Abstract
Forkhead box protein A1 (FoxA1) is a transcription factor that is involved in embryonic development and cell differentiation. In this study, we show that hydrogen peroxide (H2O2) treatment upregulated expression of FoxA1 and UCP2 in the A549 cell line. Overexpression of FoxA1 by full-length complementary DNA reduced UCP2 expression, while silencing of FoxA1 expression by small interfering RNA significantly increased UCP2 levels. FoxA1 binds to a site from −919 to −913 bp relative to the UCP2 transcription start site. The overexpression of FoxA1 promoted the DNA binding activity and attenuated the transcription of UCP2 promoter as shown by electromobility shift, chromatin immunoprecipitation assays, and luciferase reporter assay. These data indicate an important role of FoxA1 in regulating expression of UCP2.
Keywords: Forkhead box protein A1, UCP2, Gene expression, Hydrogen peroxide
Introduction
Forkhead box (Fox) is a member of the winged-helix subgroup of the helix–loop–helix transcription factor family that exhibits homology to the fkh (forkhead) gene in Drosophila. FoxA1/Hepatocyte nuclear factor 3α (HNF3α), FoxA2/HNF3β and FoxA3/HNF3γ constitute the FoxA subfamily, which play an essential role in the development and maintenance of the endoderm-derived organs and also regulate gene transcription. Studies have shown that FoxA1 regulates many genes involved in developmental specification of not just hepatic, but several other tissues including lung. Recently, our studies have shown that FoxA1 expression is induced by H2O2 (hydrogen peroxide), lipopolysaccharide, and tumor necrosis factor-α in A549, in which it is suggested to function in A549 apoptosis (Song et al. 2009c). FoxA1 could regulate the expression of bcl-2 during H2O2-induced apoptosis in A549 cells (Song et al. 2009a). We still speculate that FoxA1 can also regulate some other apoptosis-related genes during oxidative-stress-induced apoptosis, and this idea needs further study.
Uncoupling proteins (UCPs) belong to a family of mitochondrial carrier proteins located in the inner mitochondrial membrane, a family that is currently comprised of five members. UCP2, a member of the UCP family, is expressed in various tissues including the brain, lung, spleen, kidney, liver, adipose tissues, and heart (Ricquier and Bouillaud 2000). Originally, UCP2 was postulated to decrease the production of reactive oxygen species (ROS; Goglia and Skulachev 2003). However, the role of UCP2 in cell apoptosis and in cancer was recently recognized and has attracted more attention. A growing body of evidence suggests that UCP2 could exert antiapoptosis effects by inhibiting the mitochondrial death pathway in cardiomyocytes, HepG2 cell, vascular endothelial cells, hypothalamic cells, testicular cells, and adipose cells. UCP2 has been shown to be upregulated in a number of aggressive human cancers. Increasing evidence suggests elevated UCP2 contributes not only to chemoresistance but also to early transformation (Samudio et al. 2009).
Using Matinspector Professional program at www.genomatix.de and Transcription Element Search System at www.cbil.upenn.edu, we found that the UCP2 gene contains putative FoxA1 binding sites in its promoter. However, the direct effect of FoxA1 on the expression of UCP2 remains unknown.
In this report, the expressions of FoxA1 and UCP2 in response to H2O2 were determined in A549 cells. In addition, the effects of FoxA1 on the expression of UCP2 and the mechanism of how FoxA1 regulates the UCP2 gene were also investigated. We found that the expressions of FoxA1 and UCP2 were upregulated in A549 induced by H2O2 and that FoxA1 downregulated the expression of UCP2 under normal and H2O2-induced conditions in A549 cells. Inhibition of endogenous FoxA1 with small interfering RNA increased the expression of UCP2. These results suggest that FoxA1 is a novel regulator of UCP2 expression.
Materials and methods
Cell culture and challenge
The A549 cells were maintained in Dulbecco’s modified Eagle’s medium nutrient mixture (Life Technologies), containing 10 % fetal bovine serum and 1 % penicillin-streptomycin at 37 °C and 5 % CO2. The cell was challenged with hydrogen peroxide (Sigma). At indicated time points after the treatment, cells were harvested, and messenger RNA (mRNA) and protein were extracted to assay the expression of the FoxA1 and UCP2.
RNA extraction and real-time PCR
Total RNA was extracted by TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Five micrograms of total RNA was then used as a template to synthesize complementary DNA (cDNA) using the First Strand Synthesis Kit (Invitrogen). The cDNA from this synthesis was then used in quantitative real-time PCR (RT-PCR) analysis with the TaqMan system (ABI-Prism 7700 Sequence Detection System, Applied Biosystems) using SYBR Green dye. The following primer pairs were used: FoxA1, 5′-AGGTGTGTATTCCAGACCCG-3′ and 5′-TTGACGGTTTGGTTTGTGTG-3′; UCP2, 5′-GACCTATGACCTCATCAAGG-3′ and 5′-ATAGGTGACGAACATCACCACG-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and 5′-GTGGTCGTTGAGGGCAATG-3′. RT-PCR data were normalized by measuring average cycle threshold (Ct) ratios between candidate genes and the control gene, GAPDH. The formula 2Ct(Candidate)/2Ct(Control) was used to calculate normalized ratios.
Western blot analysis
Proteins in the whole cell lysate were resolved on 12 % sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (Schleicher and Schuell). The membranes were blocked overnight in phosphate-buffered saline (PBS) containing 10 % nonfat dry milk and 0.5 % Tween-20, and incubated with primary antibodies for 2 h. Horseradish peroxidase-conjugated antirabbit or antimouse IgG was used as the secondary antibody. The immunoreactive bands were visualized using diaminobenzidine (Boster Biological Technology). Anti-GAPDH was used to normalize for equal amounts of proteins and calculate the relative induction ratio. The following antibodies were used: rabbit anti-FoxA1 polyclonal antibody (Abcam), goat anti-UCP2 polyclonal antibody (Santa Cruz Biotechnology), mouse GAPDH monoclonal antibody (Sigma), and horseradish peroxidase-conjugated antimouse and antirabbit IgG (Boster Biological Technology).
FoxA1 expression plasmid construction
Oligonucleotide primers were designed to amplify the coding sequence of human FoxA1 cDNA, yielding a 1.4-kb product. The oligonucleotide primers are as follows: FoxA1, 5′-CCG GAA TTC AGG GTG GCT CCA GGA TGT TAG-3′ (forward) and 5′-CCC AAG CTT GAA GTG TTT AGG ACG GGT ATG-3′ (reverse). The PCR product was electrophoresed onto 0.9 % agarose, and a 1.4-kb fragment was purified with the Qiagen gel purification system (Qiagen). The fragment was then inserted into the pcDNA3.1 vector (Strategene) and sequenced commercially (Invitrogen).
Lipofectamine-mediated transient transfection
Transient transfection of A549 cells was performed according to the manufacturer’s instructions (Lipofectamine 2000TM, Invitrogen). Briefly, about 5 × 105 cells per flask containing 5 ml of appropriate complete growth medium were seeded and incubated at 37 °C with 5 % CO2 until the cells were 70–80 % confluent (24 h). After the cells were rinsed with serum- and antibiotics-free medium, the cells were transfected separately with 10 μg pcDNA3.1-FoxA1 per 20 μl lipofectamine (experimental group) or 10 μg pcDNA3.1 per 20 μl lipofectamine (vector control), followed by incubation at 37 °C in a CO2 incubator for 6 h. The medium was then changed to regular medium with 10 % fetal bovine serum.
RNA interference
Transfection of FoxA1 small interfering RNA (siRNA) was performed using siPORT Amine (Ambion, Inc.), according to the siRNA transfection protocol in A549 cells. To ensure the knockdown of FoxA1 protein production, Western blot was performed with FoxA1 antibody. The short interference (si)RNAs against human FoxA1 and its control siRNA were purchased from Dharmacon (M-010319), or Qiagen (1027280), respectively.
Nuclear extract preparation and electrophoretic mobility shift assays
After cells were incubated with 0.5 mM H2O2 for 30 min, cells were harvested and washed twice with cold PBS. Briefly, the cell pellet was resuspended in 400 μl of cold buffer A [10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)]. The cells were allowed to swell on ice for 15 min, then 25 μl of a 10 % solution of Nonidet P-40 (NP-40) was added, and the tube was vortexed vigorously for 10 s. The homogenate was centrifuged at 10,000×g for 30 s, and the nuclear pellet was resuspended in 50 μl of ice-cold buffer B (20 mM Hepes, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF). After vigorously rocking at 4 °C for 15 min on a shaking platform, the nuclear extract was centrifuged at 10,000×g for 5 min in a microfuge at 4 °C, and the supernatant was frozen in aliquots at −80 °C. The protein content of the different fractions was determined by the Bradford method. Electrophoretic mobility shift assays were performed using nuclear extracts from A549 cells according to the instructions of Chemiluminescent Nucleic Acid Detection Module (Pierce). Supershift antibody for FoxA1 was incubated with nuclear extracts for 30 min at 4 °C prior to adding the biotin-labeled oligonucleotide. DNA probes were also generated according to the FoxA1 sites at positions −919 to −913 bp, −835 to −829 bp of the human UCP2 promoter as double-stranded, biotin-labeled oligonucleotides corresponding to the wild-type sequences (5′- GCCCAATTGTTGGCTCGCGT -3′, and 5′- GCCACGTGTTTGTCCCGGCC -3′, respectively) and mutant sequence for the position of −919 to −913 bp (5′- GCCACGACAAAGTCCCGGCC −3′).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was carried out with the EZ ChIP Kit (Upstate Biotechnology), as instructed by the manufacturer. In brief, cells were grown to 80–90 % confluency. After cross-linking for 10 min with 1 % formaldehyde in serum-free medium, glycine (0.125 M) was added to stop fixation, and cells were washed twice with ice-cold PBS. The chromatin lysate was sonicated on ice to an average DNA length of 600 bp. Chromatin was precleared with blocked Sepharose A, and ChIP assays were performed with either anti-FoxA1 antibody (Abcam) or FoxA2 antibody (Abcam) as the specific control, and normal rabbit IgG (Santa Cruz Biotechnology) as the negative control. Primers to amplify the proximal region of the UCP2 promoter that contained the −919 to −913 FoxA1 binding site were as follows: 5′-TGA CTG AAC GTC TTT GGG-3′ and 5′- AGC CGG GCC CAG GCC AGC TG -3′. Reaction products were analyzed on a 1.5 % agarose gel prepared using the Tris–borate EDTA buffering system, stained with ethidium bromide and visualized under UV light.
pGL-3 UCP2 promoter-reporter gene constructs and luciferase reporter gene assay
The assay was performed according to the instruction of the Dual Luciferase Reporter System (Promega). Generation of human UCP2 promoter region (−1,000 to +10 and −913 to +10) was amplified by PCR on human genomic DNA and cloned into pGL3-Basic, and authenticity was verified by sequencing. For luciferase reporter assay, exponentially growing A549 cells were seeded in 24-well culture dishes. All transfections were performed in triplicate from at least three independent experiments. Each transfection experiment contained 500 ng of pGL3-UCP2 promoter-reporter construct with 20 ng of pRL-null vector (Promega) as an internal transfection control.
Statistical analysis
Data in the figures and text were expressed as the mean ± SEM. Each experiment was performed at least three times, and statistical analysis was performed with a one-way ANOVA. Otherwise, representative data were shown. P < 0.05 was considered statistically significant.
Results
H2O2 induces the expression of FoxA1 and UCP2 in A549
Studies have shown that H2O2 could generate in tumors. In HepG2 cells and p815 mastocytoma, H2O2 concentration could reach 0.6 mM (Owada et al. 2013; Szuro-Sudol and Nathan 1982). On the other hand, previous studies have suggested that 5 × 106 cancer cells could produce 5 nM H2O2 (Szuro-Sudol and Nathan 1982; Thorne et al. 1980), so we suspected that the concentration of H2O2 could access to 0.5–1.0 mM in tumors. Thus, in this study, we used 0.25–1.0 mM H2O2 to stimulate A549 cells.
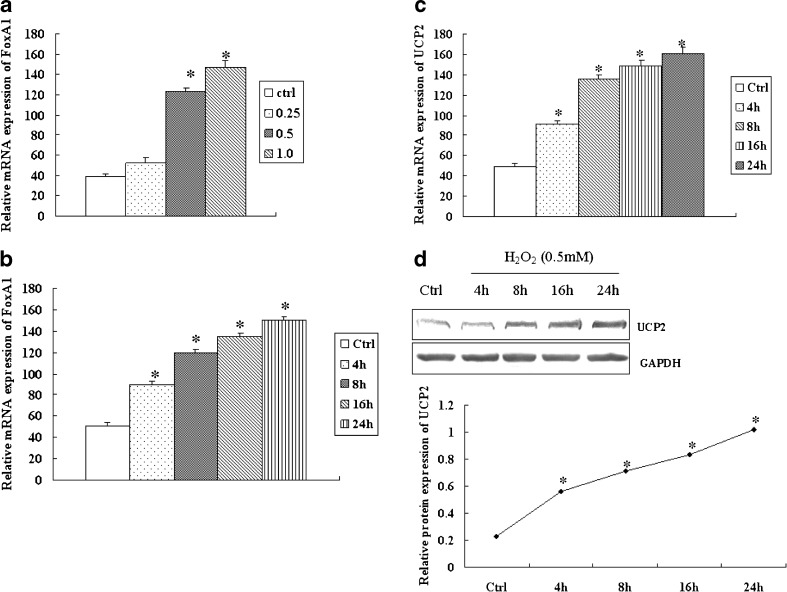
As shown in Fig. 1a and b, FoxA1 mRNA expression increased after H2O2 stimulation, and our previous studies have demonstrated that FoxA1 is upregulated sustainably in response to H2O2 in a dose- and time-dependent manner in A549 (Song et al. 2009a). To investigate the expression of UCP2, A549 cells were treated with H2O2 (0.5 mM), and the levels of UCP2 were determined. UCP2 increased 2 h after H2O2 stimulation (Fig. 1c, d). The increased levels of FoxA1 and UCP2 suggested a potential relationship between the genes.
Fig. 1.
Expression of FoxA1 and UCP2 in H2O2-stimulated A549 cells. a A549 cells were stimulated with H2O2 at the indicated dose for 4 h, and mRNA levels of FoxA1 were determined by RT-PCR. b A549 cells were stimulated with H2O2 (0.5 mM) for various periods of time and mRNA levels of FoxA1 were determined by RT-PCR. A549 cells were stimulated with H2O2 (0.5 mM) for various periods of time, and mRNA or proteins of UCP2 were determined by RT-PCR (c) and Western blot (d). The relative values of all results were determined and expressed as mean ± SEM of three experiments in duplicate. *P < 0.05, statistically significant difference versus control group (Ctrl)
FoxA1 influences the expression of UCP2 in A549
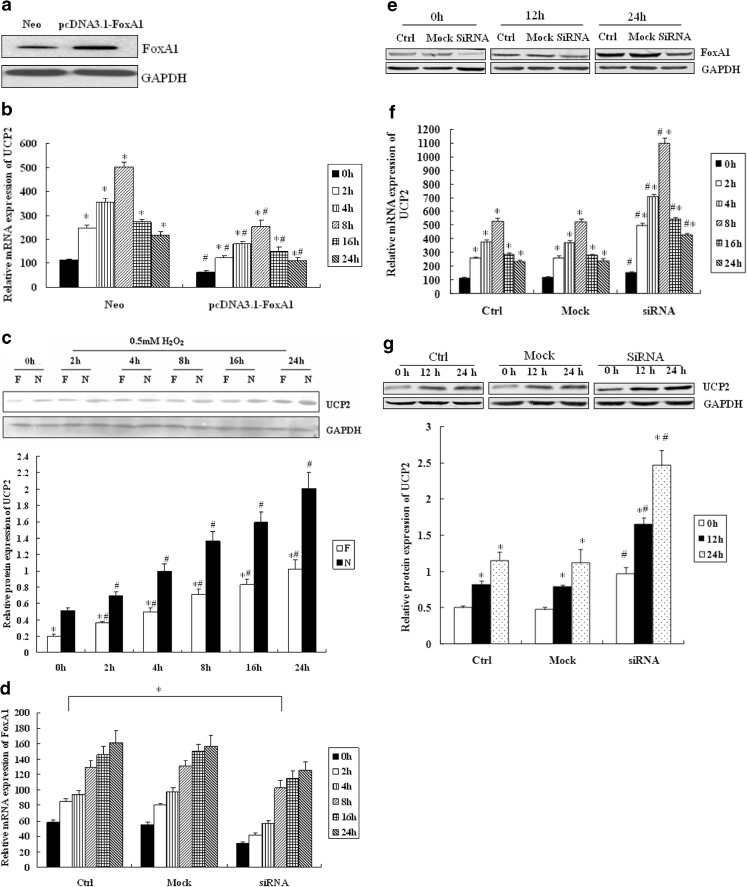
Using bioinformatics analysis, we found a putative FoxA1 binding site in the promoter sequence of UCP2. Because FoxA1 was induced by H2O2 and significantly increased in the early phase, we considered the possibility that this transcription factor may influence the expression of UCP2. We overexpressed FoxA1 in A549 using a pcDNA3.1-FoxA1 expression construct (Fig. 2a) and found that cell viability was not reduced by the transfection significantly (determined by MTT assay, data not shown). As demonstrated in Fig. 2b and c, overexpression of FoxA1 led to a decreased expression of UCP2 mRNA and protein. The basal level of UCP2 mRNA and protein after FoxA1 overexpression was decreased. H2O2 stimulation significantly increased the expression of UCP2, which was inhibited by FoxA1 overexpression.
Fig. 2.
The effect of FoxA1 on the expression of UCP2 in A549 cells. a A549 cells were transfected with pcDNA3.1-FoxA1 and the expression levels of FoxA1 were identified by Western blot analysis. b The effect of FoxA1 overexpression on UCP2 in A549 cells was determined by RT-PCR and Western blot analysis (c). Neo (N) the vector control group; pcDNA3.1-FoxA1 (F) FoxA1 overexpression group. *P < 0.05, statistically significant difference from 0 h. # P < 0.05, statistically significant difference from relevant Neo group. d A549 cells were transfected with short interference (si)RNAs against human FoxA1, and expressions of FoxA1 in response to H2O2 were detected by RT-PCR and Western blot analysis (e) for identification of basal FoxA1 inhibition. *P < 0.05, statistically significant difference from control group (Ctrl). f Effect of FoxA1 inhibition on the levels of UCP2 was measured by RT-PCR and western blot analysis (g), respectively. *P < 0.05, statistically significant difference from 0 h. # P < 0.05, statistically significant difference from the control group (Ctrl). The relative values of all results were determined and expressed as mean ± SEM of three experiments in duplicate. Cells were treated with H2O2 (0.5 mM) for indicated durations
In order to observe the effect of FoxA1 inhibition on the expression of UCP2, we transfected (si)RNAs against human FoxA1 into A549. RT-PCR and Western blot were used to assess basal and H2O2-induced FoxA1 expression (Figs. 2d, e). Following the inhibition of basal FoxA1 expression, the expression of UCP2 was determined by RT-PCR and Western blot. As shown by Fig. 2f and g, after FoxA1 inhibition, the basal and H2O2-induced (0.5 mM) expression of UCP2 increased compared to that of the control group.
FoxA1 regulates UCP2 promoter in A549
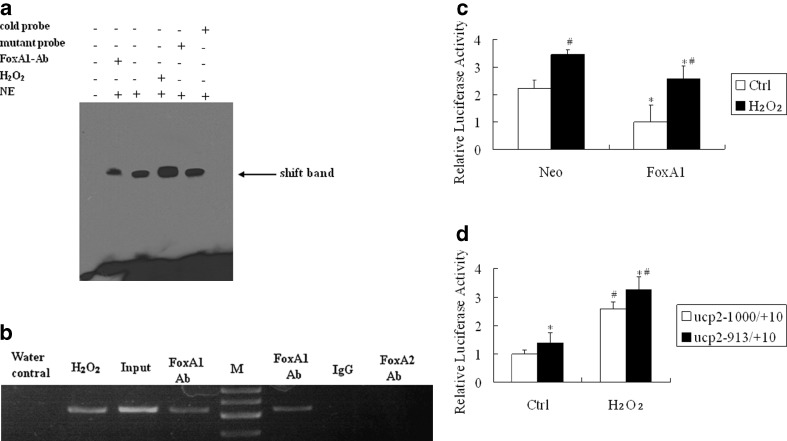
To determine whether the potential FoxA1 binding sites on the UCP2 promoter are capable of binding FoxA1, we performed electrophoretic mobility shift assays (EMSAs). Figure 3a shows that the biotin-labeled probe designed according to the UCP2 promoter (site at −919 to −913 bp) can bind to the FoxA1 protein in the nuclear extract of A549, and H2O2 stimulation induced the further binding of DNA to FoxA1. Specificity was verified using mutant oligonucleotides, which failed to compete for binding with FoxA1, and by supershift studies with FoxA1 antibody. The site at −835 to −829 bp had no binding activity with FoxA1 protein in either normal or H2O2 stimulation condition (data not shown).
Fig. 3.
DNA binding activity and transcription activity of FoxA1 to the FoxA1 binding element of UCP2 promoter in A549 cells. a FoxA1 bound to the FoxA1 binding element in the region −919 to −913 bp in the UCP2 promoter, as shown by EMSA. Cold probe Competition with cold probe (200-fold excess concentration); mutant probe competition with mutant cold probe (200-fold excess concentration); FoxA1 Ab supershift group by Foxa1 antibody; H 2 O 2 cells were stimulated by H2O2 (0.5 mM) for 30 min; NE nuclear extract. b Association of FoxA1 with UCP2 promoter shown by ChIP. Chromatin was extracted, and binding of FoxA1 to the UCP2 promoter was analyzed by ChIP using a pair of primers that contained the FoxA1 binding site at −919 to −913 bp, which specifically targeted the human UCP2 proximal promoter region. The cross-linked protein–DNA complexes were immunoprecipitated with the anti-FoxA1 antibody (lanes 2, 4, and 6), or with a purified rabbit IgG as a negative control (lane 7), or with the anti-FoxA2 antibody as a specific control (lane 8). PCR of the input (sample representing PCR amplification from a 1:25 dilution of total input chromatin from the ChIP experiment) is shown in lane 3. The PCR control represents the PCR amplification in the absence of DNA (lane 1). Lane M marker; lane Water control negative control; lane H 2 O 2 H2O2 treatment (0.5 mM for 30 min) plus FoxA1 antibody; lane Input positive control; lane FoxA1 Ab untreated cells plus FoxA1 antibody; lane IgG control negative control for FoxA1 antibody; lane FoxA2 Ab untreated cells plus FoxA2 antibody. The image is representative of three independent experiments. c A549 cells were transiently co-transfected with an expression plasmid of FoxA1 (500 ng) and a reporter driven by UCP2 promoter (500 ng). All transfections were performed at least three times in triplicate. *P < 0.05, statistically significant difference versus the vector control group (Neo). # P < 0.05, statistically significant difference from the control group (Ctrl). d Transient co-transfection studies were performed in A549 cells using full-length FoxA1 and a reporter driven by each of the truncate UCP2 promoter (500 ng). *P < 0.05, statistically significant difference from the UCP2 –1,000/+10 group. #P < 0.05, statistically significant difference from the control group (Ctrl). Ctrl cells were untreated with H2O; H 2 O 2 cells were treated with H2O2 for 30 min; Neo vector control group; FoxA1 full-length FoxA1 group
To investigate the endogenous relevance of FoxA1 with UCP2 promoter, a ChIP assay was used to determine whether FoxA1 can bind to the UCP2 promoter. Figure 3b showed the PCR product after immunoprecipitation of the cross-linked chromatin with the FoxA1 antibody. As a specific control, purified rabbit IgG performed in parallel did not yield detectable PCR product. Input DNA, also obtained from cross-linked chromatin, served as a positive control for PCR effectiveness. Collectively, these data agree that FoxA1 binds to the UCP2 promoter and H2O2 treatment increased FoxA1 binding to the UCP2 promoter.
In order to understand how FoxA1 can regulate UCP2, we assessed its effect on UCP2 promoter activity. A strong trans-inhibition effect of FoxA1 on the UCP2 promoter both on a basic and H2O2-stimulated situation is shown in Fig. 3c. To identify the FoxA1 binding region in the UCP2 promoter, the promoter activity of −913/+10 was examined. Cotransfection of pcDNA3.1-FoxA1 with the UCP2 −913/+10 region revealed that the promoter activity of −913/+10 regions was higher than that of −1,000/+10 regions both on basic and H2O2-stimulated situation, indicating that the −919 to −913 bp region contained the potential FoxA1 binding sites (Fig. 3d).
Discussion
FoxA1 is a winged-helix transcription factor and has been widely investigated in both normal development and carcinogenesis. High expression of FoxA1 has been reported in various tumors, including lung, esophageal, breast cancer, bladder cancer, and prostate cancer. In bladder cancer, DeGraff et al. (2012) reported that knockdown of FoxA1 resulted in significantly increased cell proliferation, while overexpression of FoxA1 significantly decreased cell growth and invasion in RT4 bladder cancer cells. Study has shown that FoxA1 plays an important role as a lineage-specific oncogene in proliferation of cancer cells derived from mammary luminal cells (Yamaguchi et al. 2008). It has also been indicated that FoxA1 silencing increases migration and invasion of luminal cancer cells, which suggests that this protein could be a novel, potential prognostic factor in breast cancer (Bernardo et al. 2013; Wolf et al. 2007) Studies (Abe et al. 2012; Mehta et al. 2012) also showed that FoxA1 functions as a tumor suppressor in endometrial cancer through modulation of proliferation and migration of endometrial cancer cells. Lin et al. (2002) and Deutsch et al.(2012) reported that FoxA1 is amplified and overexpressed in lung adenocarcinomas, which may suggest a potential oncogenic role for FoxA1 in tumorigenesis. Indeed, our previous study (Ricquier and Bouillaud 2000) indicated that FoxA1 expression was upregulated in H2O2-induced apoptosis of A549 and that FoxA1 could induce A549 apoptosis, which suggests FoxA1 might have a role of in apoptosis in lung cancer. However, the expression of FoxA1 and its function in lung cancer remains poorly understood.
Abundant evidence has also shown that overexpression of UCP2 plays an antiapoptotic role by modulating the generation of intracellular ROS. Sanming et al. (Deng et al. 2012) reported that UCP2 inhibits ROS-mediated apoptosis in A549 under hypoxic conditions. Our results also show that the expression of UCP2 resulted in a dose- and time-dependent increase under H2O2-stimulated conditions in A549 cells. The increased levels of FoxA1 and UCP2 suggested a potential relationship between the genes that related to the H2O2-induced A549 cell apoptosis. In further experiments, we found a basal decrease in UCP2 upon FoxA1 overexpression, and the subsequent expression changes of UCP2 induced by H2O2 after FoxA1 overexpression or inhibition. In response to H2O2 (0.5 mM) treatment, the upregulation of UCP2 was suppressed in FoxA1-overexpressed group. After FoxA1 inhibition, the H2O2-induced UCP2 expression increased compared to the control group. The results reveal that FoxA1 might induce A549 cell apoptosis by decreasing UCP2 expression.
As a transcriptional factor, many target genes of FoxA1 have been demonstrated, including bcl2 and HSP72 (Song et al. 2009a, b). FoxA1 regulates its target genes by binding to the potential FoxA1 binding elements in their promoters. We have shown that FoxA1 decreased transcription of the UCP2 gene by binding to the −919 to −913 binding element. Our studies provide further elucidation of the molecular mechanism by which FoxA1 exerts its regulatory effects on the expression of UCP2. H2O2 and FoxA1 overexpression promoted the binding of FoxA1 to its binding site in the UCP2 promoter.
Recent findings have shown that expression of UCP2 is associated with various human cancers, and inhibition of UCP2 by FoxA1 may have important implications for various cancers. There is abundant evidence showing that UCP2 might be important for tumor growth, invasiveness, metastasis, and resistance to chemotherapy. For example, in HCT116, HT29, and HepG2, among other cell types, overexpression of UCP2 plays an antiapoptotic role by modulating the generation of intracellular ROS after tumor cells are exposed to chemotherapeutic agents (Derdak et al. 2008; Santandreu et al. 2010; Mailloux et al. 2010). Most studies point to UCP2 as being associated with the development of colon and breast cancer. Studies have demonstrated that the levels of both UCP2 mRNA and protein were higher in colon cancer tissue samples, and it may be involved in colon cancer metastasis (Horimoto et al. 2004; Kuai et al. 2010). Sayeed et al.(2010) and Won and Kim (2010) independently reported a significant association of UCP2 with tumor grade in primary breast cancer. Wang et al. (2010) demonstrated that cell transformation was suppressed when UCP2 was downregulated using a siRNA approach, suggesting that UCP2 may serve as a tumor promoter during early tumorigenesis.
In light of the important roles of FoxA1 and UCP2 in cell apoptosis and in many human cancers, we predict that FoxA1 is a regulator of human cancers by downregulating the expression of UCP2, although our data have only shown the regulation of FoxA1 on the expression of UCP2 in the A549 human lung cancer cell line. In summary, our study demonstrates that FoxA1 represses UCP2 expression in A549 cells by interaction with the FoxA1 element in the UCP2 promoter. It is conceivable that FoxA1 and UCP2 may participate in modulating tumor progression. In order to understand the exact functions of these genes, further investigations are needed.
Acknowledgments
This work was support by funding from the Nature Science Foundation of China (81000026 and 81202451), the National Science Foundation for post-doctoral Scientists of Cnina (2012M511731), the Research Fund for the Doctoral program of Higher Education of China (20114323120002), and the National Nature Science Foundation of Hunan, China (12JJ4029).
Footnotes
L. Song and Z. Xu contributed equally to this study.
References
- Abe Y, Ijichi N, Ikeda K, Kayano H, Horie-Inoue K, Takeda S, Inoue S. Forkhead box transcription factor, forkhead box A1, shows negative association with lymph node status in endometrial cancer, and represses cell proliferation and migration of endometrial cancer cells. Cancer Sci. 2012;103(4):806–812. doi: 10.1111/j.1349-7006.2012.02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo GM, Bebek G, Ginther CL, Sizemore ST, Lozada KL, Miedler JD, Anderson LA, Godwin AK, Abdul-Karim FW, Slamon DJ, Keri RA. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32(5):554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGraff DJCP, Cates JM, Yamashita H, Robinson VL, Yu X, Smolkin ME, Chang SS, Cookson MS, Herrick MK, Shariat SF, Steinberg GD, Frierson HF, Wu XR, Theodorescu D, Matusik RJ. Loss of the urothelial differentiation marker FOXA1 is associated with high grade, late stage bladder cancer and increased tumor proliferation. PLoS One. 2012;7(5):e36669. doi: 10.1371/journal.pone.0036669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Yang Y, Han Y, Li X, Wang X, Li X, Zhang Z, Wang Y. UCP2 inhibits ROS-mediated apoptosis in A549 under hypoxic conditions. PLoS One. 2012;7(1):e30714. doi: 10.1371/journal.pone.0030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdak Z, Mark NM, Beldi G, Robson SC, Wands JR, Baffy G. The mitochondrial uncoupling protein-2 promotes chemoresistance in cancer cells. Cancer Res. 2008;68(8):2813–2819. doi: 10.1158/0008-5472.CAN-08-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch L, Wrage M, Koops S, Glatzel M, Uzunoglu FG, Kutup A, Hinsch A, Sauter G, Izbicki JR, Pantel K, Wikman H. Opposite roles of FOXA1 and NKX2-1 in lung cancer progression. Genes, chromosomes & cancer. 2012;51(6):618–629. doi: 10.1002/gcc.21950. [DOI] [PubMed] [Google Scholar]
- Goglia F, Skulachev VP. A function for novel uncoupling proteins: antioxidant defense of mitochondrial matrix by translocating fatty acid peroxides from the inner to the outer membrane leaflet. FASEB J. 2003;17(12):1585–1591. doi: 10.1096/fj.03-0159hyp. [DOI] [PubMed] [Google Scholar]
- Horimoto M, Resnick MB, Konkin TA, Routhier J, Wands JR, Baffy G. Expression of uncoupling protein-2 in human colon cancer. Clin Cancer Res. 2004;10(18 Pt 1):6203–6207. doi: 10.1158/1078-0432.CCR-04-0419. [DOI] [PubMed] [Google Scholar]
- Kuai XY, Ji ZY, Zhang HJ. Mitochondrial uncoupling protein 2 expression in colon cancer and its clinical significance. World J Gastroenterol. 2010;16(45):5773–5778. doi: 10.3748/wjg.v16.i45.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, Glover TW, Beer DG. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62(18):5273–5279. [PubMed] [Google Scholar]
- Mailloux RJ, Adjeitey CN, Harper ME. Genipin-induced inhibition of uncoupling protein-2 sensitizes drug-resistant cancer cells to cytotoxic agents. PLoS One. 2010;5(10):e13289. doi: 10.1371/journal.pone.0013289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RJJR, Leung S, Choo J, Nielsen T, Huntsman D, Nakshatri H, Badve S. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res Treat. 2012;131(3):881–890. doi: 10.1007/s10549-011-1482-6. [DOI] [PubMed] [Google Scholar]
- Owada S, Shimoda Y, Tsuchihara K, Esumi H. Critical role of H2O2 generated by NOX4 during cellular response under glucose deprivation. PLoS One. 2013;8(3):e56628. doi: 10.1371/journal.pone.0056628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricquier D, Bouillaud F. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem J. 2000;345(Pt 2):161–179. doi: 10.1042/0264-6021:3450161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudio I, Fiegl M, Andreeff M. Mitochondrial uncoupling and the Warburg effect: molecular basis for the reprogramming of cancer cell metabolism. Cancer Res. 2009;69(6):2163–2166. doi: 10.1158/0008-5472.CAN-08-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santandreu FM, Roca P, Oliver J. Uncoupling protein-2 knockdown mediates the cytotoxic effects of cisplatin. Free Radic Biol Med. 2010;49(4):658–666. doi: 10.1016/j.freeradbiomed.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Sayeed AMZ, Luciani G, Chen LC, Bennington JL, Dairkee SH. Negative regulation of UCP2 by TGFβ signaling characterizes low and intermediate-grade primary breast cancer. Cell Death Dis. 2010;2010(1):e53. doi: 10.1038/cddis.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Wei X, Zhang B, Luo X, Liu J, Feng Y, Xiao X. Role of Foxa1 in regulation of bcl2 expression during oxidative-stress-induced apoptosis in A549 type II pneumocytes. Cell stress & chaperones. 2009;14(4):417–425. doi: 10.1007/s12192-008-0095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Xu Z, Zhang C, Qiao X, Huang C. Up-regulation of the HSP72 by Foxa1 in MCF-7 human breast cancer cell line. Biochem Biophys Res Commun. 2009;386(1):30–34. doi: 10.1016/j.bbrc.2009.05.120. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang B, Feng Y, Luo X, Wei X, Xiao X. A role for forkhead box A1 in acute lung injury. Inflammation. 2009;32(5):322–332. doi: 10.1007/s10753-009-9139-x. [DOI] [PubMed] [Google Scholar]
- Szuro-Sudol A, Nathan CF. Suppression of macrophage oxidative metabolism by products of malignant and nonmalignant cells. J Exp Med. 1982;156(4):945–961. doi: 10.1084/jem.156.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne KJ, Svvennsen RJ, Franks D. Role of hydrogen peroxide in the cytotoxic reaction of T lymphocytes. Clin Exp Immunol. 1980;39(2):486–495. [PMC free article] [PubMed] [Google Scholar]
- Wang F, Fu X, Chen X, Chen X, Zhao Y. Mitochondrial uncoupling inhibits p53 mitochondrial translocation in TPA-challenged skin epidermal JB6 cells. PLoS One. 2010;5(10):e13459. doi: 10.1371/journal.pone.0013459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP. FOXA1: Growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer. 2007;120(5):1013–1022. doi: 10.1002/ijc.22389. [DOI] [PubMed] [Google Scholar]
- Won KYKG, Kim YW. Uncoupling Protein 2 (UCP2) and p53 expression in invasive ductal carcinoma of breast. Korean J Pathol. 2010;44:565–570. doi: 10.4132/KoreanJPathol.2010.44.6.565. [DOI] [Google Scholar]
- Yamaguchi N, Ito E, Azuma S, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Tatsuta K, Inoue J, Semba K, Watanabe S (2008) FoxA1 as a lineage-specific oncogene in luminal type breast cancer. Biochem Biophys Res Commun 365(4):711–717 [DOI] [PubMed]