Abstract
Peptide-based proteomic data sets are ever increasing in size and complexity. These data sets provide computational challenges when attempting to quickly analyze spectra and obtain correct protein identifications. Database search and de novo algorithms must consider high-resolution MS/MS spectra and alternative fragmentation methods. Protein inference is a tricky problem when analyzing large data sets of degenerate peptide identifications. Combining multiple algorithms for improved peptide identification puts significant strain on computational systems when investigating large data sets. This review highlights some of the recent developments in peptide and protein identification algorithms for analyzing shotgun mass spectrometry data when encountering the aforementioned hurdles. Also explored are the roles that analytical pipelines, public spectral libraries, and cloud computing play in the evolution of peptide-based proteomics.
Introduction
Peptide based, or shotgun, mass spectrometry is one of the most fundamental methods in the field of proteomics. In shotgun proteomics, complex protein samples are enzymatically digested or chemically fragmented to peptides and introduced to a mass analyzer through separation and ionization techniques (e.g. liquid chromatography and electrospray ionization). Peptides observed through precursor survey scans are selectively isolated and fragmented along their peptide backbone, typically using collision induced dissociation (CID). The peptide fragment masses are recorded in tandem (MS/MS) spectra. These spectra are then analyzed computationally to identify the peptide sequences, in what is known as a peptide spectrum match (PSM). The PSMs are then used to infer which protein(s) are in the sample (see [1–3] for more detailed reviews).
With modern mass spectrometers, it is possible to acquire MS/MS spectra on tens of thousands of peptides in a few hours, enabling the study of thousands of proteins simultaneously from a single sample. However, the task of going from spectrum to result is far from trivial, and requires the use of several software algorithms. Recently, the routine acquisition of MS/MS spectra using electron transfer dissociation (ETD) and high-energy collisional dissociation (HCD) have enabled researchers to collect different spectra of the same peptides [4–8]. However, spectra obtained from the use of ETD and HCD have challenged existing algorithms and analytical paradigms, prompting the interest in new software solutions.
On the computational forefront, the availability of fast computers and free software has enabled more thorough data analysis than ever before. Multiple algorithms can be combined into robust pipelines for high-throughput analysis. Data and results can be stored and shared in large public repositories. Cloud computing offers cheap and efficient processing power to solve some of the most daunting analytical tasks. These advances will help to make whole proteome analysis both fast and routine for a multitude of organisms in the near future.
This review explores the process of going from spectrum to result in proteomics with focus on recent algorithm development and publicly available computational resources (summarized in Table 1). Generally speaking, the process involves identifying peptide sequences from spectra, statistically validating those sequences, and then using those sequences to infer which proteins are in a sample (Figure 1). More specifically, this review will focus on the new wave of algorithm development for database searching and de novo sequencing resulting from adoption of ETD and HCD. Software solutions for combining and validating results from multiple analyses will be presented. New developments in protein inference will also be investigated, as well as software tools that allow the assembly, customization, and interaction of these algorithms into versatile pipelines for data analysis. Interspersed among these developments will be the roles that shared data repositories, spectral libraries, and cloud computing will play in shotgun proteomics in the coming years.
Table 1.
Summary of software tools featured in this review
| Software tool | Purpose |
|---|---|
| Andromeda | Database search |
| MS-GFDB | Database search |
| pFind | Database search |
| pNovo | De novo sequencing |
| PeptideProphet | PSM validation |
| Percolator | PSM validation |
| FDRAnalysis | Multiple search combination |
| iProphet | Multiple search combination |
| Pepitome | Spectral library searching |
| PeptideAtlas | Annotated spectrum repository |
| Fido | Protein inference |
| IDPicker 2.0 | Protein inference |
| MAYU | Protein inference |
| ProteinProphet | Protein inference |
| MaxQuant | Analytical software suite |
| Trans-Proteomic Pipeline | Analytical software suite |
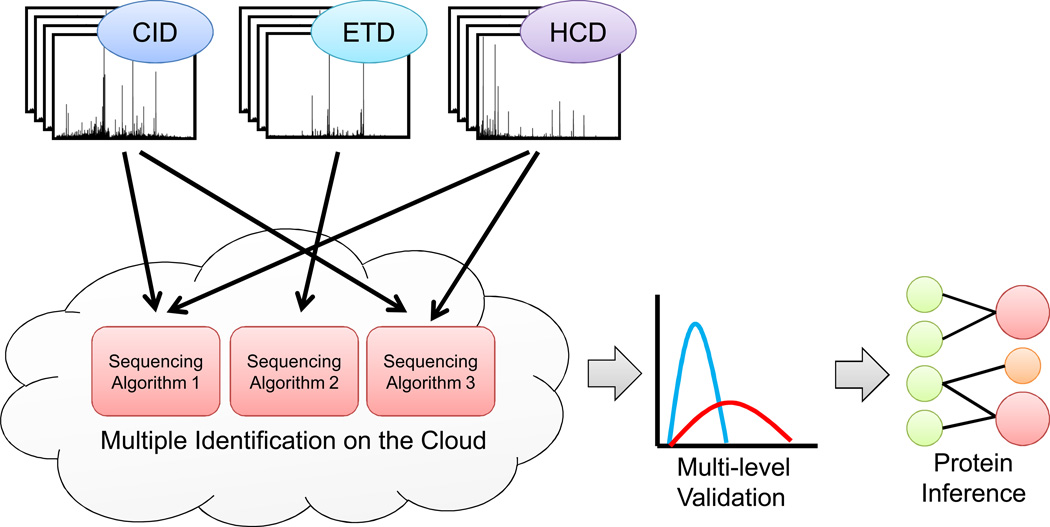
Figure 1.
A robust computational workflow for shotgun proteomics. Spectra acquired from several complementary fragmentation techniques are simultaneously analyzed with multiple sequencing algorithms, utilizing cloud-based computing resources. Multi-level peptide validation algorithms assign probabilities to peptide sequences from the combined sequencing results. The most likely proteins in the sample are inferred from the confident peptide sequences.
Peptide sequence identification from MS/MS spectra
Peptide based proteomics can be performed using a variety of instruments, fragmentation methods, and analytical techniques; however, historically those methods have predominantly consisted of low-resolution CID spectra acquisition (via quadrupole and ion trap mass analyzers) and database search or de novo identification. The consistency in instrumentation and technique produced a general analytical workflow and robust computational tools (reviewed in [2,3,9]). Alternative methods of generating MS/MS spectra include, but are not limited to, electron-capture dissociation (ECD)[10], ETD, and HCD. A brief overview and comparison of these different fragmentation methods can be found in Box 1. Recently, these alternative modes of fragmentation have gained widespread adoption for their complementarity to the CID paradigm. For example, ETD has shown superiority over CID for larger, higher charged (>2+) peptides [6,11]. Higher-energy collisional activation (HCD) prior to Orbitrap acquisition produces a more complete ion series in the low mass region [12]. Additionally, modern instrumentation makes acquisition of fragmentation spectra at high-resolution fast enough to keep pace with low-resolution data acquisition [12–14]. Because of the complementarity of these different approaches for peptide identification, data are now routinely collected that combine all these acquisition methods [5,6]. CID-centric algorithms are not optimal for analysis of these new data [15]. This limitation has spurred a new wave of algorithmic development for peptide identification.
Box 1: Overview of gas phase ion fragmentation methods discussed in this review.
Collision induced dissociation (CID): Ions are accelerated to a high kinetic energy using an electrical potential. The ions then collide with neutral gas molecules (usually helium, argon, or nitrogen), where the kinetic energy is converted to internal energy resulting in bond breakage and fragmentation [62]. Peptide fragmentation usually occurs along the amide backbone to produce b and y ions.
High-energy collisional dissociation (HCD): Ions are fragmented in the same manner as CID using higher degrees of kinetic energy. This process is can be performed in a dedicated collision cell coupled to a C-trap in an oribtrap mass spectrometer. Ions are then analyzed at high resolution in the orbitrap mass analyzer [63].
Electron capture dissociation (ECD): Multiply protonated ions are trapped in the gas phase and exposed to low-energy electrons. The neutralization of the molecule cations with the low-energy elections causes fragmentation [10]. This method differs from CID and HCD in that it does not require kinetic energy redistribution to cause fragmentation. Peptide fragmentation predominantly results in c and z type ions.
Electron transfer dissociation (ETD): This method is similar to ECD, but uses anions as vehicles for delivering electrons to multiply protonated ions trapped in the gas phase [64].
ETD has gained traction due to its complementarity to CID and ease of use. ETD is well suited for fragmentation of large peptides, thus improving sequence coverage over use of CID alone [16]. Additionally, ETD is advantageous for the study of certain post-translational modifications (PTMs), such as phosphorylation [4]. However, CID-based database search algorithms often underperform when analyzing ETD spectra, thus necessitating ETD-specific algorithms [17,18]. For example, while CID fragmentation results in predictable b and y ion series, ETD fragmentation produces the more complex z’, c, a’, and y ion series. Numerous additional fragmentation artifacts occur, such as 1) frequent neutral losses, 2) charge-reduced precursor observation, 3) side chain losses on histidine and alkylated cysteine residues, and 4) observation of c-H ions from doubly charged peptides [19]. Recently, the pFind database search algorithm was updated to incorporate these artifacts and showed improvement over the popular CID-centric Mascot algorithm for ETD spectra. Interestingly, the greatest improvement was in the identification of 2+ peptides, which are generally considered poor candidates for ETD fragmentation [19]. Similar results were observed using probabilistic scoring models trained for ETD spectra with the algorithm, MS-GFDB [20]. MS-GFDB goes even further to improve peptide identification when both CID and ETD spectra are acquired on the same peptide. The two fragmentation scans are combined in silico into a single spectrum that shows improved sequence identification over CID or ETD alone.
With the ever increasing prevalence of modern Orbitrap mass analyzers, HCD is fast becoming another common method of fragmentation. Although similar to CID (such as generation of b and y fragment ions), HCD offers two advantages: 1) improved detection of low mass fragment ions, and 2) high mass accuracy measurements. These features have shown HCD to be a powerful tool for PTM identification, such as phosphorylation [21–23]. Several, but not all, database search algorithms can be optionally configured to take advantage of the improved resolution and mass accuracy [7,24]. Where the benefit of HCD is particularly useful is in the development of de novo peptide sequencing algorithms.
De novo sequencing algorithms have advantages over database search algorithms, such as that they can be used to identify peptides not contained in the database. This capability is especially important for identification of protein variants not represented in the database. For example, one approach to identify protein variants with a database search is using RNA-Seq data, if available [25]. De novo algorithms are not limited by the availability of transcriptome data. However, database search algorithms have historically outperformed de novo algorithms where sample proteomes are known [9]. Incomplete ion series, noisy spectra, and poor fragment ion mass accuracy contribute much to the inaccuracy of de novo algorithms. These difficulties have limited de novo algorithms to identification of small peptides [26], or in conjunction with database search for improved accuracy and efficiency [27–31]. The emergence of HCD has since breathed new life into de novo identification strategies. pNovo is one of the first algorithms devoted to de novo sequencing of HCD spectra [32]. The algorithm also contains an interesting feature: low mass HCD spectra (containing dominant fragment ions < 500 m/z) can be merged with normal HCD spectra to the same precursor ion to improve sequencing of the N- and C-termini. These regions can be highly problematic when performing de novo identifications. The increased success rate of de novo algorithms when presented with HCD data may highlight a paradigm shift in shotgun proteomics towards integrating more de novo identification strategies in the coming years.
Evaluation of peptide spectrum matches
Identifying the most likely peptide sequence for any given spectrum does not mean the peptide was correctly identified in the sample. Many spectra do not provide adequate information to produce sequence identifications, yet sequences (likely false) are still reported for these spectra. To help separate true from false identifications, each algorithm provides a metric to evaluate the likelihood a given PSM is correct. The metric may differ, and usually does, from algorithm to algorithm, and the appropriate score cutoff may also differ from dataset to dataset. For this reason, different database search algorithms can result in a widely varying array of PSMs for the same set of spectra. As a way of addressing this uncertainty, target-decoy searches are frequently performed when using database search algorithms to give an estimate of false discovery rate (FDR) at a given score threshold (Figure 2). Target-decoy searches involve scoring all spectra against a database that contains reversed or shuffled protein sequences (decoys) in addition to true protein sequences (targets). PSMs to decoy sequences are known to be false, and can be exploited to determine the FDR at a given score threshold. In this manner, the different scoring metrics between algorithms can be normalized to a fixed FDR (e.g. 1% or 5%), and the PSMs reported from the different algorithms can be compared at all FDR thresholds [33]. Few database search algorithms perform FDR calculation as part of their operation. Thus, stand-alone algorithms have been created to make use of the target-decoy approach to estimating FDR, including PeptideProphet [34,35] and Percolator [36,37]. Although target-decoy searches are routine, PeptideProphet can alternatively estimate FDR using a Baysian mixture modeling approach for spectra searched against a target-only database.
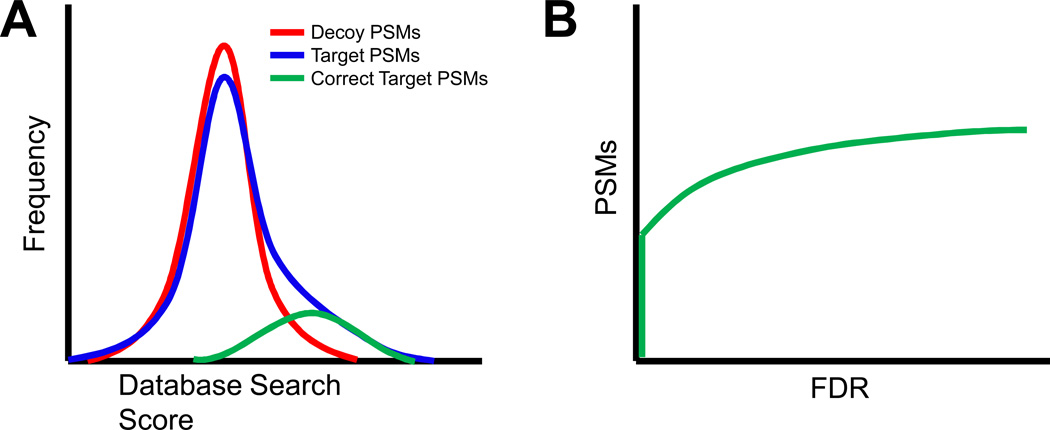
Figure 2.
Estimating false discovery rate (FDR) using target-decoy searches. (A) The score distribution of target PSMs (blue line) shows a right tail when compared to the decoy distribution (red line), representing correct PSMs (green line). (B) The distribution of scores for the target and decoy PSMs are used to estimate the FDR at any score threshold.
Even at a low FDR, any algorithm used for peptide identification produces different PSMs for the same data [33]. This is true for any shotgun proteomics analytical tool, and despite the abundance of algorithms available to researchers [38], there is no perfect solution for any given set of MS/MS spectra. One algorithm may be able to produce a correct PSM for a spectrum where another failed, and vice versa. Rather than limit an analysis to a single software tool, it is now possible to combine multiple results of the same data using different database search and de novo algorithms [39–41].
One such algorithm to take advantage of multiple search algorithms is FDRAnalysis [42]. Based on the combined FDRScore approach [43], this web-based algorithm reclassifies peptide sequence identifications from three different database search tools in terms of FDR. By doing so, the different scoring metrics employed by each tool can be comparatively analyzed. A combined FDR is then computed for each peptide. Peptides observed in all three search tools receive much lower combined FDR values than those observed in only one search tool. Initial studies using the combined FDRScore algorithm showed an average 35% increase in peptide identifications over individual database search results [43]. Although easily accessible, FDRAnalysis is limited in the database search algorithms supported.
Of recent note, the iProphet algorithm supports virtually any peptide sequence identification scheme [44]. After calculating the posterior probability of a PSM from any peptide identification algorithm, refined posterior probabilities are calculated in iProphet using a series of models related to identification with multiple peptide sequencing tools. The authors showed a 30% increase in PSMs combining six different database search algorithm results [44]. These results highlight that the limitations of a single search algorithm may be overcome by combining the results of multiple algorithms. Given the amount of readily available processing power from modern computers, especially when considering the accessibility of cloud computing, multiple algorithm analysis may soon be routine for peptide identification.
Spectral libraries
The spectrum in a PSM can be considered a type of barcode for that peptide. If acquired again under similar conditions, the spectrum for a given peptide will look nearly identical in terms of observed fragments and their peak intensities. This reproducibility can be exploited to quickly and confidently identify peptides that have been seen in previous experiments through a process called spectral library searching [45]. Accurate PSMs are compiled into spectra libraries, and newly acquired spectra can be matched against those libraries to identify known peptides using dot products [46–48]. More recently, the Pepitome algorithm adopted probabilistic scoring metrics [49]. Spectral library search algorithms use only a fraction of the time of database search algorithms; however, spectral library searching can only identify previously observed peptides. Furthermore, ETD and HCD spectra are fundamentally different than their CID spectra counterparts. Thus, independent spectral libraries must be compiled for each fragmentation method. These caveats have somewhat limited rate of adoption of spectral library search algorithms.
The increasing availability of large shotgun proteomics data repositories will help lend weight to spectral library searching. At the time of this writing, the PeptideAtlas [50] project contains approximately one-half million peptide annotations from nearly 30 million PSMs across twelve different species. As spectra repositories continue to grow in coverage, organisms represented, and types of spectral acquisitions, it will soon be possible to search entire proteomes using spectral library matching.
Improved methods for protein inference
Confident PSMs are used to infer which proteins are in a sample. At first glance, a PSM that maps to a protein would indicate that the protein is in the sample. This may be true for PSMs that are undoubtedly correct and proteotypic, meaning the peptide sequence matches to only a single protein. If the PSM can be mapped to two or more proteins, then which of those proteins is in the sample, or all of them, is difficult to determine (Figure 3). Several algorithms attempt to solve this issue using both non-probabilistic and probabilistic methods. The non-probabilistic approach returns the simplest set of proteins that explains the observed high-scoring PSMs [51]. The probabilistic approach, on the other hand, uses additional metrics, such as the quality of a PSM, to give a probability that a protein is present in a sample. For example, ProteinProphet [52] may infer a protein with high confidence from multiple low scoring PSMs because such an occurrence is highly unlikely by chance. Recent developments in improving protein inference are explored in detail below.
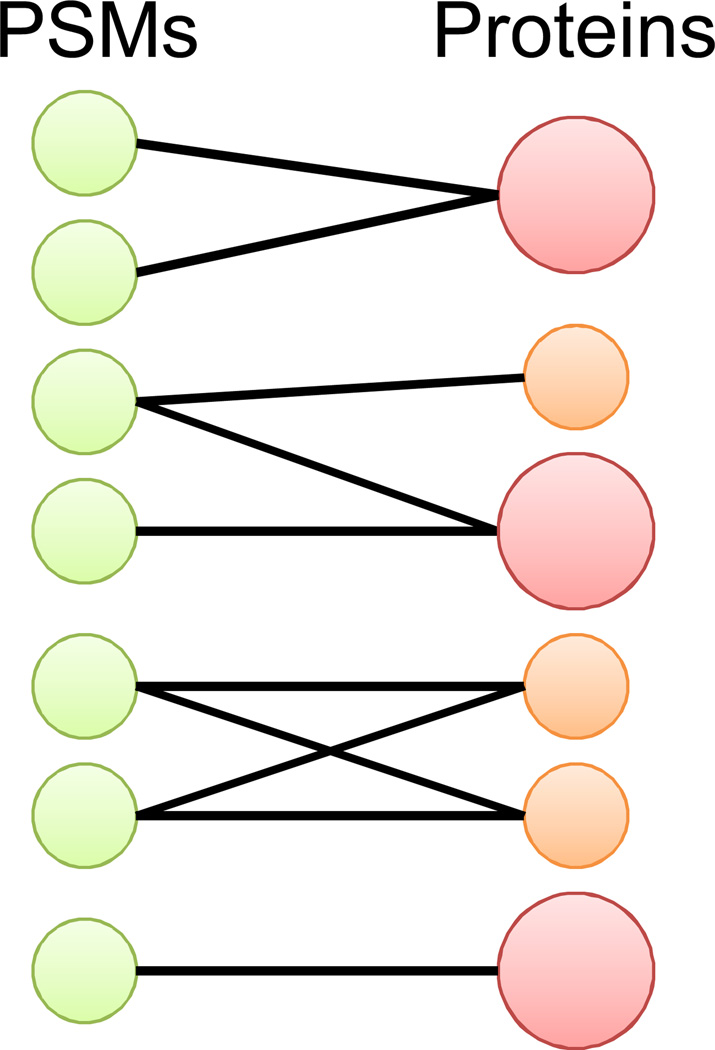
Figure 3.
Protein inference from PSMs. Peptide sequences from PSMs are used to infer proteins in the sample. Peptide degeneracy, or when a PSM can be matched to two or more proteins, makes correct protein identification difficult. Some algorithms use probabilistic models, represented by large red circles versus small orange circles, to discriminate correct from incorrect protein identifications.
A common strategy for protein inference is parsimony, or the smallest number of proteins that result in the observed PSMs. Consider, though, using a large database (e.g. NCBI NR) which contains homologous sequences across multiple species. Even with parsimony, degenerate peptides may return far more proteins than are truly in the sample. IDPicker 2.0 attempts to solve this problem non-probabilistically by allowing the user to set a specific number of distinct peptide identifications for a protein that are not explained by other proteins [41]. That number may be set interactively by the user, including a value of zero to disable parsimony. Tests identifying human serum against a multispecies database showed how setting even low counts of two and three distinct peptide identifications dramatically reduced the number of orthologous proteins reported with little loss of human proteins reported.
Probabilistic methods for protein inference attempt to qualify protein identifications with, for example, an FDR. This is not a straightforward process because incorrectly identified peptides are randomly sampled among all proteins in the database and correct peptides tend to cluster on a limited protein set. Thus peptide-level FDR does not directly translate to protein-level FDR, and special care must be taken to avoid inflating the FDR when generating a final protein list. The MAYU algorithm was developed specifically to address this issue, which becomes more difficult as data set size increases [53]. Here a target-decoy model is applied to the protein identifications, with decoy protein identifications derived from decoy PSMs present in the data. Additional complications often arise to challenge the computation of accurate protein probabilities, for instance, when considering degenerate peptides. The Fido algorithm uses a Bayesian method to compute protein posterior error probabilities when given a set of PSMs [54]. Peptide degeneracy is resolved by rewarding protein sets that have additional PSM evidence in addition to the degenerate peptide sequences. Of interest, with Fido it is possible for a protein inferred from a high-scoring PSM to receive a poor probability. With large data sets, there is increased likelihood a protein could be associated with an erroneous high-scoring PSM. Few other algorithms, however, consider the possibility that the best scoring PSMs might have an error among them. These examples highlight the challenges faced by protein inference algorithms as data sets continue to increase in both size and proteome coverage.
Assembling multiple algorithms into pipelines
As described above, the software algorithms for getting from spectra to protein identification are constantly adapting in the rapidly evolving field of shotgun proteomics. No single tool is sufficient to perform all analytical steps necessary to obtain a result. Thus, software suites and pipelines have been developed to help researchers perform the required steps for streamlined data analysis. The different types of suites can be broken into two major groups: all-in-one packages or interchangeable pipelines. There are strengths and weaknesses to each format. MaxQuant [55] is an excellent example of the former. With the recent completion of its Andromeda database search algorithm [56], it can now be used to perform data analysis from spectra to protein identification inside a single interface without the need for third party software. The caveat is that the software is not open source or customizable beyond what its developers have been able to provide for users. In contrast, the Trans-Proteomic Pipeline (TPP) [57] offers the set of tools needed to perform complete shotgun analysis in an open-source, customizable interface. Admittedly, the tradeoff for this platform is the difficulty for non-tech savvy scientists to integrate their own algorithms into the TPP; however, the TPP comes with a set of tools specifically for migrating data to open standards [58]. Additionally, the open-source interface enables integration of emerging algorithms with existing ones [44], whether provided with the TPP or from third parties. This customizability helps ensure rapid adoption of new algorithms to keep pace with evolving mass spectrometry technology.
Looking ahead
Shotgun mass spectrometry is constantly evolving to make the goal of complete proteome analysis a reality. To achieve this goal, shotgun proteomics algorithms are expanding and adapting to new technology. This development in proteomics algorithms creates challenges for researchers in terms of processing power and software interoperability. One of the biggest challenges in shotgun proteomics is simply how to perform such large data analysis efficiently, reproducibly, and easily.
The combination of multiple peptide identification algorithms is fast becoming routine in shotgun proteomics. Simultaneously, the size and complexity of shotgun data sets have grown significantly. Together, these factors contribute to considerable strain on computational systems for data analysis. Computer clusters are one option to provide additional processing power, but due to their expense, difficulty to maintain, and limited lifespan, alternative strategies are being explored. A graphics processing unit can be configured to provide parallel computing and has recently been shown to improve efficiency for database search and spectral matching [59,60]. Alternatively, database search algorithms can be adapted to operate on the internet through cloud computing services [61]. Cloud computing offers several advantages for shotgun proteomics analysis, with foremost being access to vast computational resources without the difficulty and expense of maintaining them on site. Whereas any given cluster has a limit to the amount of data it can analyze at a time, cloud resources can be expanded on demand to meet the needs of large data sets. These cloud resources are rapidly becoming available from a variety of vendors (e.g. Amazon, Microsoft, and Google). Cloud computing may soon be the preferred method for shotgun data analysis.
Software interoperability is critically important to shotgun proteomics. Without pipelines in place, many researchers would have difficulty in efficiently and correctly analyzing their data. However, there is a lag in the development of new algorithms and their widespread adoption in analytical pipelines. This lag might be overcome with a robust development environment for creating algorithms and pipelines. Perhaps it will soon be possible to download algorithms, much like apps for a smartphone, into an environment where their interoperability is already established. Thus, in a matter of seconds, researchers would be able to build custom analytical pipelines specific to their experimental design. Certainly, shotgun proteomics algorithms are evolving at a rapid pace, and researchers are constantly finding creative ways to go from spectrum to result.
Acknowledgements
This work has been funded in part with federal funds from the National Science Foundation MRI (grant No. 0923536), the Luxembourg Centre for Systems Biomedicine, and the University of Luxembourg. The authors would like to thank David Shteynberg for review of this manuscript.
References
- 1.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 2.Käll L, Vitek O. Computational mass spectrometry-based proteomics. PLoS Comput Biol. 2011;7:e1002277. doi: 10.1371/journal.pcbi.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eng JK, Searle BC, Clauser KR, Tabb DL. A face in the crowd: recognizing peptides through database search. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.R111.009522. R111 009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frese CK, Altelaar AF, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJ, Mohammed S. Improved peptide identification by targeted fragmentation using CID HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res. 2011;10:2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 6.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAlister GC, Phanstiel D, Wenger CD, Lee MV, Coon JJ. Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal Chem. 2010;82:316–322. doi: 10.1021/ac902005s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Tolic N, Xie F, Zhao R, Purvine SO, Schepmoes AA, Moore RJ, Anderson GA, Smith RD. Effectiveness of CID, HCD, and ETD with FT MS/MS for degradomic-peptidomic analysis: comparison of peptide identification methods. J Proteome Res. 2011;10:3929–3943. doi: 10.1021/pr200052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma B, Johnson R. De novo sequencing and homology searching. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.014902. O111 014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 11.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized Fast and Sensitive Acquisition Methods for Shotgun Proteomics on a Quadrupole Orbitrap Mass Spectrometer. J Proteome Res. 2012 doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- 14.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. M111 011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coon JJ. Collisions or electrons? Protein sequence analysis in the 21st century. Anal Chem. 2009;81:3208–3215. doi: 10.1021/ac802330b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina H, Matthiesen R, Kandasamy K, Pandey A. Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal Chem. 2008;80:4825–4835. doi: 10.1021/ac8007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Shan B, Xin L, Ma B. Better score function for peptide identification with ETD MS/MS spectra. BMC Bioinformatics. 2010;11(Suppl 1):S4. doi: 10.1186/1471-2105-11-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadygov RG, Good DM, Swaney DL, Coon JJ. A new probabilistic database search algorithm for ETD spectra. J Proteome Res. 2009;8:3198–3205. doi: 10.1021/pr900153b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun RX, Dong MQ, Song CQ, Chi H, Yang B, Xiu LY, Tao L, Jing ZY, Liu C, Wang LH, et al. Improved peptide identification for proteomic analysis based on comprehensive characterization of electron transfer dissociation spectra. J Proteome Res. 2010;9:6354–6367. doi: 10.1021/pr100648r. [DOI] [PubMed] [Google Scholar]
- 20. Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJ, Pevzner PA. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010;9:2840–2852. doi: 10.1074/mcp.M110.003731. Combines CID and ETD for improved peptide identification.
- 21.Nagaraj N, D'Souza RC, Cox J, Olsen JV, Mann M. Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J Proteome Res. 2010;9:6786–6794. doi: 10.1021/pr100637q. [DOI] [PubMed] [Google Scholar]
- 22.Nagaraj N, D'Souza RC, Cox J, Olsen JV, Mann M. Correction to Feasibility of Large-Scale Phosphoproteomics with Higher Energy Collisional Dissociation Fragmentation. J Proteome Res. 2012 doi: 10.1021/pr100637q. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Ficarro SB, Li S, Marto JA. Optimized Orbitrap HCD for quantitative analysis of phosphopeptides. J Am Soc Mass Spectrom. 2009;20:1425–1434. doi: 10.1016/j.jasms.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Guthals A, Bandeira N. Peptide identification by tandem mass spectrometry with alternate fragmentation modes. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.R112.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Slebos RJ, Wang D, Halvey PJ, Tabb DL, Liebler DC, Zhang B. Protein identification using customized protein sequence databases derived from RNA-Seq data. J Proteome Res. 2012;11:1009–1017. doi: 10.1021/pr200766z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitzer E, Masselot A, Colinge J. Assessing peptide de novo sequencing algorithms performance on large and diverse data sets. Proteomics. 2007;7:3051–3054. doi: 10.1002/pmic.200700224. [DOI] [PubMed] [Google Scholar]
- 27.Jeong K, Kim S, Bandeira N, Pevzner PA. Gapped spectral dictionaries and their applications for database searches of tandem mass spectra. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002220. M110 002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Gupta N, Bandeira N, Pevzner PA. Spectral dictionaries: Integrating de novo peptide sequencing with database search of tandem mass spectra. Mol Cell Proteomics. 2009;8:53–69. doi: 10.1074/mcp.M800103-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 30.Tabb DL, Saraf A, Yates JR., 3rd GutenTag: high-throughput sequence tagging via an empirically derived fragmentation model. Anal Chem. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabb DL, Ma ZQ, Martin DB, Ham AJ, Chambers MC. DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring. J Proteome Res. 2008;7:3838–3846. doi: 10.1021/pr800154p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi H, Sun RX, Yang B, Song CQ, Wang LH, Liu C, Fu Y, Yuan ZF, Wang HP, He SM, et al. pNovo: de novo peptide sequencing and identification using HCD spectra. J Proteome Res. 2010;9:2713–2724. doi: 10.1021/pr100182k. [DOI] [PubMed] [Google Scholar]
- 33.Balgley BM, Laudeman T, Yang L, Song T, Lee CS. Comparative evaluation of tandem MS search algorithms using a target-decoy search strategy. Mol Cell Proteomics. 2007;6:1599–1608. doi: 10.1074/mcp.M600469-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Ding Y, Choi H, Nesvizhskii AI. Adaptive discriminant function analysis and reranking of MS/MS database search results for improved peptide identification in shotgun proteomics. J Proteome Res. 2008;7:4878–4889. doi: 10.1021/pr800484x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 36.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 37.Spivak M, Weston J, Bottou L, Kall L, Noble WS. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J Proteome Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alves G, Wu WW, Wang G, Shen RF, Yu YK. Enhancing peptide identification confidence by combining search methods. J Proteome Res. 2008;7:3102–3113. doi: 10.1021/pr700798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Searle BC, Turner M, Nesvizhskii AI. Improving sensitivity by probabilistically combining results from multiple MS/MS search methodologies. J Proteome Res. 2008;7:245–253. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- 41.Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, et al. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wedge DC, Krishna R, Blackhurst P, Siepen JA, Jones AR, Hubbard SJ. FDRAnalysis: a tool for the integrated analysis of tandem mass spectrometry identification results from multiple search engines. J Proteome Res. 2011;10:2088–2094. doi: 10.1021/pr101157s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones AR, Siepen JA, Hubbard SJ, Paton NW. Improving sensitivity in proteome studies by analysis of false discovery rates for multiple search engines. Proteomics. 2009;9:1220–1229. doi: 10.1002/pmic.200800473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. M111 007690. Software capable of integrating virtually any peptide identification algorithm. Easily integrated into existing software pipelines.
- 45.Lam H. Building and searching tandem mass spectral libraries for peptide identification. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.R111.008565. R111 008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal Chem. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- 47.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 48.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J Proteome Res. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- 49. Dasari S, Chambers MC, Martinez MA, Carpenter KL, Ham AJ, Vega-Montoto LJ, Tabb DL. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. J Proteome Res. 2012;11:1686–1695. doi: 10.1021/pr200874e. Uses probabilistic algorithm to evaluate spectral library searches.
- 50.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 53.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Serang O, MacCoss MJ, Noble WS. Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. J Proteome Res. 2010;9:5346–5357. doi: 10.1021/pr100594k. Probabilistic approach to protein inference. Not all high-scoring PSMs are guaranteed to infer a protein identification.
- 55.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 56.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 57. Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. Complete guide to peptide-based proteomics data analysis and interpretation.
- 58.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100024. 2005 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baumgardner LA, Shanmugam AK, Lam H, Eng JK, Martin DB. Fast parallel tandem mass spectral library searching using GPU hardware acceleration. J Proteome Res. 2011;10:2882–2888. doi: 10.1021/pr200074h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milloy JA, Faherty BK, Gerber SA. Tempest: GPU-CPU Computing for High-Throughput Database Spectral Matching. J Proteome Res. 2012 doi: 10.1021/pr300338p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pratt B, Howbert JJ, Tasman NI, Nilsson EJ. MR-Tandem: parallel X!Tandem using Hadoop MapReduce on Amazon Web Services. Bioinformatics. 2012;28:136–137. doi: 10.1093/bioinformatics/btr615. Use of cloud computing to perform rapid peptide identification.
- 62.Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. J Mass Spectrom. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- 63.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 64.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]