Abstract
Brucella melitensis causes the most severe and acute symptoms of all Brucella species in human beings and infects hosts primarily through the oral route. The epithelium covering domed villi of jejunal-ileal Peyer's patches is an important site of entry for several pathogens, including Brucella. Here, we use the calf ligated ileal loop model to study temporal in vivo Brucella-infected host molecular and morphological responses. Our results document Brucella bacteremia occurring within 30 min after intraluminal inoculation of the ileum without histopathologic traces of lesions. Based on a system biology Dynamic Bayesian Network modeling approach (DBN) of microarray data, a very early transient perturbation of the host enteric transcriptome was associated with the initial host response to Brucella contact that is rapidly averted allowing invasion and dissemination. A detailed analysis revealed active expression of Syndecan 2, Integrin alpha L and Integrin beta 2 genes, which may favor initial Brucella adhesion. Also, two intestinal barrier-related pathways (Tight Junction and Trefoil Factors Initiated Mucosal Healing) were significantly repressed in the early stage of infection, suggesting subversion of mucosal epithelial barrier function to facilitate Brucella transepithelial migration. Simultaneously, the strong activation of the innate immune response pathways would suggest that the host mounts an appropriate protective immune response; however, the expression of the two key genes that encode innate immunity anti-Brucella cytokines such as TNF-α and IL12p40 were not significantly changed throughout the study. Furthermore, the defective expression of Toll-Like Receptor Signaling pathways may partially explain the lack of proinflammatory cytokine production and consequently the absence of morphologically detectable inflammation at the site of infection. Cumulatively, our results indicate that the in vivo pathogenesis of the early infectious process of Brucella is primarily accomplished by compromising the mucosal immune barrier and subverting critical immune response mechanisms.
Introduction
Brucellosis is a worldwide anthropozoonotic infectious disease caused by small aerobic, non-motile, Gram negative coccobacilli belonging to the genus Brucella. The traditional classification of Brucella species (B. melitensis, B. abortus, B. suis, B. canis, B. ovis and B. neotomae) is based on host preference [1]. Recent isolates from human (B. inopinata), aquatic mammals (B. pinnipedialis and B. ceti) and a common vole (B. microti) have been recognized as new species [2], [3], [4], bringing the current number to ten species in the genus. In susceptible hosts, Brucella spp. produce chronic infections with persistent or recurrent bacteremia, and in middle to late gestation abortion in pregnant animals. With the exception of B. ovis and B. neotomae that are exclusively pathogenic in their primary hosts (sheep and desert rat wood, respectively), and the newest Brucella species whose host specificity has yet to be fully evaluated, brucellae are able to infect other susceptible animals with similar pathogenic effect and clinical disease [5].
The preferred hosts for B. melitensis are goats and sheep. However, B. melitensis can also infect cattle, among which it can be transmitted under specific epidemiological conditions [6], [7], [8], and it causes the most severe and acute symptoms in human beings [9]. The predominant route for B. melitensis penetration after natural exposure is the alimentary tract [1], [10]. Susceptible hosts are primarily infected by contact with aborted fetuses and placental membranes or ingestion of contaminated milk products. Usually B. melitensis enter through the oral mucosa and colonize the lymph nodes that drain the eye, nose and mouth [11], however several studies have isolated Brucella from different sections of the alimentary tract [12] and feces [13] revealing that brucellae survive under the different environmental conditions of the alimentary canal and invade in multiple sites of the gastrointestinal tract.
The epithelium covering domed villi of jejunal-ileal Peyer's patches is an important site of entry for several pathogens, including Brucella [12], [14]. The calf ligated ileal loop model has demonstrated to be a very useful model to study in vivo host:agent molecular and morphological initial interaction [15], [16], [17], [18], a field of study that has been somewhat neglected in brucellosis. We hypothesize that in the early phase of infection B. melitensis actively modulates host responses to avert pathological lesions and immune-based inflammatory cellular pathways to rapidly establish bacteremia and colonize reticular-endothelial and reproductive systems. Here, we describe the temporal in vivo transcriptional profile of the bovine jejunal-ileal Peyer's patches after 0.25, 0.5 1, 2 and 4 h post-B. melitensis infection based on a systems biology Dynamic Bayesian Network modeling approach (DBN) of microarray data. Our results document Brucella bacteremia occurring within 30 min after intraluminal ileum inoculation without histopathologic traces of lesions and only a transient, very early perturbation of the host enteric transcriptome associated to the initial host:pathogen interactions that was rapidly averted later by the pathogen. These data identify major perturbations of pathways/GO and mechanistic regulatory points modulating critical cellular elements at the onset of the Brucella infectious process.
Materials and Methods
Bacterial strain, media and culture conditions
Smooth virulent Brucella melitensis 16M Biotype 1 (ATCC 23456) (American Type Culture Collection, Manassas, VA), originally isolated from an aborted fetal goat was maintained as frozen glycerol stocks. An aliquot of a saturated culture was inoculated into a 50 ml of cell culture media [F12K medium (ATCC)] supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS) (ATCC)], and incubated at 37°C with 5% CO2 with shaking (200 rpm) until reaching the late-log growth phase (OD600 = 0.4) [19]. The concentration and purity of the inoculum was confirmed by plating a serial dilution on tryptic soy agar (TSA) (BD, Franklin Lakes, NJ) and incubated at 37°C with 5% CO2.
Experimental animals and ligated ileal loop surgeries
Four unrelated, clinically healthy 3 to 4-week old, brucellosis-free, male Holstein calves weighing 45–55 kg, maintained on antibiotic-free milk replacer twice daily up to 24 h and water ad libitum up to 12 h prior to the surgery, were used in these experiments. To minimize the interference that other enteropathogens may have on the host gene expression profile, all calves were tested for fecal excretion of Salmonella spp. and Eimeria spp. oocysts twice as previously explained [15], [17], and only negative animals were used. All animal experiments were approved by the Texas A&M University Institutional Animal Care and Research Advisory Committee (AUP 2003-178). Surgeries were performed under biosecurity laboratory 3 (ABSL3) conditions in CDC approved isolated buildings located in the Veterinary Medical Park, Texas A&M University (College Station, TX). The surgical procedures were previously described by our laboratory [20]. Briefly, the calves were fasted for 12 hours prior to the surgery. The calves were pre-sedated with propofol (Propoflo; Abbot Laboratories, Chicago, IL) followed by placement of an endotracheal tube and maintenance with isofluorane (Isoflo; Abbott Laboratories) for the duration of the experiment. Throughout the experimental procedure (12 h), the calves were monitored for vital signs (blood pressure, heart rate, hydration status, depth of anesthesia and temperature). The abdominal wall was incised, the distal jejunum and ileum exteriorized, and segments ranging in length from 6 to 8 cm were ligated with umbilical tape leaving 1-cm interloops between them. Seven loops were inoculated intraluminally with 3 ml of a suspension containing 1×109 CFU of B. melitensis 16 M/ml (B. melitensis inoculated loops) using a 26 gauge 3/8 inch needle, and the seven other loops (media inoculated control) were injected with 3 ml of sterile cell culture media (F12K media supplemented with 10% HI-FBS). The loops were replaced into the abdominal cavity, the incision temporarily closed, and reopened for collecting samples beginning at 15 min and continuing through 12 h. Calves were euthanized with an intravenous bolus of sodium pentobarbital at the completion of the procedures. One infected and one control loop were collected at 7 time points (0.25, 0.5, 1, 2, 4, 8 and 12 h post-inoculation), and the samples were processed for quantification of tissue-associated bacteria, morphology and host gene expression profiling. Samples from the surgery room were transported in triple container to an approved BSL3 laboratory for immediate processing.
Bacteriology
For quantification of Peyer's patch-associated B. melitensis, two-6 mm biopsy punches (0.1 g) were excised from every infected loop, intensely washed three times in PBS to reduce extracellular bacteria, macerated and diluted in 1 ml of distilled water. Similar procedures were followed in control loops to identify any viable Brucella, cross contamination during material processing, or Brucella dissemination via lymphatic or blood vessels. To determine the number of viable CFU of B. melitensis in tissues, lysates were serially diluted and cultured on selective Farrell's selective media [TSA (BD) supplemented with Brucella selective supplement (Oxoid Limited, Hampshire, UK) according to manufacture's instructions] [21].
To address the possibility of B. melitensis systemic invasion and dissemination, 5 ml of blood were collected by aseptic venipuncture of the jugular vein into 0.75 ml of acid-citrate-dextrose (ACD) at T0 (pre-inoculation), 0.5, 1, 2, 4, 8 and 12 h time, points, and samples from mesenteric lymph nodes and liver were immediately extracted after the calves were euthanized. One ml of blood from every time point was cultured in a non-selective biphasic medium (also known as Castañeda's media) [21]. Briefly, TSA (BD) +1% agar (Difco, Lawrence, KS) cooled to 56°C after autoclaving before adding the Brucella selective supplement (Oxoid). The molten medium was well mixed and 20 ml dispensed into 75 cm2 cell culture flask (Corning, Corning, NY). Flasks were laid down to allow the media to solidify along one side. The following day, tryptic soy broth (TSB) (BD) was autoclaved, cooled and Brucella selective supplement (Oxoid) added according to manufacture's instructions. Fifteen ml were dispensed aseptically in each flask already containing the solid phase, and the sterility of the media was checked by overnight incubation at 37°C. Blood cultures were incubated for at least 1 month at 37°C and examined for growth of Brucella twice a week. To quantify the number of viable CFU of B. melitensis in mesenteric LN and liver, 0.1 g of tissue were lysed, serially diluted in distilled water and cultured on selective Farrell's selective media.
Morphologic analysis
For light microscopy observation, full cross-sections of each loop always including Peyer's patch were fixed in buffered 10% formalin, processed according to the standard procedures for paraffin embedding, sectioned at 5-µm thickness, stained with hematoxylin and eosin, and examined with light microscopy.
Isolation of total RNA from intestinal loops
Six to ten 6 mm biopsy punches were excised from every loop. The mucosa of the samples was immediately dissected, macerated and homogenized in TRI-Reagent® (Ambion, Austin, TX) (2 biopsy punches/1 ml of reagent) with a hand-held mechanical tissue grinder equipped with a RNase, DNase free plastic disposable pestle. RNA was extracted according to TRI-Reagent manufacturer's instructions. The pellet was re-suspended in DEPC-treated water (Ambion) with 2% DTT and 1% RNase inhibitor (Promega). Contaminant genomic DNA was removed by RNase-free DNase I treatment (Ambion) according to the manufacture's instructions, and samples were stored at −80°C until used. RNA concentration was quantified by NanoDrop® ND-1000 (NanoDrop, Wilmington, DW), and the quality was determined using a Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Sample preparation and slide hybridization
Four biological replicates from every time point (T0.25, T0.5, T1, T2 and T4) and every condition (B. melitensis- inoculated loops and media-inoculated control loops) were labeled and hybridized as previously described [22]. Briefly, 10 µg of total RNA from each experimental sample (n = 40) were reverse transcribed to cDNA using random hexamer primers (Invitrogen), labeled with Cy5 (Amersham Pharmacia Biosciences) and co-hybridized against Cy3 labeled cDNA generated from the bovine reference RNA sample to a custom 13K bovine 70mer oligoarray [23]. Slides were hybridized at 42°C for approximately 40 h in a dark humid hybridization chamber (Corning).
Microarray data acquisition, normalization and analysis
Immediately after washing, the slides were scanned using a commercial laser scanner (GenePix 4100; Axon Instruments Inc., Foster City, CA). Scans were performed using the autoscan feature with the percentage of saturated pixels set at 0.03%. The genes represented on the arrays were adjusted for background and normalized to internal controls using image analysis software (GenePixPro 6.0; Axon Instruments Inc.). Genes with fluorescent signal values below background were disregarded in all analyses. Arrays were initially normalized against the bovine reference RNA, and the resulting data were analyzed and modeled using an integrated platform termed the BioSignature Discovery System (BioSignatureDS™) (Seralogix, LLC, Austin, TX; www.seralogix.com) explained in detail elsewhere [15], [17], [24], [25], [26]. To achieve a rigorous analysis, genes were ranked and ordered according to their expression magnitudes and gene variance was computed using a Bayesian predicted variance value. The Bayesian variance was determined by using a sliding window algorithm that averages 50 variances directly on the ascending and descending ordered sides of each gene of interest [27]. Significantly changed genes were determined with the Bayesian z-test (p<0.025). BiosignatureDS tools for statistical Z-score gene thresholding, Bovine pathway and GO activation scoring, Mechanistic gene identification and Genetic network system model were used for the comprehensive analysis performed in this study. Microarray data are deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) Accession # GSE41835.
Microarray results validation
Five immunity-related genes, which had differential expression by microarray results, were analyzed at every time point by quantitative RT-PCR (qRT-PCR) following the protocol described elsewhere [15], [17]. Briefly, two micrograms of RNA were reverse transcribed using TaqMan® Reverse Transcription reaction (Applied Biosystems, Foster City, CA). For relative quantitation of target cDNA, samples were analyzed in individual tubes in SmartCycler II (Cepheid, Sunnyvale, CA). Primers (Sigma Genosys) of tested genes were designed by Primer Express Software v2.0 (Applied Biosystems) ( Table 1 ). For each gene tested, the individual calculated threshold cycles (Ct) were averaged among each condition and normalized to the Ct of the bovine glyceraldehyde phosphate dehydrogenase (GAPDH) from the same cDNA samples before calculating the fold change using the ΔΔCt method [28]. Statistical significance was determined by Student's t-test and expression differences considered significant when P<0.05. As gene expression by microarray and qRT-PCR were based on z-score and fold-change, respectively, array data were considered valid if the fold change of each gene tested by qRT-PCR was expressed in the same direction as determined by microarray analysis.
Table 1. Primers for Real Time – PCR Analysis of Genes in B. melitensis-Infected Bovine Peyer's Patch.
| GENEBANK ACCESSION # | GENE SYMBOL | GENE NAME | FORWARD PRIMERS (5′-3′) | REVERSE PRIMERS (5′-3′) |
| NM_173895 | BPI | Bactericidal/permeability-increasing protein | CCTCCGAAACTCACCATGAAG | TGTCCAATCTGAGCTCTCCAATAA |
| NM_175793 | MAPK1 | Mitochondrial-activated protein kinase 1 | GGCTTGGCCCGTGTTG | GGAAGATGGGCCTGTTGGA |
| NM_174006 | CCL2 | Chemokine (C-C motif) ligand 2 | TCCTAAAGAGGCTGTGATTTTCAA | AGGGAAAGCCGGAAGAACAC |
| NM_173925 | IL8 | Interleukin 8 | TGCTTTTTTGTTTTCGGTTTTTG | AACAGGCACTCGGGAATCCT |
| NM_001033608 | MIF | Macrophage migration inhibitory factor | CTGCAGCCTGCACAGCAT | TTCATGTCGCAGAAGTTGATGTAG |
| NM_001034034 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | TTCTGGCAAAGTGGACATCGT | GCCTTGACTGTGCCGTTGA |
Results and Discussion
B. melitensis Colonize and Invade Bovine Host through Peyer's patches Without Inducing Morphologic Lesions in the First 12 h p.i
While it is accepted that the alimentary tract is the main route of invasion for B. abortus and B. melitensis [1], [10], however the specific portal(s) of entry are not well defined. Although Brucella spp. are not considered to be enteric pathogens [29], B. abortus was isolated from the small intestine of calves 5 h after being fed with infected milk [12] and from feces of coyotes orally infected [13], which indicates that under natural conditions viable Brucella are able to reach the intestinal Peyer's patches.
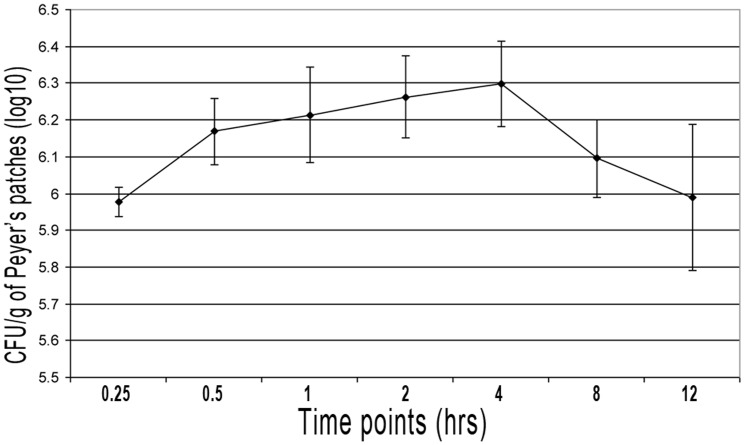
We assessed the kinetics of B. melitensis 16 M infection after intraluminal inoculation by quantifying the CFU present in the Peyer's patches at different time points. The intestinal loops were intraluminally inoculated with 3×109 CFU of B. melitensis 16 M and 0.1 g of Peyer's patches collected beginning at 15 min and continuing through 12 h. Fifteen minutes post inoculation (p.i.), a few less than 106 CFU/g of tissue were recovered. The number of tissue-associated B. melitensis rose rapidly, reaching the peak (2×106 CFU/g of tissue) by 4 h p.i., and decreasing at later time points ( Figure 1 ). No differences in histopathological features were detected between control loops and B. melitensis-infected loops of the four calves at any time during the 12 h p.i. experiments. The inverse relation between Brucella invasion and time post-infection was also observed in B. abortus S19-infected bovine ileal loops [30]. The decreasing uptake over the time was attributed to the degeneration of Brucella in the intestinal lumen and consequent lack of adhesion due to enteric surface modifications. Another proposed explanation was that Brucella saturated the surface receptors on the epithelium, as it was reported in Salmonella [31]. A third possible explanation for the decreasing number of tissue-associated Brucella over time may be the very rapid translocation from the lumen through the tissue into lymph and ultimately into systemic circulation.
Figure 1. Kinetics of Peyer's Patch Infection with B. melitensis 16 M.
Jejunal-ileal loops were intraluminally inoculated in 3 ml containing 1×109 CFU of B. melitensis 16 M/ml. Tissue (Peyer's patch) samples of 0.1 g of mucosal tissue were extracted at 0.25, 0.5, 1, 2, 4, 8 and 12 h from every infected loop, intensively washed 3 times in PBS, macerated and diluted in 1 ml of distilled water. To evaluate the kinetics of the infection, macerated samples were serially diluted and cultured on Farrell's medium. Numbers of CFU recovered from bovine Peyer's patches are the average of 4 calves. Bars represent standard deviation.
To assess Brucella systemic invasion and dissemination, blood from the jugular vein was collected and cultured. Blood samples taken from the first calf were contaminated and not considered in the final analysis of the data. Blood from the second calf was collected as early as 1 h time point while samples from the two other animals were taken from 30 min p.i. through the end of the procedure. All blood samples before the inoculation (T0, control) were Brucella-free, but B. melitensis were isolated (but could not be quantified) by using Casteñeda's media from all samples from 30 min p.i. through 12 h p.i. B. melitensis were also isolated from mesenteric lymph nodes and liver at 12 h p.i. (an average of 3.2×103 and 1×102 B. melitensis CFU/g of tissue, respectively) from all inoculated calves, and also from control loops of one animal at 8 and 12 h time points (2×102 and 8×102 CFU of B. melitensis/g of Peyer's patch, respectively). These results indicate a very rapid penetration of B. melitensis through Peyer's patches followed by systemic dissemination and organ colonization via blood and lymphatic vessels without traces of associated histopathologic lesions.
To elucidate host and Brucella-induced host mechanisms responsible for this very rapid invasion and translocation, we studied global gene expression using microarray analysis to host gene expression coupled with a Bayesian inference analysis modeling.
Host gene expression response is perturbed immediately after infection but rapidly returns to normal state
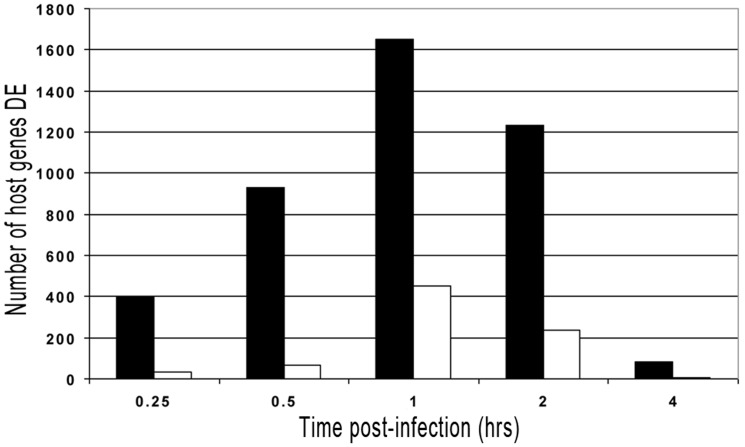
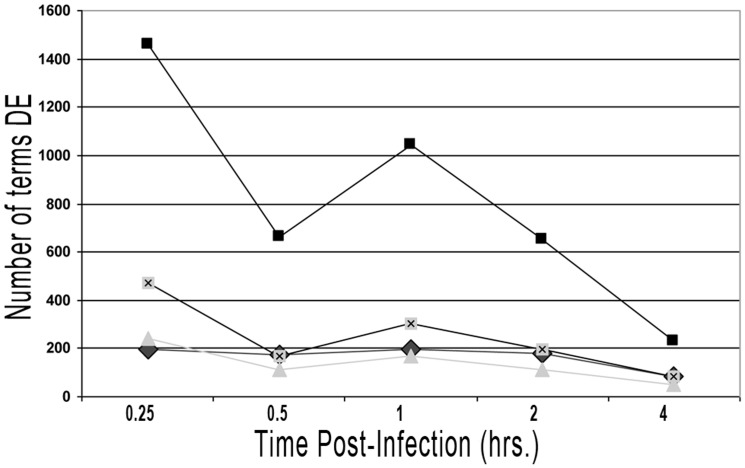
Previous work in our lab indicated that four is the lowest number of biological replicates (i.e., calves) to detect significant differences between the treatments used [15], [17], [25]. Considering that B. melitensis was isolated from control intestinal loops of one animal at 8 and 12 h p.i., only RNA samples taken from 15 min to 4 h p.i. were considered for further analysis. Bayesian inference identified a progressive host gene, pathway and gene ontology (GO) terms expression modification with the highest activity at one hour p.i. in infected intestinal loop tissues compared with control tissues, which decreased at later time points ( Figure 2 and 3). More details are shown in File S1, Supplementary Figure 1 and Supplementary Table 1 through 23 (TableS1 – S23 in File S1). Overall, these results indicate that host molecular response is markedly perturbed at a very early time post-Brucella infection, with a strong tendency to rapidly return to a normal state at later time points. These results are in concordance with results from our previous experiments [26] and other studies [36] that had analyzed host gene expression at two different time points after Brucella infection.
Figure 2. Graphic Representation of Bovine Genes Differentially Expressed throughout the Experiment.
(DE = differentially expressed). Solid bars represent genes up-regulated; open bars represent genes down-regulated.
Figure 3. Graphic Representation of Number of Pathways and GO Terms Differentially Expressed at each time point from 15 min to 4 h p.i.
Squares = GO Biological processes; Squares with X = GO Molecular functions; Triangle = Cellular components; Diamond = Pathways.
Focusing our analysis on host gene expression alterations involved in response to Brucella spp. infection [22], [26], [32], [33], [34], [35], [36] has enabled us to follow the behavior of pathways for Cell communications, Cell growth and death, Cell motility, Immune system, Infectious disease, Membrane transport, Signal transduction and Signaling molecules and interaction categories. These categories create a subgroup of 56 pathways.
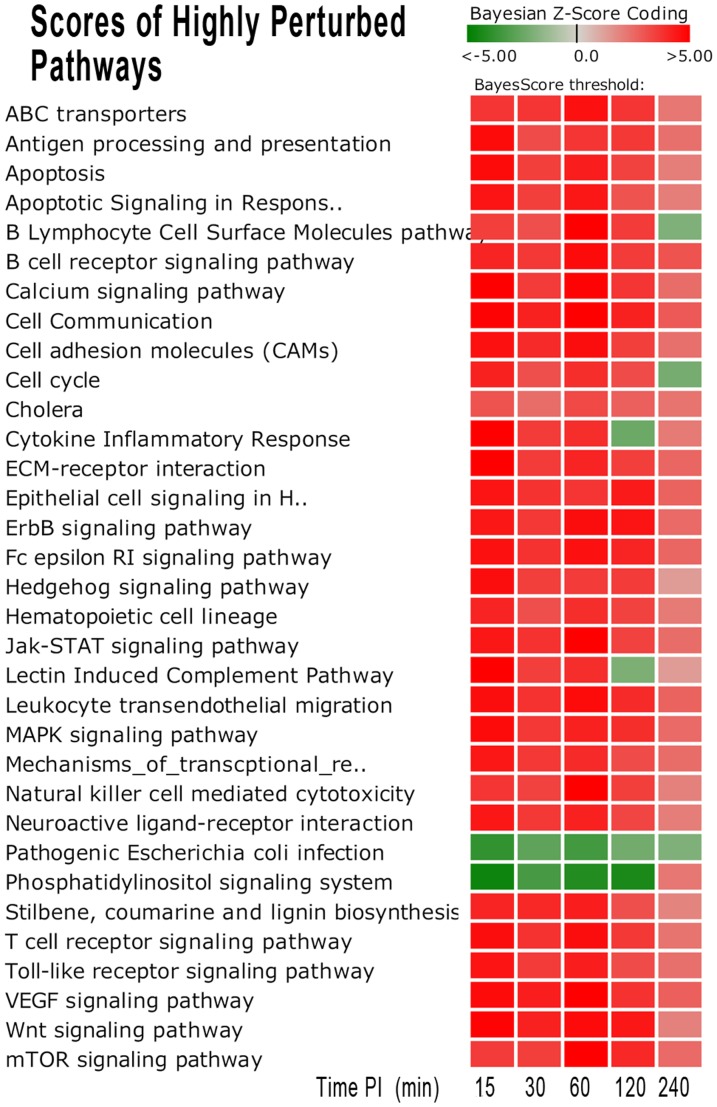
Among them, there were 37 pathways that were highly perturbed at earlier time points (either activated or repressed) and then had decreased differential expression (DE) at later time points to near or non-statistical differences in comparison with control tissues ( Table 2 , Figure 4). Within these perturbed (either activated or repressed) pathways, there were several pathways that have been extensively studied in host-response manipulation by Brucella infection such as Apoptosis [37], [38], Antigen processing and presentation [39], [40], Toll like receptor signaling [14], Leukocyte transendothelial migration [41] and MAPK signaling [26], [42].
Table 2. Bayesian z-score of 56 Pathways in Eight categories post- B. melitensis Infection Compared to Controls.
| Name | Description | Category* | 0.25 h | 0.5 h | 1 h | 2 h | 4 h |
| hsa04510 | Integrin-mediated cell adhesion | CC | 4.48 | 3.68 | 4.3 | 4.35 | 2.31 |
| hsa04520 | Adherens junction | CC | 4.06 | −3.13 | 4.01 | 3.84 | 1.84 |
| hsa04530 | Tight junction | CC | −4.35 | −3.68 | −4.74 | 4.27 | 2.23 |
| hsa04540 | Gap junction | CC | 3.74 | 2.88 | 3.05 | 3.12 | 2.18 |
| hsa04110 | Cell cycle | CGD | 4.18 | 3.05 | 3.86 | 3.19 | −2.21 |
| hsa04210 | Apoptosis | CGD | 4.72 | 3.44 | 4.25 | 3.42 | 2.02 |
| hsa04810 | Regulation of actin cytoskeleton | CM | 4.51 | 3.43 | 4.4 | 4.16 | 2.55 |
| hsa04610 | Complement and coagulation cascades | IS | 4.85 | −3.23 | 4.56 | −2.39 | 2.28 |
| hsa04612 | Antigen processing and presentation | IS | 4.72 | 3.15 | 3.69 | 3.67 | 2.27 |
| hsa04620 | Toll-like receptor signaling pathway | IS | 4.48 | 3.55 | 4.28 | 3.13 | 2.29 |
| hsa04640 | Hematopoietic cell lineage | IS | 4.11 | 3.1 | 3.87 | 3.38 | 2.07 |
| hsa04650 | Natural killer cell mediated cytotoxicity | IS | 3.71 | 3.36 | 5.13 | 3.5 | 1.9 |
| hsa04660 | T cell receptor signaling pathway | IS | 4.66 | 3.82 | 4.61 | 3.6 | 2.2 |
| hsa04662 | B cell receptor signaling pathway | IS | 4.13 | 3.63 | 4.72 | 3.55 | 2.98 |
| hsa04664 | Fc epsilon RI signaling pathway | IS | 4.56 | 3.75 | 4.56 | 4.08 | 2.53 |
| hsa04670 | Leukocyte transendothelial migration | IS | 4.65 | 3.77 | 4.73 | 3.91 | 2.58 |
| hsa99103 | Apoptotic DNA fragmentation and homeostasis pathway | IS | −1.72 | −2.16 | −2.17 | −1.83 | −2.18 |
| hsa99104 | Apoptotic Signaling Response to DNA Damage pathway | IS | 4.52 | 3.47 | 4.41 | 3.01 | 1.98 |
| hsa99105 | BRC signaling pathway | IS | 3.8 | −2.48 | 3.72 | 2.19 | −1.47 |
| hsa99106 | B Cell Receptor Complex pathway | IS | 2.39 | 4.05 | 4.56 | 0.95 | 2.9 |
| hsa94660 | T cell receptor signaling pathway Antigen Processing | ID | 4.71 | 4.17 | 4.94 | 3.73 | 2.15 |
| hsa00626 | Nitrobenzene degradation | ID | 4.11 | 2.21 | 4.86 | 2.7 | 1.69 |
| hsa00940 | Stilbene, coumarine and lignin biosynthesis | ID | 4.13 | 4 | 4.28 | 3.07 | 1.8 |
| hsa01430 | Cell Communication | ID | 4.92 | 4.19 | 5.77 | 4.16 | 2.82 |
| hsa05110 | Cholera | ID | 2.98 | 2.42 | 3.22 | 2.67 | 2.26 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | ID | 4.47 | 3.79 | 3.69 | 4.35 | 2.58 |
| hsa05130 | Pathogenic Escherichia coli infection | ID | −3.77 | −2.77 | −3.12 | −2.7 | −2.03 |
| hsa99020 | Lectin Induced Complement Pathway | ID | 5.39 | 3.48 | 3.87 | −2.06 | 1.27 |
| hsa99100 | Activation of Csk through the T-Cell Receptor pathway | ID | 3.29 | 1.51 | 2.45 | 1.75 | 0.91 |
| hsa99101 | Activation of cAMP-dependent protein kinase PKA pathway | ID | 2.47 | −1.03 | 1.22 | 2.29 | 1.79 |
| hsa99102 | Anthrax Toxin Mechanism of Action pathway | ID | 2.85 | 1.39 | 3.15 | 3.87 | 0.99 |
| hsa99107 | B Lymphocyte Cell Surface Molecules pathway | ID | 3.51 | 3.06 | 5.2 | 3.57 | −2.01 |
| hsa99108 | Blockade Neurotransmitter Release by Botulinum Toxin path | ID | −0.96 | 0 | 0 | 0.63 | 0 |
| hsa99109 | CCR3 signaling in Eosinophils pathway | ID | 4.78 | −2.89 | 5.06 | −4.63 | 3.29 |
| hsa99110 | CD40L Signaling Pathway | ID | 5.71 | 4.32 | 3.64 | −4.24 | 3.28 |
| hsa99111 | Calcium Signaling by HBx of Hepatitis B virus pathway | ID | −1.23 | −0.97 | −2.21 | −0.1 | 0 |
| hsa99112 | Cytokine Inflammatory Response | ID | 5.43 | 3.58 | 3.84 | −2.49 | 2.08 |
| hsa99113 | Trefoil Factors Initiate Mucosal Healing | ID | −4.46 | −3.07 | 4.37 | 3.56 | −2.57 |
| hsa99114 | Transcptional_repression_by_DNA_methylation | ID | 4.39 | 3.61 | 3.91 | 3.15 | 2.35 |
| hsa02010 | ABC transporters | MT | 3.74 | 3.72 | 4.55 | 3.7 | 2.18 |
| hsa04010 | MAPK signaling pathway | ST | 4.75 | 3.63 | 4.17 | 3.89 | 2.43 |
| hsa04012 | ErbB signaling pathway | ST | 4.39 | 3.66 | 4.61 | 4.53 | 2.47 |
| hsa04020 | Calcium signaling pathway | ST | 4.97 | 3.59 | 4.86 | 3.68 | 2.37 |
| hsa04070 | Phosphatidylinositol signaling system | ST | −4.62 | −3.35 | −4.21 | −4.3 | 2.18 |
| hsa04150 | mTOR signaling pathway | ST | 3.55 | 3.46 | 5.24 | 4.01 | 2.45 |
| hsa04310 | Wnt signaling pathway | ST | 4.85 | 4.15 | 4.73 | 4.41 | 1.93 |
| hsa04330 | Notch signaling pathway | ST | −4.43 | 3.61 | 3.56 | 4.11 | 2.6 |
| hsa04340 | Hedgehog signaling pathway | ST | 4.66 | 3.49 | 3.55 | 3.52 | 1.27 |
| hsa04350 | TGF-beta signaling pathway | ST | 4.47 | −2.88 | −3.65 | 4.02 | 1.96 |
| hsa04370 | VEGF signaling pathway | ST | 4.75 | 4.24 | 5.11 | 3.82 | 2.68 |
| hsa04630 | Jak-STAT signaling pathway | ST | 4.43 | 3.76 | 4.93 | 3.37 | 2.39 |
| hsa04060 | Cytokine-cytokine receptor interaction | SM | 4.34 | 3.38 | 4.12 | −3.06 | 2.04 |
| hsa04080 | Neuroactive ligand-receptor interaction | SM | 4.44 | 3.62 | 4.17 | 3.31 | 2.02 |
| hsa04512 | ECM-receptor interaction | SM | 5.03 | 3.55 | 4.07 | 3.47 | 2.52 |
| hsa04514 | Cell adhesion molecules (CAMs) | SM | 4.55 | 3.95 | 4.64 | 3.48 | 2.31 |
Categories - CC = cell communication; CGD = cell growth and death; CM = cell motility; IS = immune system; ID = infectious disease; MT = membrane transport; ST = signal transduction; SM = signaling molecule measured time points 15, 30, 60, 120, and 240 minutes p.i. The Bayeian z-scores represent the degree of perturbation of the group of pathway genes versus the controls. Positive z-scores represent activation of the pathway (pathway score is dominated by more up-regulated genes), while the negative z-score represents pathway repression (pathway score is dominated by down-regulated genes). Bolded pathways represent pathways highly perturbed at earlier time points (either activated or repressed) with decreased differential expression at later time points to near or non-statistical differences in comparison with control tissues.
Figure 4. Heatmap of Significantly Perturbed Pathways Scores in Bovine Peyer's Patch Infected with B. melitensis.
Thirty-seven pathways were identified as significantly perturbed (97.5% confidence) from the early stage of infection (15–60 m.p.i.). The darker red gradients indicate higher Bayesian activation scores (more up-regulated gene expression within the pathway) while the darker green gradients indicate more repressed pathway activity (more down-regulated gene expression).
We will now focus the manuscript on the most relevant findings detected by our analysis related to the host response during the early onset of Brucella infection, i.e., bacterial adherence, invasion, colonization and dissemination within the host [43], [44].
Brucella adhere and invade by compromising the mucosal barrier
The bacterial adhesion to cell membrane is the essential first step in colonization, followed by invasion. According to previous publications, Brucella attach to cultured epithelial cell lines via receptor molecules containing sialic acid or sulphated residues and also bind extracellular matrix proteins (ECM), which probably contribute to the spread of the pathogen and tissue colonization [45]. A very recent publication indicates that Brucella attach to the cellular prion protein on the M cells surface before internalizing [46]. Our analysis identified two adherence-related pathways significantly activated throughout the experiment in infected samples compared to control ones: the Cell Molecules Adhesion (CAM) and the ECM receptor interaction pathways. Deeper analysis of both pathways revealed several genes differentially expressed (Bayesian z-score >|2.24|) at different times post-infection (Tables 3 and 4). Some of these genes showed an erratic expression, probably due to the several different cell types analyzed simultaneously; however other genes were consistently up or down-regulated through the study and may be strong candidates involved in Brucella adherence. Among them, there are specifically four gene products (SDC2 –syndecan 2, ITGAL –integrin alpha L, ITGB2 –integrin beta 2, and IBSP – integrin binding sialoprotein-) that commonly are expressed on cell surfaces and their products are involved in bacterial pathogenesis. Syndecans comprise a major family of cell surface heparan sulfate proteoglycans (HSPGs) and their role in microbial infections is currently an active area of research. Neisseria gonorrhoeae, Pseudomona aeuruginosa and several enterobacteria attach and invade hosts by interacting to sydecans molecules [47], [48], [49]. Particularly, syndecan-2 (SDC2) expressed on the surface of dendritic cells has been shown to bind to HIV and facilitate viral transmission to CD4-positive T cells [50]. Until now, nothing was known about the role of SDC in Brucella pathogenesis, but this finding may illustrate a fruitful avenue of future investigation. Integrins are a group of mammalian cell surface molecules which can function as receptors for bacterial ligands, promoting bacterial adherence and internalization into mammalian cells [51]. ITGAL combines with ITGB2 to form the integrin LFA-1 (lymphocyte function-associated antigen-1), which is expressed on all leukocytes and resting Langerhan's dendritic cells as well [52]. We had demonstrated previously that blocked α-chain (ITGAL) of the integrin LFA-1, decrease Brucella abortus binding to mononuclear phagocytes [53]. Our studies surprisingly discovered IBSP gene as being mechanistic in nature. IBSP is a major structural protein of the bone matrix, and the only known extraskeletal site of its synthesis is the trophoblast, a major target cell for the pathogenesis of Brucella induced abortion. Little is known about the potential roles of the IBSP gene product in the microbial pathogenesis of brucellosis particularly in Peyer's patch, however given the uniqueness of this finding IBSP deserves special emphasis.
Table 3. Significantly Altered Genes (Bayesian z-score |2.24|) in the Cell Molecule Adhesion (CAM) pathway.
| Symbol | Description | 0.25 h | 0.5 h | 1 h | 2 h | 4 h |
| Cd28 | CD28 antigen | 2.12 | 0.69 | −3.13 | 0 | −0.22 |
| CD34 | CD34 molecule | 0 | −1.61 | −2.02 | −2.35 | 0 |
| CD58 | CD58 molecule | −3.47 | −2.7 | −2.81 | −3.13 | 0 |
| CD99 | CD99 molecule | −0.99 | 0.86 | 0.82 | 2.5 | 0 |
| CDH1 | cadherin 1, type 1, E-cadherin (epithelial) | −1.88 | −2.82 | 0 | −1.83 | 0.52 |
| CDH15 | cadherin 15, M-cadherin (myotubule) | 2.42 | 0.66 | −0.59 | 2.99 | 1.63 |
| CDH3 | cadherin 3, type 1, P-cadherin (placental) | 2.85 | 1.75 | 0 | 0 | −0.99 |
| CDH5 | cadherin 5, type 2 (vascular endothelium) | −3.36 | −2.08 | −2.54 | −2.65 | −1.97 |
| CLDN1 | claudin 1 | −2.49 | −1.73 | −2.39 | −1.76 | −1.03 |
| Cldn16 | claudin 16 | 2.42 | 1.46 | 1.05 | 0.28 | 2.09 |
| CLDN3 | claudin 3 | 2.71 | 0 | 0 | 0.42 | 2.27 |
| CLDN4 | claudin 4 | 1.33 | 2.18 | 2.43 | 0.95 | 1.29 |
| CLDN5 | claudin 5 (transmembrane protein deleted in velocardiofacial) | 1.79 | 2.14 | 2.95 | 0 | 1.94 |
| CLDN7 | claudin 7 | 1.16 | −0.65 | −3.48 | −2.05 | 0 |
| CNTN1 | contactin 1 | −2.86 | −1.18 | −1.43 | −0.68 | −0.21 |
| Cntnap2 | contactin associated protein-like 2 | 0.51 | 2.59 | 3.41 | 2.75 | 1.26 |
| VCAN | versican | 2.7 | 3.98 | 5.16 | 4.14 | 2.58 |
| ESAM | endothelial cell adhesion molecule | 2.94 | 4.05 | 1.81 | 1.85 | 2.87 |
| VCAM1 | vascular cell adhesion molecule 1 | −2.33 | −2.02 | −3.27 | −3.89 | −1.26 |
| Pecam1 | platelet/endothelial cell adhesion molecule 1 | 2.87 | 0.8 | −1.18 | −1.76 | −0.64 |
| NCAM1 | neural cell adhesion molecule 1 | −3.03 | 0 | 0 | 1.22 | −0.1 |
| F11R | F11 receptor | −0.74 | −0.92 | −0.3 | −1.37 | 2.42 |
| GLG1 | golgi apparatus protein 1 | 0.51 | 1.04 | 2.22 | 0.54 | 2.72 |
| HLA-A | major histocompatibility complex, class I, A | −3.44 | −2.13 | −2.14 | −2.92 | 0 |
| HLA-DMA | major histocompatibility complex, class II, DM alpha | 1.35 | 3.65 | 2.3 | 0.75 | 0 |
| HLA-DOA | major histocompatibility complex, class II, DO alpha | 2.37 | 0 | 1.95 | 0 | 0.61 |
| HLA-DOB | major histocompatibility complex, class II, DO beta | 0.97 | 0.98 | 2.45 | 0.08 | 1.25 |
| HLA-DRA | major histocompatibility complex, class II, DR alpha | −3.07 | −1.86 | −1.32 | −3.09 | −1.57 |
| ICAM1 | intercellular adhesion molecule 1 | 0.25 | 0 | 2.25 | 0 | 0 |
| ICAM2 | intercellular adhesion molecule 2 | 3.06 | 4.5 | 6.19 | 0.44 | 2.82 |
| ITGA4 | integrin, alpha 4 (antigen CD49D, alpha 4 of VLA-4 receptor) | 2.04 | 2.18 | 3.61 | −0.63 | 0 |
| ITGA8 | integrin, alpha 8 | 0.47 | 1.12 | 3.18 | 0.22 | 0.9 |
| ITGA9 | integrin, alpha 9 | −4.84 | −3.42 | −2.55 | −2.27 | −2.02 |
| ITGAL | integrin, alpha L (antigen CD11A (p180), LFA 1; alpha peptide | 1.74 | 2.73 | 4.28 | 2.81 | 0.81 |
| ITGAV | integrin, alpha V (vitronectin receptor, alpha peptide, CD51) | −2.7 | −2.42 | −0.24 | −1.82 | 0 |
| ITGB2 | integrin, beta 2 (complement component 3 receptor 3 and 4 unit) | 3.13 | 2.8 | 4.15 | 4.04 | 2.34 |
| NRXN3 | neurexin 3 | −2.85 | 0 | −1.88 | 1.53 | −2.39 |
| PTPRM | protein tyrosine phosphatase, receptor type, M | 4.37 | −3.96 | −3.08 | −1.19 | −0.24 |
| PVRL1 | poliovirus receptor-related 1 (herpesvirus entry mediator C) | 1.12 | 1.24 | 1.05 | 2.17 | 2.59 |
| PVRL2 | poliovirus receptor-related 2 (herpesvirus entry mediator B) | 0.84 | 1.87 | 5.18 | 1.33 | 1.19 |
| SDC2 | syndecan 2 | 4.05 | 3.81 | 3.75 | 3.27 | 2.16 |
The table lists significantly perturbed (activated or repressed) genes at any time among the 5 different time points (0.25 h through 4 h p.i.).
Table 4. Significantly Altered Genes (Bayesian z-score |2.24|) in the Extra-Cellular Membrane (ECM) pathway.
| Symbol | Description | t = 15 | t = 30 | t = 60 | t = 120 | t = 240 |
| Agrn | agrin | −0.89 | −2.48 | 1.26 | 2.28 | 1.83 |
| Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | −3.3 | −3.8 | −4.64 | −1.05 | 0 |
| Chad | chondroadherin | −2.65 | −0.08 | −2.44 | −1.6 | −2.62 |
| Col5a1 | collagen, type V, alpha 1 | 4.03 | 3.59 | 4.81 | 3.31 | 2.5 |
| Col5a2 | collagen, type V, alpha 2 | −2.89 | −1.59 | −1.73 | −2.45 | −1.94 |
| COL6A2 | collagen, type VI, alpha 2 | 0.13 | 2.88 | 2.43 | 1.84 | 0.65 |
| COL6A3 | collagen, type VI, alpha 3 | 3.13 | 0 | 0 | −0.34 | 1.59 |
| OI4 | collagen, type I, alpha 2 | 0 | 0 | 0 | −2.95 | 0 |
| SEDC | collagen, type II, alpha 1 | 2.35 | 0 | −2.18 | −1.74 | 3.21 |
| DAG1 | dystroglycan 1 (dystrophin-associated glycoprotein 1) | −2.29 | −0.68 | −0.5 | 0 | −0.03 |
| Fndc1 | fibronectin type III domain containing 1 | 4.22 | −2.3 | −2.69 | −0.87 | −2.14 |
| Ibsp | integrin binding sialoprotein | −2.82 | −2.03 | −3.84 | −2.43 | 3.32 |
| ITGA10 | integrin, alpha 10 | 2.86 | 0.93 | 4.74 | 2.05 | 1.85 |
| ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 3.18 | 0.61 | 1.92 | 2.26 | 2.27 |
| ITGA2B | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | 0.99 | 2.01 | 4.05 | 2.51 | 2.81 |
| ITGA4 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | 1.73 | 1.66 | 2.88 | −0.68 | 0 |
| ITGA8 | integrin, alpha 8 | 0.36 | 1.15 | 3.1 | 0 | 0.86 |
| ITGA9 | integrin, alpha 9 | −3.58 | −2.82 | −2.38 | −1.94 | −0.62 |
| ITGB4 | integrin, beta 4 | 1.31 | 0 | 2.78 | 2.22 | 1.1 |
| LAMA1 | laminin, alpha 1 | 0.38 | 1.27 | 3.41 | 1.28 | 1.09 |
| Lama5 | laminin, alpha 5 | −1.32 | 2.6 | 0.78 | 2.42 | 2.43 |
| LAMB2 | laminin, gamma 1 (formerly LAMB2) | 0.88 | 2.67 | 2.89 | 2.24 | 2.48 |
| LOC131873 | no description | 2.26 | 1.12 | 0.12 | 0.49 | 0.34 |
| Npnt | nephronectin | 3.04 | 0.04 | −0.36 | −2.2 | 0.83 |
| SDC2 | syndecan 2 | 3.71 | 3.82 | 3.01 | 2.91 | 1.95 |
| THBS1 | thrombospondin 1 | −3.07 | −0.85 | 0 | 1.07 | 1.88 |
| THBS2 | thrombospondin 2 | 2.82 | −0.14 | −2.71 | −2.49 | −0.07 |
| Tnn | trophinin | 2.89 | 1.64 | 0.74 | 0 | 0.64 |
| VTN | vitronectin | 0 | 2.25 | 0 | 0 | −1.33 |
The table lists significantly perturbed (activated or repressed) genes at any time among the 5 different time points (0.25 h through 4 h p.i.).
A few minutes after adhesion, Brucella are quickly internalized in in vitro studies. In non-professional phagocytic cells, Brucella are able to auto-induce internalization by activating small GTPases of the Rho subfamily (i.e. Rho, Rac, Cdc42) and modulate rearrangements of the host cell actin cytoskeleton and microtubules [26], [54]. In phagocytic cells, entry and survival of Brucella requires functional lipid rafts on cell membrane [55]. In vivo studies using the ileal loop model demonstrated that transepithelial migration of Brucella occur mainly through endocytosis by the follicle associated epithelium (FAE) of Peyer's patches and uptake by dendritic cells-penetrating the FAE [14], [30], [46]. In addition to enterocytes, a critical component of the intestinal barrier is the intercellular junction complexes between adjacent enterocytes that form a semi-permeable diffusion barrier. Several pathogens have developed strategies to interact and manipulate junctional complexes, in order to disrupt and cross the epithelial barriers [56]. Our pathway analysis revealed two intestinal barrier-related pathways (Tight Junction (TJ) and Trefoil Factors Initiated Mucosal Healing (TFIMH)) significantly repressed in the early stage of infection (15–60 minutes p.i.) (Table 2). The repressed expression of these two pathways associated with Brucella host invasion suggest subversion of mucosal epithelial barrier function as previously shown that activation or repression of gene expressions of junction pathways may lead to strengthening or weakening of the primary intestinal barrier, respectively [57], [58]. These results are in concordance with our previous publication where we reported that Brucella-infected epithelioid like cells down-regulate GO biological processes related with cell-cell adhesion, such as positive regulation of cell adhesion (GO:0045785), regulation of cell-cell adhesion (GO:0022407), cell-cell junction organization (GO:0045216), regulation of cell adhesion (GO:0030155) and cell junction organization (GO:0034330) among others [26]. Of note is the fact that other cell communication/junction pathways that include Gap Junction (GJ), Adherents Junction (AJ) and Integrin-mediated Cell Adhesion (IMCA) were not scored as repressed as shown in the heat map of Figure 5 .
Figure 5. Pathway Score Heat Map for Cell Communication Related Pathways.
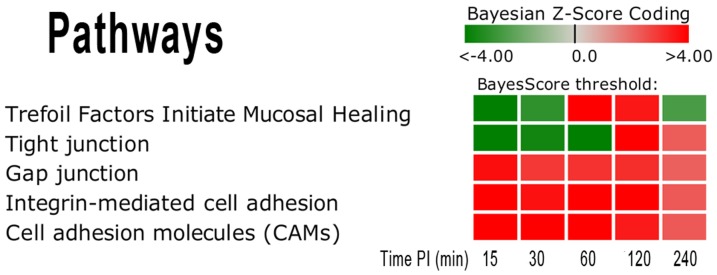
This heat map shows the repressed state of activity for the Tight Junction (TJ) and Trefoil Factors (TF) pathway in comparison to the Gap Junction, Integrin-mediated Cell Adhesion and Cell Adhesion Molecules pathways. Note that the TJ and TF pathways have a complex activation pattern. The TJ is tri-phasic in that it is highly repressed in the early stage, becomes moderately activated at 60 and 120 min p.i, and then becomes repressed at 240 min p.i. The TF pathway is bi-phasic in that it is highly repressed in the early stage but becomes moderately activated at 120 and 240 min p.i. The darker red gradients indicate higher Bayesian activation scores (more up-regulated gene expression within the pathway) while the darker green gradients indicate more repressed pathway activity (more down-regulated gene expression).
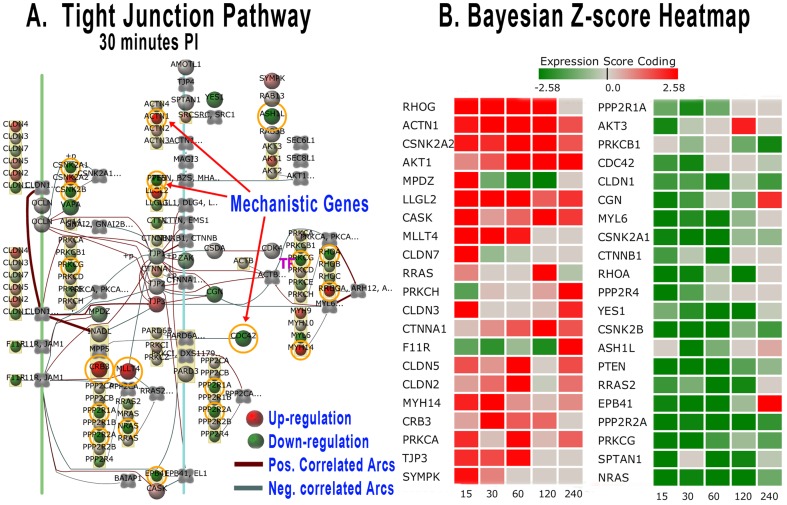
Tight junction pathway analysis
The tight junctions are the most apical intercellular complex and are responsible for controlling the permeability of the paracellular conduits between epithelia cells. The molecular interactions between genes of this pathway were visualized by the BioSignatureDS software as shown in Figure 6(A) , while Figure 6(B) provides a heat map of genes on this pathway found significantly expressed (Bayesian z-score |>2.24|). The repressed state of the TJ pathway in the first hour p.i. is evident by the numerous down-regulated genes in both the Bayesian network model and the gene score heat map. Some of the more dominant down-regulated genes in the early stage of infection are NRAS, SPTAN1, PRKCG, PPP2R2A, EPB41, PTEN, CSNK2B, YES1, RHOA, CSNK2A1, MYL5, CGN, CLDN1, CDC42, and AKT3. The biological roles of these subverted genes are described in Supplemental Table 24 (Table S24 in File S1). Interestingly, NRAS, RHOA, CDC42, and AKT3 were previously identified as mechanistic genes having intersecting points across several pathways (Table S22 in File S1). Of special note are the genes SPTAN1, EPB41, RHOA, MYL5, CGN, and CLDN1 that are involved in maintaining the integrity of the epithelial layer and permeability. Additional insight into the regulatory relationships within the Tight Junction pathway was found by interrogating the Bayesian network model of Figure 6(A). The model suggests that RHOA (Ras homolog family member A) has significant correlated influence over MYL6 (myosin, light chain 6, alkali, smooth muscle and non-muscle), which is involved in actin binding. Additionally, it was found that CGN (Cingulin) has regulatory influence on the genes MYL6, ACTB (actin, beta), MYH9, MYH10, and MYH14 (myosin, heavy chain 9, 10 and 14), all of which are involved in cell motility, cell shape, integrity and cell attachments.
Figure 6. Tight Junction Pathway Bayesian Network Model and Gene Score Heat Map.
(A) Tight Junction pathway Bayesian network representation at 30 min post-inoculation. Gene nodes with orange circles on the network are those defined as mechanistic genes that surpass a threshold Bayesian z-score| >2.24|. The network shows gene nodes with gradient colors representing the level of expression (deeper red for higher up-regulated genes and deeper green for down-regulated). (B) The Bayesian score heat map for the gene expression of Brucella infected host Peyer's patch versus non-infected controls. The heat map is colorized and corresponds to the gene node expression levels. Grey color represents little to no expression difference between Brucella-infected and control loops. The heat map columns are by time post-infection in minutes.
Trefoil factors initiated mucosal healing (TFIMH) pathway analysis
Epithelial continuity can also depend on a family of small, yet abundant, secreted proteins, namely the trefoil factors. The trefoil factors maintain the integrity of the gastrointestinal tract, despite the continual presence of microbial flora and injurious agents [59]. The immune related TFIMH pathway was suppressed in the early stage post infection ( Figure 5 ). Unfortunately, not all the trefoil factors gene probes were included on the bovine microarray employed during this study. However, the TFIMH pathway suppression (as determined by other observed gene expressions) implies impaired trefoil factor gene expression, and consequently, a possible invasion mechanism of Brucella by subverting mucosal healing. Genes that dominate the suppressed pathway activity are PTK2, GHR, RHOA, CTNNB1, MAPK3, and SHC1. The biological roles of these genes are described in Supplemental Table 25 (Table S25 in File S1) except for RHOA, which was described in Table S24 in File S1. The gene PTK2 (Protein tyrosine kinase 2) is strongly down regulated across all time points and was identified previously as a mechanistic gene intersecting several pathways (Table S22 in File S1). PTK2 encodes a protein-tyrosine kinase that plays an essential role in regulating cell migration, adhesion, spreading, reorganization of the actin cytoskeleton, formation and disassembly of focal adhesions and cell protrusions, cell cycle progression, cell proliferation and apoptosis. It is found concentrated in the focal adhesions that form between cells growing in the presence of extracellular matrix constituents. Even though Brucella are not normally regarded as an enteric pathogens and the paracellular route of entry has not been previously described in its pathogenesis, our analysis indicates that alteration of genes involved in maintenance of the intestinal integrity may constitute an important and novel target associated with the successful invasion by Brucella through the host's mucosal barrier.
Brucella establish and disseminate in the host by subverting the immune response
Upon crossing the first level of defense at the mucosal epithelium, Brucella are transported to lymph nodes by phagocytic cells and phagosomes act as intracellular niche for Brucella [5]. Based on this premise, it's expected that pathways involved in phagocytosis are activated during Brucella infection. Our study confirms that the Lectin pathway, which is initiated by the binding of mannose-binding lectin to carbohydrates found on bacterial cell surfaces and leads to phagocyte recruitment, was significantly activated during the early stage of Brucella infection (with a Bayesian z-score of >2.24 at 15 to 60 min p.i. [ Table 2 ]). This result is in agreement with Fernandez-Prada et al. who demonstrated that Lectin pathway was involved in complement deposition and complement-mediated killing of Brucella [60]. The Lectin pathway (or mannose-binding pathway) is part of the Complement and Coagulation Cascades (CCC), a nonspecific defense mechanism involved in triggering the innate immune response against pathogens. Of the seven genes in the lectin induced complement pathway, MASP2, C9 and C5 are up-regulated and C6 is down-regulated at 15 min p.i.; and C5 and C9 are up-regulated at 30 min p.i. The biological roles of these four genes are shown in Supplemental Table 26 (Table S26 in File S1 ). Coincidently, Lectin and CCC pathways were found to be up-regulated in Brucella- infected HeLa cells at 4 h post-infection, and MASP2 gene was indicated as a key mechanistic gene in both pathways [26]. Though the Lectin pathway is significantly activated in the early stages, however, the combined CCC pathway is observed to have temporal cyclic response (activation and repression) during the first 4 h p.i. Activation of the Lectin pathway may lead to phagocytosis, which plays an important role in establishment and dissemination of Brucella in the host. The lack of a phagocytic pathway in KEGG, lead us to identify the significant genes for “phagocytosis” using GO terms. Of the 34 genes representing phagocytosis, 13 were significantly perturbed in the first 4 h post-Brucella infection. CD47, CDC42SE2, and DOCK1 were down-regulated across all time points while SIRPA was strongly down-regulated in the early stage of infection. GATA2 and AHSG were up-regulated at 15 min p.i. and ELMO1, ELMO2, FCER1G, AZU1, SCARB1, SFTPD and CORO1A were up-regulated between 15–60 min p.i. Their biological roles for phagocytosis GO Group are summarized in Supplemental Table 27 (Table S27 in File S1 ). CD47 plays an important role in both cell adhesion and in the modulation of integrins, and is a receptor for SIRPA, binding to which prevents maturation of immature dendritic cells and inhibits phagocytosis and cytokine production. As previously stated, SIRPA was repressed during early Brucella-host interaction, but is later activated, suggesting a novel mechanism of Brucella manipulation of dendritic cells maturation.
From a different perspective of phagocytosis, Starr et al. demonstrated that Brucella abortus subvert the autophagy machinery of host cells to establish an intracellular niche favorable for its replication [61]. Regulation of autophagy pathway is repressed as indicated by a Bayesian z-score <−2.24 at 15 and 30 min p.i. ( Table 2 ) which supports Brucella's ability of taking advantage of the repressed autophagy machinery for its own survival instead of being engulfed and killed.
Pathways related to the immune system i.e. Leukocyte transendothelial migration, Toll-like receptor signaling, Hematopoietic cell lineage, Fc-epsilon RI signaling, B cell receptor signaling, B cell receptor complex, T cell receptor signaling, Apoptotic signaling in response to DNA damage, Antigen processing and presentation, JAK-STAT signaling and Natural killer cell mediated cytotoxicity were activated as indicated by a Bayesian z-score of >2.24 across all the time points ( Table 2 ). The strong activation of these innate immune response pathways should suggest that the host is mounting an appropriate protective immune response. However, Brucella are known to successfully evade the host's response suggesting that Brucella may be selectively subverting critical signaling mechanisms. For instance, expression of the two key innate immunity anti-Brucella cytokines [62] such as TNF-α and IL12p40 (or IL12B) were not significantly changed (Bayesian z-score of >|2.24|) throughout the study. These results are in agreement with previous publications that had identified several Brucella's strategies to avoid activation of innate immune system during the onset of the infection [39], [41], [63], [64], [65]. As examples, the Toll-Like Receptor and Cytokine-cytokine Receptor Interaction pathways were examined in more detail to determine if any important signaling events were disrupted following Brucella invasion as described next.
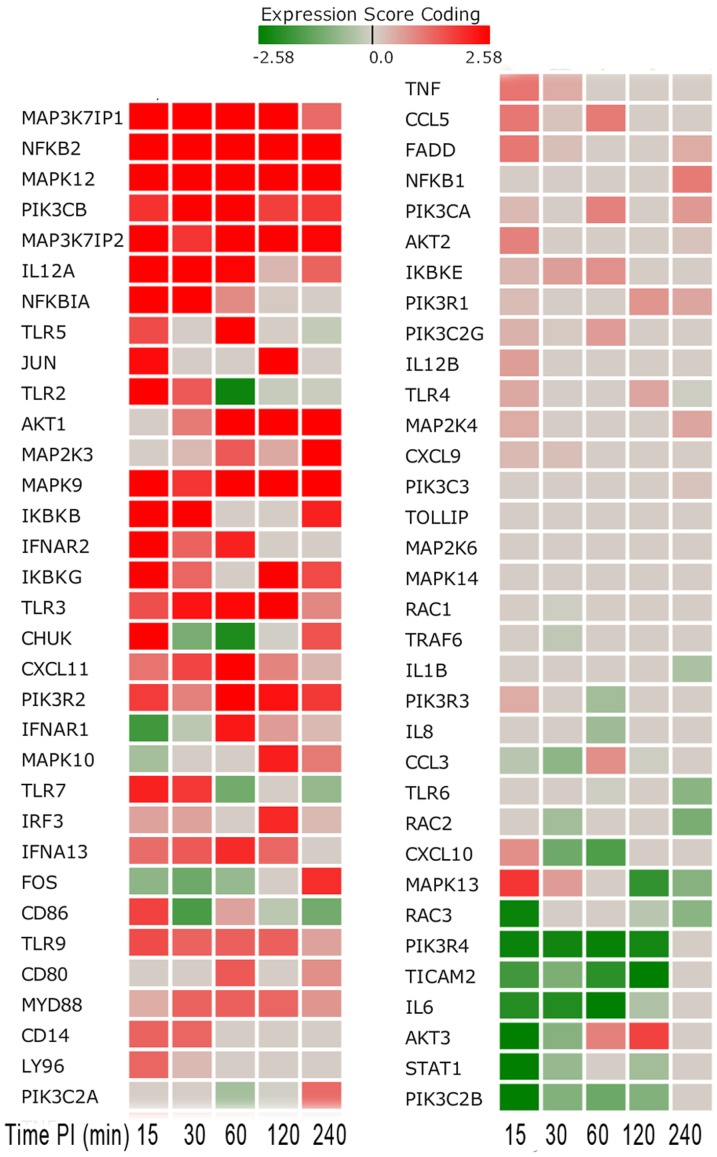
Toll-like receptor signaling (TLRS) pathway subversion. TLRs are crucial components of the innate immune system that recognize conserved microbial components and trigger antimicrobial responses. With the triggering of the TLRS pathway, it could be presumed that the host had initiated an effective immune response. Examining this pathway at the network and gene expression level indicated that the source of pathway perturbation comes from genes that are both highly up-regulated and interestingly, a number of repressed genes over the complete time course. Figure 7 shows the Bayesian z-score gene expression heat map for all genes on the TLRS pathway, and Table 5 shows z-scores of those genes significantly expressed in the pathway. Our analysis reveals that the only toll-like receptors differentially expressed during the study, were TLR2 (activated at 15 min p.i. and repressed at 1 h p.i.), TLR3 (activated at 0.5, 1 and 2 h p.i.) and TLR5 (significantly up-regulated only at 15 min p.i.), but not for any other toll-like receptors, which is consistent with the hypothesis that Brucella modify, reduce and/or hide PAMP-bearing molecules to reach its replication niche before host immune system's detection [39]. In addition, Brucella protein Btp1 (recently re-named BtpA) and BtpB interfere with TLR signaling by producing inhibitory homologues of the TIR (Toll/Interleukin 1 receptor) domain that block MyD88-induced activation of NFkβ or NFkβ nuclear translocation [14], [66], [67]. The persistent inhibition of NFkβ1 has been linked to inappropriate immune cell development and continues to provide evidence that supports the lack of inflammatory response through subversion of key signaling events in the TLRS pathway. Furthermore, the toll-like receptor signaling appears defective in that it is not producing the expected expression pattern for proinflammatory cytokines. For instance, proinflammatory cytokines-encoded genes such as IL-1β, TNF-α, IL-6 and IL8 are either not differentially expressed or are down-regulated. Additionally, chemokines CCL3 (MIP-1α), CCL5 (RANTES), CXCL9, CXCL10, and CXCL11 -encoded genes were not significantly expressed and suggesting a potential disruption of monocyte and natural killer cell stimulation and T-cell migration. This lack of expression of inflammatory mediators may partially explain the absence of morphologically detectable inflammation at the site of infection [5].
Figure 7. Toll-like Receptor Related Gene Score Heat Map by Time Point Post-Inoculation.
The heat map shows a number of up-regulated genes occurring in the early stage (15–60 minutes post-inoculation) as indicated by the darker red boxes as well as down-regulated genes as indicated by the darker green boxes.
Table 5. Genes Significantly Expressed in the TLRS Pathway (z-score |2.24|).
| Symbol | Description | 0.25 | 0.5 | 1 h | 2 h | 4 h |
| MAP3K7IP1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | 3.15 | 4.67 | 5.83 | 5.27 | 1.28 |
| NFKB2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | 3.46 | 3.92 | 5.44 | 3.83 | 3.04 |
| MAPK12 | mitogen-activated protein kinase 12 | 3.6 | 4.28 | 3.78 | 4.67 | 5.32 |
| PIK3CB | phosphoinositide-3-kinase, catalytic, beta polypeptide | 1.96 | 2.6 | 4.86 | 1.81 | 1.89 |
| MAP3K7IP2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | 3.3 | 1.92 | 4.32 | 3.79 | 2.52 |
| IL12A | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, | 3.5 | 3 | 2.51 | 0.34 | 1.37 |
| NFKBIA | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 3.28 | 2.59 | 0.88 | 0.09 | 0 |
| TLR5 | toll-like receptor 5 | 1.64 | 0 | 3.23 | 0 | −0.21 |
| JUN | jun oncogene | 2.42 | 0 | 0 | 3.22 | 0 |
| TLR2 | toll-like receptor 2 | 3.17 | 1.48 | −2.41 | −0.17 | −0.15 |
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | 0 | 1.05 | 3.14 | 3.15 | 2.82 |
| MAP2K3 | mitogen-activated protein kinase kinase 3 | 0 | 0.29 | 1.48 | 0.49 | 3.09 |
| MAPK9 | mitogen-activated protein kinase 9 | 3.05 | 1.9 | 3.08 | 2.95 | 2.68 |
| IKBKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | 2.96 | 2.55 | 0 | 0 | 2.17 |
| IFNAR2 | interferon (alpha, beta and omega) receptor 2 | 2.92 | 1.36 | 2.17 | 0 | 0 |
| IKBKG | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | 2.69 | 1.31 | 0.04 | 2.84 | 1.69 |
| TLR3 | toll-like receptor 3 | 1.59 | 2.33 | 2.46 | 2.78 | 0.89 |
| CHUK | conserved helix-loop-helix ubiquitous kinase | 2.77 | −1.12 | −2.21 | −0.02 | 1.54 |
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 1.16 | 1.7 | 2.67 | 0.94 | 0.34 |
| PIK3R2 | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | 1.82 | 0.99 | 2.59 | 2.34 | 1.86 |
| IFNAR1 | interferon (alpha, beta and omega) receptor 1 | −1.79 | −0.31 | 2.26 | 0.68 | 0.32 |
| RAC3 | ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | −2.42 | 0 | 0 | −0.34 | −0.91 |
| PIK3R4 | phosphoinositide-3-kinase, regulatory subunit 4 | −2.44 | −2.34 | −2.48 | −2.3 | 0 |
| TICAM2 | toll-like receptor adaptor molecule 2 | −1.79 | −1.05 | −2.05 | −2.55 | 0 |
| IL6 | interleukin 6 (interferon, beta 2) | −2.12 | −2.16 | −2.58 | −0.48 | 0 |
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) | −2.88 | −0.93 | 0.97 | 1.76 | 0 |
| STAT1 | signal transducer and activator of transcription 1, 91 kDa | −3.04 | −0.75 | 0 | −0.61 | 0 |
| PIK3C2B | phosphoinositide-3-kinase, class 2, beta polypeptide | −3.5 | −0.98 | −1.27 | −0.99 | 0 |
The table lists significantly perturbed (activated or repressed) genes at any time among the 5 different time points (0.25 h through 4 h p.i.).
Cytokine-cytokine receptor interactions (CCRI) pathway subversion
The CCRI pathway was activated in the early stage of infection with several genes dominating the activation. These genes were involved in extracellular membrane receptor interaction that included chemokines (CC and CXC), interleukins (ILs) and platelet-derived growth factors (PDGFs). Chemokines and their receptors are important for the migration of various cell types into inflammatory sites. With regard to the chemokines, only CCL21, CXCL10, and BLR1 were highly up-regulated at 15 min p.i.; BLR1, CCL14, CXCL16, CXCL11 and CCR4 were significantly expressed at 1 h p.i.; and only CCL11 and CXCL16 became significantly up-regulated at 4 h p.i. The other remaining chemokines were either down-regulated or minimally expressed as shown in Figure 8 .
Figure 8. Chemokine Gene Score Heat Map by Time Point Post-Inoculation.
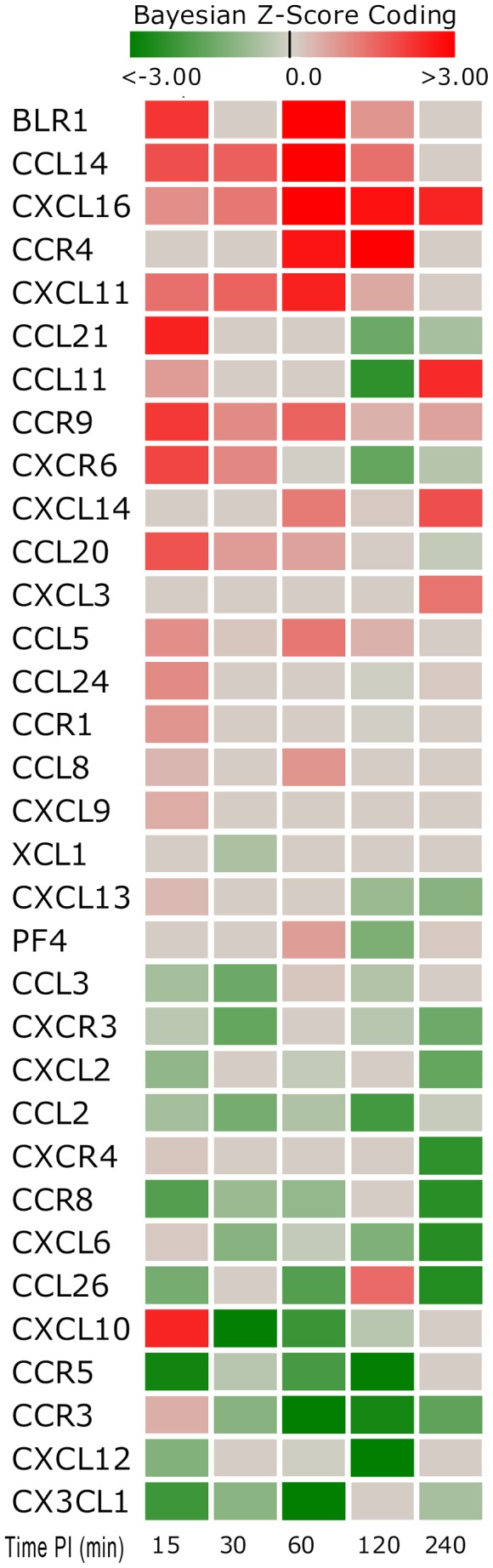
The heat map shows a number of both up-regulated and down-regulated genes (15–240 minutes post-inoculation). Note that there were large numbers of down-regulated and non-expressed chemokine genes. Red indicates an activated state while green indicates repression and grey is no expression change from control.
The function of the immune system depends in a large part on interleukins (ILs) that are predominately synthesized by helper CD4+ T lymphocytes, as well as through monocytes, macrophages, and endothelial cells. Interleukins promote the development and differentiation of T, B and hematopoietic cells. The strongly expressed genes encoded by interleukins or their receptors at 15 min post-Brucella infection include IL-1RAP, IL-12A, IL-4, IL-3, IL-15, IL-28RA, IL-6R, IL-7, IL-15RA, IL-2RG, IL-5, IL-2, and IL-10RB. However, the up-regulation was short-lived since many of these genes reversed direction of expression or become minimally expressed in later time points as shown in Figure 9 . Their biological roles are summarized in Supplemental Table 28 (Table S28 in File S1). IL-1RAP, IL-2RG, IL-10RB, IL-15RA reversed expression at 30 min p.i. and later became significantly down-regulated. IL-1RAP induces synthesis of acute phase and proinflammatory proteins during infection or tissue damage; the IL-2RG gene is an important signaling component of many interleukin receptors, including those of interleukin −2, −4, −7 and −21; the gene IL-10RB encodes a cell surface receptor required for the activation of five cytokines: IL10, IL22, IL26, IL28 and IL29; and IL-15RA encodes a receptor that is reported to enhance cell proliferation and expression of apoptosis inhibitor BCL2L1/BCL2-XL and BCL2. This down-regulation of such genes at 30 min p.i. suggested that Brucella manipulate the host's immune response for survival and proliferation after its initial invasion. Interestingly, the soluble epithelial factors, IL-7 and IL-15, maintained higher expression levels from 15 to 60 min p.i. The proteins encoded by these genes differentially regulate homeostasis of intraepithelial lymphocytes and other mucosal leukocytes. IL7 can be produced locally by intestinal epithelial and epithelial goblet cells, and may serve as a regulatory factor for intestinal mucosal lymphocytes.
Figure 9. Interleukin Gene Score Heat Map by Time Point Post Inoculation.
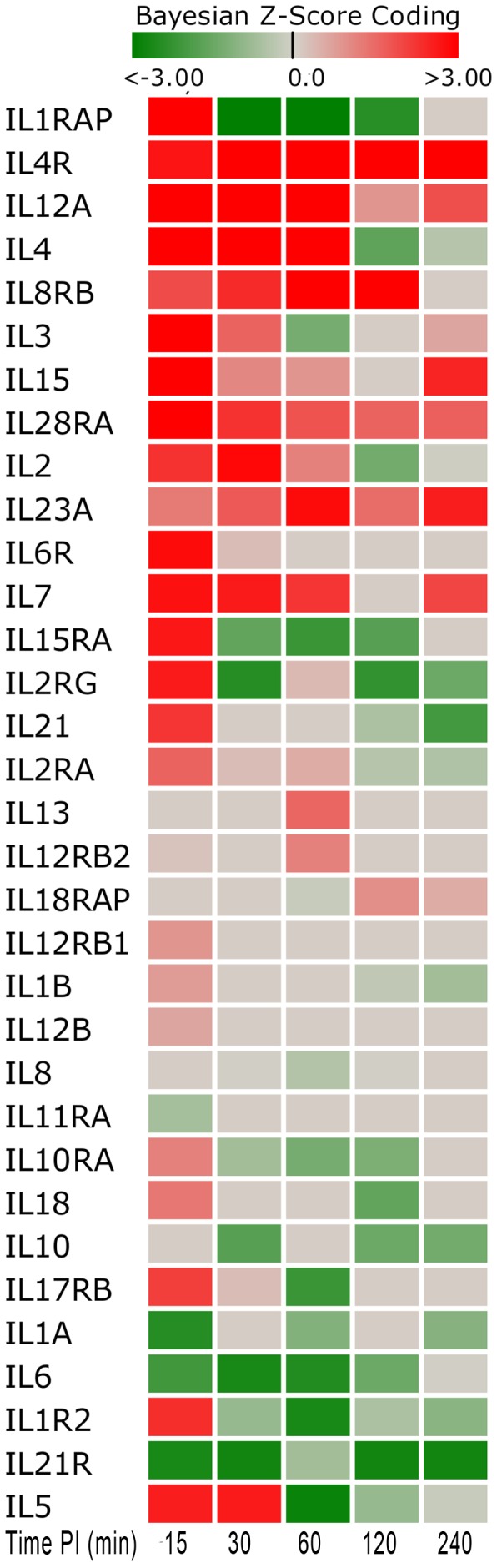
The heat map shows a number of both up-regulated and down-regulated genes (15–240 minutes post-inoculation). The heat map shows that much of the up-regulation is short-lived for many of these genes and reversed expression direction or minimally expressed in later time points. Red indicates an activated state while green indicates repression and grey is no expression change from control.
The elevated expression of platelet-derived growth factors (PDGF) has been linked to early signaling events for infection by intracellular pathogens. Only two PDGF pathway genes, VEGFB and CSF1 were strongly up-regulated at some time points in the early stage (15–60 min p.i.). VEGFB signals via the endothelial receptor FLT1 which encodes a receptor tyrosine-kinase that plays a key role in vascular development and regulation of vascular permeability. However, FLT1 was strongly down-regulated throughout the time course. CSF1 encodes a cytokine that plays an essential role in the regulation of survival, proliferation and differentiation of hematopoietic precursor cells, especially mononuclear phagocytes, such as macrophages and monocytes, the in vivo refuge of Brucella. The KIT gene was strongly down-regulated across all time points. KIT encodes a tyrosine-protein kinase that acts as cell-surface receptor for the cytokine KITLG/SCF and plays an essential role in the regulation of cell survival and proliferation, hematopoiesis, stem cell maintenance, gametogenesis, mast cell development, migration and function, and in melanogenesis. The down regulation of KIT may lead to the disruption of a number of important signaling events and may be a novel survival/proliferation mechanism exploited by Brucella.
Conclusions
In summary, our systems biology pathway and GO analyses of the in vivo host intestinal transcriptome revealed that in the early phase of infection, B. melitensis actively modulated host responses to avert pathological lesions and immune-based inflammatory cellular pathways to rapidly invade Peyer's patch, metastasize to mesenteric lymph nodes and quickly establish bacteremia. As expected, the analysis identified genes and pathways previously known to have roles in Brucella infection and also detected new infection-related genes, pathways and GO terms. Molecular analysis provided evidence that the enteric mucosal barrier is compromised during early time post-infection. We identified cell molecule adhesion (CAM) and ECM receptor interaction pathways that were perturbed, more specifically SDC2, ITGAL, ITGB2, and IBSP genes. Additionally, several of the pathways of the enteric Tight Junction of the mucosa were significantly repressed in the early stages of infection, especially NRAS, SPTAN1, PRKCG, PPP2R2A, EPB41, PTEN, CSNK2B, YES1, RHOA, CSNK2A1, MYL5, CGN, CLDN1, CDC42, and AKT3 genes. Likewise, the Trefoil Factor Initiated Mucosal Healing pathways were significantly suppressed in early stage of infection, more specifically, PTK2, GHR, RHOA, CTNNB1, MAPK3, and SHC1 genes. Our data confirmed prior observations that Toll-Like Receptor Signaling pathways were largely subverted, apparently by Brucella reducing or hiding PAMP-bearing molecules to reach its niche before host immune detection. While the Cytokine-Cytokine Receptor Interactions (CCRI) pathways were initially up-regulated very early in the infection, this response was very short-lived and quickly down-regulated within the first hour post-infection. In summary, our data indicate that the pathogenesis of the early infectious process of B. melitensis consists of compromising the mucosal immune barrier and subverting critical immune response mechanisms.
Supporting Information
Supplemental figures and tables. Figure A in File S1. Validation of Bovine Microarray Results by Quantitative Real Time-PCR. cDNA was synthesized from the same RNA samples used for microarray hybridization. Five randomly selected genes (A = BPI; B = MAPK1; C = MIF; D = CCL2; E = IL8.) that were differentially expressed by microarrays in B. melitensis-infected bovine Peyer's patch between 15 min and 4 h p.i. as compared to non-infected tissues (control) extracted at the same time points, were validated by quantitative RT-PCR. Fold changes was normalized to the expression of GAPDH and calculated using the ΔΔCt method. All tested genes at all time points had fold-changes altered in the same direction in microarray and qRT-PCR. White bars represent fold-change by microarray analysis and black bars represent fold-change by qRT-PCR. Table S1 in File S1. Detailed List of Host Genes with Differential Expression (z-score >|2.24|) in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch in at least one time point. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S2 in File S1. Bayesian z-score for All Host Pathways in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S3 in File S1. List of All Biological Process-Related Host Genes Differentially Expressed in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S4 in File S1. List of All Cellular Component-Related Host Genes Differentially Expressed in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S5 in File S1. List of All Molecular Function-Related Host Genes Differentially Expressed in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S6 in File S1. Group 1 - GO Biological Process Terms Significantly Perturbed Early (Activated or Repressed) but Not Later. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S7 in File S1. Group 2 - GO Biological Process Terms Consistently Perturbed (Activated or Repressed) Throughout the Experiment; Not Significantly Perturbed Early but Later; or Consistently Altered Perturbation (Activated to Repressed or Vice Versa). Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S8 in File S1. Group 1 - GO Cellular Components Terms Significantly Perturbed Early (Activated or Repressed) but Not Later. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S9 in File S1. Group 2 - GO Cellular Component Terms Consistently Perturbed (Activated or Repressed) Throughout the Experiment; or Terms Not Significantly Perturbed Early but Later; or Consistently Altered Perturbation (Activated to Repressed or Vice Versa). Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S10 in File S1. Group 1 - GO Molecular Function Terms Significantly Perturbed Early (Activated or Repressed) but Not Later. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S11 in File S1. Group 2 - GO Molecular Function Terms Consistently Perturbed (Activated or Repressed) Throughout the Experiment; or Terms Not Significantly Perturbed Early but Later; or Consistently Altered Perturbation (Activated to Repressed or Vice Versa). Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S12 in File S1. Comparative Table of Group 1 and Group 2 Gene Ontology Terms Clustered to the Top 15 Primary Immune Related Biological Process Categories. This table shows lists the mapping of perturbed GO term gene groups to their higher level GO category definitions. The number of perturbed GO terms that mapped to each category is provided in the Count column. Table S13 in File S1. Group 1 - Detailed Listing of the Top 15 Highly Perturbed Gene Ontology Terms Mapped to Major Immune Related Biological Process. This table lists each GO term that was mapped (clustered) to a main category related to an immune response. There were 108 main categories to which 686 perturbed GO term sets of genes resulted in mapping to 55 categories. Group 1 represents those GO terms that were significantly perturbed at earlier time points (either activated or repressed) but not at later time points. Table S14 in File S1. Group 2 - Detailed Listing of the Top 15 Highly Perturbed Gene Ontology Terms Mapped to Major Immune Related Biological Process. This table lists each GO term that was mapped (clustered) to a main category related to an immune response. There were 108 main categories to which 114 perturbed GO term sets of genes resulted in mapping to 23 categories. Group 2 are GO terms consistently perturbed (up- or down-regulated) throughout the experiment; or GO terms not significantly perturbed at earlier time points but at later times; or GO terms that consistently changed their state of perturbation (from activated- to repressed or vice versa). Table S15 in File S1. Comparative Table of Group 1 and Group 2 Gene Ontology Terms Clustered to Primary Cellular Component Categories. The mapping of terms in this table is to major categories related to the cellular component categories. Table S16 in File S1. Group 1 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Cellular Component Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 12 main categories to which 112 perturbed GO term sets of genes resulted in mapping to 9 categories. Definition of Group 1 is same as described in Table S13. Table S17 in File S1. Group 2 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Cellular Component Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 12 main categories to which 26 perturbed GO term sets of genes resulted in mapping to 6 categories. Definition of Group 2 is same as described in Table S14. Table S18 in File S1. Comparative Table of Group 1 and Group 2 Gene Ontology Terms Clustered to Primary Molecular Function Categories. The mapping of terms in this table is to major categories related to the primary molecular function categories. Table S19 in File S1. Group 1 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Molecular Function Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 17 main categories to which 220 perturbed GO term sets of genes resultedin mapping to 10 categories. Definition of Group 1 is same as described in Table S13. Table S20 in File S1. Group 2 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Molecular Function Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 17 main categories to which 38 perturbed GO term sets of genes resulted in mapping to 6 categories. Definition of Group 1 is same as described in Table S14. Table S21 in File S1. Mechanistic Genes with Overlapping Intersections among Multiple Pathways. This table lists 476 unique mechanistic genes associated with the list of pathways in Table 2. The table shows the number of pathways for which each gene has an intersection. The Bayesian z-score by time point is provided for each gene. Table S22 in File S1. Significantly Down-Regulated Mechanistic Genes with Bayesian z-score <−2.24 in the Early Stage of Infection (15, 30, & 60 minutes post-infection). The table lists significantly perturbed down-regulated genes with a high degree of overlap (intersection or cross-talk) across multiple pathways (5 or more intersections) associated with those pathways listed in Table 2. Significant down-regulated Bayesian z-scores in the first hour p.i. are bolded. Table S23 in File S1. Significantly Up-Regulated Mechanistic Genes with Bayesian z-score >2.24 in the Early Stage of Infection (15, 30, & 60 minutes post-infection). The table lists significantly perturbed up-regulated genes with a high degree of overlap (intersection or crosstalk) across multiple pathways (5 or more intersections) associated with those pathways listed in Table 2. Significant up-regulated Bayesian z-scores in the first hour p.i. are bolded. Table S24 in File S1. Description of the Biological Role of Dominant Down-Regulated Mechanistic Genes in Tight Junction Pathway. Table S25 in File S1. Description of the Biological Role of Dominant Down-Regulated Mechanistic genes in Trefoil Factors Initiated Mucosal Healing. Table S26 in File S1. Significant Perturbed Genes and Their Biological Roles for the Lectin Pathway. Table S27 in File S1. Significant Perturbed Genes and Their Biological Roles for Phagocytosis Gene Ontology Group. Table S28 in File S1. Interleukin Mechanistic Genes Significantly Perturbed at 15 minutes post-infection.
(ZIP)
Funding Statement
LGA and HRG were supported by grants from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases Western Regional Center of Excellence 1U54 AI057156-01. LGA is also supported by the U.S. Department of Homeland Security National Center of Excellence for Foreign Animal and Zoonotic Disease Defense ONR-N00014-04-1-0 grant. CAR was supported by an I.N.T.A.-Fulbright Argentina Fellowship. CLG received support from an NIH cardiology fellowship, Cardiology Department, University of Texas Southwestern Medical Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Olsen SC, Thoen CO, Cheville NF (2004) Brucella; GylesCL, PrescottJF, SongerJG, ThoenCO, editors. AmesIowa: Blackwell Publishing Ltd. 309–319p. [Google Scholar]
- 2. Foster G, Osterman BS, Godfroid J, Jacques I, Cloeckaert A (2007) Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int J Syst Evol Microbiol 57: 2688–2693. [DOI] [PubMed] [Google Scholar]
- 3. Scholz HC, Hubalek Z, Sedlacek I, Vergnaud G, Tomaso H, et al. (2008) Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int J Syst Evol Microbiol 58: 375–382. [DOI] [PubMed] [Google Scholar]
- 4. Scholz HC, Nockler K, Gollner C, Bahn P, Vergnaud G, et al. (2010) Brucella inopinata sp. nov., isolated from a breast implant infection. Int J Syst Evol Microbiol 60: 801–808. [DOI] [PubMed] [Google Scholar]
- 5.Enright FM (1990) The pathogenesis and pathobiology of Brucella infection in domestic animals. In: Nielsen K, Duncan JR, editors. Animal Brucellosis. Boca Raton, Florida: CRC Press, Inc. pp. 301–320. [Google Scholar]
- 6.Verger JM (1985) B. melitensis infection in cattle. Brucella melitensis: November 14–15, 1984–85, Brussells, Belgium. Dordrecht: Martinus Niihoff Publishers. pp. 197–203.
- 7.Alton GG (1990) Brucella melitensis. In: Nielsen K, Duncan JR, editors. Animal Brucellosis. Boca Raton, Florida: CRC Press, Inc. pp. 383–409. [Google Scholar]
- 8. Kahler SC (2000) Brucella melitensis infection discovered in cattle for first time, goats also infected. J Am Vet Med Assoc 216: 648. [PubMed] [Google Scholar]
- 9.Young EJ (1995) An overview of human brucellosis. Clin Infect Dis 21: : 283–289; quiz 290. [DOI] [PubMed] [Google Scholar]
- 10. Adams LG (2002) The pathology of brucellosis reflects the outcome of the battle between the host genome and the Brucella genome. Vet Microbiol 90: 553–561. [DOI] [PubMed] [Google Scholar]
- 11.Plommet M (1977) Brucellosis in domestic animals; Crawford RP, Hidalgo RJ, editors. College Station and London: Texas A&M University Press. 116–134p. [Google Scholar]
- 12. Carpenter CM (1924) Bacterium abortem invasion of the tissues of calves from the ingestion of infected milk. The Cornell Veterinarian 14: 16–31. [Google Scholar]
- 13. Davis DS, Heck FC, Williams JD, Simpson TR, Adams LG (1988) Interspecific transmission of Brucella abortus from experimentally infected coyotes (Canis latrans) to parturient cattle. J Wildl Dis 24: 533–537. [DOI] [PubMed] [Google Scholar]
- 14. Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, et al. (2008) Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog 4: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khare S, Lawhon SD, Drake KL, Nunes JE, Figueiredo JF, et al. (2012) Systems Biology Analysis of Gene Expression during In Vivo Mycobacterium avium paratuberculosis Enteric Colonization Reveals Role for Immune Tolerance. PLoS One 7: e42127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khare S, Nunes JS, Figueiredo JF, Lawhon SD, Rossetti CA, et al. (2009) Early phase morphological lesions and transcriptional responses of bovine ileum infected with Mycobacterium avium subsp. paratuberculosis. Vet Pathol 46: 717–728. [DOI] [PubMed] [Google Scholar]
- 17. Lawhon SD, Khare S, Rossetti CA, Everts RE, Galindo CL, et al. (2011) Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica Serovar Typhimurium: a systems biology analysis approach. PLoS One 6: e26869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nunes JS, Lawhon SD, Rossetti CA, Khare S, Figueiredo JF, et al. (2010) Morphologic and cytokine profile characterization of Salmonella enterica serovar typhimurium infection in calves with bovine leukocyte adhesion deficiency. Vet Pathol 47: 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossetti CA, Galindo CL, Lawhon SD, Garner HR, Adams LG (2009) Brucella melitensis global gene expression study provides novel information on growth phase-specific gene regulation with potential insights for understanding Brucella:host initial interactions. BMC Microbiol 9: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santos RL, Tsolis RM, Baumler AJ, Smith R 3rd, Adams LG (2001) Salmonella enterica serovar typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect Immun 69: 2293–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alton GG, Jones LM, Angus RD, Verger JM (1988) Techniques for the brucellosis laboratory. ParisFrancs: Institut National de la Recherche Agronomique. 174 p. [Google Scholar]
- 22. Rossetti CA, Galindo CL, Everts RE, Lewin HA, Garner HR, et al. (2011) Comparative analysis of the early transcriptome of Brucella abortus--infected monocyte-derived macrophages from cattle naturally resistant or susceptible to brucellosis. Res Vet Sci 91: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loor JJ, Everts RE, Bionaz M, Dann HM, Morin DE, et al. (2007) Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics 32: 105–116. [DOI] [PubMed] [Google Scholar]
- 24. Adams LG, Khare S, Lawhon SD, Rossetti CA, Lewin HA, et al. (2011) Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: linking systems biology with vaccine development. Vaccine 29: 7197–7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams LG, Khare S, Lawhon SD, Rossetti CA, Lewin HA, et al. (2011) Multi-comparative systems biology analysis reveals time-course biosignatures of in vivo bovine pathway responses to B.melitensis, S.enterica Typhimurium and M.avium paratuberculosis. BMC Proc 5 Suppl 4S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossetti CA, Drake KL, Adams LG (2012) Transcriptome analysis of HeLa cells response to Brucella melitensis infection: a molecular approach to understand the role of the mucosal epithelium in the onset of the Brucella pathogenesis. Microbes Infect 14: 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldi P, Hatfield GW (2002) DNA Microarrays and Gene Expression: From Experiments to Data Analysis and Modeling: Cambridge University Press, 1st Edition, p. 107–115.
- 28. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. Gorvel JP, Moreno E, Moriyon I (2009) Is Brucella an enteric pathogen? Nat Rev Microbiol 7: 250. [DOI] [PubMed] [Google Scholar]
- 30. Ackermann MR, Cheville NF, Deyoe BL (1988) Bovine ileal dome lymphoepithelial cells: endocytosis and transport of Brucella abortus strain 19. Vet Pathol 25: 28–35. [DOI] [PubMed] [Google Scholar]
- 31. Bolton AJ, Osborne MP, Wallis TS, Stephen J (1999) Interaction of Salmonella choleraesuis, Salmonella dublin and Salmonella typhimurium with porcine and bovine terminal ileum in vivo . Microbiology 145: 2431–2441. [DOI] [PubMed] [Google Scholar]
- 32. Zheng K, Chen DS, Wu YQ, Xu XJ, Zhang H, et al. (2012) MicroRNA expression profile in RAW264.7 cells in response to Brucella melitensis infection. Int J Biol Sci 8: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galindo RC, Munoz PM, de Miguel MJ, Marin CM, Blasco JM, et al. (2009) Differential expression of inflammatory and immune response genes in rams experimentally infected with a rough virulent strain of Brucella ovis. Veterinary Immunology and Immunopathology 127: 295–303. [DOI] [PubMed] [Google Scholar]
- 34. Wang F, Hu S, Liu W, Qiao Z, Gao Y, et al. (2011) Deep-sequencing analysis of the mouse transcriptome response to infection with Brucella melitensis strains of differing virulence. PLoS One 6: e28485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carvalho Neta AV, Stynen APR, Paixao TA, Miranda KL, Silva FL, et al. (2008) Modulation of the bovine trophoblastic innate immune response by Brucella abortus . Infection and Immunity 76: 1897–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He Y, Reichow S, Ramamoorthy S, Ding X, Lathigra R, et al. (2006) Brucella melitensis triggers time-dependent modulation of apoptosis and down-regulation of mitochondrion-associated gene expression in mouse macrophages. Infection and Immunity 74: 5035–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gross A, Terraza A, Ouahrani-Bettache S, Liautard JP, Dornand J (2000) In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect Immun 68: 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galdiero E, Romano Carratelli C, Vitiello M, Nuzzo I, Del Vecchio E, et al. (2000) HSP and apoptosis in leukocytes from infected or vaccinated animals by Brucella abortus . New Microbiology 23: 271–279. [PubMed] [Google Scholar]
- 39. Forestier C, Deleuil F, Lapaque N, Moreno E, Gorvel JP (2000) Brucella abortus lipopolysaccharide in murine peritoneal macrophages acts as a down-regulator of T cell activation. J Immunol 165: 5202–5210. [DOI] [PubMed] [Google Scholar]
- 40. Lapaque N, Forquet F, de Chastellier C, Mishal Z, Jolly G, et al. (2006) Characterization of Brucella abortus lipopolysaccharide macrodomains as mega rafts. Cellular Microbiology 8: 197–206. [DOI] [PubMed] [Google Scholar]
- 41. Barquero-Calvo E, Chaves-Olarte E, Weiss DS, Guzmán-Verri C, Chacon-Diaz C, et al. (2007) Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. Plos One 2: e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dimitrakopoulos O, Liopeta K, Dimitracopoulos G, Paliogianni F (2013) Replication of Brucella melitensis inside primary human monocytes depends on mitogen activated protein kinase signaling. Microbes and Infection 15: 450–460. [DOI] [PubMed] [Google Scholar]
- 43. Finlay BB, Falkow S (1997) Common themes in microbial patogenicity revisited. Microbiology and Molecular Biology Reviews 61: 136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liautard JP, Gross A, Dornand J, Kohler S (1996) Interactions between professional phagocytes and Brucella spp. Microbiologia 12: 197–206. [PubMed] [Google Scholar]
- 45. Castaneda-Roldán EI, Avelino-Flores F, Dall'Agnol M, Freer E, Cedillo L, et al. (2004) Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cellular Microbiology 6: 435–445. [DOI] [PubMed] [Google Scholar]
- 46. Nakato G, Hase K, Suzuki M, Kimura M, Ato M, et al. (2012) Cutting Edge: Brucella abortus exploits a cellular prion protein on intestinal M cells as an invasive receptor. Journal of Immunology 189: 1540–1544. [DOI] [PubMed] [Google Scholar]
- 47. Haynes A 3rd, Ruda F, Oliver J, Hamood AN, Griswold JA, et al. (2005) Syndecan 1 shedding contributes to Pseudomonas aeruginosa sepsis. Infection and Immunity 73: 7914–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henry-Stanley MJ, Hess DJ, Erlandsen SL, Wells CL (2005) Ability of the heparan sulfate proteoglycan syndecan-1 to participate in bacterial translocation across the intestinal epithelial barrier. Shock 24: 571–576. [DOI] [PubMed] [Google Scholar]
- 49. Freissler E, Meyer auf der Heyde A, David G, Meyer TF, Dehio C (2000) Syndecan-1 and syndecan-4 can mediate the invasion of OpaHSPG-expressing Neisseria gonorrhoeae into epithelial cells. Cell Microbiol 2: 69–82. [DOI] [PubMed] [Google Scholar]
- 50. Bobardt MD, Saphire AC, Hung HC, Yu X, Van der Schueren B, et al. (2003) Syndecan captures, protects, and transmits HIV to T lymphocytes. Immunity 18: 27–39. [DOI] [PubMed] [Google Scholar]
- 51. Hauck CR, Borisova M, Muenzner P (2012) Exploitation of integrin function by pathogenic microbes. Current Opinion in Cell Biology 24: 637–644. [DOI] [PubMed] [Google Scholar]
- 52. De Panfilis G, Manara GC, Ferrari C, Torresani C (1991) Adhesion molecules on the plasma membrane of epidermal cells. III. Keratinocytes and Langerhans cells constitutively express the lymphocyte function-associated antigen 3. Journal of Investigative Dermatology 96: 512–517. [DOI] [PubMed] [Google Scholar]
- 53. Campbell GA, Adams LG, Sowa BA (1994) Mechanisms of binding of Brucella abortus to mononuclear phagocytes form cows naturally resistant or susceptible to brucellosis. Veterinary Immunology and Immunopathology 41: 295–306. [DOI] [PubMed] [Google Scholar]
- 54. Guzman-Verri C, Chaves-Olarte E, von Eichel-Streiber C, Lopez-Goni I, Thelestam M, et al. (2001) GTPases of the Rho subfamily are required for Brucella abortus internalization in nonprofessional phagocytes: direct activation of Cdc42. J Biol Chem 276: 44435–44443. [DOI] [PubMed] [Google Scholar]
- 55. Watarai M, Makino SI, Fujii Y, Okamoto K, Shirahata T (2002) Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication. Cellular Microbiology 4: 341–355. [DOI] [PubMed] [Google Scholar]
- 56. Nikitas G, Cossart P (2012) Adherens junctions and pathogen entry. Sub-Cellular Biochemistry 60: 415–425. [DOI] [PubMed] [Google Scholar]
- 57. Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, et al. (2011) Regulation of tight junction permeability by intestinal bacteria and dietary components. Journal of Nutrition 141: 769–776. [DOI] [PubMed] [Google Scholar]
- 58. Roxas JL, Koutsouris A, Bellmeyer A, Tesfay S, Royan S, et al. (2010) Enterohemorrhagic E. coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin independent manner. Laboratory Investigation 90: 1152–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taupin D, Podolsky DK (2003) Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol 4: 721–732. [DOI] [PubMed] [Google Scholar]
- 60. Fernandez-Prada CM, Nikolich M, Vemulapalli R, Sriranganathan N, Boyle SM, et al. (2001) Deletion of wboA enhances activation of the lectin pathway of complement in Brucella abortus and Brucella melitensis. Infect Immun 69: 4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Starr T, Child R, Wehrly TD, Hansen B, Hwang S, et al. (2012) Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baldwin CL, Goenka R (2006) Host immune responses to the intracellular bacteria Brucella: Does the bacteria instruct the host to facilitate chronic infection Critical Review in Microbiology. 26: 407–442. [DOI] [PubMed] [Google Scholar]
- 63. Martirosyan A, Moreno E, Gorvel JP (2011) An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunological Reviews 240: 211–234. [DOI] [PubMed] [Google Scholar]
- 64. Billard E, Dornand J, Gross A (2007) Brucella suis prevents human dendritic cell maturation and antigen presentation through regulation of tumor necrosis factor alpha secretion. Infection and Immunity 75: 4980–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dornand J, Gross A, Lafont V, Liautard J, Oliaro J, et al. (2002) The innate immune response against Brucella in humans. Veterinary Microbiology 90: 383–394. [DOI] [PubMed] [Google Scholar]
- 66. Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, et al. (2008) Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nature Medicine 14: 399–406. [DOI] [PubMed] [Google Scholar]
- 67. Salcedo S, Marchesini MI, Degos C, Terwagne M, Von Bargen K, et al. (2013) BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Frontiers in Cellular and Infection Microbiology 3: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables. Figure A in File S1. Validation of Bovine Microarray Results by Quantitative Real Time-PCR. cDNA was synthesized from the same RNA samples used for microarray hybridization. Five randomly selected genes (A = BPI; B = MAPK1; C = MIF; D = CCL2; E = IL8.) that were differentially expressed by microarrays in B. melitensis-infected bovine Peyer's patch between 15 min and 4 h p.i. as compared to non-infected tissues (control) extracted at the same time points, were validated by quantitative RT-PCR. Fold changes was normalized to the expression of GAPDH and calculated using the ΔΔCt method. All tested genes at all time points had fold-changes altered in the same direction in microarray and qRT-PCR. White bars represent fold-change by microarray analysis and black bars represent fold-change by qRT-PCR. Table S1 in File S1. Detailed List of Host Genes with Differential Expression (z-score >|2.24|) in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch in at least one time point. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S2 in File S1. Bayesian z-score for All Host Pathways in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S3 in File S1. List of All Biological Process-Related Host Genes Differentially Expressed in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S4 in File S1. List of All Cellular Component-Related Host Genes Differentially Expressed in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S5 in File S1. List of All Molecular Function-Related Host Genes Differentially Expressed in B. melitensis Infected vs. Control Bovine Jejunal-Ileal Peyer's Patch. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S6 in File S1. Group 1 - GO Biological Process Terms Significantly Perturbed Early (Activated or Repressed) but Not Later. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S7 in File S1. Group 2 - GO Biological Process Terms Consistently Perturbed (Activated or Repressed) Throughout the Experiment; Not Significantly Perturbed Early but Later; or Consistently Altered Perturbation (Activated to Repressed or Vice Versa). Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S8 in File S1. Group 1 - GO Cellular Components Terms Significantly Perturbed Early (Activated or Repressed) but Not Later. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S9 in File S1. Group 2 - GO Cellular Component Terms Consistently Perturbed (Activated or Repressed) Throughout the Experiment; or Terms Not Significantly Perturbed Early but Later; or Consistently Altered Perturbation (Activated to Repressed or Vice Versa). Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S10 in File S1. Group 1 - GO Molecular Function Terms Significantly Perturbed Early (Activated or Repressed) but Not Later. Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S11 in File S1. Group 2 - GO Molecular Function Terms Consistently Perturbed (Activated or Repressed) Throughout the Experiment; or Terms Not Significantly Perturbed Early but Later; or Consistently Altered Perturbation (Activated to Repressed or Vice Versa). Black numbers in the body of the table indicate differentially expressed (activated: (+) numbers; repressed: (−) numbers) while red numbers represent non-differentially expressed genes. Table S12 in File S1. Comparative Table of Group 1 and Group 2 Gene Ontology Terms Clustered to the Top 15 Primary Immune Related Biological Process Categories. This table shows lists the mapping of perturbed GO term gene groups to their higher level GO category definitions. The number of perturbed GO terms that mapped to each category is provided in the Count column. Table S13 in File S1. Group 1 - Detailed Listing of the Top 15 Highly Perturbed Gene Ontology Terms Mapped to Major Immune Related Biological Process. This table lists each GO term that was mapped (clustered) to a main category related to an immune response. There were 108 main categories to which 686 perturbed GO term sets of genes resulted in mapping to 55 categories. Group 1 represents those GO terms that were significantly perturbed at earlier time points (either activated or repressed) but not at later time points. Table S14 in File S1. Group 2 - Detailed Listing of the Top 15 Highly Perturbed Gene Ontology Terms Mapped to Major Immune Related Biological Process. This table lists each GO term that was mapped (clustered) to a main category related to an immune response. There were 108 main categories to which 114 perturbed GO term sets of genes resulted in mapping to 23 categories. Group 2 are GO terms consistently perturbed (up- or down-regulated) throughout the experiment; or GO terms not significantly perturbed at earlier time points but at later times; or GO terms that consistently changed their state of perturbation (from activated- to repressed or vice versa). Table S15 in File S1. Comparative Table of Group 1 and Group 2 Gene Ontology Terms Clustered to Primary Cellular Component Categories. The mapping of terms in this table is to major categories related to the cellular component categories. Table S16 in File S1. Group 1 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Cellular Component Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 12 main categories to which 112 perturbed GO term sets of genes resulted in mapping to 9 categories. Definition of Group 1 is same as described in Table S13. Table S17 in File S1. Group 2 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Cellular Component Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 12 main categories to which 26 perturbed GO term sets of genes resulted in mapping to 6 categories. Definition of Group 2 is same as described in Table S14. Table S18 in File S1. Comparative Table of Group 1 and Group 2 Gene Ontology Terms Clustered to Primary Molecular Function Categories. The mapping of terms in this table is to major categories related to the primary molecular function categories. Table S19 in File S1. Group 1 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Molecular Function Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 17 main categories to which 220 perturbed GO term sets of genes resultedin mapping to 10 categories. Definition of Group 1 is same as described in Table S13. Table S20 in File S1. Group 2 - Detailed Listing of Highly Perturbed Gene Ontology Terms Mapped to Major Molecular Function Categories. This table lists each GO term that was mapped (clustered) to a main category related to its cellular component association. There were 17 main categories to which 38 perturbed GO term sets of genes resulted in mapping to 6 categories. Definition of Group 1 is same as described in Table S14. Table S21 in File S1. Mechanistic Genes with Overlapping Intersections among Multiple Pathways. This table lists 476 unique mechanistic genes associated with the list of pathways in Table 2. The table shows the number of pathways for which each gene has an intersection. The Bayesian z-score by time point is provided for each gene. Table S22 in File S1. Significantly Down-Regulated Mechanistic Genes with Bayesian z-score <−2.24 in the Early Stage of Infection (15, 30, & 60 minutes post-infection). The table lists significantly perturbed down-regulated genes with a high degree of overlap (intersection or cross-talk) across multiple pathways (5 or more intersections) associated with those pathways listed in Table 2. Significant down-regulated Bayesian z-scores in the first hour p.i. are bolded. Table S23 in File S1. Significantly Up-Regulated Mechanistic Genes with Bayesian z-score >2.24 in the Early Stage of Infection (15, 30, & 60 minutes post-infection). The table lists significantly perturbed up-regulated genes with a high degree of overlap (intersection or crosstalk) across multiple pathways (5 or more intersections) associated with those pathways listed in Table 2. Significant up-regulated Bayesian z-scores in the first hour p.i. are bolded. Table S24 in File S1. Description of the Biological Role of Dominant Down-Regulated Mechanistic Genes in Tight Junction Pathway. Table S25 in File S1. Description of the Biological Role of Dominant Down-Regulated Mechanistic genes in Trefoil Factors Initiated Mucosal Healing. Table S26 in File S1. Significant Perturbed Genes and Their Biological Roles for the Lectin Pathway. Table S27 in File S1. Significant Perturbed Genes and Their Biological Roles for Phagocytosis Gene Ontology Group. Table S28 in File S1. Interleukin Mechanistic Genes Significantly Perturbed at 15 minutes post-infection.
(ZIP)