Abstract
Detection of species fraud in meat products is important for consumer protection and food industries. A molecular technique such as PCR method for detection of beef, sheep, pork, chicken, donkey, and horse meats in food products was established. The purpose of this study was to identification of fraud and adulteration in industrial meat products by PCR-RFLP assay in Iran. In present study, 224 meat products include 68 sausages, 48 frankfurters, 55 hamburgers, 33 hams and 20 cold cut meats were collected from different companies and food markets in Iran. Genomic DNA was extracted and PCR was performed for gene amplification of meat species using specific oligonucleotid primers. Raw meat samples are served as the positive control. For differentiation between donkey’s and horse’s meat, the mitochondrial DNA segment (cytochrome-b gene) was amplified and products were digested with AluI restriction enzyme. Results showed that 6 of 68 fermented sausages (8.82%), 4 of 48 frankfurters (8.33%), 4 of 55 hamburgers (7.27%), 2 of 33 hams (6.6%), and 1 of 20 cold cut meat (5%) were found to contain Haram (unlawful or prohibited) meat. These results indicate that 7.58% of the total samples were not containing Halal (lawful or permitted) meat and have another meat. These findings showed that molecular methods such as PCR and PCR-RFLP are potentially reliable techniques for detection of meat type in meat products for Halal authentication.
Keywords: PCR-RFLP, Mitochondrial DNA, Meat species, Haram, Halal, Iran
Introduction
Meat is a primary source of water and fat, and contains between 20% and 35% protein, providing all essential amino acids (lysine, threonine, methionine, phenylalanine, tryptophan, leucine, isoleucine and valine), as well as good amounts of various micronutrients (Aida et al. 2005). It is an easily absorbable source of iron, zinc and selenium, as well as containing good levels of vitamins B6 and B12, and vitamin D, and significant amounts of omega-3 polyunsaturated fatty acids. Thus, it is a valuable source of some key nutrients (Ferguson 2010).
In Islam, foods containing pig, donkey and horse sources are Haram for Muslims to consume. Hence, it is an important task for food control laboratories to be able to carry out species differentiation of raw materials to be used for industrial food preparation and the detection of animal species in food products (Luo et al. 2008). Authenticity testing of the animal species present in food is important for economic, safety, legal, religious and health reasons. Food labeling regulations require that the species of meat in food products are accurately declared to the consumer. Product consumption containing nondeclared meat proteins can induce allergic reactions in predisposed individuals (Ong et al. 2007). Single and multispecies adulterations have been reported in commercial meat products (Hsieh et al. 1997). The adulteration rates of cooked meat products are higher than raw meats. Poultry consumption has been increasing because of its relatively lower saturated fat and cholesterol content than mammalian meats. The popularity of poultry meat also increases the chance of mixing mechanically deboned poultry tissue with ground or comminuted mammalian products (Hsieh et al. 1996).
This is especially crucial for Halal authentication, of food products. In order to protect consumers from fraud and adulteration several analytical approaches have been made to identify animal species in food products (Aida et al. 2005). Methods have been developed based on electrophoresis, isoelectric focusing, chromatography, DNA hybridization, polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) (Ong et al. 2007). PCR proved to be an adequate technique for detection of small amounts of DNA, specifically amplifying a target region of template DNA in a rapid and sensitive manner (Saiki et al. 1988). The aim of this study was to detection of adulteration and identification of beef’s, sheep’s, chicken’s, pork’s, donkey’s and horse’s meat using PCR and PCR-RFLP techniques in meat products of Iran.
Materials and methods
Samples
In this study, 224 meat products include 68 sausages, 48 frankfurters, 55 hamburgers, 33 hams and 20 cold cut meats were collected from different companies and food markets in Iran and 30 samples of raw meats of beef, sheep, pork, chicken, donkey and horse are used as a positive control.
DNA extraction
Genomic DNA was extracted from 25 mg of meat and meat products using DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quality of DNA was checked on agarose gel electrophoresis and quantitation done by UV-spectrophotometry.
PCR assay for gene amplification
Species-specific DNA segments of bovis, sheep, pork, chicken, donkey, and horse in addition to mitochondrial DNA segment (cytochrome-b gene) in both donkey and horse were used for amplification and detection of animal derived materials in meat products. PCR amplification was performed in a 25 μL reaction volume containing 1 μg of genomic DNA of each specie, 1 μM of each primers, 2 mM Mgcl2, 200 μM dNTP, 2.5 μL of 10X PCR buffer and 1 unit of Taq DNA polymerase (Roche applied science). Species-specific oligonucleotide primers reported by Luo et al. (2008) and Abdel-Rahman et al. (2009) were used for gene amplification are shown in Table 1 (Luo et al. 2008, Abdel-Rahman et al. 2009).
Table 1.
Species-specific oligonucleotide primers and expected lengths of amplified segments
| Primer name | Primer sequence | Product size |
|---|---|---|
| Bovis | F: 5′-GCCATATACTCTCCTTGGTGACA-3′ | 271 bp |
| Bovis | R: 5′-GTAGGCTTGGGAATAGTACGA-3′ | |
| Sheep | F: 5′-ATGCTGTGGCTATTGTC-3′ | 274 bp |
| Sheep | R: 5′-CCTAGGCATTTGCTTAATTTTA-3′ | |
| Pork | F: 5′-ATGAAACATTGGAGTAGTCCTACTATTTACC-3′ | 149 bp |
| Pork | R: 5′-CTACGAGGTCTGTTCCGATATAAGG-3′ | |
| Chicken | F: 5′-GGGACACCCTCCCCCTTAATGACA-3′ | 266 bp |
| Chicken | R: 5′-GGAGGGCTGGAAGAAGGAGTG-3′ | |
| Donkey & | F: 5′-TTCTGCTCTGGGTGTGCTACTT-3′ | 221 bp |
| Horse | R: 5′-CTACTTCAGCCAGATCAGGC-3′ |
After 5 min of initial denaturation at 94 °C, 30 cycles of denaturation at 94 °C for 1 min, annealing at 63 °C for beef, 59 °C for sheep, 69 °C for chicken, 60 °C for pork, 58 °C for donkey and horse, for 1 min, extension at 72 °C for 1 min and final extension at 72 °C for 6 min were performed for PCR reaction by thermal cycler (Mastercycler Gradient, Eppendrof, Germany).
PCR products detection
The products of PCR amplification were separated on 1% agarose gels in 1X TBE buffer at 100 V for 30 min, stained with Ethidium Bromide, and images were obtained in a UVIdoc gel documentation systems (UK).
RFLP analysis
For restriction analysis, 10 μL of PCR product (359 bases of mitochondrial cytochrome-b gene) in donkey and horse were digested overnight at 37 °C with AluI restriction enzyme (Fermentas, Germany), according to the manufacturer’s instructions. Digested DNA was separated on 2% agarose gels in IX TBE buffer and Ethidium Bromide staining.
Results and discussion
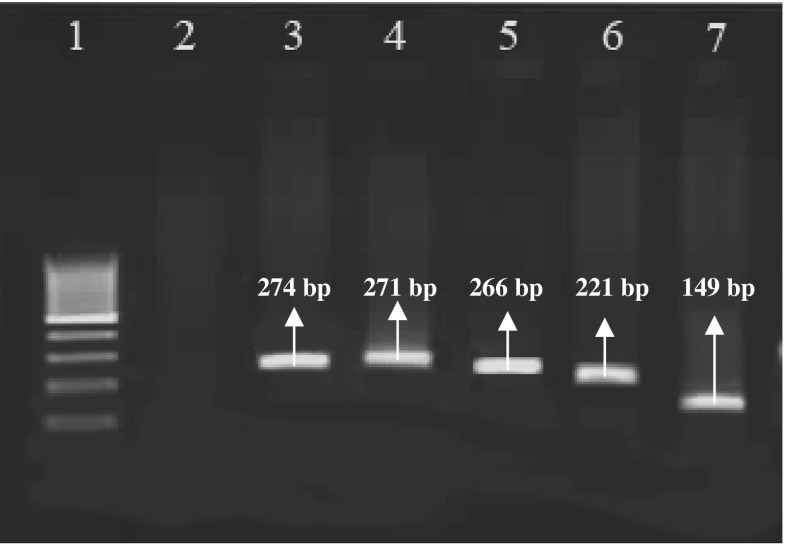
Amplification with species-specific oligonucleotide primers revealed a 271, 274, 149, 266, and 221 bp from bovine, sheep, pork, chicken, and both donkey and horse genomic DNA, respectively (Fig. 1).
Fig. 1.
The electrophoresis of PCR products was generated by species-specific oligonucleotide primers. Line 1 is a 100 bp molecular weight marker (Fermentas, Germany). Line 2 is negative control, lines 3–7 are sheep, bovine, chicken, both donkey and horse, and pork amplified fragments, respectively
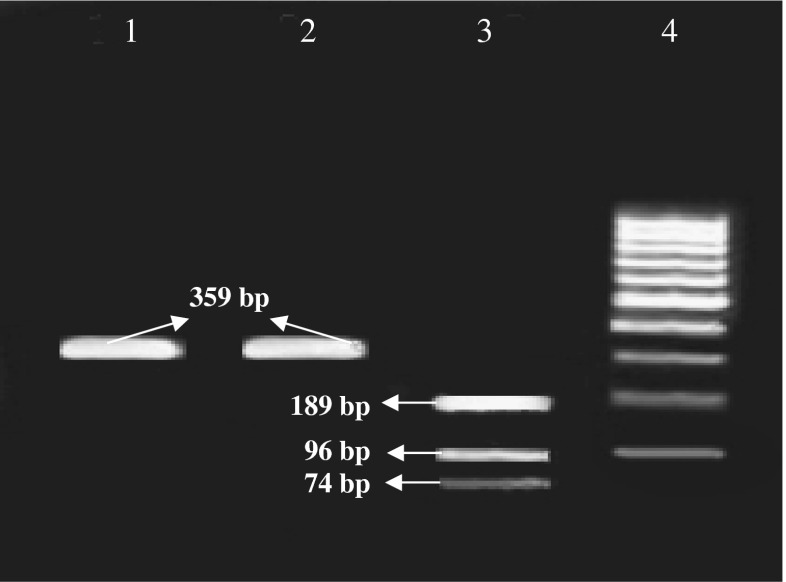
The amplification of mitochondrial DNA segment (cytochrome-b gene) in both donkey and horse yielded the same amplicon with a size of 359 bp (Fig. 2). The PCR amplification size and the position of the PCR with species-specific oligonucleotide primers and the mitochondrial DNA segment (221 bp and 359 bp) with both donkey and horse are exactly same (Figs. 1 and 2).
Fig. 2.
Agarose gel electrophoresis of amplified cytochrome-b gene following digestion with AluI generated three fragments with sizes of 189, 96 and 74 bp (lane 1: uncut fragment, lane 2: donkey, and lane 3: horse). Lane 4 is a 100 bp DNA ladder (Fermentas, Germany)
For differentiation between donkey’s and horse’s meat, PCR-RFLP technique for the two PCR products (221 bp and 359 bp) was used. However, 221 bp PCR product size was treated with numerous restriction enzymes, but without success (no genetic variation was found). The amplified region of the gene encoding cytochrome-b (359 bp) in both donkey and horse was also treated with many restriction enzymes. Eventually, three different patterns only in horse were generated after the AluI restriction enzyme digestion with sizes; 189, 96 and 74 bp, while in donkey no digestion was obtained (359 bp) allowing an identification of donkey’s and horse’s meat (Fig. 2).
The results showed that 6 of 68 fermented sausages (8.82%), 4 of 48 frankfurters (8.33%), 4 of 55 hamburgers (7.27%), 2 of 33 hams (6.6%), and 1 of 20 cold cut meat (5%) were found to contain Haram meat. These results showed that 7.58% of the total samples were not containing Halal (pure) meat and have Haram meat. The detail of the range of Haram meat in meat products of Iran is shown in Table 2.
Table 2.
The range of Haram meat in meat products of Iran
| Animal type meat | |||||
|---|---|---|---|---|---|
| Meat product | Fermented sausages (percent) | Frankfurters (percent) | Hamburgers (percent) | Hams (percent) | Cold cut meat (percent) |
| Pork | 0 | 0 | 1 (1.81%) | 0 | 0 |
| Donkey | 5 (7.35%) | 4 (8.33%) | 2 (3.63%) | 2 (6.06%) | 1 (5%) |
| Horse | 1 (1.47%) | 0 (0%) | 1 (1.81%) | 0 (0%) | 0 (0%) |
Preventing adulteration of meat foods with less desirable or objectionable meat species is important for economic, religious and health reasons (Ayaz et al. 2006). In addition, determination of the species of origin of the meat components in meat products is an important task in food hygiene, food codex, food control and veterinary forensic medicine (Ayaz et al. 2006). Adulteration of meat species is important for people whose religious practices limit the types of meat they eat, and for people who have allergies to certain types of meat proteins (Hsieh et al. 1997). Several methods have been developed to identify different species of meat. Each method has advantages and disadvantages. The conventional methodology used for the determination of species origin in meat products had been predominantly based on immunosorbent assay (ELISA), immunochemical and electrophorectic analysis of protein. Electrophoresis requires several hours and presents low reproducibility. Additionally, through the acquisition of sequence data, DNA can potentially provide more information than type of protein content, due to the degeneracy of the genetic code and the presence of many non-coding regions (Partis et al. 2000). DNA hybridization) Jain et al. 2007) and PCR methods (Chikuni et al. 1994) have been used for the identification of meats and meat products. PCR is a helpful technique for meat species identification. The present study is focused on the use of PCR and PCR-RFLP techniques for a rapid detection and identification of meat species in meat products of companies and food markets in Iran.
The results of these two techniques showed good evidence for molecular markers linked to genetic identification of beef’s, sheep’s, chicken’s, pork’s, donkey’s and horse’s meat. For discrimination between donkey’s and horse’s meat, using AluI restriction enzyme, three fragments (189 bp, 96 bp and 74 bp) from the amplified gene encoding cytochrome-b gene (359 bp) were obtained in horse, whereas in donkey no fragments were obtained (Fig. 2). This finding allowed us a direct and rapid identification and detection of adulteration of beef’s, sheep’s, chicken’s, pork’s, donkey’s and horse’s meat in meat products.
In current study from a total of 224 meat products, including 68 sausages, 48 frankfurters, 55 hamburgers, 33 hams and 20 cold cut meat 7.58% were contained Haram meat. Fermented sausage, hamburgers, frankfurter, and hams in generally have beef meat but in fraud meat products contained poultry, pig, donkey, and horse meat. Beef tissue was not found in the pork products samples. Since beef is more expensive than pork, there is no apparent economic reason for the addition of beef to pork products. Results indicated that the primary problem centers on the meat grinding operation. Companies and food markets readily admitted that they did not routinely clean grinders when changing from ground beef to another meat. On the other hand, poultry, donkey, and horse meat is cheaper than sheep and beef meat in Iran and indicating the possibility of adulteration for economic reasons. Pork meat is rare in Muslims countries such as Iran but sometimes through smuggling, pork meat may use in meat products of some companies.
Hsieh et al. (1995) reported that beef or lamb meat was found to be the contaminating species in ground turkey sold in retail markets. The reasons for substituting more expensive meat such as beef and lamb with cheaper meat such as poultry include the use of the unmarketable trimmings from expensive meats and improper cleaning of the grinder between each change of meat species prior to grinding (Hsieh et al. 1995). Meyer et al. detected 0.5% pork in beef using the duplex PCR technique. Their results revealed that PCR was the method of choice for identifying meat species in muscle foods (Meyer et al. 1994). Furthermore, Meyer et al. in 1995 detected 0.01% soy protein in processed meat products using the nested-PCR technique (Meyer et al. 1995). Partis et al. detected 1% pork in beef using RFLP (Partis et al. 2000) whereas Hopwood et al. detected 1% chicken in lamb using PCR (Hopwood et al. 1999). The study of Aida et al. in Malaysia showed PCR-RFLP is a potentially reliable technique for detection of pig meat and fat from other animals for Halal authentication (Aida et al. 2005). Shally et al. were used multiplex PCR technique for detection of meat species via tracing of cytochrome-b gene (Jain et al. 2007). Ong et al. in 2007 were used three restriction enzymes in PCR-RFLP using the mitochondrial cytochrome b region to establish a differential diagnosis which detect and discriminate between three meat species and they were showed this technique can be applied to food authentication for the identification of different species of animals in food products and as a same to the results of present study (Ong et al. 2007). Luo et al. in 2008 were showed the application of a PCR for detection of beef, sheep, pig, and chicken derived materials in feedstuff and indicated that high sensitivity and specificity of PCR technique with a minimum detection level of 0.1% (Luo et al. 2008). Abdel-Rahman et al. in 2009 were detected adulteration in cat’s, dog’s, donkey’s and horse’s meats using species-specific primers and PCR-RFLP technique (Abdel-Rahman et al. 2009).
Conclusion
The present study confirms previous findings and showed low adulteration in meat products of Iran. Since, the results of this study might be useful for effective control of adulterated consumer and fraud in meat products and violations of labeling requirements for meat products. Furthermore, PCR-RFLP technique is a useful method for meat species identification in meat products.
Acknowledgements
The authors would like to thank all the staff of Biotechnology Research Center of Islamic Azad University of Shahrekord Branch in southwest Iran for their sincere support.
References
- Abdel-Rahman SM, El-Saadani MA, Ashry KM, Haggag AS. Detection of adulteration and identification of cat’s, dog’s, donkey’s and horse’s meat using species-specific PCR and PCR-RFLP techniques. Aust J Basic Appl Sci. 2009;3(3):1716–1719. [Google Scholar]
- Aida AA, Che Man YB, Wong CMVL, Raha AR, Son R. Analysis of raw meats and fats of pigs using polymerase chain reaction for Halal authentication. Meat Sci. 2005;69:47–52. doi: 10.1016/j.meatsci.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Ayaz Y, Ayaz ND, Erol I. Detection of species in meat and meat products using enzyme-linked immunosorbent assay. J Muscle Foods. 2006;17:214–220. doi: 10.1111/j.1745-4573.2006.00046.x. [DOI] [Google Scholar]
- Chikuni K, Tabata T, Kosugiyama M, Monma M, Saito M. Polymerase chain reaction assay for detection of sheep and goat meats. Meat Sci. 1994;37:337–345. doi: 10.1016/0309-1740(94)90051-5. [DOI] [PubMed] [Google Scholar]
- Ferguson LR. Meat and cancer. Meat Sci. 2010;84:308–313. doi: 10.1016/j.meatsci.2009.06.032. [DOI] [PubMed] [Google Scholar]
- Hopwood AJ, Fairbrother KS, Lockley AK, Bardsley RG. An actin gene-related polymerase chain reaction (PCR) test for identification of chicken in meat mixtures. Meat Sci. 1999;53:227–231. doi: 10.1016/S0309-1740(99)00060-1. [DOI] [PubMed] [Google Scholar]
- Hsieh YHP, Woodward BB, Ho SH. Detection of species substitution in raw and cooked meats using immunoassays. J Food Prot. 1995;58:555–559. doi: 10.4315/0362-028X-58.5.555. [DOI] [PubMed] [Google Scholar]
- Hsieh YHP, Johnson MA, Wetzstein CJ, Green NR. Detection of species adulteration in pork products using agar gel immunodiffusion and enzyme linked immunosorbent assay. J Food Qual. 1996;19:1–9. doi: 10.1111/j.1745-4557.1996.tb00401.x. [DOI] [Google Scholar]
- Hsieh YHP, Chen FC, Sheu SC. AAES research developing simple, inexpensive tests for meat products. Highlights Agric Res. 1997;44(2):19–20. [Google Scholar]
- Jain S, Brahmbhait MN, Rank DN, Joshi CG, Solank JV. Use of cytochrome b gene variability in detecting meat species by multiplex PCR assay. Indian J Anim Sci. 2007;77(9):880–888. [Google Scholar]
- Luo J, Wang J, Bu D, Li D, Wang L, Wei H, Zhou L. Development and application of a PCR approach for detection of beef, sheep, pig, and chicken derived materials in feedstuff. Agr Sci China. 2008;7(10):1260–1266. doi: 10.1016/S1671-2927(08)60173-X. [DOI] [Google Scholar]
- Meyer R, Candrian U, Luthy J. Detection of pork in heated meat products by the polymerase chain reaction. J AOAC Int. 1994;77:617–622. [PubMed] [Google Scholar]
- Meyer R, Hofelein C, Luthy J, Candrian U. Polymerase chain reaction-restriction fragment length polymorphism analysis: a simple method for species identification in food. J AOAC Int. 1995;78:1542–1551. [PubMed] [Google Scholar]
- Ong SB, Zuraini MI, Jurin WG, Cheah YK, Tunung R, Chai LC, Haryani Y, Ghazali FM, Son R. Meat molecular detection: sensitivity of polymerase chain reaction-restriction fragment length polymorphism in species differentiation of meat from animal origin. ASEAN Food J. 2007;14(1):51–59. [Google Scholar]
- Partis L, Croan D, Guo Z, Clark R, Coldham T, Murby J. Evaluation of a DNA fingerprinting method for determining the species origin of meats. Meat Sci. 2000;54:369–376. doi: 10.1016/S0309-1740(99)00112-6. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]