Abstract
Beckwith-Wiedemann syndrome (BWS) is a human loss-of-imprinting syndrome primarily characterized by macrosomia, macroglossia, and abdominal wall defects. BWS has been associated with misregulation of two clusters of imprinted genes. Children conceived with the use of assisted reproductive technologies (ART) appear to have an increased incidence of BWS. As in humans, ART can also induce a similar overgrowth syndrome in ruminants which is referred to as large offspring syndrome (LOS). The main goal of our study is to determine if LOS shows similar loss-of-imprinting at loci known to be misregulated in BWS. To test this, Bos taurus indicus × Bos taurus taurus F1 hybrids were generated by artificial insemination (AI; control) or by ART. Seven of the 27 conceptuses in the ART group were in the > 97th percentile body weight when compared with controls. Further, other characteristics reported in BWS were observed in the ART group, such as large tongue, umbilical hernia, and ear malformations. KCNQ1OT1 (the most-often misregulated imprinted gene in BWS) was biallelically-expressed in various organs in two out of seven overgrown conceptuses from the ART group, but shows monoallelic expression in all tissues of the AI conceptuses. Furthermore, biallelic expression of KCNQ1OT1 is associated with loss of methylation at the KvDMR1 on the maternal allele and with downregulation of the maternally-expressed gene CDKN1C. In conclusion, our results show phenotypic and epigenetic similarities between LOS and BWS, and we propose the use of LOS as an animal model to investigate the etiology of BWS.
Keywords: BWS, LOS, KvDMR1, KCNQ1OT1, epigenetics, genomic imprinting
Introduction
Beckwith-Wiedemann syndrome (BWS) (OMIM 130650) is a pediatric overgrowth condition with an occurrence of 1 in 13,700 natural births.1,2 BWS is a complex syndrome and has highly variable clinical features.1,2 The primary features of BWS include macrosomia (overgrown bodyweight > 97th percentile), macroglossia (enlarged tongue), and abdominal wall defects (umbilical hernia).1,2 Secondary characteristics such as ear malformations, visceromegaly, neonatal hypoglycemia, and nevus flammeus are less frequently observed in BWS patients.1,2 In addition, BWS is associated with increased risk of childhood tumors (rate ranges from 4% to 21%), with Wilms? tumor of kidney and hepatoblastoma being the two most commonly observed.2,3
Genomic imprinting is a series of epigenetic processes that lead to parental-allele-specific gene expression in mammals.4-6 Because of genomic imprinting, both maternal and paternal genomes are required for embryonic growth and development. Of the identified imprinted genes, most are found in clusters containing two or more imprinted genes in an imprinting domain which is in turn regulated by a differentially methylated region of DNA known as the imprinting control region (ICR).4-6 The parental-allele-specific DNA methylation of the ICRs is erased in primordial germ cells and re-established during prospermatogenesis in male and oocyte growth in female.4-6
The molecular alterations responsible for BWS have been mapped to chromosome region 11p15 (syntenic to mouse chromosome 7) which has two imprinting clusters: imprinting center 1 (IC1) and IC2.1,2 In humans, IC2 contains one paternally-expressed non-coding RNA (ncRNA) and at least six maternally-expressed protein-coding genes,7 and this cluster is regulated by the ICR referred to as KvDMR1. The KvDMR1 is unmethylated on the paternal chromosome. In mice, it has been shown that unmethylated KvDMR1 permits the transcription of long non-coding RNA Kcnq1ot1, which recruits the Polycomb group proteins (such as Ezh2 and Rnf2) and repressive histone marks (such as H3K27me3 and H2AK119U1) to create a repressive chromatin conformation where maternally- expressed genes are located and repressed on the paternal allele.8,9 However, on the maternal allele, methylation of the KvDMR1 prevents the transcription of the Kcnq1ot1 gene; therefore, flanking maternally-expressed genes such as Cdkn1c, Kcnq1, and Phlda2 are transcribed.10-12 About 50% of naturally occurring BWS cases present with loss of methylation at KvDMR1 on the maternal allele, which is coupled with biallelic expression of KCNQ1OT1 and downregulation of CDKN1C.1,2 Reports estimated a 3- to 9- fold increased likelihood of BWS in children conceived with the use of assisted reproductive technologies (ART).13-18 Currently, determination of the methylation status of the KvDMR1 is the most often used procedure to associate the syndrome to a molecular lesion.
IC1 contains the maternally-expressed ncRNA H1919 and the paternally-expressed fetal growth factor, IGF2.20 The imprinting mechanism of this locus has been elucidated in a series of mouse experiments. On the maternal allele, H19/Igf2 ICR is unmethylated, allowing for the binding of the insulator protein CTCF (CCCTC binding factor). This binding results in a chromosome boundary which prevents the interaction of the Igf2 promoter with the downstream enhancers. Thus, H19 has access to the enhancer and is transcribed. However, on the paternal allele, H19/Igf2 ICR is methylated which avoids the binding of CTCF protein,21 therefore, allowing the Igf2?s promoter to interact with the downstream enhancers. Approximately 2?7% of naturally-conceived BWS cases have a gain of methylation on H19/IGF2 ICR, which is associated with the increased expression level of IGF2.1,2
Large offspring syndrome is an overgrowth disorder in ruminants which is phenotypically similar to BWS.22 The features of LOS include: excessive birth weight, large tongue, umbilical hernia, hypoglycemia, and visceromegaly.22-28 As in BWS, LOS can result from ART.22-28 Previous studies observed hypomethylation of the KvDMR1 and biallelic expression of KCNQ1OT1 in somatic nuclear transfer (SCNT) and ART-produced bovine conceptuses.29,30 However, ascription of parental origin to the alleles during methylation studies has been difficult as a result of the polymorphic nature of cattle, which is similar to the situation in humans. A previous study performed in our laboratory showed that allelic expression of KCNQ1OT1, CDKN1C, and H19 and DNA methylation of the KvDMR1 and H19/IGF2 ICR in day 65 bovine conceptuses is conserved to humans.31
Given the similarities between BWS and LOS, and together with previous studies29-31 we hypothesized that bovine conceptuses with the overgrowth phenotype would have similar misregulation of imprinted loci as those reported for the human overgrowth condition BWS. In the present study, we used B. t. indicus × B. t. taurus F1 hybrid conceptuses produced by ART. We determined the allele-specific DNA methylation and expression of imprinted genes in IC1 and IC2 by using the identified31 fixed polymorphisms between the two subspecies of cattle. We show that LOS conceptuses at day ~105 resemble the phenotype of BWS. Most importantly, two LOS conceptuses display biallelic expression of the ncRNA KCNQ1OT1, which is coupled with loss of methylation of KvDMR1 and downregulation of CDKN1C.
Results
Generation of LOS conceptuses
To determine if the IC2 and IC1 are misregulated in LOS as in BWS, we generated LOS conceptuses with the use of ART procedures known to induce the syndrome in bovine.22-28 Based on published observations27,28 we expected that 16% of our ART conceptuses would be of the overgrown phenotype. LOS conceptuses were collected at day ~105; when features of LOS can be first characterized.32 We used B. t. indicus × B. t. taurus F1 hybrid conceptuses which allowed us to distinguish paternal and maternal alleles based on fixed polymorphisms at these loci between the two subspecies of cattle.31 B. t. indicus × B. t. taurus F1 hybrid control conceptuses were produced by artificial insemination (AI). Twenty-seven conceptuses were collected from the ART group and nine from the AI group (Fig. 1A). Average bodyweight, organ weight (liver, heart, lung, kidney, spleen, and tongue), crown-rump length, foreleg length, and head width were not significantly different (p > 0.05) between groups (data not shown). Body weight did not differ between singletons and twins. However, heart girth (an indirect measure of body weight) of ART conceptuses was significantly larger than AI conceptuses (p < 0.03; means ± SEM = 16.42 ± 0.19 vs. 15.53 ± 0.32 cm, for ART and AI group, respectively).
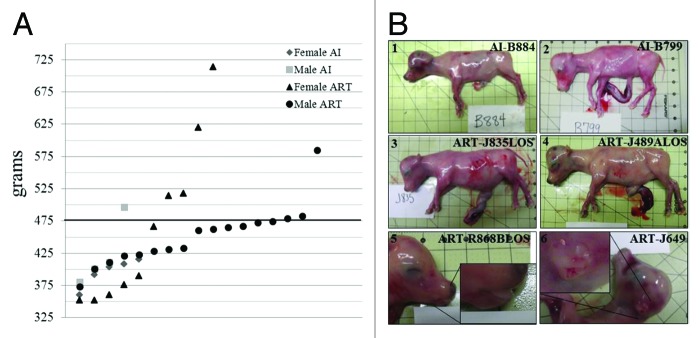
Figure 1. LOS bovine fetuses have similar phenotype characteristics as those reported in BWS patients. (A). Fetal weight at day ~105 gestation. Y axis represents the weight in grams. X axis has no actual implication and is used to scatter the spots representing each fetus for ease of visualization. The sex of the fetuses and the way they were generated is shown at the top-right side. The bold line represents the 97th percentile of control weight (i.e. 476.8 g). (B). Primary and secondary characteristics of BWS can be observed in LOS; B.1 = AI-B884 (control female weighing 400 g) and B.2 = AI-B799 (control male weighing 408 g which is the approximate average weight of the control fetuses). B.3 and B.4 show fetuses with macrosomia (ART-J835LOS ? female weighing 714 g and ART-J489ALOS? female weighing 514 g). B.5 shows an example of macroglossia in a female weighing 620 g and B.6 shows an ear malformation in a female weighing 320 g. Each square on the background = 2.54 cm2. LOS, large offspring syndrome; BWS, Beckwith-Wiedemann syndrome; AI, artificial insemination; ART, assisted reproductive technologies.
Children with at least three primary features or two primary features and one or more secondary features are diagnosed as BWS patients.1,2 In the present study, we used the overgrown feature (bodyweight > 97th percentile) as a major criterion to diagnose fetuses with LOS. The 97th percentile was calculated based on the bodyweight of AI conceptuses, and this explains why one AI conceptus (AI-C010) was also above the bodyweight 97th percentile (Fig. 1A). For the ART conceptuses, seven out of 27 (26%) were above 97th percentile, and females showed a greater variability in bodyweight than males (range ? male = 372?584 g and female = 352?714 g; Fig. 1A). Besides increased bodyweight, other features of LOS were also observed in the ART conceptuses (Fig. 1B) including enlarged tongue (macroglossia; n = 3), umbilical hernia (n = 2; data not shown), and ear malformation (n = 1).
Expression analysis of imprinted genes
To test if BWS-associated imprinted genes are similarly misregulated in LOS, we determined allelic expression of six imprinted genes in liver, muscle, brain, tongue, heart, lung, kidney, and placenta (Fig. 2). Four of these genes, CDKN1C, KCNQ1, PHLDA2, and H19 are expressed from the maternal chromosome, whereas KCNQ1OT1 and IGF2 are expressed from the paternal chromosome. Fifty percent of naturally-conceived BWS patients show loss of methylation on the maternal allele of the differentially methylated region known as KvDMR1, and this loss-of-imprinting is correlated with biallelic expression of KCNQ1OT1.1,2 KCNQ1OT1 was biallelically-expressed in several tissues in two (ART-J835LOS and ART-J489ALOS) of the seven overgrown conceptuses from the ART group, but showed monoallelic expression in all tissues of the AI conceptuses (Fig. 3, Fig. S1 and Table S1.1). H19, IGF2, and CDKN1C were imprinted for both ART and AI groups in liver, muscle, tongue, heart, lung, kidney, and placenta. However, CDKN1C and IGF2 were expressed from both parental alleles in the brain of fetuses from both groups (Table S1.2, S1.4?S1.5). PHLDA2 only showed monoallelic expression in liver and placenta in both groups, but was biallelically-expressed in other tissues in both conditions (Table S1.4). KCNQ1 showed global biallelic expression with a bias toward the maternal allele in both groups (Table S1.3).
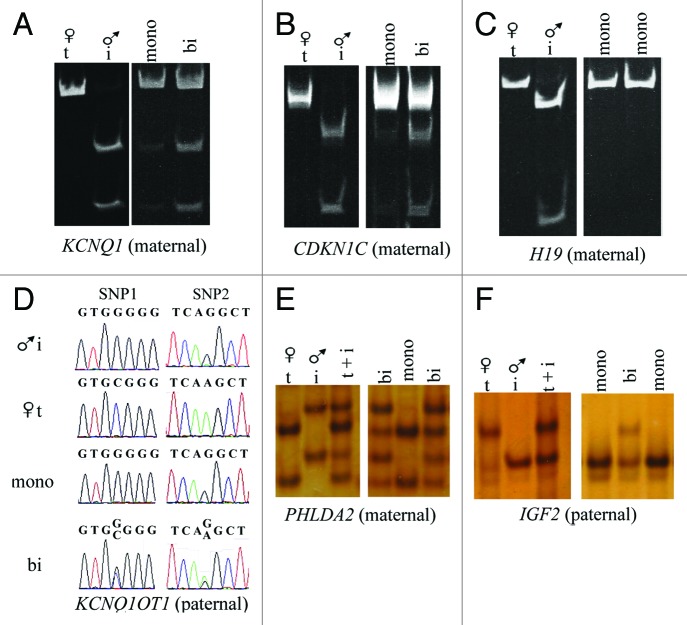
Figure 2. Example of assays used to determine allelic expression in tissues from B. t. indicus × B. t. taurus F1 hybrid conceptuses. Shown are examples of allelic determination by RT-PCR followed by RFLP and PAGE (A-C), Sanger sequencing (D) or SSCP analysis (E-F). The left portion of the panels (A-C, E and F) shows the band pattern of B. t. taurus and B. t. indicus control tissues (liver) which was used as reference to determine parental expression of imprinted gene in tissues from B. t. indicus × B. t. taurus F1 hybrids. The right portion of the panels shows examples of monoallelic and biallelic expression of several imprinted genes in ~d105 conceptus. (D) is an example of the Sanger sequencing allelic assay for KCNQ1OT1, a paternally-expressed gene. Two SNPs were used in this assay; double peaks demonstrate biallelic expression. The contribution of each parental allele to the total expression was determined by the use of Image J (NIH). Only samples with at least 10% expression from the repressed allele were considered to be biallelically-expressed. T, B. t. taurus; i, B. t. indicus; mono, monoallelic; bi, biallelic; SNP, single nucleotide polymorphism; RFLP, restriction fragment length polymorphism; SSCP, single strand conformation polymorphism; PAGE, polyacrylamide gel electrophoresis.
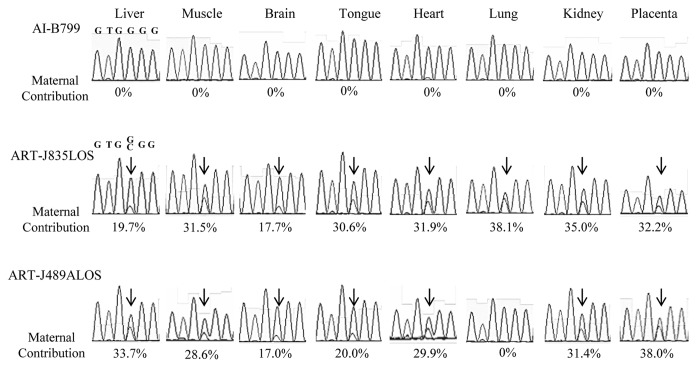
Figure 3. Biallelic expression of KCNQ1OT1 in LOS fetuses. Shown is Sanger sequencing data of KCNQ1OT1 RT-PCR product in tissues analyzed in AI-B799 (control), ART-J835LOS and ART-J489ALOS fetuses. The columns show the chromatograph for each tissue of each fetus. Values below the chromatograph are the percentage of KCNQ1OT1 expressed from the maternal allele. Arrows show the double peaks of SNP1 site (refer to Fig. 2) in ART-J835LOS and ART-J489ALOS fetuses. For clarity of depiction, SNP2 site is shown here (Fig. S1). LOS, large offspring syndrome; RT-PCR, reverse transcription-polymerase chain reaction; SNP, single nucleotide polymorphism.
Since biallelic expression of KCNQ1OT1 is associated with the repressed expression of CDKN1C from the maternal allele, we then performed quantitative RT-PCR to determine CDKN1C mRNA levels in tissues with biallelic expression of KCNQ1OT1. We compared CDKN1C expression level in each of the two LOS conceptuses (ART-J835LOS and ART-J489ALOS) with the average level of transcript of eight AI conceptuses. To get a better understanding of whether CDKN1C?s expression is directly affected by the expression of KCNQ1OT1 from the maternal allele or if it is an artifact of the ART procedures we compared the CDKN1C expression of ART-J835LOS and ART-J489ALOS to the five remaining LOS conceptuses. We found that the level of the CDKN1C RNA of ART-J835LOS and ART-J489ALOS was lower when compared with the average expression of the AI controls and the average expression of the monoallelic LOS group (Fig. 4A). CDKN1C expression level in the AI group and LOS group with correct imprinting of KCNQ1OT1 was comparable in most tissues except in the placenta where the LOS group is higher (Fig. 4A; p < 0.05). It should be noted that, the AI-C010 fetus (bodyweight > 97 percentile) was included in the eight fetuses analyzed and its CDKN1C expression distributed randomly among the AI controls and never occupied an extreme position. We next queried about the level of CDKN1C expression between tissues in control fetuses and we found that this gene is expressed at lower levels in brain and heart when compared with other tissues (Fig. 4B).We also analyzed the expression level of PHLDA2, which is a maternally-expressed gene and expected to be regulated similar to CDKN1C. ART-J835LOS showed downregulation of PHLDA2 in all tissues except muscle, while expression of this gene was only downregulated in liver, heart and lung in ART-J489ALOS (Fig. S2A). In contrast to what we observed for CDKN1C, PHLDA2 showed different expression level in the various tissues analyzed with highest expression observed in the kidney and placenta (Fig. S2B). As 2?7% of BWS cases are associated with biallelic expression of IGF2, which leads to increased mRNA level of IGF2,1,2 we also determined IGF2 expression level in LOS conceptuses. IGF2 transcript levels were similar between LOS and the AI control group (data not shown).
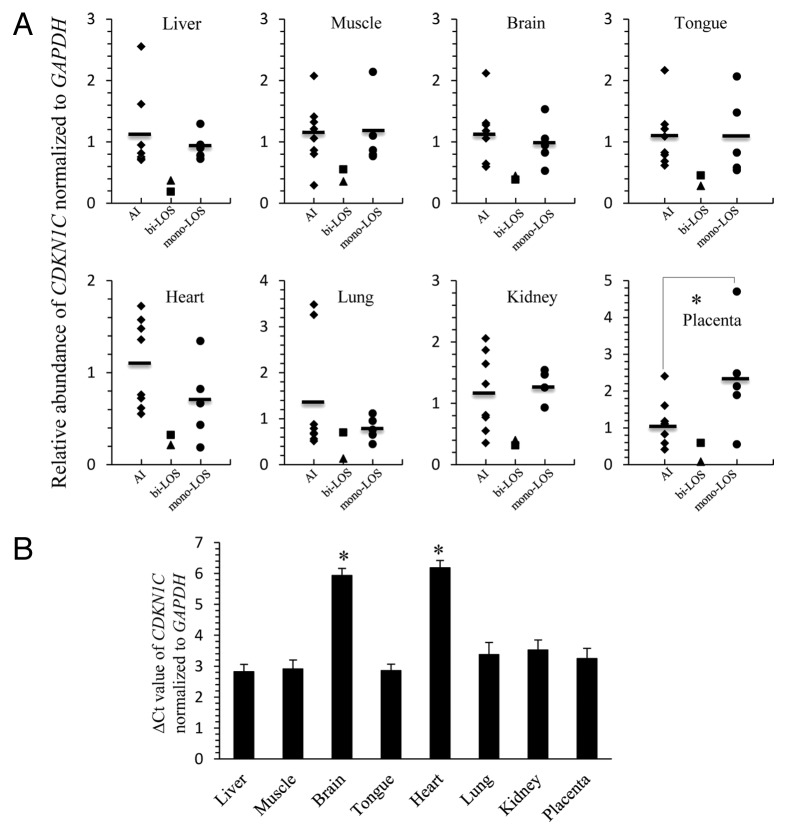
Figure 4. LOS fetuses with biallelic expression of KCNQ1OT1 show downregulation of CDKN1C. (A) The CDKN1C RNA was determined by quantitative RT- PCR in several tissues from eight AI fetuses (diamonds; AI), two LOS fetuses with biallelic expression of KCNQ1OT1 (triangle = ART-J835LOS; square = ART-J489ALOS; bi-LOS), and five LOS fetuses with correct imprinting of KCNQ1OT1 (circles; mono-LOS). The short line among the diamonds and circles represents the average level of the individuals. CDKN1C level was normalized to the expression of GAPDH. Note that for each tissue analyzed CDKN1C level in ART-J835LOS is at least 2-fold lower than the average level found in AI fetuses and LOS fetuses with correct imprinting of KCNQ1OT1. It should be also noted that lung from ART-J489ALOS which had correct imprinting of KCNQ1OT1 (Fig. 3) exhibits the comparative CDKN1C level with controls. (B) The threshold cycle (Ct) of CDKN1C was normalized to the reference gene GAPDH in each tissue from the eight AI fetuses. The data are expressed as mean ± SEM. *p < 0.05 between the brain, heart and other tissues analyzed. LOS, large offspring syndrome.
Methylation analyses of KvDMR1, H19/IGF2 ICR and CDKN1C exon 2
Loss of methylation of KvDMR1 is the most common epimutation in BWS.1,2 Sodium bisulfite mutagenesis was used to investigate the methylation status of the KvDMR1 in the tissues of fetuses that had maternal KCNQ1OT1 expression. We determined methylation status of a 385 bp region containing 37 CpGs. We show that in most tissues, loss of methylation on the maternal allele was coupled with biallelic expression of KCNQ1OT1 in these fetuses (Fig. 5 and Fig. S3). Interestingly, the placental tissue of AI-C010, the largest AI conceptus in the control group, also showed reduced methylation of the KvDMR1 on the maternal allele (Fig. 5).
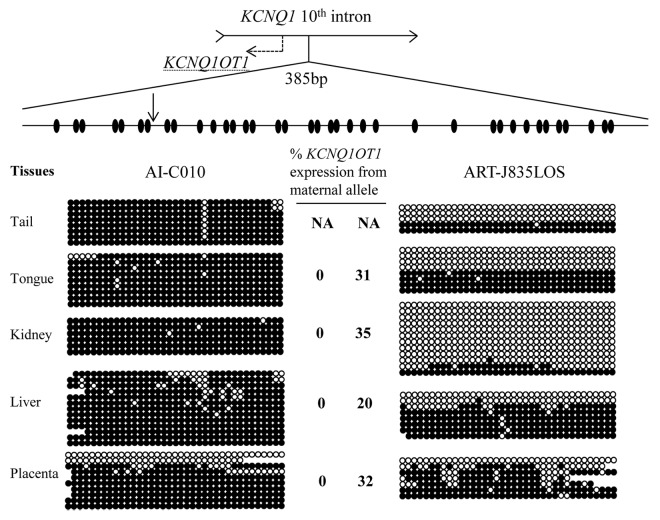
Figure 5. Loss of methylation of KvDMR1 on the maternal allele is associated with biallelic expression of KCNQ1OT1 in LOS fetuses. DNA was treated with sodium bisulfite prior to PCR, and PCR product was cloned before sequencing. Sequencing data was used to determine the DNA methylation status at the KvDMR1. Shown on top is a depiction of the 10th intron of the maternally-expressed gene KCNQ1 and its direction of transcription is shown with an arrow. The region harbors the promoter of the antisense long ncRNA KCNQ1OT1 (shown as dashed arrow), which is also an imprinting control region known as KvDMR1. A 385 bp region of the KvDMR1 was used to determine the DNA methylation status of 37 CpG sites (ovals). A SNP (vertical arrow) between B. t. indicus and B. t. taurus was used to determine the parental origin of the alleles and only maternal alleles are shown here. Five tissues from two fetuses are shown. Filled and open circles represent methylated and unmethylated CpG dinucleotides, respectively. Missing circles are sequencing data of low quality. Each line denotes an individual DNA strand. The level of maternal KCNQ1OT1 expression is shown in the center and next to the strands. Tail tissues were collected for the purpose of DNA analysis, precluding its use for gene expression determinations. NA, not available.
The H19/IGF2 ICR is normally unmethylated on the maternal allele but methylated on the paternal allele. We then asked if the biallelic expression of IGF2 in the brain of the fetuses studied was associated with gain of methylation of H19/IGF2 ICR on the maternal alleles. Here we show that H19/IGF2 ICR had differential methylation in brain samples where IGF2 was biallelically-expressed (Fig. S5).
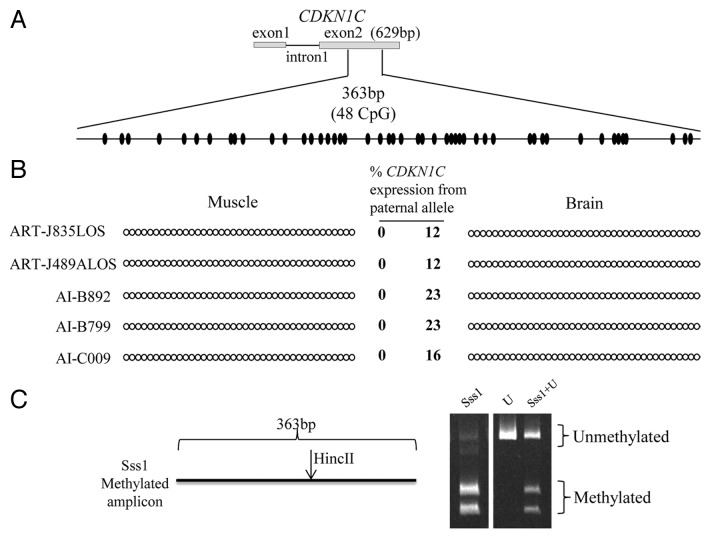
In mice, differential methylation of Cdkn1c region was observed from -600 bp from the transcription start site to exon 2 33. Cdkn1c DMR is a somatic imprint,33 and therefore is established after implantation in mice.33,34 However, the homologous region in humans is unmethylated on both alleles.35,36 In the present study, biallelic expression of CDKN1C was observed in brain in both ART and AI conditions. We then asked if DNA methylation is involved in the regulation of CDKN1C imprinting. Currently, DNA sequence information in the upstream region of CDKN1C in bovine (GenBank accession number NW_003104648.1: 2774900?2775500) harbors a sequencing gap, and we were unable to amplify the 5? end of CDKN1C. Therefore, we focused on the CDKN1C region encompassing exon 1 to exon 2. This region in the bovine (GenBank accession number NW_003104648.1: 2775692?2776645) has 78% identity to the locus in humans (GenBank accession number NT 009237.18: 2847003?2846299). We determined methylation status of a 363 bp region located in the second exon which contains 48 CpG dinucleotides. Since no SNPs between B. t. indicus and B. t. taurus are available at this locus,31 we used Sanger sequencing for direct sequencing of the PCR product in order to investigate the potential of differential methylation which would be identifiable as a double peak (i.e. one for C and one for T) in the chromatograph. Sequencing results showed hypomethylation in both muscle, in which CDKN1C is normally imprinted, and brain, where CDKN1C is biallelically-expressed (Fig. 6A?B and Table S1.2). It is conceivable that only the unmethylated maternal alleles were amplified because of a bias introduced during PCR amplification. To exclude this possibility, we co-incubated genomic DNA with the methyltransferase Sss137 prior to performing bisulfite mutagenesis. Sss1 treatment combined with COBRA showed that no bias was introduced during PCR amplification as both methylated and unmethylated DNA were similarly amplified (Fig. 6C).
Figure 6. Imprinted expression of CDKN1C is not regulated by somatic DNA methylation. (A) Shown on top is a depiction of the first two exons and the first intron of the maternally-expressed gene CDKN1C. No DNA polymorphisms were identified in this region. Bisulfite converted specific primers for a 363 bp region of the exon 2 were used to determine the DNA methylation status of 48CpG sites (ovals). (B) The PCR product was sequenced without cloning. Two tissues from five fetuses are shown. Filled and open circles represent methylated and unmethylated CpG dinucleotides, respectively. Each line denotes the DNA methylation pattern of each sample. Only the first 39 CpGs are shown here because of the low quality sequencing of last 9 CpGs due to primer binding. The level of paternal CDKN1C expression is shown in the center and next to the strands (based on a SNP in exon 4). (C) COBRA performed to ensure ability of primers to equally amplify methylated and unmethylated alleles. The restriction enzyme HincII recognized and cleaved the methylated amplicon. Sss1 = genomic DNA co-incubated with the methyltransferase Sss1 prior to bisulfite conversion, U = bisulfite converted DNA with no Sss1 treatment, Sss1 + U = equal portions of methylated and unmethylated DNA were used for PCR amplification. The digested products were resolved by PAGE.
Discussion
In the present study we show that the bovine model of LOS has extensive similarities with BWS. Phenotypically, LOS exhibited macrosomia, macroglossia, and umbilical hernia, which are primary characteristics of BWS.1,2 Additionally, a secondary feature of BWS, namely ear malformations, was also observed in LOS.1,2 At the epigenetic level, two LOS conceptuses showed loss of methylation at the KvDMR1 on the maternal allele. This epimutation is the major molecular signature of BWS1,2 and is observed in 50% of individuals afflicted with this syndrome.1,2 Furthermore, downregulation of CDKN1C was observed in conceptuses with biallelic expression of KCNQ1OT1. This is a finding similar to what have been reported using skin fibroblasts from BWS patients with loss of methylation at the KvDMR1.38
It should also be noted that in our study, female conceptuses showed greater variability in bodyweight when compared with males. In humans, it has been observed that monozygotic female twins have a higher frequency of BWS cases than monozygotic male twins.39 Likewise, the monozygotic female twins with BWS were generally associated with loss of methylation of the KvDMR1 on the maternal allele.39 Weksberg and others39 suggested that the higher frequency of BWS in monozygotic female twins might be partially explained by the overlap in timing of X-inactivation and global DNA methylation reprogramming in the preimplantation period.
IGF2 was exclusively expressed from the paternal chromosome except in brain where it had biallelic expression. This is in accordance with previous studies where IGF2 showed biallelic expression in brain in both mice and cattle.40,41 Similarly, in our study, KCNQ1 was biallelically-expressed in all tissues analyzed which is consistent with the situation in the mouse, where even though Kcnq1 is maternally-expressed at midgestation, it is globally biallelically-expressed at birth.42,43
Even though it is accepted1,2 that BWS is associated with misregulation of one or more imprinted genes in one or more imprinting clusters, several pivotal questions remain unanswered about this overgrowth syndrome. First, what are the cellular and molecular alterations causing loss-of-imprinting at the specified loci? Second, how does misregulation of imprinted gene expression translates into the highly variable and complex phenotypes of BWS?
At present, only associations exist between loss of methylation at specific imprinting centers and BWS. No evidence exists that points to any particular genomic region which when epimutated, triggers the overgrowth phenotype and associated developmental errors in humans. It is not known if loss-of-imprinting is the cause or a symptom of BWS. Currently, no animal model exists that faithfully recapitulates the various phenotypic and epigenetic singularities of BWS. Several mouse models for this syndrome have been generated by introducing genetic mutations into IC1 and/or IC2. The genetic mutations include: maternal mutation of Cdkn1c,44 double mutation including H19?13 (deletion of H19/Igf2 ICR and H19) and Cdkn1c,45 maternal H19?13 which leads to higher expression level of Igf2,46,47 and overexpression of Igf2.48 All these mouse models provided fundamental understanding of the essential function of imprinted genes in embryonic development as well as mechanisms of genomic imprinting regulation. However, these models did not phenocopy the overgrowth as well as other the primary and secondary characteristics of BWS. For example, maternal mutant Cdkn1c mice exhibited 20% overgrowth during prenatal period, but the increased bodyweight was not seen at birth which can probably be explained by intrauterine competition for maternal nutrients in litter bearing species.44
Several reasons exist to propose the use of LOS as an adequate animal model to study BWS. First, only ruminants and humans have been reported to display the overgrowth and excessive weight at birth as a result of minimal ART manipulations.13-18,22-28,49-51 Second, loss of methylation at KvDMR1 and biallelic expression of KCNQ1OT1 were observed in both human1,2 and LOS. Third, here we show lack of DNA methylation at the bovine CDKN1C exon 2 which is in accordance to what has been reported for humans35,36 but is in stark contrast to the situation in the mouse where differential methylation is evident.33 Fourth, females of both species carry primarily singleton pregnancies (monotocous). It has been suggested that variance for growth regulation exist between litter bearing and non-litter bearing species.44 Fifth, both human and bovine have a nine month gestation period. This is important because sequential events that lead to molecular lesions resulting in the overgrown phenotype or other features of BWS may occur at similar times during pregnancy and the potential exists to evaluate the timing of intervention strategies.
BWS has more recently been associated with misregulation at loci other than the IC2 and IC1 such as MEST (PEG1), PLAGL1 (ZAC1) and GNAS 52,53. Misregulation of multiple imprinting clusters in BWS speculates that highly variable clinical features of BWS may result from diverse combinations of epimutation of each imprinting center. Future work is planned to determine if the same is observed in the LOS model.
In conclusion, our results show phenotypic and epigenetic similarities between LOS and BWS, and we propose the use of LOS as an animal model to investigate the etiology of BWS.
Materials and Methods
Animals
We used B. t. indicus and B. t. taurus, two subspecies of cattle, to produce F1 hybrid progenies. The use of B. t. indicus × B. t. taurus F1 individuals allowed us to determine allele-specific expression and DNA methylation of imprinted genes by the use of polymorphisms between the two subspecies. A previous study in our laboratory identified DNA polymorphisms between the two subspecies in IC1 and IC2 imprinting domains.31
Experimental groups
Control conceptuses
The estrous cycle of B. t. taurus (Holstein breed) females was synchronized and the females were artificially inseminated (AI) with semen from one B. t. indicus bull (Nelore breed; ABS CSS MR N OB 425/1 677344 29NE0001 97155). Four males and five female B. t. indicus × B. t. taurus F1 conceptuses (fetus + placenta) were collected on day ~105 (104?106). This time was chosen because phenotypic characteristics of LOS can be recognized at this stage.32 Conceptuses were retrieved from the gravid uterus at caesarean section in order to preserve nucleic acid integrity. At collection, crown-rump length, heart girth, foreleg length and head width were measured,54 as well as body and organ weight. The following tissues were collected: liver, muscle, brain, tongue, heart, lung, kidney, spleen, reproductive tract, intestine, skin and placenta. Tissues were diced and mixed at collection and were snap frozen in liquid nitrogen and stored at -80°C until use.
ART conceptuses
In vitro production of bovine embryos was performed as previously described by us55 and http://www.animal.ufl.edu/hansen/ivf/. All media (Hepes-TL, IVF-TL, SP-TL) were purchased from Caisson Laboratorys. All chemicals used to prepare media were purchased from Sigma. Briefly, B. t. taurus (Holstein) cumulus-oocyte complexes (COCs) were shipped overnight in maturation medium from TransOva Genetics. At receipt, the oocytes were rinsed in Hepes-TALP (Tyrode?s Albumin Lactate Pyruvate) and immediately placed in IVF-TALP. Semen from the same B. t. indicus bull used to generate control conceptuses was used for IVF. The semen in one straw was thawed at 37.0°C. The semen straw contents were added into a 15 ml centrifuge tube containing 13 ml SP-TALP and centrifuged for 15 min at 200 × g. The sperm pellet was suspended in 300 ?l of IVF-TALP and this volume was pipetted into a glass wool column in order to separate live sperm cells from debris and dead sperm. Note: this bull does not perform well in vitro when purified by Percoll gradient. The COCs and the sperm cells were co-incubated in IVF-TALP at 38.5°C in humidified air containing 5% CO2 for 18 h.55 Putative zygotes were cultured in KSOM-BE55 in groups of 25?30. The embryos were cultured at 38.5°C in humidified atmosphere containing 90% N2, 5% CO2 and 5% O2. On day 5 after fertilization, embryo culture drops were supplemented with 10% estrus cow serum. On day 6, 20?30 early blastocysts were placed in 2.0 ml tubes containing 1.8 ml KSOM-BE supplemented with 180 ?l (10%) estrus cow serum and covered with mineral oil. The embryos were shipped overnight to TransOva Genetics in a portable incubator maintained at 38.5°C. Upon receipt, the embryos were transferred into synchronized B. t. taurus recipients (2 blastocyst per recipient; one/uterine horn). Embryo transfers and conceptus retrievals were performed by theriogenologists at TransOva Genetics. On day ~105 (104?106), conceptuses were collected and processed as described for the control conceptuses. Twenty-seven conceptuses were collected from 19 recipients (8 recipients had twins).
All animal procedures were performed at TransOva Genetics by veterinarians, and all procedures were approved by TransOva?s animal care and use committee.
RNA isolation, cDNA synthesis and reverse transcriptase polymerase chain reaction
RNA was isolated from fetal tissues with the use of Trizol Reagent (Invitrogen) according to the manufacturer?s instructions. RNA amount and quality were confirmed by spectrometry and agarose gel electrophoresis, respectively. RNA was treated with DNase (Fisher Scientific) before cDNA synthesis. 100 ng RNA was used as template to synthesize cDNA in a 20 ?l reaction with: 10 mM DTT (Invitrogen), 1x First Strand Buffer (Invitrogen), 0.5 ?g random primer (Promega), 1mM dNTPs (Fisher Scientific), 100 U SuperscriptII reverse transcriptase (Invitrogen), and 20 U of Optizyme RNase Inhibitor (Fischer Scientific). The samples were incubated at 42°C for 1 h, at 95°C for 10 min, and stored at -20°C until use. To ensure no genomic DNA contamination, a minus reverse-transcriptase control was also included. For PCR amplification, 2 ?l cDNA (~10ng) was added into the PCR mix [1 × colorless GoTaq Flexi Buffer (Promega), 0.3 ?M forward and reverse primer (IDT), 2.5 mM MgCl2 (Promega), 200 ?M dNTP (Fisher Scientific) and 0.5 U GoTaq Hot Start polymerase (Promega)]. For each assay, at least 5 sets of primers were tested to get specific amplification. The PCR conditions were as follows: denaturation at 94°C for 2 min 15 sec, then 35 cycles of 94°C for 15 sec, 59.5?62.8°C for 20 sec and 72°C for 30 sec, and final extension at 72°C for 5 min (Table S2.1).
Allele-specific expression analysis of imprinted genes
The six imprinted genes analyzed in this study were the maternally-expressed genes: CDKN1C, PHLDA2, H19, and KCNQ1 and the paternally-expressed genes: KCNQ1OT1 and IGF2. These genes were analyzed in eight tissues; namely, liver, muscle, brain, tongue, heart, lung, kidney, and placenta. Expression of KCNQ1OT1 was determined by RT-PCR followed by Sanger sequencing. The samples were sequenced at the University of Missouri?s DNA core using the 96-capillary applied Biosystems 3730 DNA analyzer with Big Dye Terminator. Sequencing data was aligned to a reference with the use of MacVector software (Cary). Allelic expression of CDKN1C, H19, and KCNQ1 were determined by RT-PCR followed by allele-specific restriction enzyme digests (Table S2.1). The digested PCR products were resolved by polyacrylamide gel electrophoresis (PAGE). The assay used to determine allele-specific expression of H19 was previously described.31 Allelic expression of PHLDA2 and IGF2 were determined by RT-PCR-SSCP (single strand conformation polymorphism) because restriction enzymes that recognized the sequence of interest were not available. Briefly, SSCP was conducted on an 8% polyacrylamide gel and run at 110V overnight (~14 h). The SSCP gel was then subjected to silver staining (Bio-Rad) and dried by Gel-Dry (Invitrogen). The contribution of each parental allele to the total expression was determined by Image J (NIH). Only samples with at least 10% expression from the repressed allele were considered biallelic.56
Quantitative RT-PCR of CDKN1C, PHLDA2 and IGF2
Taqman gene expression assays (Applied Biosystems; Table S2.1) were used to determine if CDKN1C, PHLDA2, and IGF2 showed different expression levels among control conceptuses, the LOS conceptuses with biallelic expression of KCNQ1OT1, and the LOS conceptuses with monoallelic expression of KCNQ1OT1. The assay was conducted in the eight tissues described in the allele-specific expression analysis section. The CDKN1C level of expression of the eight tissues of the two LOS conceptuses with biallelic expression of KCNQ1OT1 were compared with the tissues of the eight control conceptuses (5 females and 3 males) and five LOS conceptuses (2 females and 3 males) with monoallelic KCNQ1OT1 The samples were analyzed in triplicates, and the threshold cycle was normalized to the housekeeping gene GAPDH using an ABI Real-time 7500 system. The expression level for each gene in each tissue was calculated using the comparative CT method. The expression levels of our bovine samples were plotted as described before in human.38
DNA isolation and bisulfite conversion
DNA from B. t. indicus × B. t. taurus F1 individuals was isolated using phenol-chloroform. Bisulfite mutagenesis was conducted with the Imprint DNA Modification Kit (Sigma) according to manufacturer?s instructions. During this procedure, unmethylated cytosines are converted into uracils, but methylated cytosines remain cytosines. After PCR amplification, uracils are replaced by thymines. Primers for the bisulfite-converted DNA were designed for KvDMR1, H19/IGF2 ICR, and CDKN1C exon 2 (Table S2.2). The PCR conditions were as follows: denaturation at 94°C for 2 min15 sec, then 45 cycles at 94°C for 30 sec, 53.5?62.1°C for 45s and 72°C for 1 min 30 sec, and final extension at 72°C for 5 min (Table S2.2). Note that 1M Betaine was necessary for amplification of the H19/IGF2 ICR.
DNA methylation analysis of KvDMR1 and H19/IGF2 ICR
The PCR product of the bisulfite-converted regions of interest was isolated from a 1% agarose gel with Wizard SV gel and PCR Clean-Up System (Promega). KvDMR1 (385 bp containing 37 CpGs; GenBank accession number NW_003104648.1: 2960086?2960470) and H19/IGF2 ICR (318 bp containing 20/21 CpGs; GenBank accession number NW_003104648.1: 3556002?3556319) amplicons were inserted into pCC1 vector with chloramphenicol resistance gene and cloned using CopyControl PCR cloning kit with TransforMax EPI300 electrocompetent E. coli cells (Epicenter Biotechnologies)56 according to the manufacturer?s instructions except that all the cloning incubation procedures were done at 25°C. Note: it took approximately 2 d to form visible colonies at this temperature. The individual clones were sequenced and analyzed as described for KCNQ1OT1 sequencing.
DNA methylation analysis of CDKN1C exon 2
In mice, Cdkn1c DMR starts from 600 bp upstream of transcription start site of Cdkn1c and extends through exon 2 33. The homologous region in humans is, however, unmethylated.35,36 A 363 bp region of bisulfite-converted exon 2 (containing 48 CpGs and no SNPs; GenBank accession number NW_003104648.1: 2776175?2776537) was amplified by PCR. The PCR product was processed and sequenced as described above. Primer information can be found in Table S2.2.
Sequencing data showed hypomethylation of CDKN1C exon 2. To ensure that the primers used were equally able to amplify bisulfite converted methylated and unmethylated DNA, we did the following; (1) an aliquot of DNA was bisulfite converted with no Sss1 treatment; (2) another aliquot of DNA was treated with Sss1 methyltransferase (New England BioLabs) prior to bisulfite conversion; (3) samples were mixed with a 1:1 ratio. The three types of template were analyzed separately by combined bisulfite restriction analysis (COBRA). The enzyme used for COBRA was HincII (New England BioLabs) which only cuts the methylated amplicons.
Statistical analysis
Bodyweight, organ weight, crown-rump length, heart girth, foreleg length and head width were analyzed by using standard General Linear Model procedure of SAS with fixed factors: ART/AI and sex. The significance level is p < 0.05.
Funding
This work was supported by the National Institutes of Health (grant number 5R21HD062920); the Reproductive Biology Group Food for the 21st Century program at the University of Missouri, and the University of Missouri Research Board (grant number CB000384).
Supplementary Material
Acknowledgments
We would like to acknowledge Mr. Matthew Sepulveda, Ms. Angela Schenewerk, and Ms. Verónica Negrón-Pérez, for their invaluable assistance with the collection of the B. t. indicus × B. t. taurus day 105 conceptuses. We would also like to thank TransOva Genetics for being so accommodating to our needs of space and timing during conceptus collections. We need to thank Mr. Brian Brace from ABS Global for donating several straws of various B. t. indicus breeds which were used to optimize in vitro production procedures as well as for identification of sub-species specific fixed polymorphisms. Finally, we also thank Dr. X. Cindy Tian from the University of Connecticut for her assistance in developing the SSCP assay.
Glossary
Abbreviations:
- BWS
Beckwith-Wiedemann syndrome
- LOS
large offspring syndrome
- ART
assisted reproductive technologies
- AI
artificial insemination
- PAGE
polyacrylamide gel electrophoresis
- PCR
Polymerase Chain Reaction
- SSCP
single strand conformation polymorphism
- SNP
single nucleotide polymorphism
- RFLP
restriction fragment length polymorphism
- B. t. taurus
Bos taurus taurus
- B. t. indicus
Bos taurus indicus
- COBRA
combined bisulfite restriction analysis
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24655
References
- 1.Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2010;18:8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:343–54. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- 3.Rump P, Zeegers MP, van Essen AJ. Tumor risk in Beckwith-Wiedemann syndrome: A review and meta-analysis. Am J Med Genet A. 2005;136:95–104. doi: 10.1002/ajmg.a.30729. [DOI] [PubMed] [Google Scholar]
- 4.Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–59. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 5.Schulz R, Woodfine K, Menheniott TR, Bourc?his D, Bestor T, Oakey RJ. WAMIDEX: a web atlas of murine genomic imprinting and differential expression. Epigenetics. 2008;3:89–96. doi: 10.4161/epi.3.2.5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–9. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Weksberg R, Shuman C, Smith AC. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2005;137C:12–23. doi: 10.1002/ajmg.c.30058. [DOI] [PubMed] [Google Scholar]
- 8.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–79. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Horike S, Mitsuya K, Meguro M, Kotobuki N, Kashiwagi A, Notsu T, et al. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum Mol Genet. 2000;9:2075–83. doi: 10.1093/hmg/9.14.2075. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32:426–31. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 12.Thakur N, Kanduri M, Holmgren C, Mukhopadhyay R, Kanduri C. Bidirectional silencing and DNA methylation-sensitive methylation-spreading properties of the Kcnq1 imprinting control region map to the same regions. J Biol Chem. 2003;278:9514–9. doi: 10.1074/jbc.M212203200. [DOI] [PubMed] [Google Scholar]
- 13.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–60. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, et al. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003;40:62–4. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halliday J, Oke K, Breheny S, Algar E, J Amor D. Beckwith-Wiedemann syndrome and IVF: a case-control study. Am J Hum Genet. 2004;75:526–8. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, et al. Assisted reproductive therapies and imprinting disorders--a preliminary British survey. Hum Reprod. 2006;21:1009–11. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- 17.Lim D, Bowdin SC, Tee L, Kirby GA, Blair E, Fryer A, et al. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod. 2009;24:741–7. doi: 10.1093/humrep/den406. [DOI] [PubMed] [Google Scholar]
- 18.Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–41. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 20.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–59. doi: 10.1016/0092-8674(91)90513-X. [DOI] [PubMed] [Google Scholar]
- 21.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 22.Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3:155–63. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- 23.Farin PW, Piedrahita JA, Farin CE. Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology. 2006;65:178–91. doi: 10.1016/j.theriogenology.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Miles JR, Farin CE, Rodriguez KF, Alexander JE, Farin PW. Angiogenesis and morphometry of bovine placentas in late gestation from embryos produced in vivo or in vitro. Biol Reprod. 2004;71:1919–26. doi: 10.1095/biolreprod.104.031427. [DOI] [PubMed] [Google Scholar]
- 25.Bertolini M, Anderson GB. The placenta as a contributor to production of large calves. Theriogenology. 2002;57:181–7. doi: 10.1016/S0093-691X(01)00665-3. [DOI] [PubMed] [Google Scholar]
- 26.Bertolini M, Mason JB, Beam SW, Carneiro GF, Sween ML, Kominek DJ, et al. Morphology and morphometry of in vivo- and in vitro-produced bovine concepti from early pregnancy to term and association with high birth weights. Theriogenology. 2002;58:973–94. doi: 10.1016/S0093-691X(02)00935-4. [DOI] [PubMed] [Google Scholar]
- 27.Hiendleder S, Mund C, Reichenbach HD, Wenigerkind H, Brem G, Zakhartchenko V, et al. Tissue-specific elevated genomic cytosine methylation levels are associated with an overgrowth phenotype of bovine fetuses derived by in vitro techniques. Biol Reprod. 2004;71:217–23. doi: 10.1095/biolreprod.103.026062. [DOI] [PubMed] [Google Scholar]
- 28.Hiendleder S, Wirtz M, Mund C, Klempt M, Reichenbach HD, Stojkovic M, et al. Tissue-specific effects of in vitro fertilization procedures on genomic cytosine methylation levels in overgrown and normal sized bovine fetuses. Biol Reprod. 2006;75:17–23. doi: 10.1095/biolreprod.105.043919. [DOI] [PubMed] [Google Scholar]
- 29.Hori N, Nagai M, Hirayama M, Hirai T, Matsuda K, Hayashi M, et al. Aberrant CpG methylation of the imprinting control region KvDMR1 detected in assisted reproductive technology-produced calves and pathogenesis of large offspring syndrome. Anim Reprod Sci. 2010;122:303–12. doi: 10.1016/j.anireprosci.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Couldrey C, Lee RS. DNA methylation patterns in tissues from mid-gestation bovine foetuses produced by somatic cell nuclear transfer show subtle abnormalities in nuclear reprogramming. BMC Dev Biol. 2010;10:27. doi: 10.1186/1471-213X-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robbins KM, Chen Z, Wells KD, Rivera RM. Expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 imprinting control regions is conserved between human and bovine. J Biomed Sci. 2012;19:95. doi: 10.1186/1423-0127-19-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breukelman SP, Reinders JM, Jonker FH, de Ruigh L, Kaal LM, van Wagtendonk-de Leeuw AM, et al. Fetometry and fetal heart rates between Day 35 and 108 in bovine pregnancies resulting from transfer of either MOET, IVP-co-culture or IVP-SOF embryos. Theriogenology. 2004;61:867–82. doi: 10.1016/j.theriogenology.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Bhogal B, Arnaudo A, Dymkowski A, Best A, Davis TL. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics. 2004;84:961–70. doi: 10.1016/j.ygeno.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 34.John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81:270–80. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Chung WY, Yuan L, Feng L, Hensle T, Tycko B. Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms? tumors. Hum Mol Genet. 1996;5:1101–8. doi: 10.1093/hmg/5.8.1101. [DOI] [PubMed] [Google Scholar]
- 36.Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A. 2006;103:6623–8. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo K, Silke J, Gramatikoff K, Schaffner W. The CpG-specific methylase SssI has topoisomerase activity in the presence of Mg2+ Nucleic Acids Res. 1994;22:5354–9. doi: 10.1093/nar/22.24.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiesa N, De Crescenzo A, Mishra K, Perone L, Carella M, Palumbo O, et al. The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet. 2012;21:10–25. doi: 10.1093/hmg/ddr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weksberg R, Shuman C, Caluseriu O, Smith AC, Fei YL, Nishikawa J, et al. Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum Mol Genet. 2002;11:1317–25. doi: 10.1093/hmg/11.11.1317. [DOI] [PubMed] [Google Scholar]
- 40.Hemberger M, Redies C, Krause R, Oswald J, Walter J, Fundele RH. H19 and Igf2 are expressed and differentially imprinted in neuroectoderm-derived cells in the mouse brain. Dev Genes Evol. 1998;208:393–402. doi: 10.1007/s004270050195. [DOI] [PubMed] [Google Scholar]
- 41.Curchoe C, Zhang S, Bin Y, Zhang X, Yang L, Feng D, et al. Promoter-specific expression of the imprinted IGF2 gene in cattle (Bos taurus) Biol Reprod. 2005;73:1275–81. doi: 10.1095/biolreprod.105.044727. [DOI] [PubMed] [Google Scholar]
- 42.Paulsen M, Davies KR, Bowden LM, Villar AJ, Franck O, Fuermann M, et al. Syntenic organization of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5. Hum Mol Genet. 1998;7:1149–59. doi: 10.1093/hmg/7.7.1149. [DOI] [PubMed] [Google Scholar]
- 43.Gould TD, Pfeifer K. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet. 1998;7:483–7. doi: 10.1093/hmg/7.3.483. [DOI] [PubMed] [Google Scholar]
- 44.Tunster SJ, Van de Pette M, John RM. Fetal overgrowth in the Cdkn1c mouse model of Beckwith-Wiedemann syndrome. Dis Model Mech. 2011;4:814–21. doi: 10.1242/dmm.007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caspary T, Cleary MA, Perlman EJ, Zhang P, Elledge SJ, Tilghman SM. Oppositely imprinted genes p57(Kip2) and igf2 interact in a mouse model for Beckwith-Wiedemann syndrome. Genes Dev. 1999;13:3115–24. doi: 10.1101/gad.13.23.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–9. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 47.Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–42. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun FL, Dean WL, Kelsey G, Allen ND, Reik W. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 1997;389:809–15. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 49.Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–4. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- 50.Blondin P, Farin PW, Crosier AE, Alexander JE, Farin CE. In vitro production of embryos alters levels of insulin-like growth factor-II messenger ribonucleic acid in bovine fetuses 63 days after transfer. Biol Reprod. 2000;62:384–9. doi: 10.1095/biolreprod62.2.384. [DOI] [PubMed] [Google Scholar]
- 51.Kuentz P, Bailly A, Faure AC, Blagosklonov O, Amiot C, Bresson JL, et al. Child with Beckwith-Wiedemann syndrome born after assisted reproductive techniques to an human immunodeficiency virus serodiscordant couple. Fertil Steril. 2011;96:e35–8. doi: 10.1016/j.fertnstert.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 52.Rossignol S, Steunou V, Chalas C, Kerjean A, Rigolet M, Viegas-Pequignot E, et al. The epigenetic imprinting defect of patients with Beckwith-Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet. 2006;43:902–7. doi: 10.1136/jmg.2006.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bliek J, Verde G, Callaway J, Maas SM, De Crescenzo A, Sparago A, et al. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2009;17:611–9. doi: 10.1038/ejhg.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O?rourke PK, Entwistle KW, Arman C, Esdale CR, Burns BM. Fetal development and gestational changes in Bos taurus and Bos indicus genotypes in the tropics. Theriogenology. 1991;36:839–53. doi: 10.1016/0093-691X(91)90350-M. [DOI] [PubMed] [Google Scholar]
- 55.Rivera RM, Kelley KL, Erdos GW, Hansen PJ. Alterations in ultrastructural morphology of two-cell bovine embryos produced in vitro and in vivo following a physiologically relevant heat shock. Biol Reprod. 2003;69:2068–77. doi: 10.1095/biolreprod.103.020347. [DOI] [PubMed] [Google Scholar]
- 56.Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17:1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.