT-3′; reverse, 5′-CTTTCAGGGGCTACCTCGGCTTT-3′. GAPDH (NM_001034034.2): forward, 5′-TCTGCACCTTCTGCCGATG-3′; reverse, 5′-AGCAGTTGGTGGTGCAGGA-3′. A melting curve analysis was performed to ensure a single product was amplified from each primer pair at a temperature consistent with the expected amplification product.
In situ hybridization
Luteal tissue slices were quickly frozen in Optimal Cutting Temperature compound (OCT) on nitrogen liquid and stored at −80°C until sectioning. Tissues were sectioned 12 μm and placed on superfrost plus slides (VWR Scientific) and stored at −80°C. Tissue sections were sent to Affymetrix, Inc. to detect ATF3 and GAPDH gene expression by in situ hybridization using protocol QG ViewRNA-Fresh Frozen Tissue. Briefly, fresh frozen tissues samples were fixed for 16–18 hours in 4% formaldehyde at room temperature followed by protease digestion, target hybridization, and signal amplification in which labeled probe oligonucleotides conjugated to alkaline phosphate hybridize to each amplifier molecule. Then Fast Red Substrate was added to form a precipitate (red dots) that indicates the presence of the target RNA molecule. Finally, sections were counterstained with Gill's Hematoxylin.
Western blot analysis
For in vivo tissue samples, corpora lutea were weighed and then put into Pierce RIPA buffer (Thermo Scientific) with protease and phosphatase inhibitor cocktails and brought to the laboratory in dry ice. Tissues were then homologized with tissue homogenizer (OMNI) and were sonicated for five 2-second bursts with a cell homogenizer (Sonics & Materials Inc.). For in vitro samples, luteal cell cultures were harvested into lysis buffer (20 mM Tris, 1 mM EDTA, 0.2 mM EGTA, 150 mM NaCl, and 1% Triton X-100 and protease and phosphatase inhibitor cocktails). Cells were then lysed by sonication. Lysates were centrifuged at 18 350 × g for 15 minutes at 4°C, and the supernatant was collected for SDS-PAGE analysis. Protein concentrations were determined by protein assay dye reagent (Bio-Rad; catalog no.500–0006). Aliquots of each sample (40–50 μg of protein) were resolved on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. After transfer, the membranes were blocked in TBST (10 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20; pH 7.5) with 5% fat-free milk for 1 hour at room temperature. Membranes were incubated overnight with the primary antibody diluted in TBST with 5% nonfat milk or BSA at 4°C. After three 5-minutes washes with TBST, membranes were incubated for 1 hour at room temperature with antirabbit (1:20 000) or mouse (1:2000) HRP-conjugated IgG diluted in TBST with 5% nonfat milk. After three 5-minute washes, protein bands were detected with ECL reagent (Thermo Science or PerkinElmer). Signals were visualized using x-ray film or a Kodak Digital Sciences Image Station 440 (Eastman Kodak). Densitometry was performed using Image J software (http://rsbweb.nih.gov/ij/).
Immunohistochemistry
Corpora lutea were excised and fixed by immersion in 10% formalin for 24 hours and then changed into 70% ethanol until embedded in paraffin. Tissues were cut into 4-μm sections and mounted onto polylysine-coated slides. Slides were deparaffinized through 2 changes of xylene and through graded alcohols to water. Slides were microwaved in citrate-based unmasking solution (Vector Laboratories) for antigen retrieval. Endogenous peroxidase was quenched with 0.3% hydrogen peroxide in methanol for 30 minutes. Sections were incubated without (negative control) or with anti-ATF3 (1:500) and subsequently an antirabbit VECTASTAIN ABC Kit, and stained using a 3,3′diaminobenzidine detection kit. Slides were counterstained with hematoxylin, dehydrated through graded alcohols, and coverslipped with VectaMount.
Fluorescence immunocytochemistry
Bovine luteal cells were cultured in 4-well Lab-Tek glass chamber slides (Nalge Nunc International). After treatment with 100 nM PGF for 2 hours, the cells were washed with PBS and fixed with 4% paraformaldehyde for 15 minutes. Cells were then blocked with 10% donkey serum, 0.1%Triton X-100, and 0.1% sodium azide in PBS (pH 7.4) for 30 minutes at room temperature. Sections were then incubated with anti-ATF3 (1:100) at 4°C in a humidified chamber overnight. Immune reactive proteins were detected by incubating with a Texas red-X goat antirabbit antibody (1:200, red fluorescence) and Alexa Fluor 488-conjugated antimouse IgG (1:500, green fluorescence) for 1 hour at room temperature. Nuclei were stained simultaneously with 4′,6-diamidino-2-phenylindole (1:100, blue fluorescence). Sections were washed with PBS and then mounted with Vectashield. Two hours later, sections were examined under epifluorescence in an Olympus instruments equipped with a DP71 microscope digital camera (Olympus), and the images were captured using MicroSuite Basic Edition image analysis software (Olympus America).
Treatment with adenoviruses
The adenoviruses expressing ATF3 (Ad.ATF3) and β-galactosidase (Ad.GLB1) were prepared by Chris Wolford (Ohio State University, Columbus, Ohio) as previously described (29). Preliminary experiments established the efficiency of adenoviral transduction to be greater than 90%. Prior to adenovirus infection, the cells were preequilibrated with serum-free medium for 2 hours. Viral stocks were diluted in serum-free medium and were added to the cells for 1.5 hours incubation. Cells were incubated for an additional 20 hours with medium containing 10% FBS before further treatment. Initial experiments were performed to determine viral titers needed for ATF3 expression.
Cell viability assays
MTT was added to the media at a final concentration of 500 μg/mL and after 3 hours incubation, the MTT working solution was removed. DMSO was added to dissolve the converted dye and samples read at 570 nm using a SpectraMax Plus Spectrophotometer. Caspase 3/7 activity was detected using a kit (Promega Corp.) following the manufacturer's instructions and luminescence was detected using a FLUOstar OPTIMA (BMG LABTECH) plate reader. Cleaved-caspase 3 was detected by Western blot.
In order to determine whether expression of ATF3 affected cell survival signaling in response to IGF1, cells infected with Ad.ATF3 or Ad.GLB1 were treated with IGF-1 (50 ng/mL) for 15 and 30 minutes, and Western blot was used to detect the phosphorylation of AKT (Thr308) and AKT (Ser473).
CRE luciferase activity
To determine the effect of ATF3 on transcription driven by the cAMP response element (CRE), we employed an adenoviral CRE-driven luciferase (Luc) reporter construct (Ad.CRE-Luc firefly luciferase), and the corresponding adenoviral control constructs Ad.MCS-Luc (firefly luciferase) and internal control construct Ad.pRL-Luc (Renilla luciferase). All of these adenoviral constructs were purchased from Vector Biolabs. Briefly, cells were mixed with 2 × 106 pfu/mL of Ad.CRE-Luc plus Ad.pRL-Luc or Ad.MCS-Luc plus Ad.pRL-Luc in serum-free medium, after which cells were seeded into 48-well plates. FBS (10% final concentration) was added to the media 1.5 hours later, and cells were incubated overnight. The cells expressing various Luc constructs were then infected with either Ad.ATF3 or Ad.GLB1 for 20 hours. Cells were then treated with control media or LH for 4 hours. The Dual-luciferase reporter assay system kit (Promega) was used to detect the luciferase signal according to the manufacturer's instructions. Briefly, cells were washed with cold PBS, and passive lysis buffer was added to lyse cells by shaking for 15 minutes. Lysates were transferred into white opaque tissue culture plates (BD Biosciences), and LARII (firefly luciferase) and STOP & GLO (Renilla luciferase) solutions were added to read luminescence using a FLUOstar OPTIMA (BMG LABTECH) plate reader. The firefly luciferase reporter activity was normalized to the Renilla luciferase signal.
Progesterone RIA
Cell culture-conditioned medium was collected for progesterone determination using a progesterone RIA kit (Diagnostic Systems Laboratories, Inc) according to the manufacturer's instructions.
Statistical analysis
The data are presented as the means ± SEM. The differences in means were analyzed by one-way ANOVA followed by Tukey's multiple comparison tests to evaluate multiple responses, or by t tests to evaluate paired responses. Two-way ANOVA was used to evaluate repeated measures with Bonferroni posttests to compare means. Statistical analysis was performed using GraphPad Prism software from GraphPad Software, Inc. *, P < .05; **, P < .01; ***, P < .001.
Results
PGF induces ATF3 mRNA and ATF3 protein expression in vivo
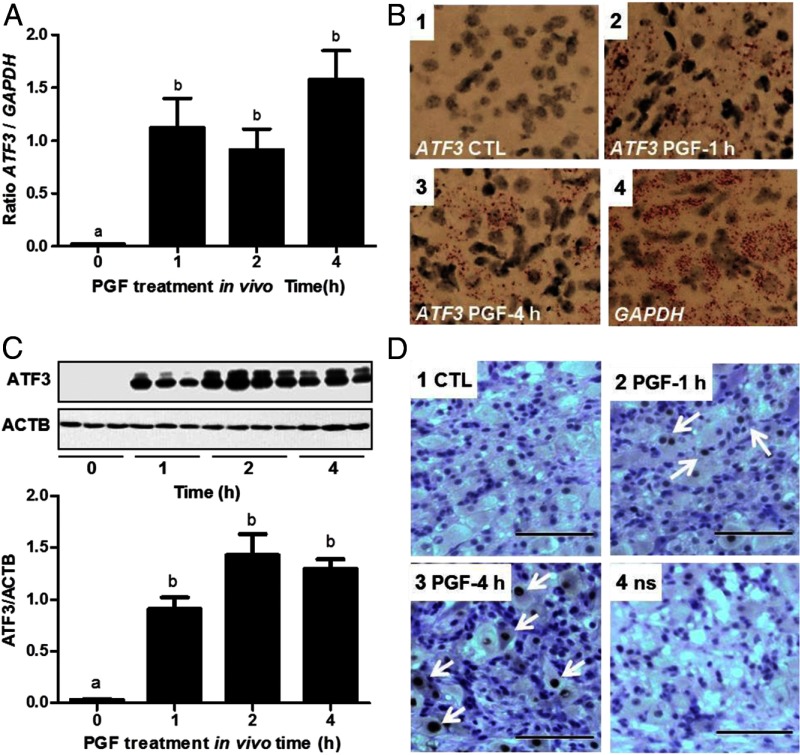
To determine whether PGF could induce ATF3 expression in vivo, corpora lutea were collected from saline-injected controls (0 hours) or 1, 2, and 4 hours after injection with a luteolytic dose of a PGF analog. Very little ATF3 mRNA or ATF3 protein was detected in corpora lutea from untreated control cows (Figure 1). Real-time PCR (Figure 1A) and in situ hybridization (Figure 1B) analysis revealed that PGF increased ATF3 mRNA expression. The increase in ATF3 mRNA was observed within 1 hour of treatment (56-fold increase, P < .001) and remained elevated for at least 240 minutes after treatment with PGF. ATF3 protein was elevated within 1 hour following PGF treatment and remained elevated after 4 hours of PGF treatment (Figure 1C). The expression and subcellular localization of ATF3 in tissue samples were evaluated by immunohistochemistry. Figure 1D shows representative immunostaining for ATF3 after PGF treatment compared with the control. Little ATF3 staining of the cytoplasm or nucleus was apparent in the luteal tissues sections of saline-treated cows. In response to PGF treatment, ATF3 was present in nuclei of LLCs. It appeared that the nuclei of some of the SLCs were also stained after 4 hours, but the nuclei of most other cell types (eg, endothelial cells) were not stained. A larger representation of the immunostaining is provided as Supplemental Figure 1 published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org.
Figure 1.
PGF treatment in vivo induces ATF3 mRNA and ATF3 protein expression. Postpubertal cattle of composite breeding were treated at midcycle with saline (n = 3) or PGF (Lutalyse; n = 9). Ovariectomies were performed after 0, 1, 2, or 4 hours. A, Corpora lutea were processed for RNA and ATF3 expression was measured by quantitative real-time PCR. Data are shown as means ± SEM. Statistical difference was determined by ANOVA followed by Tukey's test. Bars with different letters are significantly different, P < .001. B, Tissues were processed for in situ hybridization to detect ATF3 mRNA (magnification, ×200). 1) Control (CTL); 2) Treatment with PGF for 1 hour; 3) Treatment with PGF for 4 hours; 4) GAPDH in Control. The mRNA is represented as red dots. C, Corpora lutea were processed for protein to analyze ATF3 expression by Western blot (upper panel). Quantitative analysis of the Western blot data is shown in the lower panel. Results are shown as means + SEM of the ratio of ATF3 to GAPDH. Statistical difference was determined by ANOVA and Tukey's test. Bars with different letters are significantly different, P < .001. D, Tissues were processed for immunohistochemistry to detect ATF3 protein (black staining). White arrows indicate representative cells with positive ATF3 nuclear staining. 1) Control (CTL); 2) Treatment with PGF for 1 hour; 3) Treatment with PGF for 4 hours; 4) Negative control; ns, nonspecific staining. Scale bar, 100 μm. ACTB, β-actin.
PGF induces expression of ATF3 in LLCs
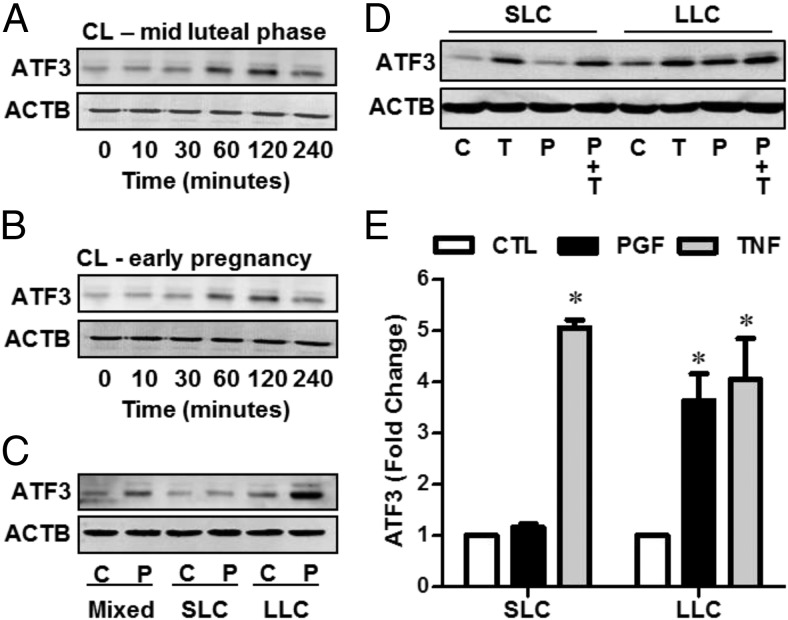
Bovine luteal cells were isolated from corpora lutea obtained at the midluteal phase and during early pregnancy. PGF increased ATF3 within 1–2 hours after treatment in luteal cells prepared from corpora lutea of the midluteal phase (Figure 2A) and early pregnancy (Figure 2B). To further identify the cell type responsible for the increase in ATF3 observed in response to PGF, SLCs and LLCs were prepared by centrifugal elutriation. Results showed that PGF induced ATF3 expression in mixed cell preparations and in highly enriched preparations of LLCs, but not in enriched preparations of SLCs (Figure 2C). The expression and subcellular localization of ATF3 in isolated luteal cells treated with PGF were also evaluated by fluorescence immunocytochemistry. ATF3 was expressed in the nucleus after PGF treatment in vitro (Supplemental Figure 2).These results correspond well with the immunohistochemistry results showing that ATF3 was expressed mostly in the nucleus (Figure 1D).
Figure 2.
PGF induces ATF3 expression in LLCs. Bovine luteal cells prepared from the midluteal phase (A) and early pregnancy (B) were treated with PGF (100 nM) for 10, 30, 60, 120, or 240 minutes after which cells were processed for ATF3 expression by Western blot. β-Actin (ACTB) served as a loading control. C, Mixed luteal cells or highly purified preparations of SLCs and LLCs were treated without (C) or with PGF (P, 100 nM) for 2 hours and ATF3 expression was analyzed by Western blot. D and E, SLCs and LLCs were treated without (C) or with PGF (P, 100 nM), or TNF (T, 50 ng/mL) for 2 hours and ATF3 expression was analyzed by Western blot. E, Data are shown as means ± SEM from 3 experiments. Statistical difference was determined by ANOVA and Tukey's test. *, P < .05 vs CTL. CL, corpora lutea; CTL, control.
Luteal cells are known to possess receptors for and respond to TNF, a cytokine implicated in luteal regression (39). TNF stimulates ATF3 expression in other cell types such as pancreatic β-cells (40), C2C12 mouse myoblast cells (41), and human umbilical vein endothelial cells (32). We observed that whereas only LLCs responded to PGF, both SLCs and LLCs responded to TNF with significant increases in ATF3 protein expression (Figure 2, D and E). We also observed that TNF, but not PGF, stimulated ATF3 expression in cultures of luteal endothelial cells (Supplemental Figure 3).
PGF treatment in vitro induces temporal and concentration-dependent increases in ATF3 expression and phosphorylation of MAPKs in LLCs
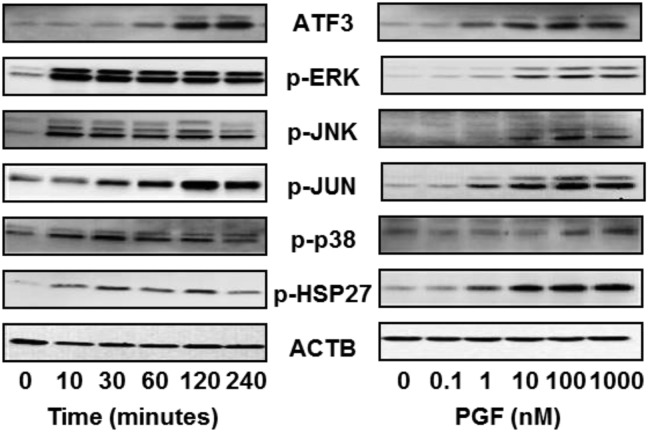
The temporal effects of PGF on ATF3 expression and activation of MAPK signaling pathways were evaluated in primary cultures of bovine LLCs (Figure 3A). ATF3 expression increased within 60 to 120 minutes following treatment with PGF (100 nM) and remained elevated for at least 240 minutes. PGF treatment also rapidly increased the phosphorylation of the MAPK family members including ERK1/2, JNK, and p38 MAPKs and their downstream substrates JUN and HSP27 (Figure 3A). ERK1/2 phosphorylation increased within 10 minutes and remained elevated throughout the 240-minute incubation with PGF. Maximal increases in JNK phosphorylation were observed as early as 10 minutes following PGF treatment and then gradually decreased throughout the 240-minute incubation. The phosphorylation of JUN, a transcription factor and known target of JNK, increased during the first 120 minutes of incubation and then remained elevated throughout the remaining 120 minutes of incubation with PGF. A rapid increase in p38 MAPK phosphorylation was accompanied by an increase in the phosphorylation of its downstream target HSP27.
Figure 3.
Treatment with PGF induces time- and concentration-dependent increases in ATF3 expression and phosphorylation of several members of the MAPK family in LLCs. A, Bovine LLCs were treated with PGF (100 nM) for 0–240 minutes. B, Bovine LLCs were treated with 0–1000 nM PGF for 2 hours. Cell lysates were analyzed by Western blot. β-Actin (ACTB) was used a loading control. Data are representative of 3 experiments.
To determine the concentration-dependent effects of PGF on ATF3 expression and activation of MAPK signaling pathways, LLCs were treated with increasing concentrations of PGF for 120 minutes, a time point established to provide near-maximal increases in ATF3 expression (Figure 3A). We observed concentration-dependent increases in ATF3 (Figure 3B) with as little as 1 nM PGF stimulating increases in ATF3, and with 10–100 nM PGF stimulating maximal increases in ATF3. Similar concentration-dependent increases in the phosphorylation of MAPK signaling components were also observed after 120 minutes of incubation (Figure 3B).
Contribution of MAPK signaling pathways to ATF3 expression in LLCs
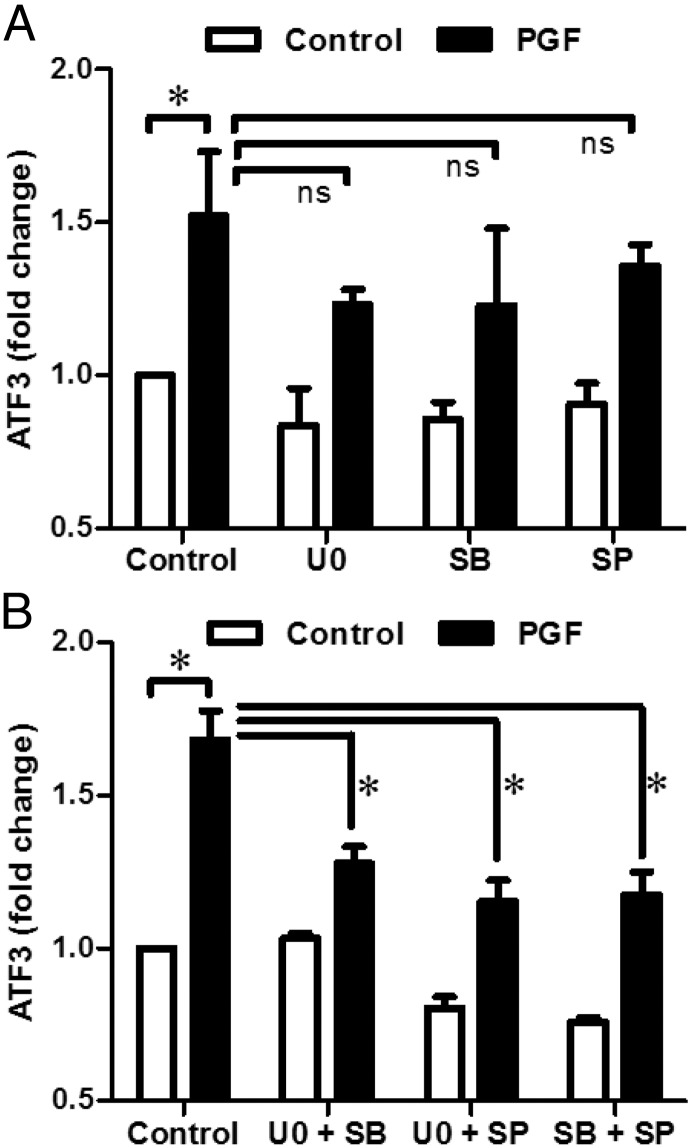
Experiments were performed to evaluate whether the inhibition of MAPK signaling would affect ATF3 expression in response to PGF. LLCs were pretreated for 1 hour with inhibitors of MEK1/2 (U0126, 20 μM), p38 MAPK (SB203580, 2 μM), or JNK (SP600125, 10 μM), after which cells were treated with PGF for 2 hours. Western blot analysis showed that inhibitors of MEK1/2, p38MAPK, and JNK, when used alone, did not significantly reduce ATF3 expression (Figure 4A). However, combinations of inhibitors were able to suppress ATF3 expression (Figure 4B), indicating that multiple converging signaling mechanisms involving several MAPKs may contribute to the induction of ATF3 by PGF in corpus luteum.
Figure 4.
Effects of MAPK pathway inhibitors on PGF-induced ATF3 expression in LLCs. A, Bovine LLCs were pretreated with vehicle alone (dimethylsulfoxide), MEK1 inhibitor U0126 (UO; 20 μM), p38 inhibitor SB203580 (SB; 2 μM), JNK inhibitor SP600125 (SP; 10 μM) for 60 minutes prior to treatment with PGF (100 nM) for 2 hours. B, LLCs were pretreated with vehicle alone or combinations of U0126 (20 μM), SB203580 (2 μM), SP600125 (10 μM) for 60 minutes prior to treatment with PGF (100 nM) for 2 hours. Cell lysates were analyzed by Western blot. Data are shown as means ± SEM from 3 or more experiments. Statistical difference was determined by ANOVA followed by Tukey's test. *, P < .05. ns, nonsignificant, P > .05.
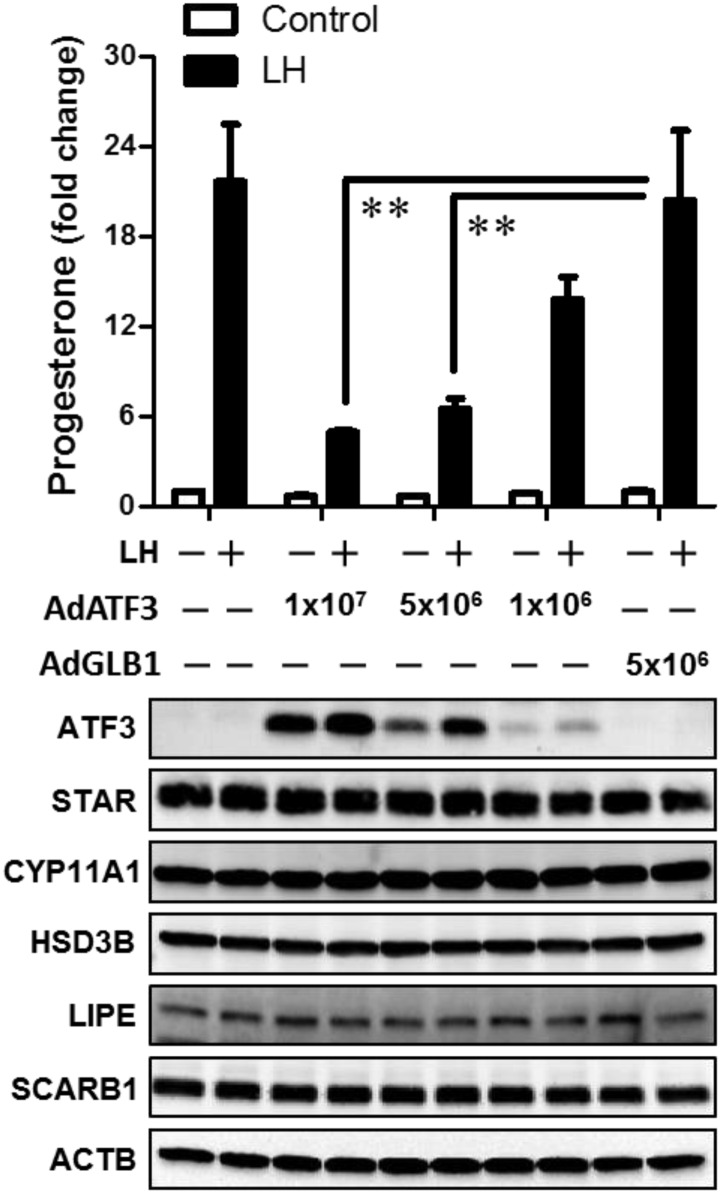
ATF3 inhibits LH-stimulated progesterone but does not alter the expression of key regulators of progesterone synthesis
To explore the possible function of ATF3 in luteal cells, we employed an adenovirus to express ATF3 in both SLCs and LLCs. Experiments were performed to determine the effects of ATF3 on cell viability, progesterone synthesis, and proteins involved in progesterone synthesis. SLCs were infected with increasing amounts of Ad.ATF3 and then treated with LH for 4 hours to stimulate progesterone secretion. As shown in Figure 5, LH significantly increased progesterone production in primary cultures of SLCs (fold increase: 21 ± 3.7, mean ± SEM, n = 3). Treatment with the control adenovirus expressing β-galactosidase (Ad.GLB1) did not alter progesterone secretion under basal conditions or in response to LH. Increasing the titer of Ad.ATF3 resulted in a concentration-dependent increase in ATF3 expression as determined by Western blot (Figure 5). It also resulted in a concentration-dependent reduction in LH-stimulated progesterone production (% inhibition: 29 ± 9, 65 ± 9, and 73 ± 6, respectively, vs Ad.GLB1, mean ± SEM, n = 3). Treatment with increasing titers of Ad.ATF3 did not statistically reduce basal progesterone (% inhibition: 15 ± 7, 31 ± 12, and 28 ± 12, respectively, vs Ad.GLB1, mean ± SEM, n = 3).
Figure 5.
Adenoviral-mediated expression of ATF3 inhibits LH-stimulated progesterone secretion in SLCs. Bovine SLCs were transduced with 1 × 106 to 1 × 107 pfu/mL of adenoviruses expressing ATF3 (Ad.ATF3) or β-galactosidase (Ad.GLB1) as control. After 20 hours the media were changed and cells were preequilibrated with serum-free medium for 2 hours prior to treatment without (Control) or with 100 ng/mL LH for 4 hours. The media were collected for analysis of progesterone by RIA (upper panel). Cells were lysed and proteins detected by Western blot analysis (lower panel). Results reflect the findings in 3 separate experiments, each of which were performed in triplicate. Statistical difference was determined by ANOVA and the Tukey's test. **, P < .01. ACTB, β-actin.
Based on the foregoing results, we infected LLCs with Ad.ATF3 prior to treatment for 4 hours with LH. LH significantly increased progesterone production in LLCs (6-fold increase, P < .01). Adenoviral-mediated expression of ATF3 significantly (P < .001) inhibited LH-stimulated progesterone in LLCs (52 ± 7% inhibition vs Ad.GLB1, mean ± SEM, n = 3) (Figure 6).
Figure 6.
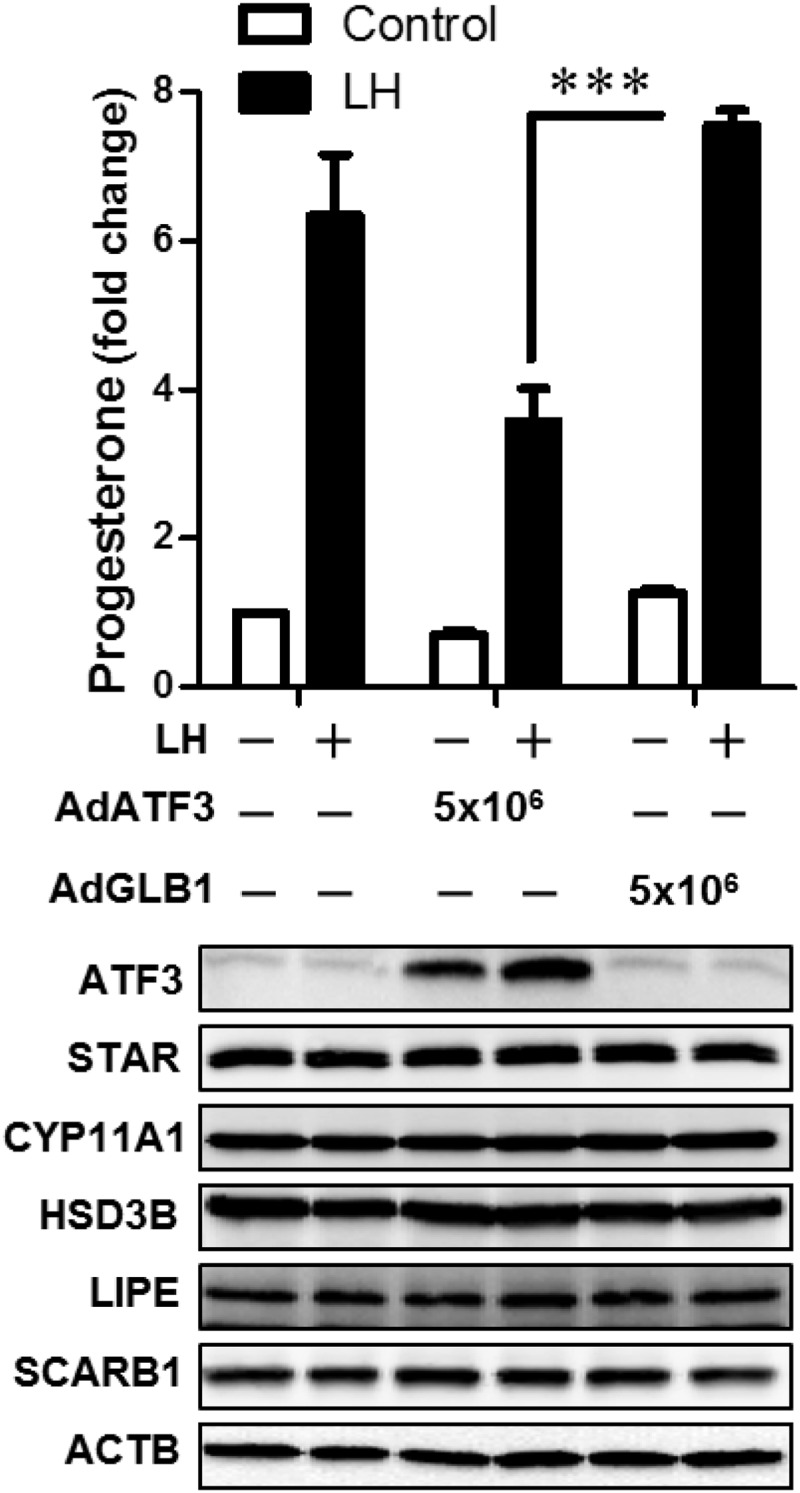
Adenoviral-mediated expression of ATF3 inhibits progesterone secretion in LLCs. Bovine LLCs were transduced with 5 × 106 pfu/mL adenovirus expressing ATF3 (Ad.ATF3) or β-galactosidase (Ad.GLB1) as control. After 20 hours the media were changed and cells were preequilibrated with serum-free medium for 2 hours prior to treatment without (Control) or with 100 ng/mL LH for 4 hours. The media were collected for analysis of progesterone by RIA (upper panel). The cells were lysed and proteins detected by Western blot analysis. Results reflect the findings in 3 separate experiments, each of which was performed in triplicate. Statistical difference was determined by ANOVA and the Tukey's test. ***, P < .001. ACTB, β-actin.
Analysis was performed to determine whether the ATF3-induced defect in LH-stimulated progesterone synthesis was the result of a reduction in the expression of proteins needed for delivery of cholesterol or the metabolism of cholesterol to progesterone. Western blot analysis performed to determine levels of scavenger receptor class B1 (SR-B1), hormone-sensitive lipase (also referred to as LIPE), StAR, cytochrome P450 side-chain cleavage (P450scc, CYP11A1), and HSD3B, and β-actin as a loading control revealed no changes in the expression of these proteins in either SLCs (Figure 5) or LLCs (Figure 6) in response to overexpression of ATF3.
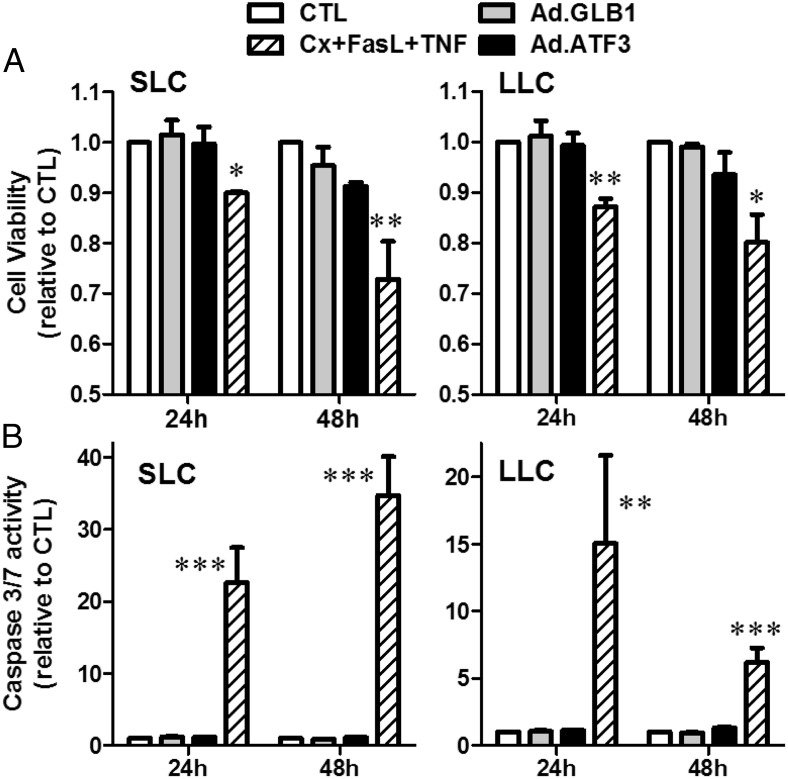
Previous reports indicate that the induction of ATF3 may contribute to cell death (29, 30, 42). In order to determine whether ATF3 expression also affected luteal cell viability, SLCs and LLCs were infected with Ad.ATF3 or the control Ad.GLB1, and caspase activity and cell viability assays were performed. As a positive control, we observed that treatment with a combination of TNF, Fas ligand, and cycloheximide effectively reduced cell viability (Figure 7A), increased caspase 3/7 activity (Figure 7B), and increased accumulation of the active cleaved-caspase 3 (Supplemental Figure 4A) within 24 hours. However, neither Ad.ATF3 nor Ad.GBL1 altered cell viability, caspase activity, or cleaved caspase 3 in SLCs or LLCs after 24 or 48 hours. Furthermore, ATF3 expression did not inhibit the stimulatory effect of IGF-1 on luteal cell survival signals, such as the phosphorylation of AKT (Ser308) and AKT (Thr473) (Supplemental Figure 4B). Taken together, the data indicate that expression of ATF3 does not affect luteal cell viability.
Figure 7.
Expression of ATF3 does not affect viability of LLCs or SLCs. Bovine SLCs and LLCs were transduced with 5 × 106 pfu/mL adenovirus expressing ATF3 (Ad.ATF3) or β-galactosidase (Ad.GLB1) as described in Figure 6. Cells were treated with TNF (50 ng/mL) + FasL (100 ng/mL) + Cycloheximide (Cx, 10 μg/mL) for 24 hours and 48 hours as a positive control for indices of cell death. MTT cell viability assays (A) and caspase 3/7 activity assays (B) were performed as described in Materials and Methods. Data are shown as means ± SEM from 3 experiments. Statistical difference was determined by ANOVA followed by Tukey's multiple-range test. *, P < .05; **, P < .01; ***, P < .001 vs control (CTL). FasL, Fas ligand.
Effect of ATF3 expression on LH-stimulated protein kinase A (PKA) signaling and CRE-mediated transcription
To determine whether treatment with Ad.ATF3 modified the early signaling response to LH, we used an antibody that detects the phosphorylation of PKA substrates as a proxy for activation of the cAMP/PKA signaling pathway. The phospho-PKA substrate (RRXphospho-S/T) antibody detects proteins containing a phospho-serine or phospho-threonine residue with arginine at the −3 and −2 positions (Cell Signaling Technology). Western blot analysis showed that LH increased PKA phospho-protein substrates harboring the PKA phosphorylation motif in response to LH in both SLCs and LLCs (Supplemental Figure 5, A and B, respectively). However, the expression of ATF3 did not alter the LH-stimulated increase in PKA phospho-protein substrates.
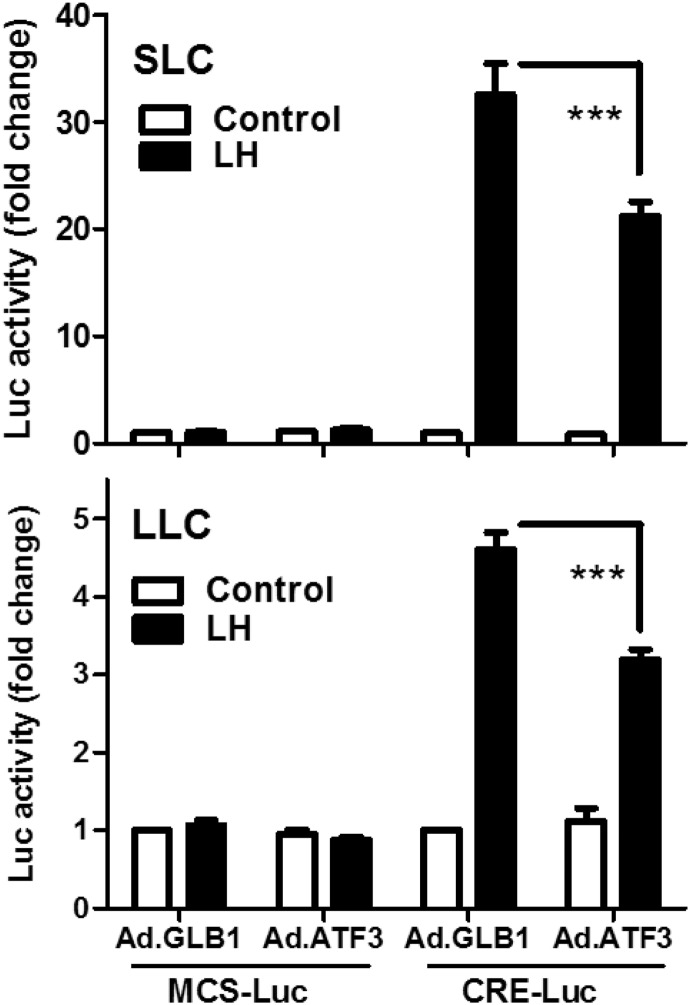
ATF3 may repress or activate gene transcription through formation of homodimers or heterodimers with other members of the ATF/CREB family (43). We employed a CREB response element (CRE) luciferase-reporter expression system to determine whether ATF3 could alter LH-stimulated CRE-mediated transcription in luteal cells. Luteal cells were infected with the reporter adenovirus construct (Ad.CRE-Luc) or control luciferase construct (Ad.MCS-Luc). As shown in Figure 8, LH significantly increased CRE-dependent luciferase activity both in SLCs (fold: 33 ± 3.0, n = 3) and LLCs (fold: 4.6 ± 0.2, n = 3), whereas it did not affect the control (MSC-Luc) luciferase activity. Importantly, we observed that expression of ATF3 significantly inhibited the LH-driven CRE-dependent transcription (% inhibition: 34 ± 2.1 in SLCs and 30 ± 5 in LLCs vs Ad.GLB1; mean ± SEM, n = 3).
Figure 8.
Expression of ATF3 inhibits LH-stimulated CRE-dependent transcription. Bovine SLCs and LLCs were mixed with Ad.MCS-Luc plus Ad.pRL-Luc, or Ad.CRE-Luc plus Ad.pRL-Luc (2 × 106 pfu/mL) and incubated overnight. The cells were then infected with either Ad.GLB1 or Ad.ATF3 (5 × 106 pfu/mL) for an additional 20 hours. After preequilibration, SLCs or LLCs were treated without (Control) or with LH (100 ng/mL) for 4 hours. A Dual-Luciferase reporter assay system was used to detect the luciferase activity. Each experiment was performed in triplicate, and 3 separate experiments were performed. Data are shown as means ± SEM from 3 experiments. Statistical difference was determined by ANOVA and the Tukey's test. ***, P < .001.
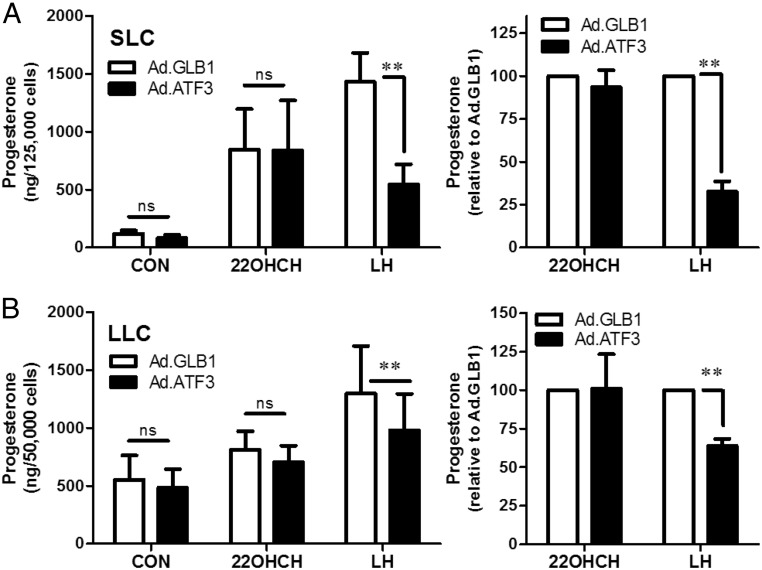
ATF3 does not inhibit progesterone production in the presence of 22R-hydroxycholesterol
Because our studies did not show a reduction in known sterol transport proteins or enzymes that convert cholesterol to progesterone, we performed an experiment to determine whether the ATF3-induced reduction in progesterone was due to a reduction in activity of the enzymes required to convert cholesterol into progesterone. Both SLCs and LLCs were infected with Ad.ATF3 or the control adenovirus Ad.GLB1 as described previously, and then treated with 22R-hydroxycholesterol (22R-OH-CH), a membrane-permeable cholesterol. We observed that treatment with 22R-OH-CH significantly increased progesterone production in SLCs (fold increase: 6.7 ± 2.2 after 4 hours, n = 4) and LLC (fold increase: 2.0 ± 0.6 after 4 hours, n = 4). Importantly, the ability of 22R-OH-CH to increase progesterone was not interrupted by prior expression of ATF3 (Figure 9) in either LLCs or SLCs. Although ATF3 did not inhibit 22R-OH-CH-stimulated progesterone production in either cell type, in this set of experiments ATF3 expression effectively inhibited LH-stimulated progesterone production in SLCs by 67 ± 6.1% after treatment for 4 hours (Figure 9A) and by 73 ± 6.0% after 24 hours. In LLCs, ATF3 inhibited LH-stimulated progesterone production by 36 ± 4.6% after 4 hours (Figure 9B).
Figure 9.
ATF3 inhibits LH but not 22-hydroxy-cholesterol (22OHCH)-induced progesterone synthesis. Bovine SLCs (panel A) and LLCs (panel B) were infected with Ad.GLB1 or Ad.ATF3 (5 × 106 pfu/mL) for 20 hours. After equilibration, the cells were treated without (control [CON]) or with LH (100 ng/mL) or 22OHCH (20 μM) for 4 hours. Progesterone in conditioned medium was measured by RIA. Each experiment was performed in triplicate, and 4 separate experiments were performed. Data are shown as means ± SEM of the amount of progesterone in the medium (left panel) and as the amount of progesterone after expressing the data relative to the control response (Ad.GLB1) in each experiment (right panel). Statistical difference was determined by two-way ANOVA, followed by Tukey's testing for differences in the means. **, P < .01. ns, nonsignificant (P > .05).
Discussion
Prostaglandin F2α initiates several signaling pathways in the corpus luteum, including elevations in intracellular calcium and activation of protein kinase C and MAPK signaling cascades (4–8). The PGF-induced changes in intracellular signaling culminate in the regulated expression of many genes in the corpus luteum (11, 34). A number of immediate early genes, including FOS, JUN, EGR1, and ATF3, have been identified but their role in PGF-induced luteolysis remains unclear (2, 9, 11, 13). In this study we report that PGF induces the expression of ATF3 mRNA and ATF3 protein in the LLCs of the bovine corpus luteum in vivo. The effect of PGF on ATF3 was limited to the LLCs in vitro, whereas the cytokine TNF induced ATF3 in both LLCs and SLCs. Overexpression of ATF3 in either LLCs or SLCs resulted in impaired progesterone secretion in response to LH. We hypothesize that the induction of ATF3 serves to modulate gonadotropin responsiveness during luteolysis.
The corpus luteum of most mammals is composed of multiple, distinctive cell types including steroidogenic cells (SLCs and LLCs) and nonsteroidogenic cells (endothelial cells, pericytes, fibroblasts, and immune cells). Endothelial cells are thought to represent about 50% of the cells in the mature bovine corpus luteum, with steroidogenic cells representing 40% and the remaining cell types about 10%. The large steroidogenic luteal cells possess most receptors for PGF (PTGFRs) whereas small steroidogenic cells have few PTGFRs (44). Recent studies provide evidence that endothelial cells may also possess the PTGFR (38, 45). Our data shows that ATF3 expression was very low to undetectable in luteal tissue from midluteal-phase cows and that PGF treatment in vivo rapidly induced the expression of ATF3 mRNA and ATF3 protein in LLCs. The size and limited number of cells responding to PGF indicates that most SLCs and endothelial cells did not express ATF3 for up to 4 hours following treatment with PGF. The cell-specific induction of ATF3 in response to PGF in the LLCs in vivo was also observed in vitro using highly purified preparations of luteal cells, in which LLCs, but neither SLCs nor corpus luteum-derived endothelial cells responded to PGF with an increase in ATF3. The lack of effect of PGF on endothelial cells derived from the bovine corpus luteum is consistent with the work of others (35, 46). The temporal increase in ATF3 observed in vivo following PGF treatment was mimicked in LLCs following PGF treatment in vitro. Furthermore, our studies showed that increases in ATF3 expression were observed at 1 nM PGF (∼0.3 ng/mL) with maximal increases observed at 10–100 nM PGF. These findings are in keeping with the established Kd for PGF binding to its receptor (47) and concentrations of PGF reported in the pulses of PGF (0.6–11 ng/ml) that reach the ovary (39, 48) during spontaneous luteal regression. Collectively, our findings demonstrate that the LLCs of the corpus luteum respond to PGF with an increase in ATF3 expression.
Although ATF3 is generally held to be a stress-inducible gene (29, 49), it is also responsive to hormones and environmental cues (50–52). The signaling mechanisms involved in induction of ATF3 are incompletely characterized; however, activated MAPK pathways have been implicated in the induction of ATF3 expression. The promoter region of the human ATF3 gene contains binding sites for a number of transcription factors, including activator protein-1, ATF/CRE, NFκB, E2F, MYC/MAX, EGR1, CCAAT enhancer binding protein, and SMAD proteins (20, 52, 53). Binder et al (50) found that GnRH, a ligand that activates a Gq-coupled receptor, specifically regulates ATF3 mRNA in murine LβT2 cells through a pathway that includes intracellular Ca2+, calcineurin, JNK, and nuclear factor of activated T cells. Lu et al (54) found that the p38 MAPK pathway plays a critical role in the induction of ATF3 by stress signals, and that ATF3 is functionally important to mediate the proapoptotic effects of p38MAPK, whereas the ERK and JNK signaling pathways are neither necessary nor sufficient to induce ATF3 in the anisomycin-induced stress paradigm. A report by Sandnes et al (55) showed that although inhibition of each of the MAPKs ERK1/2, JNK, or p38 did not affect vasopressin-induced ATF3 expression in hepatocytes, the combined inhibition of JNK and p38, and of ERK1/2 and either JNK or p38 suppressed vasopressin-induced expression of ATF3 in hepatocytes. In the present study, we found that PGF stimulates ERK1/2, p38, and JNK MAPK signaling pathways in bovine LLCs and that selective inhibition of ERK1/2, p38, and JNK signaling had very little effect on PGF-induced ATF3 expression. However, combinations of MAPK inhibitors significantly inhibited the stimulatory effect of PGF on ATF3 expression in LLCs. Although these results do not preclude the involvement of other Ca2+ and/or PKC-dependent signaling pathways, the MAPKs appear to contribute to the induction of ATF3 in a partly redundant manner in bovine LLCs. Whether other recently described PGF-induced signaling components, such as reactive oxygen species and NFκB activation, contribute to ATF3 expression in the corpus luteum requires further investigation (56).
ATF3 has been implicated in cell context-specific responses involving cellular proliferation (57), survival (58), and apoptosis (42, 59). In a study to determine the expression of genes involved in the luteolytic response to PGF, Mondal et al (34) found that PGF treatment increased ATF3 protein expression in bovine corpora lutea collected during the midluteal phase (day 11), but not in the early luteal phase (day 4), although ATF3 mRNA was abundant on day 4. This result is intriguing because the corpora lutea of both early- and midluteal phases possess PTGFR (45) and respond to PGF with rapid changes in gene expression, but the early luteal phase corpora lutea do not undergo luteolysis when challenged by PGF. This suggests that the ability of PGF to induce ATF3 expression could be required for luteal regression. Our observation that adenoviral-mediated expression of ATF3 reduced luteal progesterone synthesis supports this idea. However, our data also suggest that expression of ATF3 does not affect the induction of caspase activity or the viability of SLCs or LLCs. Therefore, it seems clear that ATF3 expression is not sufficient to induce apoptosis in steroidogenic cells. It is of interest to note that the ability of PGF to stimulate ATF3 expression was much greater in vivo when compared with in vitro results with isolated luteal cells in culture. The reasons for the differential responses are not clear but could reflect a loss of cellular responsiveness to PGF during cell isolation and the experimental conditions in vitro. It is also conceivable that optimal PGF responsiveness requires preservation of the existing 3-dimensional luteal architecture in vivo and the rapid activation of additional signaling molecules. It is appreciated that other factors like endothelins, angiotensin, and cytokines contribute to luteal regression in response to PGF (16, 17, 60, 61). That a combination of ligands could influence ATF3 expression is supported by our experiments indicating that a TNF, a cytokine known to influence luteal regression (62–65), stimulated ATF3 expression in LLCs and SLCs and endothelial cells.
ATF3 regulates transcription in a cell-specific manner, providing both stimulatory and inhibitory influences on gene transcription (43). The ATF/CRE-binding protein (CREB) family of transcription factors binds as dimers to a basic zipper (bZIP) DNA-binding domain with a 5′-TGACGTCA-3′ recognition sequence. ATF3 inhibits CREB binding to CRE sites and inhibits CRE-mediated transcription (28, 66, 67). Our experiments to determine whether ATF3 contributes in a functional way to luteal regression showed that overexpression of ATF3 significantly inhibited LH-stimulated, but not basal, progesterone production by both SLCs and LLCs. The inhibitory effect of ATF3 did not appear to alter the early signaling effects of LH because the ability of LH to increase the phosphorylation of protein kinase A (PKA) substrates was not reduced by ATF3. However, overexpression of ATF3 significantly reduced the ability of LH to stimulate CRE-mediated gene transcription as shown in experiments using a CRE-driven luciferase reporter. The inhibitory effects of ATF3 on LH-stimulated CRE-mediated transcription and progesterone synthesis were not associated with alterations in the expression of proteins important for cholesterol import (SRB1), mobilization and transport of cholesterol (LIPE, StAR), or conversion of cholesterol to progesterone (CYP11A and HSD3B). Furthermore, the activity of CYP11A and HSD3B in LLCs and SLCs was largely unaffected by ATF3 because the conversion of an exogenous source of cholesterol (22R-hydroxy cholesterol) to progesterone was similar in control and ATF3-expressing cells. Grusenmeyer and Pate (68) reported that the inhibitory effects of PGF on lipoprotein-stimulated progesterone production did not involve a reduction in cellular uptake of cholesterol nor conversion of 25-hydroxycholesterol to progesterone, suggesting an inhibition of a site subsequent to cholesterol entry into the cell but before mitochondrial cholesterol side-chain cleavage. Similar findings were reported by Wiltbank et al (69) who found that protein kinase C-mediated inhibition of progesterone synthesis in ovine LLCs was not observed when treated with 20-OH-, 22-OH-, or 25-OH-cholesterol derivatives. Because 22R-hydroxy cholesterol bypasses the first of 3 reactions mediated by the side chain cleavage enzyme CYP11A (70, 71), it is possible this initial step may be indirectly targeted by ATF3. Consequently, ATF3 appears to affect gonadotropin-stimulated progesterone secretion at a step or steps downstream of PKA signaling and before cholesterol conversion to progesterone.
Acknowledgments
This work was supported by Agriculture and Food Research Initiative Competitive Grant no. 2011–67015–20076 from the US Department of Agriculture National Institute of Food and Agriculture; by the Veterans Affairs Nebraska-Western Iowa Health Care System and is based upon work supported in part by the Department of Veterans Affairs, Office of Research and Development Biomedical Laboratory Research and Development funds; the National Institutes of Health 1 P01 AG029531; the China Scholarship Council; and The Olson Center for Women's Health, Department of Obstetrics and Gynecology, Nebraska Medical Center, Omaha, Nebraska.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
USDA is an equal opportunity provider and employer. Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of names by the USDA implies no approval of the product to the exclusion of others that may also be suitable.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ATF3
- activating transcription factor 3
- CRE
- cAMP response element
- CREB
- CRE-binding protein
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- HRP
- horseradish peroxidase
- HSD3B
- 3β-hydroxysteroid dehydrogenase
- JNK
- c-Jun N-terminal kinase
- LLC
- large luteal cell
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NFκB
- nuclear factor-κB
- PGF
- prostaglandin F2α
- PKA
- protein kinase A
- PKC
- protein kinase C
- PTGFR
- prostaglandin F receptor
- 22R-OH-CH
- 22R-hydroxycholesterol
- SLC
- small luteal cell
- StAR
- steroidogenic acute regulatory protein
- TBST
- Tris-buffered saline-Tween 20.
References
- 1. Davis JS, Rueda BR. The corpus luteum: an ovarian structure with maternal instincts and suicidal tendencies. Front Biosci. 2002;7:d1949–1978 [DOI] [PubMed] [Google Scholar]
- 2. Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr Rev. 2007;28(1):117–149 [DOI] [PubMed] [Google Scholar]
- 3. McCracken JA, Glew ME, Scaramuzzi RJ. Corpus luteum regression induced by prostaglandin F2-α. J Clin Endocrinol Metab. 1970;30(4):544–546 [DOI] [PubMed] [Google Scholar]
- 4. Davis JS, Weakland LL, Weiland DA, Farese RV, West LA. Prostaglandin F2 α stimulates phosphatidylinositol 4,5-bisphosphate hydrolysis and mobilizes intracellular Ca2+ in bovine luteal cells. Proc Natl Acad Sci USA. 1987;84(11):3728–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen DB, Westfall SD, Fong HW, Roberson MS, Davis JS. Prostaglandin F2α stimulates the Raf/MEK1/mitogen-activated protein kinase signaling cascade in bovine luteal cells. Endocrinology. 1998;139(9):3876–3885 [DOI] [PubMed] [Google Scholar]
- 6. Arvisais E, Hou X, Wyatt TA, et al. Prostaglandin F2α represses IGF-I-stimulated IRS1/phosphatidylinositol-3-kinase/AKT signaling in the corpus luteum: role of ERK and P70 ribosomal S6 kinase. Mol Endocrinol. 2010;24(3):632–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yadav VK, Sudhagar RR, Medhamurthy R. Apoptosis during spontaneous and prostaglandin F(2α)-induced luteal regression in the buffalo cow (Bubalus bubalis): involvement of mitogen-activated protein kinases. Biol Reprod. 2002;67(3):752–759 [DOI] [PubMed] [Google Scholar]
- 8. Yadav VK, Medhamurthy R. Dynamic changes in mitogen-activated protein kinase (MAPK) activities in the corpus luteum of the bonnet monkey (Macaca radiata) during development, induced luteolysis, and simulated early pregnancy: a role for p38 MAPK in the regulation of luteal function. Endocrinology. 2006;147(4):2018–2027 [DOI] [PubMed] [Google Scholar]
- 9. Chen D, Fong HW, Davis JS. Induction of c-fos and c-jun messenger ribonucleic acid expression by prostaglandin F2α is mediated by a protein kinase C-dependent extracellular signal-regulated kinase mitogen-activated protein kinase pathway in bovine luteal cells. Endocrinology. 2001;142(2):887–895 [DOI] [PubMed] [Google Scholar]
- 10. Tsai SJ, Kot K, Ginther OJ, Wiltbank MC. Temporal gene expression in bovine corpora lutea after treatment with PGF2α based on serial biopsies in vivo. Reproduction. 2001;121(6):905–913 [PubMed] [Google Scholar]
- 11. Atli MO, Bender RW, Mehta V, et al. Patterns of gene expression in the bovine corpus luteum following repeated intrauterine infusions of low doses of prostaglandin F2α. Biol Reprod. 2012;86(4):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stocco CO, Lau LF, Gibori G. A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20α-hsd genes by prostaglandin F2α in ovarian cells. J Biol Chem. 2002;277(5):3293–3302 [DOI] [PubMed] [Google Scholar]
- 13. Hou X, Arvisais EW, Jiang C, et al. Prostaglandin F2α stimulates the expression and secretion of transforming growth factor B1 via induction of the early growth response 1 gene (EGR1) in the bovine corpus luteum. Mol Endocrinol. 2008;22(2):403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berisha B, Meyer HH, Schams D. Effect of prostaglandin F2 α on local luteotropic and angiogenic factors during induced functional luteolysis in the bovine corpus luteum. Biol Reprod. 2010;82(5):940–947 [DOI] [PubMed] [Google Scholar]
- 15. Kliem H, Berisha B, Meyer HH, Schams D. Regulatory changes of apoptotic factors in the bovine corpus luteum after induced luteolysis. Mol Reprod Dev. 2009;76(3):220–230 [DOI] [PubMed] [Google Scholar]
- 16. Neuvians TP, Berisha B, Schams D. Vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) expression during induced luteolysis in the bovine corpus luteum. Mol Reprod Dev. 2004;67(4):389–395 [DOI] [PubMed] [Google Scholar]
- 17. Shirasuna K, Sasahara K, Matsui M, Shimizu T, Miyamoto A. Prostaglandin F2α differentially affects mRNA expression relating to angiogenesis, vasoactivation and prostaglandins in the early and mid corpus luteum in the cow. J Reprod Dev. 2010;56(4):428–436 [DOI] [PubMed] [Google Scholar]
- 18. Shirasuna K, Watanabe S, Nagai K, et al. Expression of mRNA for cell adhesion molecules in the bovine corpus luteum during the estrous cycle and PGF2α-induced luteolysis. J Reprod Dev. 2007;53(6):1319–1328 [DOI] [PubMed] [Google Scholar]
- 19. Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7(4–6):321–335 [PMC free article] [PubMed] [Google Scholar]
- 20. Liang G, Wolfgang CD, Chen BP, Chen TH, Hai T. ATF3 gene. Genomic organization, promoter, and regulation. J Biol Chem. 1996;271(3):1695–1701 [DOI] [PubMed] [Google Scholar]
- 21. Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr. 2010;15(1):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl). 2009;87(11):1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen-Jennings AE, Hartman MG, Kociba GJ, Hai T. The roles of ATF3 in liver dysfunction and the regulation of phosphoenolpyruvate carboxykinase gene expression. J Biol Chem. 2002;277(22):20020–20025 [DOI] [PubMed] [Google Scholar]
- 24. Kim HB, Kong M, Kim TM, et al. NFATc4 and ATF3 negatively regulate adiponectin gene expression in 3T3–L1 adipocytes. Diabetes. 2006;55(5):1342–1352 [DOI] [PubMed] [Google Scholar]
- 25. Pan Y, Chen H, Siu F, Kilberg MS. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem. 2003;278(40):38402–38412 [DOI] [PubMed] [Google Scholar]
- 26. Kang Y, Chen CR, Massagué J. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11(4):915–926 [DOI] [PubMed] [Google Scholar]
- 27. Gilchrist M, Thorsson V, Li B, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441(7090):173–178 [DOI] [PubMed] [Google Scholar]
- 28. Khuu CH, Barrozo RM, Hai T, Weinstein SL. Activating transcription factor 3 (ATF3) represses the expression of CCL4 in murine macrophages. Mol Immunol. 2007;44(7):1598–1605 [DOI] [PubMed] [Google Scholar]
- 29. Li D, Yin X, Zmuda EJ, et al. The repression of IRS2 gene by ATF3, a stress-inducible gene, contributes to pancreatic β-cell apoptosis. Diabetes. 2008;57(3):635–644 [DOI] [PubMed] [Google Scholar]
- 30. Taketani K, Kawauchi J, Tanaka-Okamoto M, et al. Key role of ATF3 in p53-dependent DR5 induction upon DNA damage of human colon cancer cells. Oncogene. 2012;31(17):2210–2221 [DOI] [PubMed] [Google Scholar]
- 31. Kawauchi J, Zhang C, Nobori K, et al. Transcriptional repressor activating transcription factor 3 protects human umbilical vein endothelial cells from tumor necrosis factor-α-induced apoptosis through down-regulation of p53 transcription. J Biol Chem. 2002;277(41):39025–39034 [DOI] [PubMed] [Google Scholar]
- 32. Nawa T, Nawa MT, Adachi MT, et al. Expression of transcriptional repressor ATF3/LRF1 in human atherosclerosis: colocalization and possible involvement in cell death of vascular endothelial cells. Atherosclerosis. 2002;161(2):281–291 [DOI] [PubMed] [Google Scholar]
- 33. Whitlock NC, Bahn JH, Lee SH, Eling TE, Baek SJ. Resveratrol-induced apoptosis is mediated by early growth response-1, Kruppel-like factor 4, and activating transcription factor 3. Cancer Prev Res (Phila). 2011;4(1):116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mondal M, Schilling B, Folger J, et al. Deciphering the luteal transcriptome: potential mechanisms mediating stage-specific luteolytic response of the corpus luteum to prostaglandin F(2)α. Physiol Genomics. 2011;43(8):447–456 [DOI] [PubMed] [Google Scholar]
- 35. Maroni D, Davis JS. TGFB1 disrupts the angiogenic potential of microvascular endothelial cells of the corpus luteum. J Cell Sci. 2011;124(Pt 14):2501–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ireland JJ, Murphee RL, Coulson PB. Accuracy of predicting stages of bovine estrous cycle by gross appearance of the corpus luteum. J Dairy Sci. 1980;63(1):155–160 [DOI] [PubMed] [Google Scholar]
- 37. Hou X, Arvisais EW, Davis JS. Luteinizing hormone stimulates mammalian target of rapamycin signaling in bovine luteal cells via pathways independent of AKT and mitogen-activated protein kinase: modulation of glycogen synthase kinase 3 and AMP-activated protein kinase. Endocrinology. 2010;151(6):2846–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mamluk R, Chen D, Greber Y, Davis JS, Meidan R. Characterization of messenger ribonucleic acid expression for prostaglandin F2α and luteinizing hormone receptors in various bovine luteal cell types. Biol Reprod. 1998;58(3):849–856 [DOI] [PubMed] [Google Scholar]
- 39. Pate JL, Johnson-Larson CJ, Ottobre JS. Life or death decisions in the corpus luteum. Reprod Domest Anim. 2012;47(Suppl 4):297–303 [DOI] [PubMed] [Google Scholar]
- 40. Gurzov EN, Barthson J, Marhfour I, et al. Pancreatic β-cells activate a JunB/ATF3-dependent survival pathway during inflammation. Oncogene. 2012;31(13):1723–1732 [DOI] [PubMed] [Google Scholar]
- 41. Park HJ, Kang YM, Kim CH, Jung MH. ATF3 negatively regulates adiponectin receptor 1 expression. Biochem Biophys Res Commun. 2010;400(1):72–77 [DOI] [PubMed] [Google Scholar]
- 42. Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27(15):2118–2127 [DOI] [PubMed] [Google Scholar]
- 43. Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273(1):1–11 [DOI] [PubMed] [Google Scholar]
- 44. Chegini N, Lei ZM, Rao CV, Hansel W. Cellular distribution and cycle phase dependency of gonadotropin and eicosanoid binding sites in bovine corpora lutea. Biol Reprod. 1991;45(3):506–513 [DOI] [PubMed] [Google Scholar]
- 45. Shirasuna K, Akabane Y, Beindorff N, et al. Expression of prostaglandin F2α (PGF2α) receptor and its isoforms in the bovine corpus luteum during the estrous cycle and PGF2α-induced luteolysis. Domest Anim Endocrinol. 2012;43(3):227–238 [DOI] [PubMed] [Google Scholar]
- 46. Cavicchio VA, Pru JK, Davis BS, Davis JS, Rueda BR, Townson DH. Secretion of monocyte chemoattractant protein-1 by endothelial cells of the bovine corpus luteum: regulation by cytokines but not prostaglandin F2α. Endocrinology. 2002;143(9):3582–3589 [DOI] [PubMed] [Google Scholar]
- 47. Anderson LE, Schultz MK, Wiltbank MC. Prostaglandin moieties that determine receptor binding specificity in the bovine corpus luteum. J Reprod Fertil. 1999;116(1):133–141 [DOI] [PubMed] [Google Scholar]
- 48. Garrett JE, Geisert RD, Zavy MT, Gries LK, Wettemann RP, Buchanan DS. Effect of exogenous progesterone on prostaglandin F2α release and the interestrous interval in the bovine. Prostaglandins. 1988;36(1):85–96 [DOI] [PubMed] [Google Scholar]
- 49. Zmuda EJ, Viapiano M, Grey ST, Hadley G, Garcia-Ocaña A, Hai T. Deficiency of Atf3, an adaptive-response gene, protects islets and ameliorates inflammation in a syngeneic mouse transplantation model. Diabetologia. 2010;53(7):1438–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Binder AK, Grammer JC, Herndon MK, Stanton JD, Nilson JH. GnRH regulation of Jun and Atf3 requires calcium, calcineurin, and NFAT. Mol Endocrinol. 2012;26(5):873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Duggan SP, Gallagher WM, Fox EJ, Abdel-Latif MM, Reynolds JV, Kelleher D. Low pH induces co-ordinate regulation of gene expression in oesophageal cells. Carcinogenesis. 2006;27(2):319–327 [DOI] [PubMed] [Google Scholar]
- 52. Huo JS, McEachin RC, Cui TX, et al. Profiles of growth hormone (GH)-regulated genes reveal time-dependent responses and identify a mechanism for regulation of activating transcription factor 3 by GH. J Biol Chem. 2006;281(7):4132–4141 [DOI] [PubMed] [Google Scholar]
- 53. Bottone FG, Jr., Moon Y, Alston-Mills B, Eling TE. Transcriptional regulation of activating transcription factor 3 involves the early growth response-1 gene. J Pharmacol Exp Ther. 2005;315(2):668–677 [DOI] [PubMed] [Google Scholar]
- 54. Lu D, Chen J, Hai T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem J. 2007;401(2):559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sandnes D, Müller KM, Akhtar K, Johansen EJ, Christoffersen T, Thoresen GH. Induction of LRF-1/ATF3 by vasopressin in hepatocytes: role of MAP kinases. Cell Physiol Biochem. 2010;25(4–5):523–532 [DOI] [PubMed] [Google Scholar]
- 56. Taniguchi K, Matsuoka A, Kizuka F, et al. Prostaglandin F2alpha (PGF2α) stimulates PTGS2 expression and PGF2α synthesis through NFκB activation via reactive oxygen species in the corpus luteum of pseudopregnant rats. Reproduction. 2010;140(6):885–892 [DOI] [PubMed] [Google Scholar]
- 57. Tamura K, Hua B, Adachi S, et al. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. EMBO J. 2005;24(14):2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang H, Park SH, Choi HJ, Moon Y. Epithelial cell survival by activating transcription factor 3 (ATF3) in response to chemical ribosome-inactivating stress. Biochem Pharmacol. 2009;77(6):1105–1115 [DOI] [PubMed] [Google Scholar]
- 59. Yin X, Wolford CC, Chang YS, et al. ATF3, an adaptive-response gene, enhances TGFβ signaling and cancer-initiating cell features in breast cancer cells. J Cell Sci. 2010;123(Pt 20):3558–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miyamoto A, Shirasuna K, Sasahara K. Local regulation of corpus luteum development and regression in the cow: impact of angiogenic and vasoactive factors. Domest Anim Endocrinol. 2009;37(3):159–169 [DOI] [PubMed] [Google Scholar]
- 61. Shirasuna K, Watanabe S, Yamamoto D, Hayashi M, Nagai K, Miyamoto A. Bovine endothelial cells interact with fully-luteinized, but not luteinizing, granulosa cells in the mRNA expression of endothelin-1 system in response to prostaglandin F2α. Reprod Domest Anim. 2007;42(6):637–642 [DOI] [PubMed] [Google Scholar]
- 62. Korzekwa A, Murakami S, Woclawek-Potocka I, Bah MM, Okuda K, Skarzynski DJ. The influence of tumor necrosis factor α (TNF) on the secretory function of bovine corpus luteum: TNF and its receptors expression during the estrous cycle. Reprod Biol. 2008;8(3):245–262 [DOI] [PubMed] [Google Scholar]
- 63. Henkes LE, Sullivan BT, Lynch MP, et al. Acid sphingomyelinase involvement in tumor necrosis factor α-regulated vascular and steroid disruption during luteolysis in vivo. Proc Natl Acad Sci USA. 2008;105(22):7670–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pru JK, Lynch MP, Davis JS, Rueda BR. Signaling mechanisms in tumor necrosis factor α-induced death of microvascular endothelial cells of the corpus luteum. Reprod Biol Endocrinol. 2003;1(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoshioka S, Acosta TJ, Okuda K. Roles of cytokines and progesterone in the regulation of the nitric oxide generating system in bovine luteal endothelial cells. Mol Reprod Dev. 2012;79(10):689–696 [DOI] [PubMed] [Google Scholar]
- 66. Kim HB, Kim WH, Han KL, et al. cAMP-response element binding protein (CREB) positively regulates mouse adiponectin gene expression in 3T3–L1 adipocytes. Biochem Biophys Res Commun. 2010;391(1):634–639 [DOI] [PubMed] [Google Scholar]
- 67. James CG, Woods A, Underhill TM, Beier F. The transcription factor ATF3 is upregulated during chondrocyte differentiation and represses cyclin D1 and A gene transcription. BMC Mol Biol. 2006;7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grusenmeyer DP, Pate JL. Localization of prostaglandin F2 α inhibition of lipoprotein use by bovine luteal cells. J Reprod Fertil. 1992;94(2):311–318 [DOI] [PubMed] [Google Scholar]
- 69. Wiltbank MC, Belfiore CJ, Niswender GD. Steroidogenic enzyme activity after acute activation of protein kinase (PK) A and PKC in ovine small and large luteal cells. Mol Cell Endocrinol. 1993;97(1–2):1–7 [DOI] [PubMed] [Google Scholar]
- 70. Chaudhuri AC, Harada Y, Shimizu K, Gut M, Dorfman RI. Biosynthesis of pregnenolone from 22-hydroxycholesterol. J Biol Chem. 1962;237:703–704 [PubMed] [Google Scholar]
- 71. Hume R, Kelly RW, Taylor PL, Boyd GS. The catalytic cycle of cytochrome P-450scc and intermediates in the conversion of cholesterol to pregnenolone. Eur J Biochem. 1984;140(3):583–591 [DOI] [PubMed] [Google Scholar]