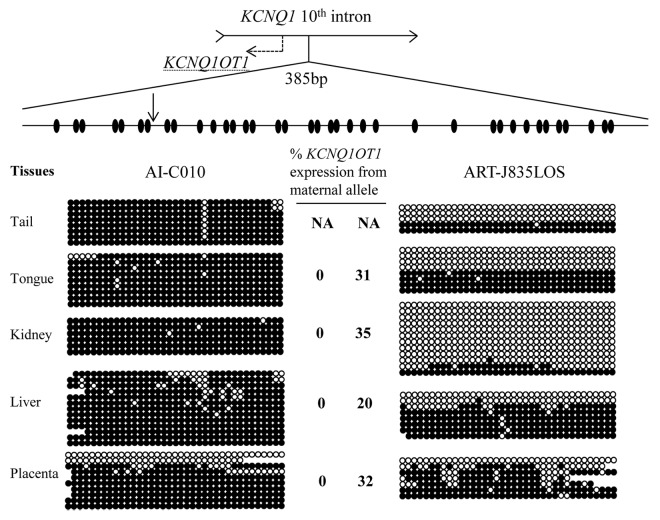
Figure 5. Loss of methylation of KvDMR1 on the maternal allele is associated with biallelic expression of KCNQ1OT1 in LOS fetuses. DNA was treated with sodium bisulfite prior to PCR, and PCR product was cloned before sequencing. Sequencing data was used to determine the DNA methylation status at the KvDMR1. Shown on top is a depiction of the 10th intron of the maternally-expressed gene KCNQ1 and its direction of transcription is shown with an arrow. The region harbors the promoter of the antisense long ncRNA KCNQ1OT1 (shown as dashed arrow), which is also an imprinting control region known as KvDMR1. A 385 bp region of the KvDMR1 was used to determine the DNA methylation status of 37 CpG sites (ovals). A SNP (vertical arrow) between B. t. indicus and B. t. taurus was used to determine the parental origin of the alleles and only maternal alleles are shown here. Five tissues from two fetuses are shown. Filled and open circles represent methylated and unmethylated CpG dinucleotides, respectively. Missing circles are sequencing data of low quality. Each line denotes an individual DNA strand. The level of maternal KCNQ1OT1 expression is shown in the center and next to the strands. Tail tissues were collected for the purpose of DNA analysis, precluding its use for gene expression determinations. NA, not available.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.