Abstract
In plants, silencing is usually accompanied by DNA methylation and heterochromatic histone marks. We studied these epigenetic modifications in different epialleles of 35S promoter (P35S)-driven tobacco transgenes. In locus 1, the T-DNA was organized as an inverted repeat, and the residing neomycin phosphotransferase II reporter gene (P35S-nptII) was silenced at the posttranscriptional (PTGS) level. Transcriptionally silenced (TGS) epialleles were generated by trans-acting RNA signals in hybrids or in a callus culture. PTGS to TGS conversion in callus culture was accompanied by loss of the euchromatic H3K4me3 mark in the transcribed region of locus 1, but this change was not transmitted to the regenerated plants from these calli. In contrast, cytosine methylation that spread from the transcribed region into the promoter was maintained in regenerants. Also, the TGS epialleles generated by trans-acting siRNAs did not change their active histone modifications. Thus, both TGS and PTGS epialleles exhibit euchromatic (H3K4me3 and H3K9ac) histone modifications despite heavy DNA methylation in the promoter and transcribed region, respectively. However, in the TGS locus (271), abundant heterochromatic H3K9me2 marks and DNA methylation were present on P35S. Heterochromatic histone modifications are not automatically installed on transcriptionally silenced loci in tobacco, suggesting that repressive histone marks and cytosine methylation may be uncoupled. However, transient loss of euchromatic modifications may guide de novo DNA methylation leading to formation of stable repressed epialleles with recovered eukaryotic marks. Compilation of available data on epigenetic modification of inactivated P35S in different systems is provided.
Keywords: transgene silencing, histone modification, DNA methylation, tobacco, dedifferentiation, callus
Introduction
Both transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS) are associated with DNA methylation, often induced by small interfering RNAs (siRNAs) in a process called RNA-directed DNA methylation (RdDM). RdDM coincides with either TGS, where promoter methylation inactivates transcription,1 or PTGS, which merely affects transcript stability and/or the translation rate and where cytosines of the transcribed region are methylated.2,3 Despite these differences, both TGS and PTGS are mechanistically related and likely evolved from a common ancestral system.4
Histone amino acid modifications represent a layer of epigenetic information that is widespread in eukaryotes, including plants.5 Repressive histone marks that are closely linked with transcriptional silencing include hypomethylation of H3K4 and dimethylation of H3K9.6 By contrast, H3 and H4 acetylation usually mark active promoter states. At the cytogenetic level, H3K4 methylation marks actively transcribing regions (euchromatin), whereas H3K9 dimethylation associate with repressed genomic territories collectively called heterochromatin.7 Genetic screens in Arabidopsis revealed several chromatin factors involved in RdDM, among which DNA methyltransferases and histone deacetylases were the most prominent.8 Cooperation of DNA methylation, H3K9 dimethylation and several other factors were shown to be needed for stable robust TGS of transgenes9 and endogenes10 in Arabidopsis. DNA methylation and histone H3K9 dimethylation are also closely interconnected at the whole genome level.11
Although tobacco has long been a classical object of genetic studies, its histone modification patterns have been rarely studied. The tobacco SET (Su(var), E(z) and Trithorax conserved) domain of protein NtSET1 binds chromatin and methylates H3K9.12 Overexpression of NtSET1 causes defects in chromosome condensation/segregation in transgenic tobacco.13 Transgenic tobacco plants expressing the Drosophila Polycomb (Pc) chromodomain show developmental abnormalities in leaves and flowers.14 Recently, additional epigenetic genes have been cloned from Nicotiana tabacum including homolog of dicer DCL2 involved in small RNA biogenesis15 and a family of DNA methytransferases.16 These studies thus suggest that tobacco may use a similar epigenetic mechanism as the model organisms to control its developmental programs although there might be differences in the setting and function of individual epigenetic tools.
The cauliflower mosaic virus 35S promoter (P35S) is the most widely used promoter for driving plant transgenes in both basic research and biotechnologies.17 Despite numerous studies showing epigenetic silencing of linked genes either at the transcriptional or posttranscriptional levels, the mechanisms of P35S inactivation are not fully understood. The epigenetic inactivation of P35S has been correlated with its increased DNA methylation, repressive histone marks and production of siRNAs (Table 1). All three characteristics seem to contribute to silencing, although each can operate in different phases, at various magnitudes and in diverse silencing systems. Posttranscriptional silencing of P35S-linked genes was associated with increased DNA methylation of transcribed regions,18-20 whereas deposition of heterochromatic histone marks was not reported.20,21 Conversely, TGS is accompanied by DNA hypermethylation, H3K9 dimethylation and overall histone deacetylation of the promoter region.22-26 Application of epigenetic inhibitors resulted in increased expression of silenced loci in most,9,27,28 but not all, cases of silenced loci.23 Generally, TGS seems to be more sensitive to chromatin factor deficiencies than PTGS, although recent reports have suggested that certain histone modifications may function in PTGS as well.29 Despite numerous transgenic lines are available, histone modifications on epigenetically inactivated 35S promoters have not been studied yet in tobacco or related Petunia species (both Solanaceae).
Table 1. Summary of the P35S epilallelic variants reported in different systems and their molecular characteristics.
| Species | Locus | Epiallele | T-DNA copy | Epilalele generation | 1Meiotic revers. | Type of silenc. |
2P35S siRNAs |
3DNA methyl. | Histone marks | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
|
Nicotiana tabacum |
435S:nptII (Locus 1) |
|
2 |
|
|
PTGS |
- |
Negligible (b.s.) |
H3K9Ac, H3K4me3 |
44
|
| |
|
Locus 1E |
2 |
Cell culture |
No |
TGS |
- |
Strong (b.s.) |
H3K9ac, H3K4me |
43
,
48
|
| |
|
Locus 1* |
2 |
siRNA signals |
No |
TGS |
- |
Strong (b.s.) |
H3K9Ac, H3K4me |
46
|
| |
35S:RIN (Locus 271) |
|
> 5 |
|
|
TGS |
+ |
Strong (s.b.) |
H3K9me2 |
26
|
| |
|
Locus 271d |
|
Rearrangement, small deletion |
No |
active |
- |
Weak (s.b.) |
n.d. |
45
|
| |
35S:hpt Locus H |
|
> 5 |
|
|
|
- |
Weak (b.s.) |
n.d. |
76
|
| |
|
Locus H* |
|
siRNA signals |
Partial |
TGS |
- |
Strong (b.s.) |
n.d. |
24
|
| |
35S:nptII (Locus 2) |
|
1 |
|
|
active |
- |
Negligible (b.s.) |
H3K9ac, H3K4me |
44
|
| |
|
Locus 2* |
1 |
siRNA signals |
Yes |
TGS |
- |
Strong (b.s.) |
H3K9ac, H3K4me |
46
|
|
Nicotiana benthamiana |
35S-GFP |
|
1 |
|
|
active |
- |
Negligible (s.b.) |
n.d. |
77
|
| |
|
35S-GFP* |
1 |
siRNA signals (VIGS) |
Partial |
TGS |
- |
Strong (s.b.) |
n.d. |
77
|
|
Petunia hybrida |
35S-CHS-A |
|
2 |
|
|
PTGS |
|
Negligible (s.b.) |
n.d. |
78
|
| |
|
35S-CHS-A* |
2 |
siRNA signals (Inverted repeat) |
n.d. |
TGS |
n.d. |
Strong (s.b.) |
n.d. |
78
|
|
Petunia hybrida |
435S-CHS-A (C001) |
|
2 |
|
|
PTGS |
- |
Weak (b.s.) |
n.d. |
28
|
| |
|
C002 |
2 |
Spontaneous conversion |
No |
TGS |
- |
Strong (b.s.) |
n.d. |
28
|
|
Gentian triflora x G. scabra |
PS |
|
1 |
|
n.d. |
active |
n.d. |
Negligible (b.s.) |
H3Kac, H3K4me3 |
23
|
| |
|
RS |
1 |
Cell culture |
n.d. |
TGS |
n.d. |
Strong (b.s.) |
H3K4me2, H3K9me2 |
23
|
|
Oryza stativa |
35S:GFP (M65) |
|
1 |
|
|
active |
- |
Negligible (b.s.) |
H3ac, H4ac |
22
|
| M65-Pi | 1 | siRNA signals | No | TGS | - | Strong (b.s.) |
H3K9me2,H3K9me2 | 22 |
1 Meiotic stability of epiallele over generations following removal of a trigger factor (callus culture, siRNA).2siRNAs produced by a transgene locus and not by a hairpin trigger.3b.s., DNA methylation determined along the ~300 bp of P35S by bisulfite sequencing, levels: < 10%, negligible, 10?20%, weak, > 20%, strong; s.b., methylation determined by methylation-sensitive restriction enzymes in combination with Southern blot hybridization. 4PTGS to TGS conversion occurred in two systems in which T-DNA was organized as an inverted repeat
Phenotypic variation known to occur in callus culture and regenerated plants (termed somaclonal variation) is likely to have a molecular background and involves an epigenetic modifications of chromatin.30 Aberrant promoter hypermethylation seems to be a ubiquitous feature of both animal and plant31 cell cultures. By contrast, some repeated sequences within the heterochromatin tend to lose heterochromatic marks in cell cultures.32 Alteration of spatial organization of chromosome territories has been noted in cytogenetic studies.33,34 Additionally, the silencing potential of hairpin constructs seems to be less efficient in calli than in the differentiated leaf.35 Although cell culture-induced epialleles do not necessarily persist in regenerated plants,33,36 there are several examples of their transmission to regenerated plants and even transgeneration inheritance.30,37-40 Alterations of DNA methylation patterns seem to be the most stable modification, probably due to the inheritance of symmetrical CG motifs.41,42 In previous reports, we characterized epiallelic variants of tobacco PTGS transgenic locus 1 that arose at high frequency among cell culture regenerants.43 The meiotically stable TGS variant (locus 1E) maintained inactive hypermethylated P35S over generations without detectable siRNA signals.
Epialleles represent an excellent system to study the correlation of chromatin modification with the expression state and inheritance of the silencing. Here, we studied chromatin histone marks imposed on tobacco transgene loci during the PTGS to TGS conversion induced by RNA signals or arising spontaneously during dedifferentiation of cells. Using chromatin immunoprecipitation (ChIP), we analyzed the distribution of histone marks along different regions of transgenes, addressing the relationships between expression activity, DNA methylation and histone modification.
Results
Organization of transgenic loci and experimental set up
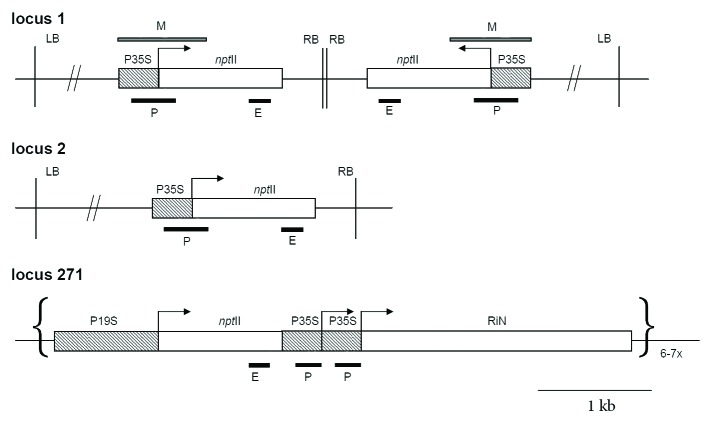
Locus 1 (Lo1; Fig. 1) and locus 2 (Lo2) were described in detail previously.19 T-DNA contain the neomycin phosphotransferase II reporter transgene driven by the 35S promoter (P35S:nptII) together with a non-silenced hygromycine resistence gene (hpt) close to the left border under the control of nopaline synthase promoter (Pnos) lying about 1 kb upstream of the P35S.18 Expression of the nptII gene in Lo1 is silenced at the posttranscriptional level, DNA methylation occurs primarily at the 3? end region and the nptII-specific 21-to 25-nt siRNAs are able to induce silencing and methylation of unlinked homologous loci. The nptII gene is actively expressed and non-methylated in Lo2.44 The homologous transgenic locus 271 consists of the complex insertion of six to seven copies of the tobacco nitrite reductase (NiR) sequence in an antisense orientation (RiN) driven by two 35S promoters and the nptII transgene driven by a CaMV 19S promoter (Fig. 1). This 271 locus was shown to effectively silence in trans all transgenes driven by 19S and 35S promoters at the transcriptional level and silence the endogenous nitrite reductase gene at the posttranscriptional level.24,26 These homologous interactions were mediated by small RNA molecules.45
Figure 1. Schematic outline of T-DNA insertions in locus 1, locus 2 and locus 271. Locus 1 contains two copies of T-DNA that are arranged as an IR about the right border. Promoters are in hatched squares; P35S, CaMV 35S promoter; P19S, CAMV 19S promoter; nptII, neomycin phosphotransferase II gene; RIN, nitrate reductase gene in an antisense orientation. M bar indicates region analyzed by bisulfite genomic sequencing. P (promoter) and E (3?end) bars correspond to PCR fragments amplified after ChIP. RB, LB right and left border, respectively.
The outline of epiallele generation is schematically depicted in Figure 2 and involves (i) callus culture of PTGS-Lo1 line establishment, callus cultivation for 1 y and plant regeneration; and (ii) two generations of hybrids (F1, F2) carrying the silencer locus 271 together with locus 1 were crossed with non-transgenic tobacco plants to obtain segregants (Lo1*S1 F2, Lo1*S1 F3), containing only locus 1. Moreover, the first generation of segregants (Lo1*S1 F2) was selfed to obtain an additional generation without the silencer locus 271 (Lo1*S2 F3).46
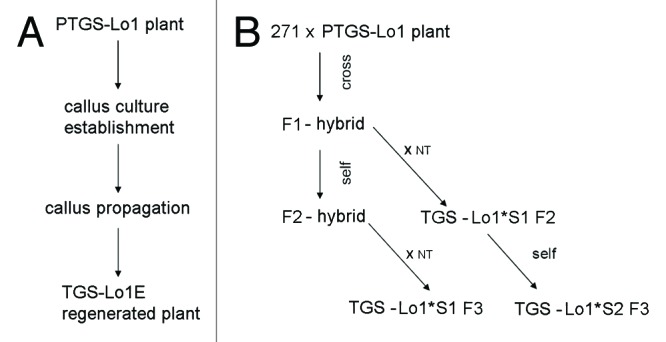
Figure 2. Schematic outline of P35S epialleles generation. (A) Epialleles generated spontaneously in callus culture. (B) Epialleles induced by siRNAs in 271 Locus 1 hybrids. S1, S2 represent plants of first or second generation after silencer locus segregation respectively. ?x NT,? cross to a non-transgenic tobacco.
Expression patterns of the nptII genes in epiallelic variants
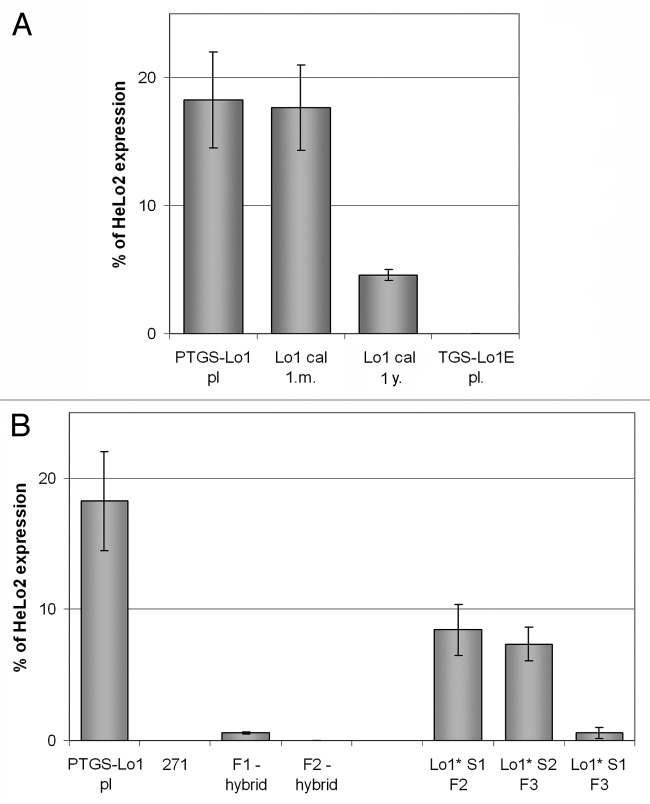
We previously reported partial loss of in trans PTGS silencing by locus 1 on locus 2 target in hybrid line exposed to callus conditions.47 To determine whether this is due to epigenetic changes at the silencer leading to TGS and the loss of the capacity to produce silencing RNA molecules, we now analyzed the nptII transgene expression level of locus 1 in a hemizygous Lo1 line, upon callus culture and upon regeneration. The parental leaf of PTGS-Lo1 plant and early callus had almost similar mRNA levels, corresponding to approximately 17% of the level of a non-silenced Lo2 plant line (Fig. 3A). By contrast, the 1-y old callus Lo1 had 4-fold lower nptII levels than the parental leaf, and virtually no transcription was found in a regenerated locus 1E plant. In conclusion, the epigenetic change imposed during callus culture lead to formation of epialleles with decreased expression of nptII genes.
Figure 3. Expression analysis of the nptII reporter genes in parental plants and derived epiallelic variants. The nptII transcripts were analyzed by quantitative RT-PCR. (A) epialleles generated by callusogenesis. Results include data from two independent experiments (3 technical replicates); (B) transcription level of epialleles generated by siRNA signals. Values are expressed as percentages of the nptII levels in a non-silenced HeLo2 line.
The TGS epialleles of locus 1 were also generated by siRNA signals in 271 Lo1 hybrids. Expression patterns of two generations of hybrids (Fig. 3B, left panel) and segregating progenies (Fig. 3B, right panel) were shown in our prior results.46 Briefly, the paramutated progeny segregating from the hybrids demonstrated reduced (Lo1*S1 F2, Lo1*S2 F3) or negligible (Lo1*S1 F3) transcription.
The silencer locus 271 failed to show substantial nptII expression, indicating TGS silencing of a P19S-driven nptII gene consistent with previous results.26
DNA methylation analysis
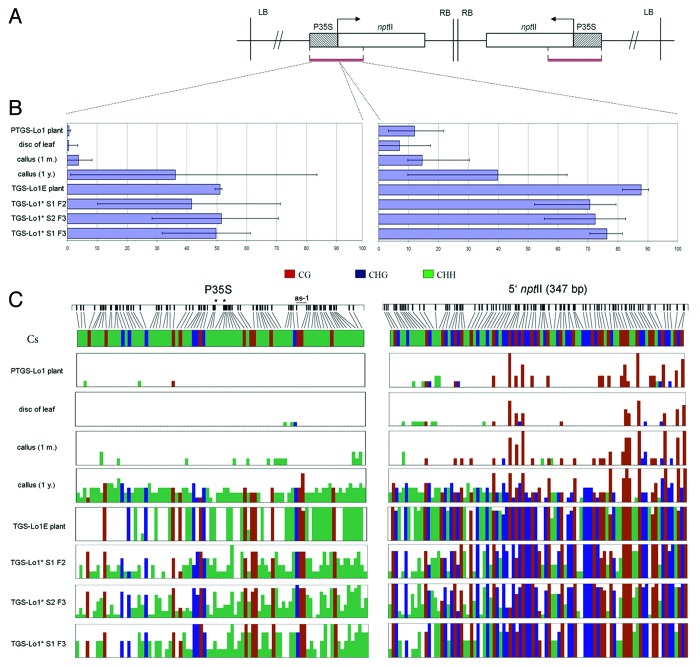
To study methylation changes accompanying formation of TGS epialleles, we performed bisulfite sequencing of locus 1. The sequenced region comprised the entire 35S promoter (344-bp) and the 347-bp subregion of the nptII 5? end (Fig. 4).
Figure 4. Detailed bisulfite methylation analysis of epiallelic variants. (A) Transgene subregions subjected to bisulfite sequencing. (B) Column graphs showing average methylation levels from 5?10 clones. Error bars represent clones with maximum and minimum level of DNA methylation. (C) Distribution of mC along the sequenced fragment. Individual vertical columns represent the average methylation at particular position. Positions of the as-1 regulatory element and Sau96I restriction sites (asterisks) are indicated.
The promoter region of PTGS-Lo1 in leaf DNA was non-methylated whereas low level of methylation occurring at the nptII 5? region was limited to the CG context cytosines (Fig. 4C). Methylation of promoter sequences slightly increased in 1-mo-old microcalli, where in several individual clones a few non-symmetrical cytosines became methylated. DNA methylation gradually increased during callus propagation and in 1-y-old calli, approximately 30?40% of cytosines in P35S and approximately 40% of cytosines at the nptII 5? end were methylated (Fig. 4B). The de novo methylation in calli was accompanied by an increase of clone to clone variability in accordance with previous results.47 Compared with the callus, a regenerated TGS-Lo1E plant showed increased methylation of the promoter (~50%) and the nptII 5? end (80?90%), whereas the clone-to-clone variability decreased compared with the callus.
The in trans silencing activity of the silencing locus 271 is connected with methylation of target promoters.24 As shown previously, promoters of PTGS-Lo1 were non-methylated (Fig. 4). Bisulfite sequencing of locus 1* revealed the inheritance of cytosine methylation in CG and non-CG motifs in both generations of segregants from Lo1 271 hybrids (Lo1*S1 F2 and Lo1*S1 F3) and their progenies (Lo1*S2 F3). Interestingly, significant de novo methylation also appeared in the 5? nptII region that was not targeted by silencing siRNAs. Contrast to locus 1* segregants, locus 2* segregants from Lo2 271 hybrids have completely lost methylation and regained expression (Fig. S1).
To validate bisulfite results we inspected methylation of restriction sites using Southern blot hybridization. The methylation status of the P35S promoter was analyzed by Sau96I enzyme, which is sensitive to the cytosine methylation in nonsymmetrical CHH context. Two recognition Sau96I sites (GGNCC) are found within the P35 promoter (Fig. S2B). The PTGS plants showed a 0.91 kb Sau96I band corresponding to a non-methylated variant while in TGS variants and callus there was an additional 1.14 kb band representing methylated molecules. Methylation of nonsymmetrical sites in the 3?region was analyzed using BamHI (GGATCC) and NcoI (CCATGG) restriction enzymes (Figs. S2C and D). While the PTGS locus 1 and TGS-Lo1* segregants displayed high level of CHH methylation, there was slight reduction of methylation in some of its epilallels which may be explained by the absence of methylation-inducing siRNAs in TGS lines.48 Nevertheless, considerable methylation of BamHI and NcoI sites was present in all locus 1 samples. In contrast, the sites in non-silenced locus 2 were unmethylated. Together the results of Southern blot and bisulfite analysis were in a good agreement.
Histone modifications in tobacco P35S-driven transgenes
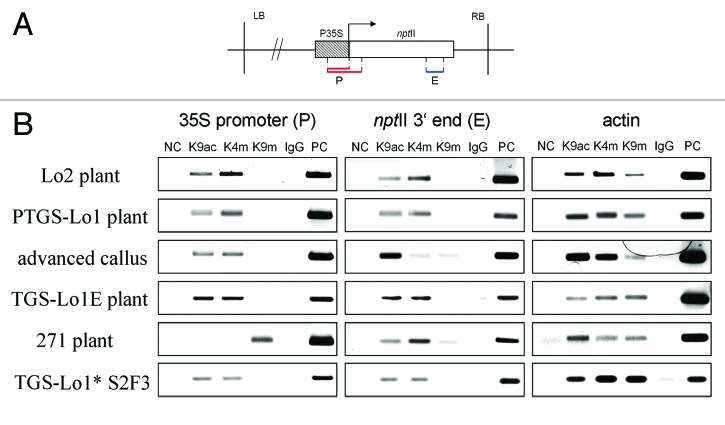
We used ChIP analysis to monitor changes in histone modification during epiallelic conversion. Chromatin was immunoprecipitated with antibodies against euchromatic and heterochromatic histone marks. Two independently established calli were analyzed as biological replicates at the promoter (~349 bp, ~212 bp in locus 271) and at the 3? end of the nptII transcribed region (~174 bp) that appear to be critical for epiallele formation49 (Fig. 1, 5A). Both regions are separated by sequence length (~700 bp), allowing sufficient resolution of ChIP. Each immunoprecipitated DNA sample was amplified with P35S-, nptII- and actin-specific primers using conventional polymerase chain reaction (PCR; Fig. 5).
Figure 5. Histone modification patterns along the transgenes. (A) Schematic outline of subregions analyzed by ChIP: P (promoter) and E (3? end of nptII). The P subregion in locus 271 was shorter (upper line) than in loci 1 and 2 (bottom line). (B) Electrophoretic profiles of PCR products obtained by amplification of immunoprecipitated DNA. Results of a repeated experiment performed on an independent callus culture are shown in Figure S3.
Histone modification changes during dedifferentiation: In a parental PTGS variant, only H3K9ac and H3K4me3 signals were visible in both the promoter and transcribed region (Fig. 5B). Similarly, both H3K9ac and H3K4me3 signals were visualized in a sample of TGS-Lo1 epialleles. The histone profiles in callus samples differed from those of parental and regenerated plants. The H3K4me3 was nearly lost from the nptII coding region, whereas faint H3K9me2 signals appeared (Fig. 5B). Semiquantitative evaluation of band intensity is shown in Figure 6. It is evident that H3K4me3 levels decreased in both analyzed calli compared with the parental leaf. Additionally, mostly repressive histone H3K27m1/K27m3 and H4K20m1 together with positive-acting H3K36m3 modifications were analyzed in parental plant and derived calli (Fig. S3A). No significant chromatin enrichment was obtained using these antibodies. The immunoprecipitation profiles of the leaf and callus were similar to results from an independently established callus (Fig. 5 and Fig. S3A).
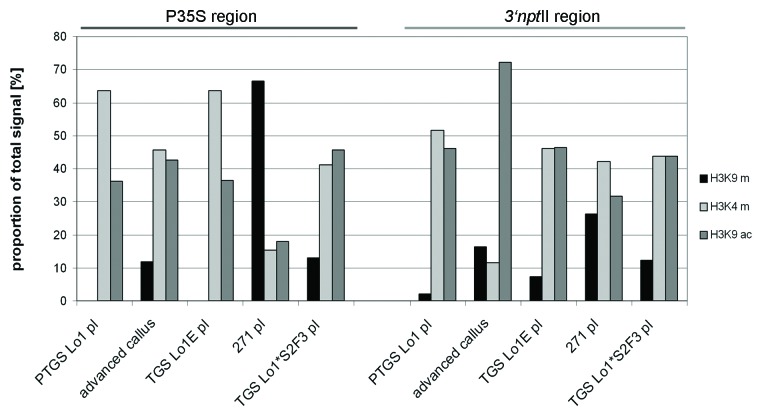
Figure 6. Semiquantitative evaluation of histone modification patterns along the transgenes. Fluorescent signals in gels (Fig. 5) were counted and the levels were expressed as percentages of total signal (a sum of H3K9me2, H3K4me3 and H3K9ac).
Paramutated locus 1* epialleles: The Lo1*S2 F3 line showed enrichment after chromatin precipitation with euchromatic H3K9ac and H3K4me3 antibodies in both subregions of the transgene (Figs. 5 and 6).
Locus 271: The anti-H3K9me2 immunoprecipitated to P35S sequences of locus 271 produced a positive signal. Conversely, no immunoprecipitation signal was obtained from the anti-H3K9ac and anti-H3K4me3 samples. The nptII coding region showed intermediate H3K9ac, H3K4me3 and H3K9me2 marks.
Locus 2: The non-silenced locus 2 carrying a similar T-DNA insertion as the silenced locus 1 showed stable euchromatic marks in both parental plant and derived callus (Fig. S3B).
Controls: No amplified product was obtained after immunoprecipitation with normal mouse IgG. The endogenous family of 5S rRNA genes was immunoprecipited with anti H3K9me2 but not H3K4me3 (Fig. S3C) consistent with their heterochromatic nature50 and heavy DNA methylation.51 Actin genes showed intermediate signals with both euchromatic and heterochromatic signals, whereas in Arabidopsis, actin genes were strictly euchromatic.52 The intermediate marks could be explained by the presence of pseudogenes and/or inactivated gene copies in allotetraploid tobacco nuclei.53 Homologous alleles inherited from both parents may be differentially imprinted in tobacco.
Discussion
Strict positive-acting histone marks at a PTGS locus despite coding region methylation
During PTGS of the nptII gene in locus 1, dense DNA methylation appeared to be restricted mostly at the 3? transcribed region.18,44,54 Consistent with this assumption, the 3? transcribed region was extensively methylated at both CG and non-CG motifs. By contrast, the 5? end of nptII was not markedly methylated at non-CG motifs, whereas some CG sites were abundantly methylated (Fig. 4C). This pattern resembles typical gene body methylation observed in endogenous genes that is only exceptionally correlated with silencing.55 Here we show that in the nptII transcribed region, only positively acting histone marks have been found, suggesting that the nptII gene-specific siRNAs cannot alter the euchromatic status. Similarly, RdDM induced during a viroid RNA infection did not increase dimethylation of lysine H3K9 or decrease acetylation of H3 in tobacco.20 It is likely that siRNAs produced during the PTGS process methylate DNA but do not change the euchromatic histone environment.
Although methylation of non-CG motifs is only rarely found in coding regions,56 a distinct class of endogenous genes seems to be regulated by this type of methylation.57 In PTGS locus 1, we hypothesize that this type of methylation may have no or only a marginal effect on transcription at least under normal physiological conditions. First, nuclear run-on patterns of a non-silenced locus 2 and PTGS locus 1 were indistinguishable.43 Second, treatment of cells with hypomethylation drugs showed only weak release of silencing.58 Similarly, the inverted repeat of the PAI genes is transcribed despite heavy cytosine and H3K9 methylation.52 The reason for why the 5? end is methylated primarily at CG, whereas the 3? end is methylated at both CG and non-CG, is not fully understood. The absence of siRNAs cannot account for these differences because experiments with transitive silencing showed that siRNAs are formed from both the central and 3? regions of the transgene.59,60 In Arabidopsis, the IBM1 (increase in BONSAI methylation 1) factor was reported to actively remove non-CG methylation from coding regions.61 One possibility is that the non-CG methylation is more actively removed from the 5? ends than from the 3? ends of the genes.
Repressive histone marks occur in some but not all TGS epialleles
TGS is usually accompanied by heterochromatic histone marks and cytosine methylation (reviewed in refs.6,62,63). However, our findings indicate that epigenetic inactivation of the 35S promoter was not associated with repressive H3K9me2 histone marks in locus 1 epialleles. Instead, euchromatic H3K9 acetylation and H3K4 trimethylation were typically present on both active and inactive 35S promoters. However, abundant heterochromatic H3K9me2 marks were found in locus 271, arguing that P35S is not refractory to heterochromatic histone modifications in tobacco. There may be several explanations. Locus 271 has a complex organization composed of several complete and incomplete T-DNA copies,26 whereas locus 1 is composed of two inverted complete T-DNA copies, each 5 kb in length.44 Perhaps heterochromatic histone modifications are more often connected with a multicopy character of the sequences rather than with the expression status (silencing). Another distinction between locus 1 and locus 271 is their capacity to produce siRNAs: locus 271 produces high levels of P35S-specific 24 nt siRNAs,45,46 while these are not detectable in TGS locus 1 epialleles.46 Although RNA signals seem to rarely induce H3K9me2 in plants,8 we cannot exclude the possibility that the deposition of H3K9me2 could be stimulated by siRNA signals in locus 271. In Arabidopsis, H3K9me2 and CHG DNA methylation are tightly interconnected, creating a self-reinforcing loop.11 It was surprising that the inactive P35S completely lacked dimethylated H3K9 despite methylation of the CHG motifs. It seems that the interplay between H3K9me2 and non-CG methylation may work at the genome-wide level, although there may be significant exceptions at the local level. In support, developmentally regulated telomerase genes were shown to possess active H3K4me3 histone mark in the presence of repressive H3K27me3 mark64 suggesting complex chromatin variants in the genomes.
Consistent DNA methylation but variable siRNA and histone marks at epigenetically inactivated 35S promoters
Because many factors including T-DNA copy number,65 insertion site,66 local features,67 and differential settings of epigenetic modifiers68 may influence transgene activity, comparisons of chromatin patterns between different loci may be difficult. Epiallelic variants of transgene loci represent an excellent system to study the relationship between gene silencing and individual epigenetic marks. Table 1 shows P35S epigenetic variants of reported in different systems. Although dense CG and non-CG methylation always accompany P35S inactivation, other repressive marks, such as dimethylation of H3K9, demethylation of H3K4, H3K9 deacetylation and silencing siRNAs, are more variable in attributes and are even dispensable for TGS, at least in the maintenance phase. In many cases including TGS epialleles of locus 1, non-symmetrical methylation was not accompanied with detectable amounts of siRNAs. The question arises regarding how the inheritance of non-CG methylation can be explained if neither heterochromatic histones nor RNA signals are present. One possibility is that specific protein complexes binding to the enhancer recruit DNA methyltransferases to the target.69
Transient histone modification changes induced by cell culture may trigger the epigenetic switch from PTGS to TGS
Previously, we showed that locus 1 bearing inverted repeated nptII transgenes reproducibly undergoes epigenetic switches from PTGS to TGS in callus culture regenerants.43,47 Here, we studied this process in detail by analyzing histone and DNA modifications in the parental plant, callus culture and regenerated plants hereof. In the dedifferentiated callus, loss of H3K4 trimethylation was accompanied by partial gain of H3K9me2. The changes were highly localized to the 3? transcribed region, whereas histone modification patterns at the promoter remained unaffected. The developmentally regulated H3K27 methylation70 was not detectable in locus 1 epialleles, confirming its mostly euchromatic nature and indicating that polycomb repressive complexes are not involved in epigenetic switches of the transgene. Significantly, a non- silenced and non-methylated locus 2 did not undergo chromatin changes in callus suggesting that chromatin alterations were associated with PTGS state and/or inverted repeat character of locus 1. As both H3K4me3 and H4K9me2 are opposing chromatin marks labeling euchromatin and heterochromatin, respectively, are present, it suggests that contradictory (active and inactive) epigenetic marks are imposed on a transgene during cell culture cultivation. The intermediate histone marks in callus cells can be explained in two ways. On the one hand, a mosaic of cells carrying the transgene with either heterochromatic or euchromatic marks may exist, reflecting known epigenetic variability in the callus. In this context, we reported cell to cell DNA methylation heterogeneity of locus 1.47 On the other hand, there may be variability at the single-nucleosome level. Under this scenario, a given histone octamer may harbor opposing modifying marks for asymmetrical distribution. Indeed, callus transgene chromatin completely lost H3K4me3 and retained high levels of H3K9ac (supporting the existence of nucleosomes with ambivalent marks).
One of the most prominent observations was a correlation between H3K4me3 demethylation at the 3? end and spreading of DNA methylation from this region into the promoter. It would be intriguing to determine whether these distally related changes are interrelated. Histone H3K4 methylation is regulated by a group of enzymes from the JmJ (Jumanji) family known to be involved in RNA-directed DNA methylation.71,72 Significantly, some JmJ14 mutants released the transgene PTGS that was correlated with an increase in promoter methylation and retardation of transcription.29 It is tempting to speculate that the activity of some JmJ proteins and perhaps other epigenetic factors, are modified in the callus, leading to a polarized (3??5?) methylation spreading over a distance that may be as long as 1 kb. Spreading of DNA methylation could be related to weakening of transcription initiation from P35S and reduced polymerase occupancy of the coding region. In support, epialleles generated by the targeting of P35S by RdDM showed similar de novo methylation of the 5? coding region to the callus-induced epialleles.
Materials and methods
Plant material, hybridization and callus culture conditions
All transgenic tobacco (Nicotiana tabacum) SR1 plants were generated by Agrobacterium-mediated transformation.18 The plants hemizygous for the PTGS locus 1 (HeLo1; Fig. 1) were obtained by crossing a plant homozygous for locus 1 with an untransformed SR1 tobacco.73 The line hemizygous for the TGS locus 1E was obtained by plant regeneration from long-term HeLo1 callus cultures.43 Seeds of the tobacco transgenic plants homozygous for the transgenic locus 271 (Fig. 1) were obtained from INRA Versailles (a gift of Dr Hervé Vaucheret). All crosses were performed by emasculating flowers manually before they opened and applying pollen to the stamen. 271 Lo1 F1 hybrids were obtained by crosses of HoLo1 to 271; the experimental strategy to obtain hybrids and relevant segregants is depicted in Figure 2. Hybrid plants and segregants were genotyped by DNA gel blot hybridization.46
Calli were established from leaf explants by hormonal treatment and grown in 0.7% agar containing B5 salts supplemented with sucrose (30 g l?1), ?-naphthaleneacetic acid (2.0 mg l?1) and 6-benzylaminopurine (0.2 mg l?1). Calli were transferred onto fresh agar medium every 30 d. To obtain regenerated plants, calli were transferred onto shoot-inducing medium (for 1 mo) containing ?-naphthaleneacetic acid (0.2 mg l?1) and 6-benzylaminopurine (2.0 mg l?1). After the rooting phase on growth medium without hormones, the plantlets were transferred into greenhouse conditions.
DNA isolation and bisulfite genomic sequencing
Total genomic DNA was isolated from lyophilized leaves or calli using the cetyltrimethylammonium bromide method as described previously.74
Bisulfite treatments were performed on purified genomic DNA using the EpiTect bisulfite kit (QIAGEN). Primers for amplification of the 35S promoter and 5? nptII region are as follows: forward primer: 5?-CATTACATCACCCATAATAAATACTTTCTC-3?; the first reverse primer: 5?-GAATAGAGAGAAAGATATATTTTTTAAGAT-3?; and the second reverse primer: 5?-GTAATAGAGATTGGAGTTTTTAAGAAAGTAG-3?. The forward primer matched the nptII coding sequence at about +410 (with respect to transcription start site). The reverse primers were located in a vector sequence at about -660 and -570, respectively. The PCR program consisted of 2 min of initial denaturation at 94°C, followed by 35 cycles of 0.5 min at 94°C, 1.5 min at 45°C and 1 min at 72°C. The program was ended with an extension step for 10 min at 72°C. The PCR products were cloned into a TA vector (pDrive, QIAGEN) and between 5 and 10 clones from each sample were sequenced (Eurofins MWG Operon). The data were processed, and the methylation density was calculated using CyMATE software.75
RNA isolation and quantitative reverse transcription (RT)-PCR analysis
Total RNA was isolated from young leaves or calli using the RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer's instructions and treated with DNaseI (TURBO DNA-free, Applied Biosystems/Life Technologies).
The cDNAs were prepared by reverse transcription of RNAs using Superscript II reverse transcriptase (Invitrogen/Life Technologies) and random nonamers (Sigma). Quantification of the nptII level related to the actin transcripts was performed using the Fast Start SYBR Green Master (Roche) by the Rotorgene 6000 (QIAGEN, Germany). The nptII gene was amplified with the forward primer 5?-CGTTACAAGAGAGAAATCGCC-3? and the reverse primer 5?-TTCTATCGCCTTCTTGACGAG-3?; actin was amplified with the forward primer 5?-CTGGATTTGCTGGTGATGAT-3?and the reverse primer 5?-CYCTCTTGGATTGAGCTT-3? in the same PCR cycle (initial denaturation at 94°C for 10 min followed by 35 cycles of 20 sec at 94°C, 20 sec at 56°C and 30 sec at 72°C). The amount of nptII transcript was determined for two to three plants/calli of each line in several technical replicates.
Chromatin immunoprecipitation (ChIP)
Immunoprecipitation of chromatin was performed using the EpiQuikTM Plant ChIP kit (Epigentek) according to the manufacturer?s instructions.
One gram of leaves or calli was cross-linked, and the isolated DNA was sonicated (6 × 10 sec; power setting 1) using the Branson Sonifier B-12 sonicator (Branson Sonic Power Company) into the 0.2- to 1-kb fragments. Immunoprecipitation was performed using commercially available antibodies anti-H3K4me3 (Abcam; catalog no. ab8580), anti-H3K9ac (Abcam; catalog no. ab10812) and anti-H3K9me2 (Millipore 07?212). Classical PCR was performed on ChIP samples. The P35S and the 5? coding sequence was amplified with the forward primer 5?-AAGGCTATCGTTCAAGATGCC-3? and the reverse primer 5?-GATTGTCTGTTGTGCCCAGTC-3?. The positions are about +140 (forward) and -210 bp (reverse) relative to the transcription start site. Because of the locus 271 arrangement it was necessary to amplify its P35S region by another reverse primer 5?-TCTCCAAATGAAATGAACTTCCTTAT -3?. The PCR program consisted of 3 min of initial denaturation at 94°C followed by 35 cycles of 20 sec at 94°C, 50 sec at 58°C and 50 sec at 72°C. The 3? nptII region and actin were amplified using the same primers as those used for quantitative RT-PCR analysis in the same PCR cycle (initial denaturation at 94°C for 2 min followed by 35 cycles of 20 sec at 94°C, 20 sec at 56°C and 30 sec at 72°C). The ChIP experiment was performed in two biological replicates
Southern blot hybridization
The standard procedures involving DNA isolation, restriction enzyme treatments and Southern hybridization were described previously46,47 with exception that the blots were washed under medium stringency conditions (2 × SSC, 55°C, 2 × 5 min and 0.6 × SSC, 55°C, 2 × 15 min). The nptII-coding sequence and the 35S promoter probes were prepared from the ~830-bp and ~980-bp inserts of the pGEMnptII and pGSJ290 plasmids, respectively.19 The hybridization bands were visualized with a PhosphorImager Typhoon (GE Healthcare) and the data were processed with the ImageQuant software (GE-Healthcare).
Supplementary Material
Acknowledgments
We thank Dr Hervé Vaucheret (INRA, Versailles, France) for providing us with a tobacco 271 line. The comments on the work from Dr Ortrun Mittelsten-Scheid (GMI, Austrian Academy of Sciences, Vienna) have been much appreciated. We thank Mrs. Jana Kubí?ková for her technical assistance. This work has been supported by the Czech Academy of Sciences, the Czech Science Foundation (P501/11/P667, P501/12/G090 and P501/13/10057S), Research Foundation-Flanders (grant no. G.0211.06N and Belgium-Czech Exchange Program).
Submitted
01/23/2013
Revised
03/28/2013
Accepted
04/08/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24613
References
- 1.Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJM, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–74. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Chawla R, Nicholson SJ, Folta KM, Srivastava V. Transgene-induced silencing of Arabidopsis phytochrome A gene via exonic methylation. Plant J. 2007;52:1105–18. doi: 10.1111/j.1365-313X.2007.03301.x. [DOI] [PubMed] [Google Scholar]
- 3.Dalmay T, Hamilton A, Mueller E, Baulcombe DC. Potato virus X amplicons in arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell. 2000;12:369–79. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel JB, Mourrain P, Béclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–4. doi: 10.1016/S0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 6.Meyer P. DNA methylation systems and targets in plants. FEBS Lett. 2011;585:2008–15. doi: 10.1016/j.febslet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Baubec T, Dinh HQ, Pecinka A, Rakic B, Rozhon W, Wohlrab B, et al. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis. Plant Cell. 2010;22:34–47. doi: 10.1105/tpc.109.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–52. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernatavichute YV, Zhang XY, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Dong AW, Shen WH. Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 2004;40:699–711. doi: 10.1111/j.1365-313X.2004.02240.x. [DOI] [PubMed] [Google Scholar]
- 13.Shen WH, Meyer D. Ectopic expression of the NtSET1 histone methyltransferase inhibits cell expansion, and affects cell division and differentiation in tobacco plants. Plant Cell Physiol. 2004;45:1715–9. doi: 10.1093/pcp/pch184. [DOI] [PubMed] [Google Scholar]
- 14.Ingram R, Charrier B, Scollan C, Meyer P. Transgenic tobacco plants expressing the Drosophila Polycomb (Pc) chromodomain show developmental alterations: possible role of Pc chromodomain proteins in chromatin-mediated gene regulation in plants. Plant Cell. 1999;11:1047–60. doi: 10.1105/tpc.11.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udriste AA, Stan V, Radu GL, Tabler M, Cucu N. Identification of a dicer homologue gene (DCL2) in Nicotiana tabacum. Plant Biol (Stuttg) 2012;14:980–6. doi: 10.1111/j.1438-8677.2012.00586.x. [DOI] [PubMed] [Google Scholar]
- 16.Fulnecek J, Matyásek R, Kovarík A. Faithful inheritance of cytosine methylation patterns in repeated sequences of the allotetraploid tobacco correlates with the expression of DNA methyltransferase gene families from both parental genomes. Mol Genet Genomics. 2009;281:407–20. doi: 10.1007/s00438-008-0420-8. [DOI] [PubMed] [Google Scholar]
- 17.Benfey PN, Chua NH. The cauliflower mosaic virus-35S promoter: combinatorial regulation of transcription in plants. Science. 1990;250:959–66. doi: 10.1126/science.250.4983.959. [DOI] [PubMed] [Google Scholar]
- 18.Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci U S A. 1994;91:10502–6. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Houdt H, Ingelbrecht I, Van Montagu M, Depicker A. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3? flanking regions. Plant J. 1997;12:379–92. doi: 10.1046/j.1365-313X.1997.12020379.x. [DOI] [Google Scholar]
- 20.Dalakouras A, Dadami E, Zwiebel M, Krczal G, Wassenegger M. Transgenerational maintenance of transgene body CG but not CHG and CHH methylation. Epigenetics. 2012;7:1071–8. doi: 10.4161/epi.21644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miki D, Shimamoto K. De novo DNA methylation induced by siRNA targeted to endogenous transcribed sequences is gene-specific and OsMet1-independent in rice. Plant J. 2008;56:539–49. doi: 10.1111/j.1365-313X.2008.03624.x. [DOI] [PubMed] [Google Scholar]
- 22.Okano Y, Miki D, Shimamoto K. Small interfering RNA (siRNA) targeting of endogenous promoters induces DNA methylation, but not necessarily gene silencing, in rice. Plant J. 2008;53:65–77. doi: 10.1111/j.1365-313X.2007.03313.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki S, Oda M, Daimon H, Mitsukuri K, Johkan M, Nakatsuka T, et al. Epigenetic modifications of the 35S promoter in cultured gentian cells. Plant Sci. 2011;180:612–9. doi: 10.1016/j.plantsci.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Park YD, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJ, et al. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 1996;9:183–94. doi: 10.1046/j.1365-313X.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- 25.Diéguez MJ, Vaucheret H, Paszkowski J, Mittelsten Scheid O. Cytosine methylation at CG and CNG sites is not a prerequisite for the initiation of transcriptional gene silencing in plants, but it is required for its maintenance. Mol Gen Genet. 1998;259:207–15. doi: 10.1007/s004380050806. [DOI] [PubMed] [Google Scholar]
- 26.Vaucheret H. Promoter-dependent trans-inactivation in transgenic tobacco plants - kinetic aspects of gene silencing and gene reactivation. Cr Acad Sci. 1994;317:310–23. [Google Scholar]
- 27.Kumpatla SP, Hall TC. Longevity of 5-azacytidine-mediated gene expression and re-establishment of silencing in transgenic rice. Plant Mol Biol. 1998;38:1113–22. doi: 10.1023/A:1006071018039. [DOI] [PubMed] [Google Scholar]
- 28.Kanazawa A, O?Dell M, Hellens RP. Epigenetic inactivation of chalcone synthase-A transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol. 2007;48:638–47. doi: 10.1093/pcp/pcm028. [DOI] [PubMed] [Google Scholar]
- 29.Le Masson I, Jauvion V, Bouteiller N, Rivard M, Elmayan T, Vaucheret H. Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell. 2012;24:3603–12. doi: 10.1105/tpc.112.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smulders MJM, de Klerk GJ. Epigenetics in plant tissue culture. Plant Growth Regul. 2011;63:137–46. doi: 10.1007/s10725-010-9531-4. [DOI] [Google Scholar]
- 31.Berdasco M, Alcázar R, García-Ortiz MV, Ballestar E, Fernández AF, Roldán-Arjona T, et al. Promoter DNA hypermethylation and gene repression in undifferentiated Arabidopsis cells. PLoS One. 2008;3:e3306. doi: 10.1371/journal.pone.0003306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanurdzic M, Vaughn MW, Jiang H, Lee TJ, Slotkin RK, Sosinski B, et al. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008;6:2880–95. doi: 10.1371/journal.pbio.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koukalova B, Fojtova M, Lim KY, Fulnecek J, Leitch AR, Kovarik A. Dedifferentiation of tobacco cells is associated with ribosomal RNA gene hypomethylation, increased transcription, and chromatin alterations. Plant Physiol. 2005;139:275–86. doi: 10.1104/pp.105.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams L, Zhao J, Morozova N, Li Y, Avivi Y, Grafi G. Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev Dyn. 2003;228:113–20. doi: 10.1002/dvdy.10348. [DOI] [PubMed] [Google Scholar]
- 35.Marjanac G, Karimi M, Naudts M, Beeckman T, Depicker A, De Buck S. Gene silencing induced by hairpin or inverted repeated sense transgenes varies among promoters and cell types. New Phytol. 2009;184:851–64. doi: 10.1111/j.1469-8137.2009.03011.x. [DOI] [PubMed] [Google Scholar]
- 36.Mittelsten Scheid O, Paszkowski J, Potrykus I. Reversible inactivation of a transgene in Arabidopsis thaliana. Mol Gen Genet. 1991;228:104–12. doi: 10.1007/BF00282454. [DOI] [PubMed] [Google Scholar]
- 37.Jaligot E, Beulé T, Rival A. Methylation-sensitive RFLPs: characterisation of two oil palm markers showing somaclonal variation-associated polymorphism. Theor Appl Genet. 2002;104:1263–9. doi: 10.1007/s00122-002-0906-4. [DOI] [PubMed] [Google Scholar]
- 38.Rhee Y, Sekhon RS, Chopra S, Kaeppler S. Tissue culture-induced novel epialleles of a Myb transcription factor encoded by pericarp color1 in maize. Genetics. 2010;186:843–55. doi: 10.1534/genetics.110.117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miguel C, Marum L. An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot. 2011;62:3713–25. doi: 10.1093/jxb/err155. [DOI] [PubMed] [Google Scholar]
- 40.Wang QM, Wang YZ, Sun LL, Gao FZ, Sun W, He J, et al. Direct and indirect organogenesis of Clivia miniata and assessment of DNA methylation changes in various regenerated plantlets. Plant Cell Rep. 2012;31:1283–96. doi: 10.1007/s00299-012-1248-6. [DOI] [PubMed] [Google Scholar]
- 41.Zeng FS, Qian JJ, Luo W, Zhan YG, Xin Y, Yang CP. Stability of transgenes in long-term micropropagation of plants of transgenic birch (Betula platyphylla) Biotechnol Lett. 2010;32:151–6. doi: 10.1007/s10529-009-0120-4. [DOI] [PubMed] [Google Scholar]
- 42.Maury S, Trap-Gentil MV, Hébrard C, Weyens G, Delaunay A, Barnes S, et al. Genic DNA methylation changes during in vitro organogenesis: organ specificity and conservation between parental lines of epialleles. Physiol Plant. 2012;146:321–35. doi: 10.1111/j.1399-3054.2012.01634.x. [DOI] [PubMed] [Google Scholar]
- 43.Fojtova M, Van Houdt H, Depicker A, Kovarik A. Epigenetic switch from posttranscriptional to transcriptional silencing is correlated with promoter hypermethylation. Plant Physiol. 2003;133:1240–50. doi: 10.1104/pp.103.023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Houdt H, Kovarík A, Van Montagu M, Depicker A. Cross-talk between posttranscriptionally silenced neomycin phosphotransferase II transgenes. FEBS Lett. 2000;467:41–6. doi: 10.1016/S0014-5793(00)01076-0. [DOI] [PubMed] [Google Scholar]
- 45.Mourrain P, van Blokland R, Kooter JM, Vaucheret H. A single transgene locus triggers both transcriptional and post-transcriptional silencing through double-stranded RNA production. Planta. 2007;225:365–79. doi: 10.1007/s00425-006-0366-1. [DOI] [PubMed] [Google Scholar]
- 46.Khaitová LC, Fojtová M, K?í?ová K, Lunerová J, Fulne?ek J, Depicker A, et al. Paramutation of tobacco transgenes by small RNA-mediated transcriptional gene silencing. Epigenetics. 2011;6:650–60. doi: 10.4161/epi.6.5.15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krizova K, Fojtova M, Depicker A, Kovarik A. Cell culture-induced gradual and frequent epigenetic reprogramming of invertedly repeated tobacco transgene epialleles. Plant Physiol. 2009;149:1493–504. doi: 10.1104/pp.108.133165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fojtová M, Bleys A, Bedrichová J, Van Houdt H, Krízová K, Depicker A, et al. The trans-silencing capacity of invertedly repeated transgenes depends on their epigenetic state in tobacco. Nucleic Acids Res. 2006;34:2280–93. doi: 10.1093/nar/gkl180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foerster AM, Dinh HQ, Sedman L, Wohlrab B, Mittelsten Scheid O. Genetic rearrangements can modify chromatin features at epialleles. PLoS Genet. 2011;7:e1002331. doi: 10.1371/journal.pgen.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Douet J, Tourmente S. Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity (Edinb) 2007;99:5–13. doi: 10.1038/sj.hdy.6800964. [DOI] [PubMed] [Google Scholar]
- 51.Fulnecek J, Matyásek R, Kovarík A, Bezdek M. Mapping of 5-methylcytosine residues in Nicotiana tabacum 5S rRNA genes by genomic sequencing. Mol Gen Genet. 1998;259:133–41. doi: 10.1007/s004380050798. [DOI] [PubMed] [Google Scholar]
- 52.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–76. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murad L, Lim KY, Christopodulou V, Matyasek R, Lichtenstein CP, Kovarik A, et al. The origin of tobacco?s T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanaceae) Am J Bot. 2002;89:921–8. doi: 10.3732/ajb.89.6.921. [DOI] [PubMed] [Google Scholar]
- 54.Lunerová-Bedrichová J, Bleys A, Fojtová M, Khaitová L, Depicker A, Kovarík A. Trans-generation inheritance of methylation patterns in a tobacco transgene following a post-transcriptional silencing event. Plant J. 2008;54:1049–62. doi: 10.1111/j.1365-313X.2008.03475.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Inagaki S, Kakutani T. Control of genic DNA methylation in Arabidopsis. J Plant Res. 2010;123:299–302. doi: 10.1007/s10265-010-0338-1. [DOI] [PubMed] [Google Scholar]
- 57.You WH, Tyczewska A, Spencer M, Daxinger L, Schmid MW, Grossniklaus U, et al. Atypical DNA methylation of genes encoding cysteine-rich peptides in Arabidopsis thaliana. BMC Plant Biol. 2012;12:51. doi: 10.1186/1471-2229-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovarík A, Van Houdt H, Holý A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Lett. 2000;467:47–51. doi: 10.1016/S0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- 59.Van Houdt H, Bleys A, Depicker A. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 2003;131:245–53. doi: 10.1104/pp.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vermeersch L, De Winne N, Nolf J, Bleys A, Kova?ík A, Depicker A. Transitive RNA silencing signals induce cytosine methylation of a transgenic but not an endogenous target. Plant J. 2013;??? doi: 10.1111/tpj.12172. [DOI] [PubMed] [Google Scholar]
- 61.Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–86. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–33. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 63.Marenkova TV, Deineko EV. Transcriptional gene silencing in plants. Russ J Genet. 2010;46:511–20. doi: 10.1134/S1022795410050017. [DOI] [PubMed] [Google Scholar]
- 64.Ogrocká A, Sýkorová E, Fajkus J, Fojtová M. Developmental silencing of the AtTERT gene is associated with increased H3K27me3 loading and maintenance of its euchromatic environment. J Exp Bot. 2012;63:4233–41. doi: 10.1093/jxb/ers107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lechtenberg B, Schubert D, Forsbach A, Gils M, Schmidt R. Neither inverted repeat T-DNA configurations nor arrangements of tandemly repeated transgenes are sufficient to trigger transgene silencing. Plant J. 2003;34:507–17. doi: 10.1046/j.1365-313X.2003.01746.x. [DOI] [PubMed] [Google Scholar]
- 66.Eike MC, Mercy IS, Aalen RB. Transgene silencing may be mediated by aberrant sense promoter sequence transcripts generated from cryptic promoters. Cell Mol Life Sci. 2005;62:3080–91. doi: 10.1007/s00018-005-5301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fischer U, Kuhlmann M, Pecinka A, Schmidt R, Mette MF. Local DNA features affect RNA-directed transcriptional gene silencing and DNA methylation. Plant J. 2008;53:1–10. doi: 10.1111/j.1365-313X.2007.03311.x. [DOI] [PubMed] [Google Scholar]
- 68.Dalakouras A, Moser M, Boonrod K, Krczal G, Wassenegger M. Diverse spontaneous silencing of a transgene among two Nicotiana species. Planta. 2011;234:699–707. doi: 10.1007/s00425-011-1433-9. [DOI] [PubMed] [Google Scholar]
- 69.Mishiba K, Yamasaki S, Nakatsuka T, Abe Y, Daimon H, Oda M, et al. Strict de novo methylation of the 35S enhancer sequence in gentian. PLoS One. 2010;5:e9670. doi: 10.1371/journal.pone.0009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011;7:e1002040. doi: 10.1371/journal.pgen.1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 2010;24:986–91. doi: 10.1101/gad.579910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol. 2008;50:886–96. doi: 10.1111/j.1744-7909.2008.00692.x. [DOI] [PubMed] [Google Scholar]
- 73.Van Houdt H, Van Montagu M, Depicker A. Both sense and antisense RNAs are targets for the sense transgene-induced posttranscriptional silencing mechanism. Mol Gen Genet. 2000;263:995–1002. doi: 10.1007/PL00008700. [DOI] [PubMed] [Google Scholar]
- 74.Kovarík A, Van Houdt H, Holý A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Lett. 2000;467:47–51. doi: 10.1016/S0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- 75.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–36. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- 76.Matzke AJM, Neuhuber F, Park YD, Ambros PF, Matzke MA. Homology-dependent gene silencing in transgenic plants: epistatic silencing loci contain multiple copies of methylated transgenes. Mol Gen Genet. 1994;244:219–29. doi: 10.1007/BF00285449. [DOI] [PubMed] [Google Scholar]
- 77.Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol. 2001;11:747–57. doi: 10.1016/S0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 78.Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JN, et al. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol. 2001;11:436–40. doi: 10.1016/S0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.