Abstract
Heat shock proteins (HSPs) are molecular chaperones and have an important role in the refolding and degradation of misfolded proteins, and these functions are related to aging. Rotifer is a useful model organism in aging research, owing to small body size (0.1–1 mm), short lifespan (6–14 days), and senescence phenotypes that can be measured relatively easily. Therefore, we used rotifer as a model to determine the role of four typical hsp genes on the aging process in order to provide a better understanding of rotifer aging. We cloned cDNA encoding hsp genes (hsp40, hsp60, hsp70, and hsp90) from the rotifer Brachionus calyciflorus Pallas, analyzed their molecular characteristics, determined its modulatory response under different temperatures and H2O2 concentrations and investigated the changes in expression of these genes during the aging process. We found that Bchsp70 mRNA expression significantly decreased with aging. In addition, we also studied the effects of dietary restriction (DR) and vitamin E on rotifer lifespan and reproduction and analyzed the changes in expression of these four Bchsp genes in rotifers treated with DR and vitamin E. The results showed that DR extended the lifespan of rotifers and reduced their fecundity, whereas vitamin E had no significant effect on rotifer lifespan or reproduction. Real-time PCR indicated that DR increased the expression of these four Bchsps. However, vitamin E only improved the expression of Bchsp60, and reduced the expression of Bchsp40, Bchsp70, and Bchsp90. DR pretreatment also increased rotifer survival rate under paraquat-induced oxidative stress. These results indicated that hsp genes had an important role in the anti-aging process.
Keywords: Aging, Brachionus calyciflorus, Dietary restriction, Heat shock proteins, Paraquat, Vitamin E
Introduction
Heat shock proteins (HSPs) are a highly conserved, ubiquitously expressed family of stress-response proteins (Hartl 1996) and are classified into six subfamilies according to their molecular weight, such as small HSPs (16–30 kDa), HSP40, HSP60, HSP70, HSP90, and the large HSP110 (Becker and CRAIG 1994; Kalmar and Greensmith 2009). HSPs can act as molecular chaperones facilitating protein folding, preventing protein aggregation, and targeting improperly folded proteins to specific degradation pathways (Kalmar and Greensmith 2009), and most of these functions are related to aging. Reactive oxygen species (ROS), such as hydrogen peroxide (H2O2) and superoxide anion (O2−), which are mainly by-products of aerobic respiration, can damage many cell macromolecules, including lipids, nucleic acids, and proteins (Finkel and Holbrook 2000; Kalmar and Greensmith 2009). The proteins damaged by ROS can easily lose their native folded conformation to become misfolded leading to functional abnormalities (Kalmar and Greensmith 2009). Some HSPs facilitate the refolding of damaged proteins and improve their impaired functions (Heikkila 2010). If the damaged proteins cannot recover, some HSPs, mainly members of the HSP70 family and its co-chaperones, play a crucial role in protein sorting and quality control by selecting and directing aberrant proteins to proteasomes or lysosomes for degradation (Mayer and Bukau 2005). Thus, HSPs regulate the quality of proteins and have an important role in the aging process. Indeed, hsp70 induced by moderate, nonlethal heat treatment is a candidate protein for longevity in the yeast Saccharomyces cerevisiae (Shama et al. 1998). More directly, the life expectancy of hsp70 transgenic Drosophila melanogaster increased by 30 % compared with the wild type when its gene expression was induced by heat treatment (Tatar et al. 1997). However, changes in hsps gene expression in rotifer aging process is very rarely reported. The rotifer, Brachionus calyciflorus, has a short lifespan (5–7 days), and it is easy to detect the changes in gene expression during the aging process. Therefore, we analyzed the changes in hsps gene expression in B. calyciflorus at different ages to explore the role of hsps genes in aging.
Dietary restriction (DR) is usually defined as a moderate (normally 20–40 %) reduction in available nutrients without causing malnutrition. It was first noted in the 1930s that food restriction significantly extended the lifespan of rodents (McCay et al. 1935). In the laboratory, DR is one of the most common lifespan-extending methods in evolutionarily distinct eukaryotes from single cell to multi-cellular organisms (Fanestil and Barrows 1965; Klass 1977; Lin et al. 2002; Lints 1985; Walker et al. 2005). It was reported by Kaneko et al. that the mRNA levels of hsp70 and glucose-regulated protein 94 (GRP94) were significantly higher in the exponential growth phase than in the stationary phase (Kaneko et al. 2002) and that rotifers in the stationary phase lived longer than those in the exponential phase (Yoshinaga et al. 2000). Nutritional deprivation also potentiates HSP60 protein induction (Wheelock et al. 2002). Therefore, we also analyzed the effects of DR on the expression of hsps genes to explore the reasons why DR extends life.
Vitamin E is an antioxidant and acts as a H2O2 radical scavenger, preventing the propagation of ROS in tissues and can affect lifespan (Traber and Stevens 2011). Previous studies have shown that vitamin E extended the lifespan of several types of rotifers (Litton 1987; Sawada and Enesco 1984; Snell et al. 2012). Thus, we hypothesized that the antioxidant effects produced by vitamin E may affect the expression of hsp genes.
The aims of this study were to: (1) clone the hsp40, hsp60, hsp70, and hsp90 cDNAs in the freshwater rotifer B. calyciflorus; (2) detect the expression of these four hsps under temperature stress and oxidative stress (produced by H2O2) conditions; (3) examine the mRNA expression levels of these four hsps during the aging process using highly sensitive quantitative real-time PCR; (4) investigate the effect of DR and vitamin E on the expression of the four hsps. These studies were undertaken to provide a better understanding of the role of hsps in rotifer aging.
Materials and methods
Rotifers
The animals used during the study, B. calyciflorus, were female neonates (0–2 h old) hatched from resting eggs in artificial freshwater medium (EPA medium, pH ∼7.8) composed of 96 mg NaHCO3, 60 mg CaSO4 · H20, 123 mg MgSO4, and 4 mg KCl in 1 L deionized water at 25 °C (ASTM 2001). This species was originally collected in Gainesville, Florida in 1983 (Snell et al. 1991) and since then, has been cultured in the laboratory continuously with periodic collection and storage of resting eggs. The females were fed by suspending the green alga Chlorella pyrenoidosa (Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China) in the culture media at ∼3 × 106 cells/mL. The feeding alga was cultured in Bold’s basal medium at 25 °C in 3-L flasks under constant fluorescent illumination at 2,000 lux.
Bchsps cloning
To clone the hsps from B. calyciflorus Pallas, the degenerate primers were designed with reference to DNA nucleotide sequences of the hsps gene reported for the rotifer Brachionus plicatilis, Brachionus ibericus, Brachionus manjavacas, the yeast S. cerevisiae, the nematode Caenorhabditis elegans, and humans.
The extraction of total RNAs from rotifer was conducted using the Trizol (Invitrogen, China) technique following the manufacturer's instructions. The total RNA concentration was determined by measuring the absorbance at OD260. RNA integrity was checked by electrophoresis. The total RNA was reverse transcribed into cDNA using the aM-MLV RTase cDNA Synthesis Kit (TaKaRa, Japan).
Polymerase chain reaction (PCR) was conducted for 3 min at 94 °C followed by 35 cycles at 94 °C for 10 s, 48 °C for 40 s, and 72 °C for 30 s. The final extension step was performed at 72 °C for 10 min. Each of the 20 μL reaction mixtures contained 1.5 mM of MgCl2, 0.2 mM of dNTP, 0.2 mM of each of the primers, 1 U of Taq DNA polymerase (TaKaRa, Japan), and 5 ng of cDNA. Amplicons of expected sizes were purified using an Agarose Gel DNA Purification Kit (TaKaRa, Japan), and then subcloned into the pMD-19T cloning vector (TaKaRa, Japan). Positive clones containing inserts of an expected size were sequenced using M13 primers and sequenced at BGI (Shanghai, China). The Bchsps partial cDNA sequence was extended using the 5′ and 3′ rapid amplification of cDNA ends (RACE; SMART™ cDNA Kit) technique following the manufacturer's instructions. A total of five gene-specific primers were designed based on the Bchsps partial cDNA sequence.
Bchsp40, Bchsp60, Bchsp70, and Bchsp90 were cloned respectively via the procedure described above. The degenerate primers for PCR and the specific primers for RACE are shown in Table 1.
Table 1.
Degenerate and specific primers used in the experiment
| Primers | Sequence (5′–3′) | Application |
|---|---|---|
| Hsp40F | TCTAGTNTGCCAGGTGAGAT | Hsp40 amplification |
| Hsp40R | TTCTNTCATAAGCTGGATCC | Hsp40 amplification |
| Hsp40-5′1 | CACCCTTTCCACTACA | Hsp40 RACE |
| Hsp40-5′2 | CACCACCACCGCCGAAGA | Hsp40 RACE |
| Hsp40-5′3 | GAAAATATCCATTGGTGAACT | Hsp40 RACE |
| Hsp40-3′1 | TCACCAGGTCTAGAACCA | Hsp40 RACE |
| Hsp40-3′2 | TGAGGGTATGCCAATG | Hsp40 RACE |
| Hsp60F | TTYGAYCGHGGMTAYATYTCNCC | Hsp60 amplification |
| Hsp60R | TCHACYTCRCTKGADCCRCCDAC | Hsp60 amplification |
| Hsp60-5′1 | TTACGTTGAGCATTGG | Hsp60 RACE |
| Hsp60-5′2 | TGGCTAATTCTAAAGCTGGT | Hsp60 RACE |
| Hsp60-5′3 | CTCTGAGATTAAGACAAAAGC | Hsp60 RACE |
| Hsp60-3′1 | GGAGCTACTGTATTCGGCGATGAAGCT | Hsp60 RACE |
| Hsp60-3′2 | GGTGTAGCCGTTTTGAAAGTCGGTGGTT | Hsp60 RACE |
| Hsp70F | GTCGANCCGCCNACCANNACAAT | Hsp70 amplification |
| Hsp70R | TGGGNGGNGGNACNTTYGACGT | Hsp70 amplification |
| Hsp70-5′1 | TTCTGATGTCTTTACCA | Hsp70 RACE |
| Hsp70-5′2 | TGTTGTCAAAATCTTCACCACC | Hsp70 RACE |
| Hsp70-5′3 | CACCATTTGTGGCTACTACTTCA | Hsp70 RACE |
| Hsp70-3′1 | ATTTGGGTGGTGAAGATTTTGACAACAGAG | Hsp70 RACE |
| Hsp70-3′2 | AAACCAGTTCAAAAGGTCTTGGAAGATGCT | Hsp70 RACE |
| Hsp70-3′3 | TCCACCAGCCCCAAGAGGTGTACCAC | Hsp70 RACE |
| Hsp70-3′4 | CCGCTGAAGACAAAGGTACTGGTAACAAA | Hsp70 RACE |
| Hsp90F | TTKGTNGARTCYTCRTGDATNGC | Hsp90 amplification |
| Hsp90R | TCHGGMACBAARGCNTTYATGGA | Hsp90 amplification |
| Hsp90-5′1 | CAACCGTATTGTGAAG | Hsp90 RACE |
| Hsp90-5′2 | CCAAACGAGTTGAAACAGTGAC | Hsp90 RACE |
| Hsp90-5′3 | CGTATTTTTCAGCATCGGATTC | Hsp90 RACE |
| Hsp90-3′1 | GTCACTGTTTCAACTCGTTTGGTCTCATC | Hsp90 RACE |
| Hsp90-3′2 | CACAATACGGTTGGTCCGCCAATATGG | Hsp90 RACE |
| Hsp40RTF | AATGGATATTTTCGAGATG | Real-time PCR |
| Hsp40RTR | TGCCTTGACAAGTTTTAC | Real-time PCR |
| Hsp60RTF | CTACTGTATTCGGCGATGAA | Real-time PCR |
| Hsp60RTR | CGGCTACACCATTACTCAAT | Real-time PCR |
| Hsp70RTF | GCCACAAATGGTGATACTCAT | Real-time PCR |
| Hsp70RTR | TGGAAGATGCTGGTTTGA | Real-time PCR |
| Hsp90RTF | CTGCTTACTTAGTCGCTGAT | Real-time PCR |
| Hsp90RTR | CTTCTTCTTCTTGTCCTTATCG | Real-time PCR |
| β-actinRTF | GAAATTGTGCGCGACATCAAGGA | Real-time PCR |
| β-actinRTR | GCAATGCCCGGGTACATGGTGGT | Real-time PCR |
Sequence analysis of Bchsps
Nucleotide and deduced amino acid sequences of hsps cDNA were analyzed using the software Vector NTI Advance 11. HSPs sequences from different organisms were obtained using the NCBI BLAST search program. The cDNA sequence and deduced amino acid sequence from B. calyciflorus were analyzed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast). Translation and protein analyses were performed using ExPASy tools (http://us.expasy.org/tools/). A multiple sequence alignment was created with ClustalW (http://www.ebi.ac.uk/clustalw/). The calculated molecular mass and the theoretical isoelectric point were predicted using the Protein Mol Wt and AA Composition Calculator (http://www.proteomics.com.cn/proteomics/pi_tool.asp). An unrooted phylogenetic tree based on the amino acid sequence alignment was constructed using the Bayesian method. A total of 39 sequences were retrieved from the GenBank databases. Multiple sequence alignment results were first converted to Nexus format and analyzed using MrBayes v3.1.2 for Windows (Huelsenbeck and Ronquist 2001). Four parallel Monte Carlo Markov chains were run for 100,000 generations to approximate the posterior probabilities, and trees were sampled every 100 generations (Rhee et al. 2011). The analysis results were visualized using the Fig Tree v1.3.1 software.
Experimental design
The effects of temperature, vitamin E and H2O2 on hsps expression
Experiments were conducted to test the effects of temperature, H2O2, and vitamin E on the expression of hsps in B. calyciflorus. In the temperature test, 100 rotifers were cultured in 50 mL EPA medium at 15, 25 (control), 30, and 37 °C for 0, 0.5, 1, 2, 3, and 6 h, respectively. The rotifers were then collected and total RNA was extracted. Three replicates were prepared for each treatment. In the H2O2 test, 100 rotifers were placed into 50 mL H2O2 (H2O2 concentration was 0.1 and 0.2 mM) medium and cultured for 0, 1, 2, 4, 6, and 12 h, respectively. The rotifers were then collected and total RNA was extracted. Three replicates were prepared for each treatment. In the vitamin E test (vitamin E concentration was 40 μM), rotifers were treated using the same procedure as in the H2O2 test described above. Vitamin E was obtained from the Sigma Chemical Company (St. Louis, MO), dissolved in DMSO and stored at −20 °C.
Age-related changes in hsps gene expression
In total, 600 neonates (0–2 h old) were collected and divided into six groups of 100 individuals, which were then cultured in 50 mL of prepared media and maintained at 25 °C for 0.5, 1, 2, 3, 4, and 5 days. Three replicates were prepared for each treatment. Rotifers were transferred into new medium every 24 h and checked every 12 h to record the number of neonates, and the neonates were removed each day. Any surviving rotifers were then collected after 0.5, 1, 2, 3, 4, and 5 days. The total RNA was then extracted and reverse transcribed into cDNA. Real-time PCR (RT-PCR) was carried out to measure the mRNA expression levels of hsps at the different time points described above. The real-time PCR method was as described below.
The effects of DR on lifespan and reproduction
Six rotifers (0–2 h old) were transferred into 12-well culture plates, which contained 3 mL culture media under different feeding regimes (AL, DR12, DR24, and DR36). In the AL group, rotifers were transferred to a newly prepared culture medium containing microalgae every 12 h. In the DR group, rotifers were alternately placed in a culture medium with and without microalgae (alternated 12, 24, and 36 h in DR12, DR24, and DR36, respectively). Six replicates were prepared for each treatment. The rotifers were checked every 12 h to record the number of neonates and remove the neonates until all the experimental animals had died.
The effects of DR on hsps expression
Rotifers hatched from resting eggs were cultured using different feeding regimes (AL, DR12, DR24, and DR36) as previously mentioned. Each treatment contained 100 rotifers in 50 mL medium. Three replicates were prepared for each treatment. Rotifers were transferred into newly prepared culture media every 12 h and checked every 12 h to remove neonates, which were then collected and the total RNA extracted and reverse transcribed into cDNA after 3 days.
The effects of vitamin E on lifespan and reproduction
Six rotifers (0–2 h old) were transferred into 24-well culture plates, which contained 3 mL of test solution and C. pyrenoidosa at 3 × 106cells/mL. The concentration of vitamin E was 40 μM. Rotifers cultured in normal medium acted as controls. Six replicates were prepared for each treatment. The rotifers were checked every 12 h to record the number of neonates and remove the neonates until all the experimental animals had died.
Survival test
One hundred twenty individuals were collected and divided into three groups, AL, DR12, and DR24. The AL group was transferred to a newly prepared culture media containing the feeding alga every 12 h, whereas the DR12 and DR24 groups were alternately placed in a culture media with and without the feeding alga every 12 and 24 h, respectively. After 2 days, 30 rotifers were transferred from each of the AL, DR12, and DR24 groups to the culture medium containing paraquat at a concentration of 15.25 mg/L. The number of surviving rotifers was recorded every 12 h after exposure. Three replicates were prepared for each treatment. Paraquat (DMSO stock) was obtained from the Jiangsu Academy of Agricultural Sciences (Nanjing, China).
Quantitative analysis of hsps gene transcription
Total RNA was extracted using the procedures described above. Total RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Japan). Real-time PCR was performed on a Mastercyclereprealplex (Eppendorf, Germany) to study gene expression. The PCR reaction was performed in a 25 μL volume with a SYBR Premix Ex Taq™Kit (TaKaRa, Japan), 2 μM of each specific primer, and 1 μL of cDNA using the following procedure: initial denaturation at 95 °C for 2 min followed by 40 cycles of amplification (95 °C for 10 s and 55 °C for 30 s). Table 1 shows the primers used in quantitative real-time PCR. Β-actin was used as the internal control for quantitative real-time PCR. The relative expression levels of different genes were calculated using the 2−ΔΔCT method (Kenneth and Thomas 2001).
Statistical analysis
All the statistical analyses were carried out using the SPSS 16.0 analytic package. Results from the lifespan experiments were analyzed using survival analysis (Kaplan–Meier log-rank test); results from the DR and vitamin E reproduction experiments were analyzed using one-way analysis of variance (ANOVA), Duncan test, and Student’s t test, respectively. The mRNA expression level was determined by one-way ANOVA with the Tukey test. Differences were considered significant when P value was <0.05. All figures were produced using Sigma Plot version 11.0.
Results
Characterization of Bchsp40, Bchsp60, Bchsp70, and Bchsp90 genes in B. calyciflorus
Bchsp40
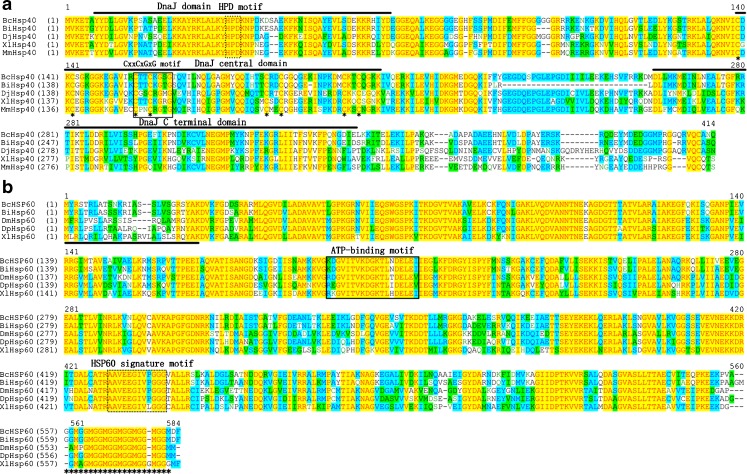
The full-length hsp40 cDNA of B. calyciflorus (Bchsp40) was 1,496 bp long, and contained 128 bp in the 5′ untranslated region (UTR), an open reading frame (ORF) of 1,212 bp that encoded 403 amino acids, and 156 bp in the 3′ UTR with a poly A signal (ATTAAA; data not shown). In a GenBank Blastx search, the cDNA sequence of BcHSP40 showed three DnaJ domains. The calculated molecular mass of the deduced mature BcHSP40 was 45.1005 kDa, and the theoretical isoelectric point was 7.09. The deduced amino acid sequence of Bchsp40 contained an N-terminal conserved domain (J domain, aa7–67), a central domain containing four highly conserved cysteine-rich repeats with a consensus sequence of CxxCxGxG where x is any amino acid (CRR domain, aa139–205), and a C-terminal domain (C domain, aa262–341) (Fig. 1a). The primary structure of BcHSP40 showed high levels of similarity to HSP40s in four other animals. It showed 80.15, 62.62, 60.11, and 58.66 % identity with orthologs of the marine rotifer B. plicatilis (GenBank accession number ADR79278), Mus musculus (NP_032324), Dugesia japonica (AES12470), and Xenopus laevis (NP_001079772), respectively. Furthermore, there were eight cysteine residues (cys-140, 142, 155, 158, 182, 185, 198, and 201) that are essential for BcHSP40 binding to zinc.
Fig. 1.
Alignment of deduced amino acid sequences of the selected HSPs. Identities are shaded yellow and similarities are shaded blue and green. Gaps (solid line) were introduced to maximize the alignment. a HSP40, three DnaJ domains are shown above the alignment. HPD motif is boxed with a dotted line. Eight cysteine residues that are essential for BcHSP40 binding to zinc are indicated with asterisks. b HSP60, the ATP-binding motif is boxed and the classical HSP60 signature motif is boxed with a dotted line. Underlined amino acids indicate the estimated transit peptides. The sequence of the GGM repeat motif conserved in mitochondrial HSP60 is indicated with asterisks. c HSP70, three HSP70 signatures and one ATP binding site are boxed. The signal peptide (position, 1–21) is underlined, and consensus signature of endoplasmic reticulum form is boxed with a dotted line. d HSP90, HSP90 family signature motifs and cytosolic signature of cytosolic form are boxed. A typical histidine kinase-like ATPases domain is boxed with a dotted line and the leucine zipper-like motifs are indicated by asterisk
Bchsp60
Full-length Bchsp60 cDNA consisted of 1940 nucleotides with a 1,740-bp ORF encoding 579 amino acids. The calculated molecular mass of the deduced mature BcHSP40 was 61.7469 kDa, and the theoretical isoelectric point was 6.66. The deduced sequence contained a presequence of 26 amino acids at the N terminus that is required for import into the mitochondria. BcHSP60 has a conserved ATP-binding/Mg2+ binding site, a hinge region, and stacking interaction sites(Marchler-Bauer et al. 2007) (Fig. 1b). BcHSP60 displayed very high homology with shrimp HSP60s from B. ibericus (ADR79280, 90.78 %), D. melanogaster (NP_511115, 70.06 %), Daphnia pulex (EFX84424, 72.09 %), and X. laevis (NP_001083970, 71.11 %).
Bchsp70
Full-length Bchsp70 cDNA consisted of 2,155 nucleotides with a 1,971-bp ORF encoding 656 amino acids. The calculated molecular mass of the deduced mature BcHSP70 was 72.3939 kDa, and the theoretical isoelectric point was 5.23. The deduced amino acid sequence of Bchsp70 also contained HSP70 family motifs and signatures, including an adenosine triphosphate/guanosine triphosphate (ATP/GTP)-binding site, signal peptide (position, 1–21), and a consensus signature of endoplasmic reticulum (ER) form KDEL motif (Fig. 1c). BcHSP70 displayed very high homology with shrimp HSP70s from Ascaris suum (ADY43887, 76.17 %), B. ibericus (ADR79282, 88.65 %), M. musculus (ABF20530, 78.10 %), and Xenopus tropicalis (XP_002941690, 78.82 %). The N-terminal two thirds of hsp70s were more conserved than the C-terminal third.
Bchsp90
The Bchsp90 cDNA contained a 2,166-bp ORF that encoded 721 amino acids with a calculated molecular weight of 82.7858 kDa and a theoretical isoelectric point of 5.07. The deduced amino acid sequence of Bchsp90 also contained HSP90 family motifs and signatures, including five signatures, a typical histidine kinase-like ATPase (HATPase) domain, and a conserved MEEVD motif (Caplan 1999; Gupta 1995). BcHSP90 exhibited high homology with HSP90s in other animals: B. ibericus (ADR79283, 93.48 %), Philodina roseola (ACC43981, 80.11 %), Harpegnathos saltator (EFN88374, 79.23 %), and Tigriopus japonicas (ACA03524, 77.32 %) (Fig. 1d). The Bchsp40, Bchsp60, Bchsp70, and Bchsp90 cDNA sequences and their deduced amino acid sequence were submitted to the NCBI GenBank under accession numbers KC176712, KC176713, KC176714, and KC176715, respectively.
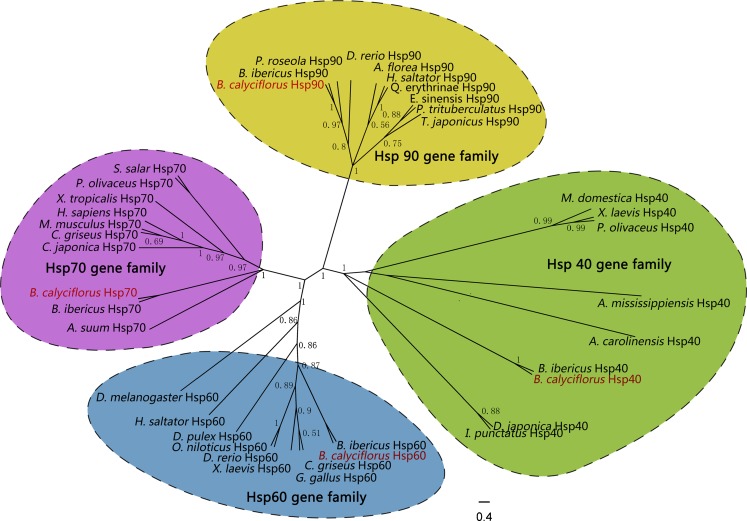
An unrooted phylogenetic tree based on the amino acid sequence alignment was constructed using the Bayesian method (Fig. 2). A total of 39 sequences were retrieved from the GenBank databases (Table 2). All HSPs can be classified into four distinct clusters: HSP40 family, HSP60 family, HSP70 family, and HSP90 family. HSP40, HSP60, HSP70, and HSP90 from B. calyciflorus clustered together with other HSP40s, HSP60s, HSP70s, and HSP90s as a subgroup, respectively.
Fig. 2.
Phylogenetic analysis of HSPs constructed by Bayesian method. The abbreviation of HSPs and the GenBank accession No. used to construct phylogenetic tree are given in Table 2. Numbers at branch nodes represent the confidence level of posterior probability
Table 2.
Species and GenBank accession numbers of HSPs sequences used for phylogenetic analysis
| Species | GenBank No. | Abbreviation type | Size (aa) |
|---|---|---|---|
| Alligator mississippiensis | BAF94139 | A. mississippiensis Hsp40 | 398 |
| Brachionus ibericus | ADR79278 | B. ibericus Hsp40 | 368 |
| Dugesia japonica | AES12470 | D. japonica Hsp40 | 411 |
| Anolis carolinensis | XP_003224682 | A. carolinensis Hsp40 | 396 |
| Ictalurus punctatus | NP_001187713 | I. punctatus Hsp40 | 408 |
| Monodelphis domestica | XP_001362945 | M. domestica Hsp40 | 423 |
| Xenopus laevis | NP_001079772 | X. laevis Hsp40 | 397 |
| Paralichthys olivaceus | ABA54277 | P. olivaceus Hsp40 | 395 |
| Brachionus calyciflorus | KC176712 | B. calyciflorus Hsp40 | 403 |
| B. ibericus | ADR79280 | B. ibericus Hsp60 | 581 |
| Cricetulus griseus | XP_003504389 | C. griseus Hsp60 | 573 |
| Danio rerio | NP_851847 | D. rerio Hsp60 | 575 |
| Daphnia pulex | EFX84424 | D. pulex Hsp60 | 576 |
| Drosophila melanogaster | NP_511115 | D. melanogaster Hsp60 | 573 |
| Gallus gallus | NP_001012934 | G. gallus Hsp60 | 573 |
| Harpegnathos saltator | EFN79769 | H. saltator Hsp60 | 608 |
| Oreochromis niloticus | XP_003456322 | O. niloticus Hsp60 | 575 |
| X. laevis | NP_001083970 | X. laevis Hsp60 | 579 |
| B. calyciflorus | KC176713 | B. calyciflorus Hsp60 | 579 |
| Ascaris suum | ADY43887 | A. suum Hsp70 | 655 |
| B. ibericus | ADR79282 | B. ibericus Hsp70 | 653 |
| Coturnix japonica | BAF38391 | C. japonica Hsp70 | 652 |
| C. griseus | NP_001233668 | C. griseus Hsp70 | 654 |
| Homo sapiens | NP_005338 | H. sapiens Hsp70 | 654 |
| Mus musculus | NP_071705 | M. musculus Hsp70 | 655 |
| P. olivaceus | ABG56392 | P. olivaceus Hsp70 | 654 |
| Salmo salar | NP_001135114 | S. salar Hsp70 | 657 |
| Xenopus tropicalis | XP_002941690 | X. tropicalis Hsp70 | 655 |
| B. calyciflorus | KC176714 | B. calyciflorus Hsp70 | 656 |
| B. calyciflorus | KC176715 | B. calyciflorus Hsp90 | 721 |
| Apis florea | XP_003694932 | A. florea Hsp90 | 717 |
| B. ibericus | ADR79283 | B. ibericus Hsp90 | 720 |
| D. rerio | AAC21566 | D. rerio Hsp90 | 724 |
| Eriocheir sinensis | ADE60732 | E. sinensis Hsp90 | 718 |
| H. saltator | EFN88374 | H. saltator Hsp90 | 723 |
| Philodina roseola | ACC43981 | P. roseola Hsp90 | 739 |
| Portunus trituberculatus | ACQ90225 | P. trituberculatus Hsp90 | 721 |
| Quadrastichus erythrinae | AFC76152 | Q. erythrinae Hsp90 | 721 |
| Tigriopus japonicus | ACA03524 | T. japonicus Hsp90 | 721 |
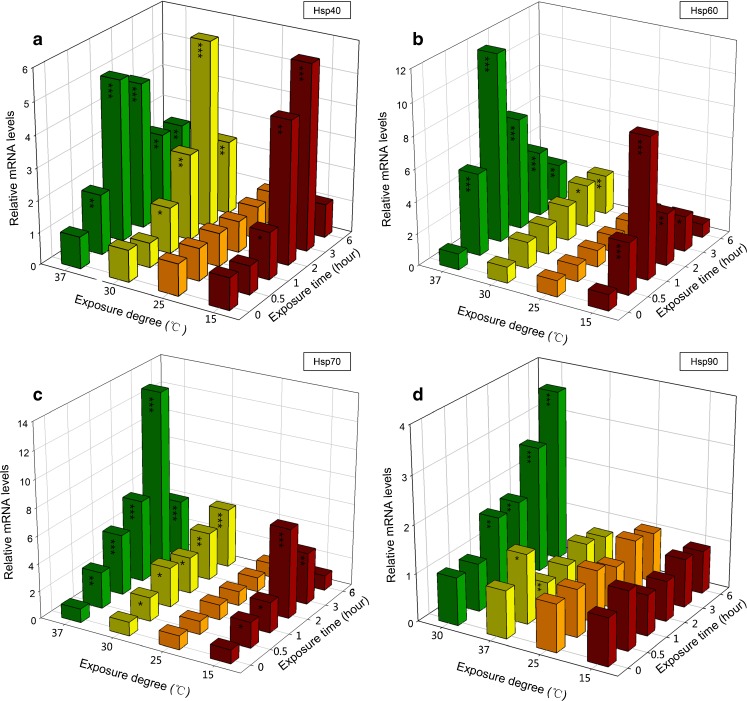
Time course of hsps expression following exposure to different temperatures
To study the modulatory effect of different temperatures on the expression of hsps, we measured expression at 15, 30, and 37 °C relative to the control (25 °C) over 6 h (Fig. 3). High temperature (37 °C) significantly increased the expression of Bchsp40, Bchsp60, and Bchsp70 at 0.5 h after exposure and showed a steady upregulation over time. However, the expression of Bchsp90 was temporarily increased after exposure to 37 °C, and then returned to normal levels (Fig. 3d). At 30 °C, the expression of Bchsp40 increased immediately (P < 0.05) (Fig. 3a). The expression of Bchsp90 increased when exposed to 30 °C for 1 h and showed a steady upregulation over time (Fig. 3d). The expression of Bchsp60 and Bchsp70 steadily increased after exposure to 30 °C. At low temperature (15 °C), the expression of Bchsp40, Bchsp60, and Bchsp70 increased significantly after 1 h of exposure, and the expression level returned to normal after 6 h of exposure. The expression of Bchsp90 was not significantly different after exposure to 15 °C (Fig. 3d).
Fig. 3.
Hsps mRNA expression of B. calyciflorus at different temperature exposures of 10, 30, and 37 °C with control (25 °C). The expression levels of hsps mRNA was statistically analyzed by one-way ANOVA with the Tukey test (*p < 0.05; **p < 0.01; ***p < 0.001): a hsp40, b hsp60, c hsp70, and d hsp90
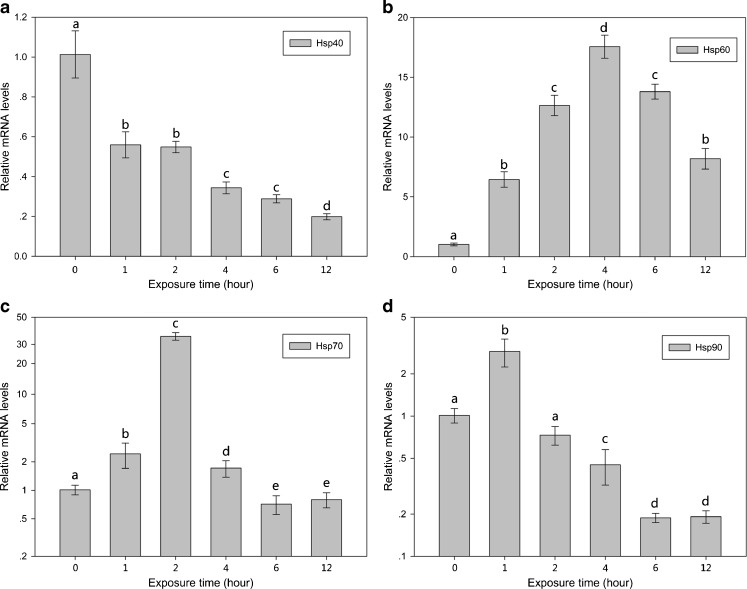
Effect of H2O2 on hsps mRNA expression
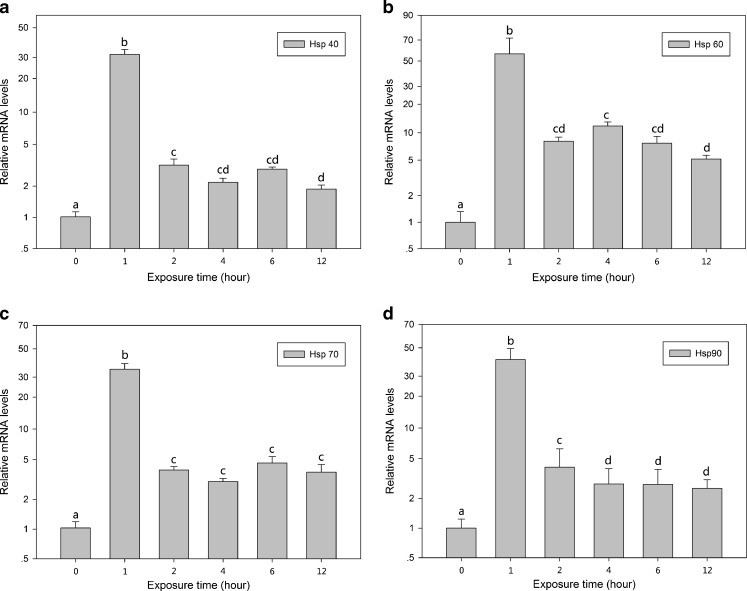
Figures 4 and 5 illustrate the effect of H2O2 on the expression of hsps mRNA. Following H2O2 (0.1 or 0.2 mM) exposure, the mRNA expression of the four hsps was upregulated to different degrees. In both the 0.1 and 0.2 mM groups, the expression of the four hsps significantly increased (30- to 90-fold) immediately after exposure to H2O2 for 1 h, and even though expression decreased slightly after 2 h, the mRNA expression levels were still significantly higher (5- to 10-fold) than the normal level (P < 0.05).
Fig. 4.
Relative mRNA levels of hsps of B. calyciflorus treated under H2O2 (0.1 mM) stress condition. The expression levels of hsps mRNA were statistically analyzed by one-way ANOVA with the Tukey test. The different letters indicate a significant difference for each time point with control (p < 0.05): a hsp40, b hsp60, c hsp70, and d hsp90
Fig. 5.
Relative mRNA levels of hsps of B. calyciflorus treated under H2O2 (0.2 mM) stress condition. The expression levels of hsps mRNA was statistically analyzed by one-way ANOVA with the Tukey test. The different letters indicate a significant difference for each time point with control (p < 0.05): a hsp40, b hsp60, c hsp70, and d hsp90
Effect of DR and vitamin E on rotifer lifespan and reproduction
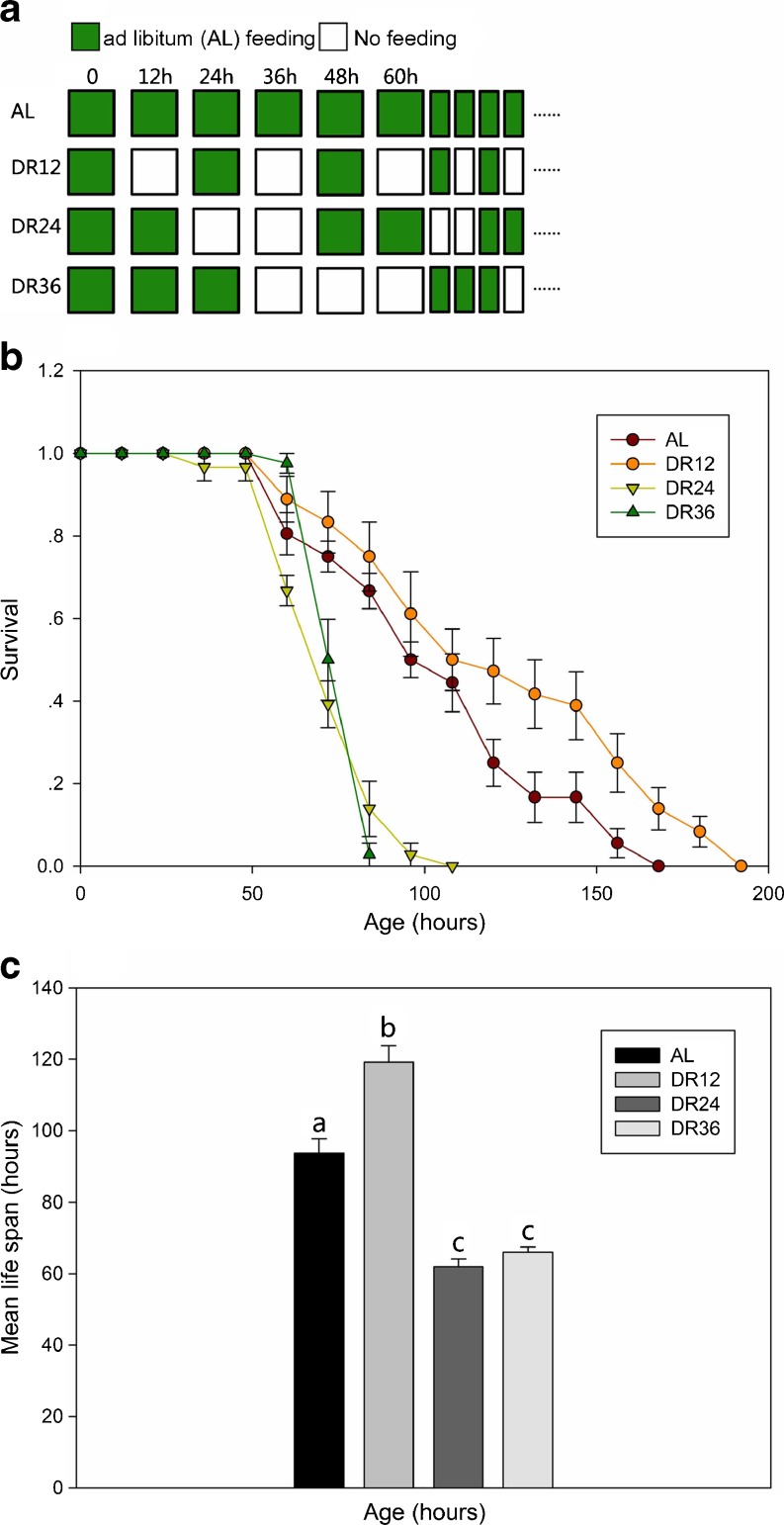
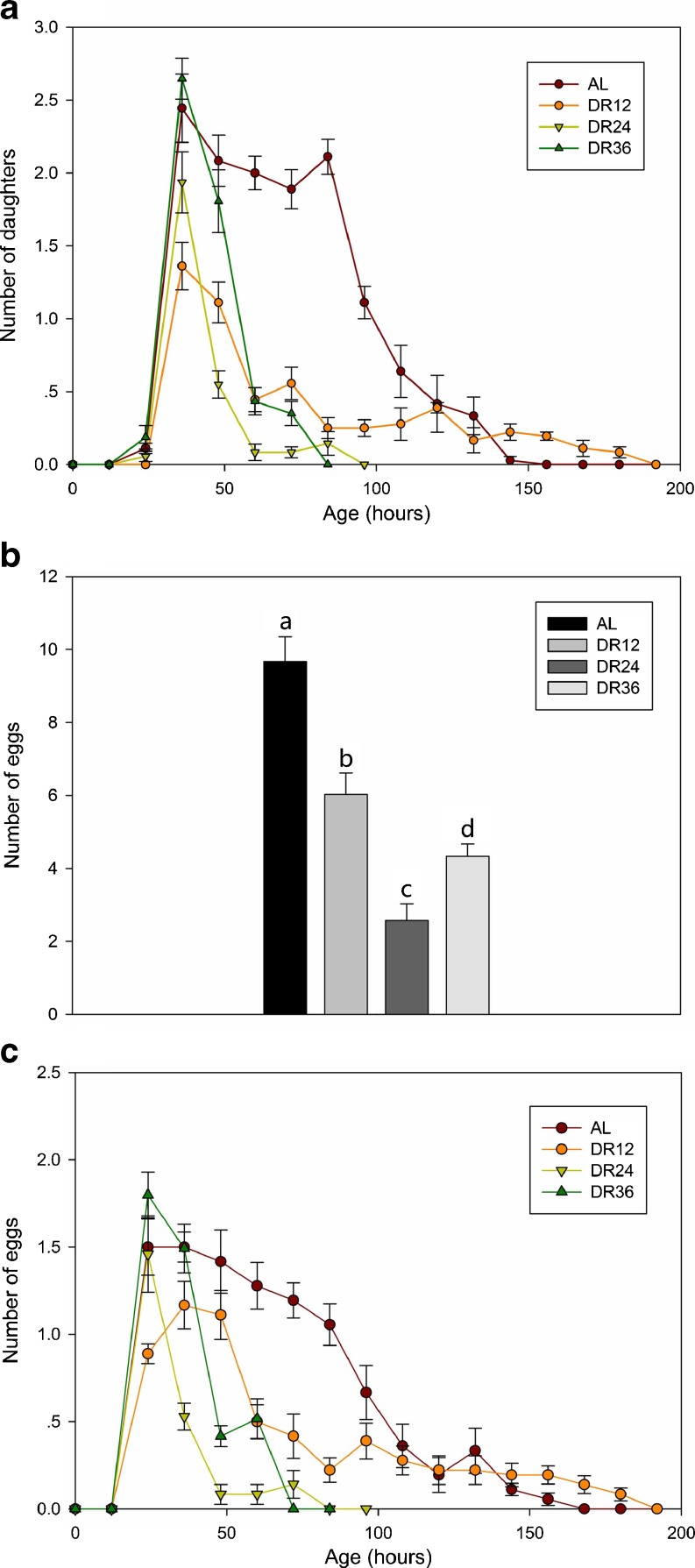
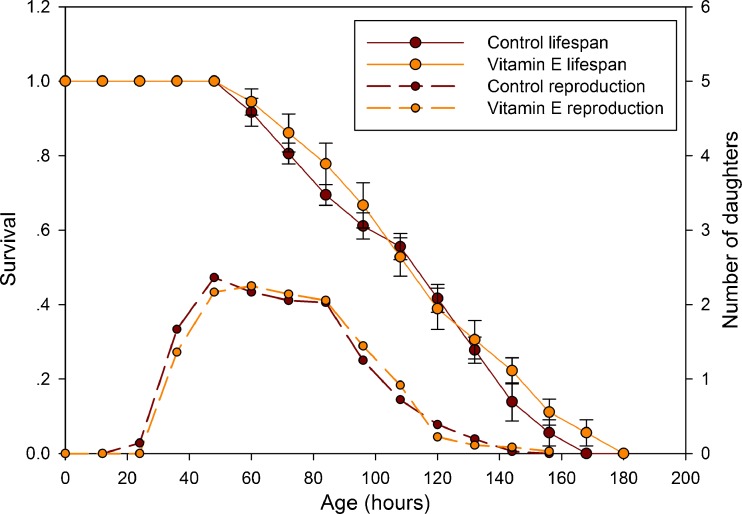
Figure 6 illustrates the effect of DR on the lifespan of B. calyciflorus. The average lifespan in the DR12 group was 120.4 ± 4.3 h, which was significantly longer (log-rank test, P < 0.0001) than that in the control group (95.0 ± 3.7 h) (Fig. 6c). However, the average lifespan in the DR24 and DR36 groups was 61.9 ± 2.3 and 66.0 ± 1.4 h, respectively and were significantly less (P < 0.05) than that in the control group (Fig. 6c). Figure 7a illustrates the fecundity of rotifers in the AL and DR groups at each time point. The average number of offspring produced per rotifer in the DR12, DR24 and DR36 groups was 5.9 ± 2.4, 2.9 ± 0.4, and 5.4 ± 0.3, respectively, significantly less (P < 0.05) than that in the control group 13.6 ± 5.5 (Fig. 7b). Figure 7c illustrates the number of egg carried by rotifers in the AL and DR groups at each time point. We also studied the effect of vitamin E on the lifespan and reproduction of B. calyciflorus and found that vitamin E had no effect on lifespan or reproduction (Fig. 8).
Fig. 6.
Effect of DR on rotifer lifespan. a Feeding regimes. b Survival rate of B. calyciflorus cultured in different feeding regimes. c Mean lifespan of B. calyciflorus cultured in different feeding regimes
Fig. 7.
Effect of DR on rotifer reproduction. a The fecundity of B. calyciflorus in the AL and DR groups at each time point. b Absolute fecundity B. calyciflorus cultured in different feeding regimes. c The number of egg carried by B. calyciflorus in the AL and DR groups at each time point
Fig. 8.
Lifespan and fecundity of B. calyciflorus cultured in vitamin E. y-axis (left) is the survival rate. y-axis (right) is the number of daughters. x-axis is survival (in hours)
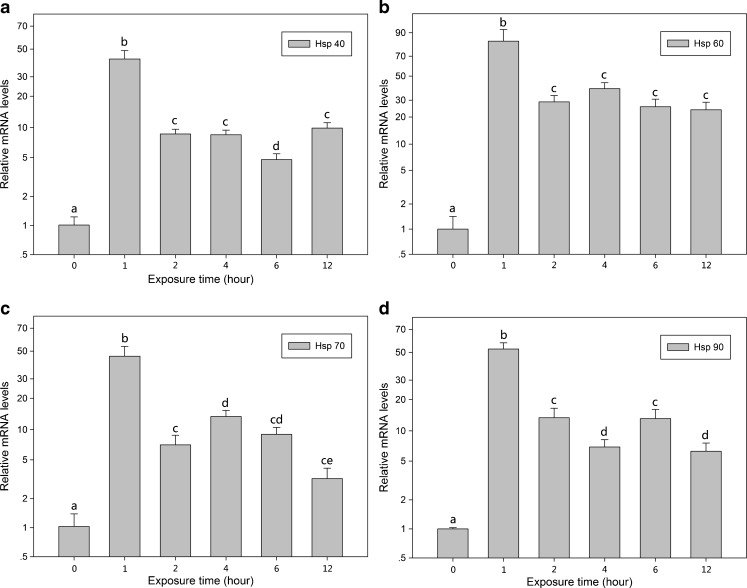
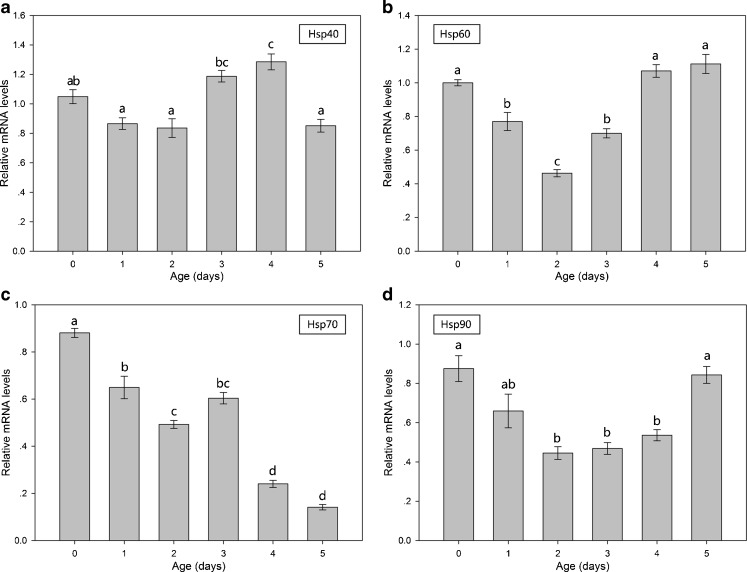
Age-related changes in hsps mRNA expression
Figure 9 shows Bchsps mRNA expression in B. calyciflorus at different ages. The expression level of Bchsp70 mRNA significantly decreased with age (P < 0.05) (Fig. 9c), whereas a small but significant (P < 0.05) increase in the mRNA levels of Bchsp40 was only observed in 4-day-old individuals (Fig. 9a). The expression of Bchsp60 mRNA significantly decreased (P < 0.05) in 1-, 2-, and 3-day-old individuals and significant decreases (P < 0.05) in the mRNA levels of Bchsp90 were observed in 2-, 3-, and 4-day-old individuals (Fig. 9b, d).
Fig. 9.
Hsps mRNA expression of B. calyciflorus of different age classes. The expression levels of hsps mRNA was statistically analyzed by one-way ANOVA with the Tukey test: a hsp40, b hsp60, c hsp70, and d hsp90
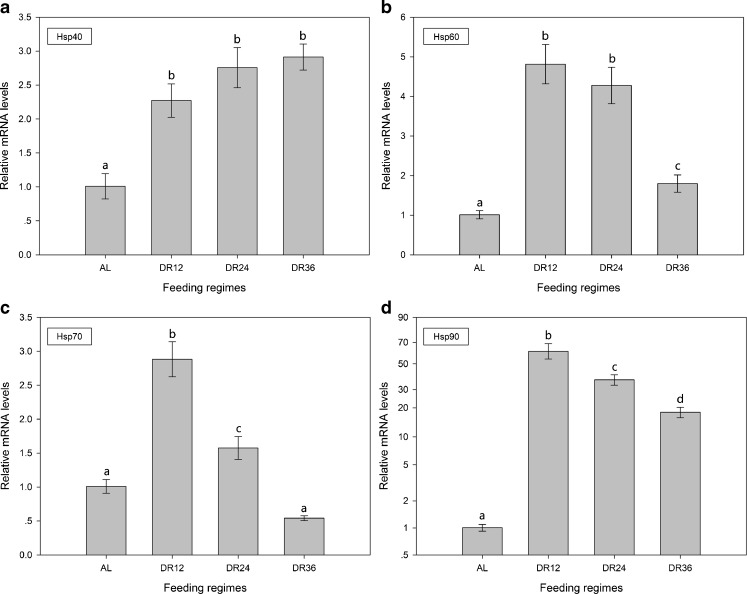
The effects DR on hsps mRNA expression
Qualitative real-time PCR showed that DR12 and DR24 treatment significantly improved (P < 0.05) the mRNA expression of Bchsp40, BcHsp60, BcHsp70, and BcHsp90 (Fig. 10), whereas DR36 treatment significantly improved (P < 0.05) the mRNA expression of Bchsp40, Bchsp60, and Bchsp90 but not Bchsp70 mRNA (P > 0.05) (Fig. 10c). The expression level of Bchsp60, Bchsp70, and Bchsp90 mRNA reached maximum levels in the DR12 group.
Fig. 10.
Hsps mRNA expression of B. calyciflorus of different feeding regimes. The expression levels of hsps mRNA was statistically analyzed by one-way ANOVA with the Tukey test. The different letters indicate a significant difference for each time point with control (p < 0.05): a hsp40, b hsp60, c hsp70, and d hsp90
Effect of vitamin E on hsps mRNA expression
Real-time quantitative RT-PCR demonstrated that vitamin E (40 μM) significantly decreased the expression of Bchsp40 (Fig. 11a). However, the expression of Bchsp70 and Bchsp90 was temporarily increased after exposure to vitamin E for 1 h, and after exposure for 4 h, the expression levels of Bchsp70 and Bchsp90 were significantly decreased compared with the control (Fig. 11c, d). The expression of Bchsp60 was significantly increased after exposure to vitamin E, and the expression level was maximal after 4 h of exposure (Fig. 11b).
Fig. 11.
Relative mRNA levels of hsps of B. calyciflorus treated with vitamin E (40 μM). The expression levels of hsps mRNA was statistically analyzed by one-way ANOVA with the Tukey test. The different letters indicate a significant difference for each time point with control (p < 0.05): a hsp40, b hsp60, c hsp70, and d hsp90
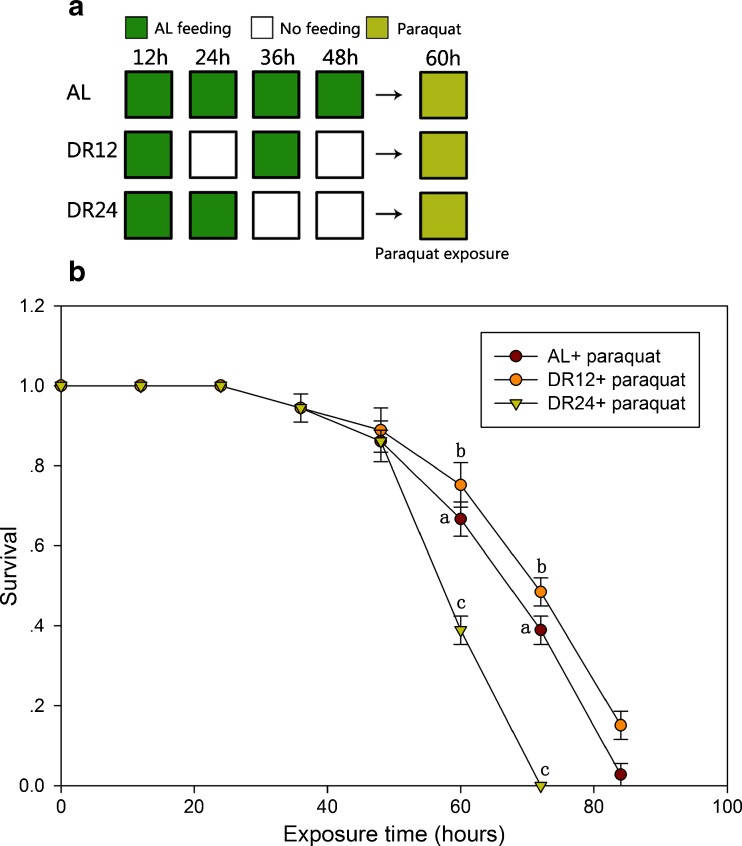
Effect of DR pretreatment on oxidative stress resistance
Figure 12 illustrates the effects of DR pretreatment on the survival rate of rotifers under oxidative stress conditions. After exposure to oxidative stress for 12 h, survival rate in the DR12 pretreatment group was 0.78 ± 0.02 which was significantly higher (P < 0.05) than that in the control group (0.60 ± 0.25). In the 24-h-exposed group, the survival rates in the DR12 pretreatment groups and control groups were 0.55 ± 0.02 and 0.38 ± 0.02, respectively (P < 0.05). However, the survival rate in the DR24 pretreatment groups were 0.39 ± 0.04 which was significantly lower (P < 0.05) than that in the control groups (0.60 ± 0.25). These results indicate that DR12 pretreatment increased rotifer survival rate under paraquat-induced oxidative stress.
Fig. 12.
Effects of DR pretreatments on the B. calyciflorus survival rate under oxidative stress conditions (paraquat). a Experimental designs. b The survival rate of B. calyciflorus in the AL and DR pretreatment groups at each time point. The survival rate was statistically analyzed by one-way ANOVA with the Duncan test
Discussion
HSPs are a highly conserved, ubiquitously expressed family of stress response proteins which are expressed at low levels under normal physiological conditions (Hartl 1996). In the present study, we cloned cDNAs encoding four typical hsp genes (Bchsp40, Bchsp60, Bchsp70, and Bchsp90) from the rotifer B. calyciflorus. Multiple sequence alignment revealed that HSPs from B. calyciflorus were highly homologous with the HSPs from other animals, which indicated that they may have a similar function to the HSPs in other species. HSPs can be induced by a variety of stressful stimuli, such as temperature, cellular energy depletion, physical injury, and various toxic substances (Feder and Hofmann 1999; Ritossa 1962), therefore, HSPs are also called “stress proteins” (Kalmar and Greensmith 2009; Wheelock et al. 2002).
In general, coordinated activation of HSP expression, called the heat shock response (HSR), is a ubiquitous adaptation mechanism in organisms ranging from bacteria to mammals, which helps them to survive and adapt to a wide range of environmental challenges (Lindquist and Craig 1988). To study the modulatory effect of different temperatures on the expression of Bchsps genes, we measured expression at 15, 30, and 37 °C relative to the control (25 °C) over 6 h and found that temperature stress (15 and 37 °C) significantly induced the expression of Bchsp40, Bchsp60, and Bchsp70 genes, indicating that Bchsp40, Bchsp60, and Bchsp70 genes were involved in the chaperoning process to protect proteins under temperature stress in rotifers. However, temperature stress (15 and 37 °C) had no effect on the expression of Bchsp90. Our findings are in accordance with previous studies where hsp40, hsp60, and hsp70 were required for rotifer thermotolerance; however, hsp90 was not essential (Smith et al. 2012). Increased hsps expression under high and low temperatures can protect cells from temperature stress (Dangi et al. 2012).
In addition to typical heat stress, oxidative stress can also induce the expression of HSPs. H2O2 is the most stable intermediate that generates intracellular nonradical ROS (Rosa et al. 2008). It can easily diffuse through cellular membranes into the cell, react with transition metals, and generate hydroxyl radicals (HO·) (Abele-Oeschger et al. 1997). As an oxidant, H2O2 can regulate the expression of many genes, for example superoxide dismutase (SOD) and the hsp20 gene in the rotifer Brachionus sp. (Rhee et al. 2011). In the present study, to analyze the modulatory effect of oxidative stress on Bchsps genes, we initially exposed rotifers to H2O2 for various time periods and found that H2O2 significantly increased the mRNA expression of these four Bchsps. The high expression levels of Bchsps contributed to activation of the HSR, and protected cells from oxidative stress. Previous studies have also shown that environmental stress (i.e., heavy metals, osmotic stress, and various chemicals) increased the expression of hsps (Rios-Arana et al. 2005; Seo et al. 2006; Ventura et al. 2007; Wheelock et al. 1999). The expression of hsps was significantly induced by typical temperature pressure and oxidative stress and reflected the characteristics of the hsps as “stress proteins.”
Aging is likely regulated by low affinity, low specificity interactions, and mediated through a complex network of overlapping pathways (Snell et al. 2012). The quality of proteins declines during the aging process, for example, in 80-year-old humans, half of all proteins may become oxidized (Stadtman and Berlett 1998). Due to these degradation defects, damaged proteins accumulate in the cells of aging organisms, and this may cause a variety of protein folding diseases (Söti and Csermely 2002). HSPs are present in the cells of all living organisms; they act as molecular chaperones and can facilitate protein folding, prevent protein aggregation, and target improperly folded proteins to specific degradation pathways (Kalmar and Greensmith 2009). For example, HSP40s are important for protein homeostasis because they stimulate the ATPase activity of the HSP70 proteins that are involved in protein translation, folding, unfolding, translocation, and degradation (Qiu et al. 2006). HSP60s is a mitochondrial chaperonin that is typically held responsible for the transportation and refolding of proteins from the cytoplasm into the mitochondrial matrix (Koll et al. 1992). HSP70s play a role in protein synthesis under normal cellular conditions, fixing denatured proteins and preventing the misfolding or aggregation of proteins (Danwattananusorn et al. 2011). HSP90 participates in the folding, maintenance of structural integrity, and the proper regulation of a subset of cytosolic proteins (Picard 2002). HSPs contribute to protein quality control and are related to aging owing to their functions mentioned above (Becker and CRAIG 1994; Calderwood et al. 2009; Nardai et al. 2002; Soti and Csermely 2003). The induced expression of HSPs in response to various stresses is dependent on the activation of specific members of a family of transcription factors, the heat shock factors (HSFs), which bind to the heat shock elements in the promoters of the genes encoding HSPs (Åkerfelt et al. 2010). Previous research has recently shown that increased protein damage during aging may be exacerbated by reduced levels of HSPs, and the resultant loss of protein quality control (Calderwood et al. 2009). In this study, we examined the effects of aging on the gene expression of four rotifer Bchsps genes to determine the relationship between aging and these four Bchsps genes. We found that Bchsp70 mRNA expression was significantly decreased with aging. HSP70 is mostly found in the cytosol and nucleus and is used for protein folding and cytoprotection as well as a molecular chaperone (Becker and CRAIG 1994). The cloned BcHSP70 contains a highly conserved typical C-terminal retention sequence (HDEL in mammalian and KDEL in yeast cells) that is common for soluble ER-localized proteins and inhibits its exit from the ER (MUNRO and PELHAM 1987). This ER-localized HSP70 protein, initially described as the mammalian glucose-regulated protein GRP78 (Lee et al. 1981; Munro and Pelham 1986) is induced by the accumulation of misfolded proteins inside the ER (Kozutsumi et al. 1988; Rose et al. 1989). Evidence has indicated that HSP70s in the ER not only aid protein folding and assembly but also may be directly involved in the import of secretory proteins (Vogel et al. 1990). Thus, the decrease in expression of Bchsp70 mRNA means that accumulation of damaged proteins is accelerated. In other organisms, the increased expression of hsp70 gene can extend the lifespan of S. cerevisiae and D. melanogaster (Shama et al. 1998; Tatar et al. 1997). On the other hand, the presence of misfolded proteins (damaged proteins) can induce activation of the HSR by activating the transcription factors, HSFs (Abravaya et al. 1992; Morimoto 1992). Age-dependent waning of the HSR is a general effect found in neuronal tissues (Hands et al. 2008; Sherman and Goldberg 2001; Winklhofer et al. 2008), skeletal muscle, cardiac muscle (Kayani et al. 2008), and liver (Gagliano et al. 2007). The decline in HSR in aging cells indicated that the capacity of the organism to resist stress diminished. The decline in the HSR may be one of the factors which cause aging.
As a natural stress, moderate starvation has been reported to enhance the expression of many genes, including SOD (Kaneko et al. 2005; Kaneko et al. 2011), catalase (CAT) (Kaneko et al. 2011), hsp70, and GRP94 (Kaneko et al. 2002). DR is the most common starvation method for prolonging life in the laboratory, and it has been reported to extend the lifespan of different organisms (Fanestil and Barrows 1965; Klass 1977; Lin et al. 2002; Walker et al. 2005). In rotifers, previous studies have also suggested that DR can extend the lifespan of different types of rotifers (Ozdemir 2009; Weithoff 2007; Yoshinaga et al. 2000). Interestingly, DR-induced longevity and other beneficial effects can be transmitted to subsequent generations in B. plicatilis (Kaneko et al. 2011). In the present study, we found that DR significantly extended the lifespan of the rotifer, B. calyciflorus, and reduced fecundity. Our findings are in accordance with previous studies where DR extended the lifespan accompanied by a reduction of infertility in D. melanogaster (Chapman and Partridge 1996; Chippindale et al. 1993; Partridge et al. 2005) and in rotifers (Ozdemir 2009; Yoshinaga et al. 2000). We also found that DR significantly increased the expression of Bchsp40, Bchsp60, Bchsp70, and Bchsp90 mRNA. The expression of HSPs is not only induced by heat stressors but also by other stressors (Feder and Hofmann 1999), and upregulated HSPs are generally described as part of the cellular stress response (Roberts et al. 2010). The increased expression of HSPs may contribute to diminish the accumulation of damaged proteins and extend the lifespan (Tatar et al. 1997). These examples confirm the hypothesis that HSPs have a better adaptation capacity to various stresses which greatly increases the chances of longevity.
Vitamin E is known to function as an antioxidant, and it has long been investigated for its effects on rotifer aging. Therefore, it was necessary for us to further determine the relationship between HSPs and aging through the anti-oxidative and anti-aging effects of vitamin E. It was reported by Sawada and Enesco that when the rotifer Asplanchna brightwelli was exposed to 25 μg/mL (58 μM) of vitamin E, its lifespan was extended by 21 % due to lengthening of the pre-reproductive and reproductive periods (Sawada and Enesco 1984). Litton reported that exposure to α-tocopherol extended the lifespan of four other rotifer species by about 10 % (Litton 1987). However, in the present study, we found that vitamin E had no effects on rotifer lifespan or reproduction. These inconsistent results may be caused by the differences in rotifer species. In recent research, Snell et al found that the lifespan of the rotifer, B. manjavacas, was extended only when antioxidants were paired with trolox, N-acetyl cysteine, or l-carnosine. Trolox treatment alone did not prolong the lifespan of rotifers (Snell et al. 2012). This also suggested that antioxidant treatment could not always extend the lifespan of rotifers. Actually, the anti-aging effect of vitamin E is also controversial in worms. For example, vitamin E (α-tocopherol) increased lifespan in the first two studies (Harrington and Harley 1988; Ishii et al. 2004) but not in the third (Adachi and Ishii 2000). Likewise, trolox, an α-tocopherol derivative, increased lifespan in Benedetti’s study (Benedetti et al. 2008) but not in Schulz’s (Schulz et al. 2007). We analyzed the expression of Bchsps in rotifers after exposure to vitamin E and found that vitamin E significantly increased the mRNA expression of Bchsp60 only and not the other three hsps. This suggests that vitamin E may not improve the HSR in rotifers. To some extent, this also explains why vitamin E does not extend the lifespan of rotifers.
Aging has been associated with a loss of stress resistance, and treatments that extend lifespan are often associated with increased stress resistance (Kirkwood and Austad 2000; Vermeulen and Loeschcke 2007). Previous studies showed that there was a close correlation between stress resistance and longevity in several long-lived C. elegans and Drosophila mutants (Lithgow and Kirkwood 1996). To assess resistance to oxidative stress in rotifers, we used paraquat, a widely used compound, to generate ROS inside the cells (Kaneko et al. 2011). Under paraquat-induced oxidative stress, moderate DR pretreatment increased rotifer survival rate. Our findings are in accordance with previous studies which showed that treatments which extended lifespan increased stress resistance. Rosa damascene is a rose hybrid commonly harvested for its oil used in perfumery and for rose water used to flavor food. It was reported by Schriner et al. that R. damascena extended the lifespan of D. melanogaster through a DR-associated mechanism (Schriner et al. 2012). The authors also found that R. damascena could improve D. melanogaster resistance to paraquat and H2O2. The same study also showed that the natural product, black tea (Camellia sinensis), blocked the age-dependent accumulation of iron, improved iron tolerance, and extended the lifespan of D. melanogaster (MASSIE et al. 1993). Enhanced stress resistance is likely to benefit from the upregulated expression of hsps. In fact, overexpression of hsp22 has been shown to increase fly lifespan, protect flies against paraquat, and improve their locomotor ability (Morrow et al. 2004). In rotifers, DR pretreatment was reported to benefit paraquat and juglone tolerance (Kailasam et al. 2011; Ozaki et al. 2010). A previous study indicated that the beneficial effects of DR may contribute to the improvement in antioxidant gene expression (SOD and CAT) (Yang et al. 2013). In the present study, we also found that inducing the expression of hsps by DR may be helpful for DR-induced extended lifespan. DR also had other beneficial effects, such as resistance to oxidative stress.
Conclusions
In this study, we successfully cloned four typical hsp genes (hsp40, hsp60, hsp70, and hsp90) from the rotifer B. calyciflorus Pallas, analyzed the molecular characteristics, and found that Bchsps was significantly induced by temperature and H2O2 stress. The mRNA expression of Bchsp70 decreased with age which indicated waning of the HSR with aging in rotifers. DR extended the lifespan and improved resistance to paraquat, and these findings may have benefitted from the upregulated expression of hsps.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No: 31272388) to JXY and the National Natural Science Foundation of China for Talents Training in Basic Science (J1103507).
Abbreviations
- ANOVA
Analysis of variance
- bp
Base pair
- DR
Dietary restriction
- ATP/GTP
Adenosine triphosphate/guanosine triphosphate
- EPA
Artificial freshwater medium
- ER
Endoplasmic reticulum
- H2O2
Hydrogen peroxide
- HSF
Heat shock factor
- HSP
Heat shock protein
- HSR
Heat shock response
- O2−
Superoxide anion
- ORF
Open reading frame
- PCR
Polymerase chain reaction
- RACE
Rapid amplification of cDNA ends
- ROS
Reactive oxygen species
- RT-PCR
Real-time PCR
- SOD
Superoxide dismutase
- UTR
Untranslated region
References
- Abele-Oeschger D, Sartoris FJ, Pörtner HO. Hydrogen peroxide causes a decrease in aerobic metabolic rate and in intracellular pH in the shrimp Crangon crangon. Comp Biochem Physiol C: Pharmacol Toxicol Endocrinol. 1997;117:123–129. [Google Scholar]
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- Adachi H, Ishii N. Effects of tocotrienols on life span and protein carbonylation in Caenorhabditis elegans. J Gerontol A: Biol Sci Med Sci. 2000;55:B280–B285. doi: 10.1093/gerona/55.6.B280. [DOI] [PubMed] [Google Scholar]
- Åkerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASTM (2001) Standard guide for acute toxicity tests with the rotifer Brachionus. In: Annual Book of ASTM Standards, Water and Environmental Technology, vol. 1105. Biological Effects and Environmental Fates. ASTM, West Conshohocken
- Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur J Biochem. 1994;219:11–23. doi: 10.1111/j.1432-1033.1994.tb19910.x. [DOI] [PubMed] [Google Scholar]
- Benedetti MG, Foster AL, Vantipalli MC, White MP, Sampayo JN, Gill MS, Olsen A, Lithgow GJ. Compounds that confer thermal stress resistance and extended lifespan. Exp Gerontol. 2008;43:882–891. doi: 10.1016/j.exger.2008.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Murshid A, Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging—a mini-review. Gerontology. 2009;55:550–558. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ. Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 1999;9:262–268. doi: 10.1016/S0962-8924(99)01580-9. [DOI] [PubMed] [Google Scholar]
- Chapman T, Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc Biol Sci. 1996;263:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Leroi AM, Kim SB, Rose MR. Phenotypic plasticity and selection in Drosophila life-history evolution. I. Nutrition and the cost of reproduction. J Evol Biol. 1993;6:171–193. doi: 10.1046/j.1420-9101.1993.6020171.x. [DOI] [Google Scholar]
- Dangi SS, Gupta M, Maurya D, Yadav VP, Panda RP, Singh G, Mohan NH, Bhure SK, Das BC, Bag S. Expression profile of HSP genes during different seasons in goats (Capra hircus) Trop Anim Heal Prod. 2012;44:1905–1912. doi: 10.1007/s11250-012-0155-8. [DOI] [PubMed] [Google Scholar]
- Danwattananusorn T, Fagutao FF, Shitara A, Kondo H, Aoki T, Nozaki R, Hirono I. Molecular characterization and expression analysis of heat shock proteins 40, 70 and 90 from kuruma shrimp Marsupenaeus japonicus. Fish Sci. 2011;77:929–937. doi: 10.1007/s12562-011-0394-z. [DOI] [Google Scholar]
- Fanestil DD, Barrows CH., Jr Aging in the rotifer. J Gerontol. 1965;20:462–469. [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Gagliano N, Grizzi F, Annoni G. Mechanisms of aging and liver functions. Dig Dis. 2007;25:118–123. doi: 10.1159/000099475. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- Hands S, Sinadinos C, Wyttenbach A. Polyglutamine gene function and dysfunction in the ageing brain. Biochim Biophys Acta (BBA)-Gene Regul Mech. 2008;1779:507–521. doi: 10.1016/j.bbagrm.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Harrington LA, Harley CB. Effect of vitamin E on lifespan and reproduction in Caenorhabditis elegans. Mech Ageing Dev. 1988;43:71–78. doi: 10.1016/0047-6374(88)90098-X. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Heikkila JJ. Heat shock protein gene expression and function in amphibian model systems. Comp Biochem Physiol A Mol Integr Physiol. 2010;156:19–33. doi: 10.1016/j.cbpa.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishii N, Senoo-Matsuda N, Miyake K, Yasuda K, Ishii T, Hartman PS, Furukawa S. Coenzyme Q10 can prolong C. elegans lifespan by lowering oxidative stress. Mech Ageing Dev. 2004;125:41–46. doi: 10.1016/j.mad.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Kailasam M, Kaneko G, Oo AKS, Ozaki Y, Thirunavukkarasu AR, Watabe S. Effects of calorie restriction on the expression of manganese superoxide dismutase and catalase under oxidative stress conditions in the rotifer Brachionus plicatilis. Fish Sci. 2011;77:403–409. doi: 10.1007/s12562-011-0334-y. [DOI] [Google Scholar]
- Kalmar B, Greensmith L. Induction of heat shock proteins for protection against oxidative stress. Adv Drug Deliv Rev. 2009;61:310–318. doi: 10.1016/j.addr.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Kaneko G, Kinoshita S, Yoshinaga T, Tsukamoto K, Watabe S. Changes in expression patterns of stress protein genes during population growth of the rotifer Brachionus plicatilis. Fish Sci. 2002;68:1317–1323. doi: 10.1046/j.1444-2906.2002.00570.x. [DOI] [Google Scholar]
- Kaneko G, Yoshinaga T, Yanagawa Y, Kinoshita S, Tsukamoto K, Watabe S. Molecular characterization of Mn-superoxide dismutase and gene expression studies in dietary restricted Brachionus plicatilis rotifers. Hydrobiology. 2005;546:117–123. doi: 10.1007/s10750-005-4107-4. [DOI] [Google Scholar]
- Kaneko G, Yoshinaga T, Yanagawa Y, Ozaki Y, Tsukamoto K, Watabe S. Calorie restriction-induced maternal longevity is transmitted to their daughters in a rotifer. Funct Ecol. 2011;25:209–216. doi: 10.1111/j.1365-2435.2010.01773.x. [DOI] [Google Scholar]
- Kayani AC, Morton JP, McArdle A. The exercise-induced stress response in skeletal muscle: failure during aging. Appl Physiol Nutr Metab. 2008;33:1033–1041. doi: 10.1139/H08-089. [DOI] [PubMed] [Google Scholar]
- Kenneth JL, Thomas DS. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Kirkwood TBL, Austad SN. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Koll H, Guiard B, Rassow J, Ostermann J, Horwich AL, Neupert W, Hartl F-U. Antifolding activity of hsp60 couples protein import into the mitochondrial matrix with export to the intermembrane space. Cell. 1992;68:1163–1175. doi: 10.1016/0092-8674(92)90086-R. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Lee AS, Delegeane A, Scharff D. Highly conserved glucose-regulated protein in hamster and chicken cells: preliminary characterization of its cDNA clone. Proc Natl Acad Sci. 1981;78:4922–4925. doi: 10.1073/pnas.78.8.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig E. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Lints FA. Non-mammalian models for research on aging. Basel: Karger; 1985. [Google Scholar]
- Lithgow GJ, Kirkwood TBL. Mechanisms and evolution of aging. Science. 1996;273:80–80. doi: 10.1126/science.273.5271.80. [DOI] [PubMed] [Google Scholar]
- Litton JR. Specificity of the α-tocopherol (vitamin E) effect on lifespan and fecundity of bdelloid rotifers. Hydrobiologia. 1987;147:135–139. doi: 10.1007/BF00025736. [DOI] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie H, Aiello V, Williams T. Inhibition of iron absorption prolongs the life span of Drosophila. Mech Ageing Dev. 1993;67:227–237. doi: 10.1016/0047-6374(93)90001-8. [DOI] [PubMed] [Google Scholar]
- Mayer M, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard L. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Morimoto RI. Transcription factors: positive and negative regulators of cell growth and disease. Curr Opin Cell Biol. 1992;4:480–487. doi: 10.1016/0955-0674(92)90015-5. [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham H. An Hsp70-like protein in the ER: identity with the 78 kD glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham H. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Nardai G, Csermely P, Söti C. Chaperone function and chaperone overload in the aged. A preliminary analysis. Exp Gerontol. 2002;37:1257–1262. doi: 10.1016/S0531-5565(02)00134-1. [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Kaneko G, Yanagawa Y, Watabe S. Calorie restriction in the rotifer Brachionus plicatilis enhances hypoxia tolerance in association with the increased mRNA levels of glycolytic enzymes. Hydrobiologia. 2010;649:267–277. doi: 10.1007/s10750-010-0269-9. [DOI] [Google Scholar]
- Ozdemir N. The effect of caloric restriction on the life span and reproduction of fresh water rotifer (Brachionus calyciflorus) J Anim Vet Adv. 2009;8:669–673. [Google Scholar]
- Partridge L, Gems D, Withers DJ. Sex and death: what is the connection? Cell. 2005;120:461–472. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci CMLS. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X-B, Shao Y-M, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci CMLS. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JS, Kim RO, Choi HG, Lee J, Lee YM, Lee JS. Molecular and biochemical modulation of heat shock protein 20 (Hsp20) gene by temperature stress and hydrogen peroxide (H2O2) in the monogonont rotifer, Brachionus sp. Comp Biochem Physiol C Toxicol Pharmacol. 2011;154:19–27. doi: 10.1016/j.cbpc.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Rios-Arana JV, Gardea-Torresdey J, Webb R, Walsh EJ. Heat shock protein 60 (HSP60) response of Plationus patulus (Rotifera: Monogononta) to combined exposures of arsenic and heavy metals. Hydrobiologia. 2005;546:577–585. doi: 10.1007/s10750-005-4308-x. [DOI] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Cell Mol Life Sci. 1962;18:571–573. doi: 10.1007/BF02172188. [DOI] [Google Scholar]
- Roberts R, Agius C, Saliba C, Bossier P, Sung Y. Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis. 2010;33:789–801. doi: 10.1111/j.1365-2761.2010.01183.x. [DOI] [PubMed] [Google Scholar]
- Rosa CE, Bianchini A, Monserrat JM. Antioxidant responses of Laeonereis acuta (Polychaeta) after exposure to hydrogen peroxide. Braz J Med Biol Res. 2008;41:117–121. doi: 10.1590/S0100-879X2006005000201. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Sawada M, Enesco H. Vitamin E extends lifespan in the short-lived rotifer Asplanchna brightwelli. Exp Gerontol. 1984;19:179–183. doi: 10.1016/0531-5565(84)90036-6. [DOI] [PubMed] [Google Scholar]
- Schriner SE, Katoozi NS, Pham KQ, Gazarian M, Zarban A, Jafari M. Extension of Drosophila lifespan by Rosa damascena associated with an increased sensitivity to heat. Biogerontology. 2012;13:105–117. doi: 10.1007/s10522-011-9357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Seo J, Park TJ, Lee YM, Park H, Yoon YD, Lee JS. Small heat shock protein 20 gene (Hsp20) of the intertidal copepod Tigriopus japonicus as a possible biomarker for exposure to endocrine disruptors. Bull Environ Contam Toxicol. 2006;76:566–572. doi: 10.1007/s00128-006-0957-3. [DOI] [PubMed] [Google Scholar]
- Shama S, Lai CY, Antoniazzi JM, Jiang JC, Jazwinski SM. Heat stress-induced life span extension in yeast. Exp Cell Res. 1998;245:379–388. doi: 10.1006/excr.1998.4279. [DOI] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/S0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- Smith HA, Burns AR, Shearer TL, Snell TW. Three heat shock proteins are essential for rotifer thermotolerance. J Exp Mar Biol Ecol. 2012;413:1–6. doi: 10.1016/j.jembe.2011.11.027. [DOI] [Google Scholar]
- Snell TW, Moffat BD, Janssen C, Persoone G. Acute toxicity tests using rotifers: IV. Effects of cyst age, temperature, and salinity on the sensitivity of Brachionus calyciflorus. Ecotoxicol Environ Saf. 1991;21:308–317. doi: 10.1016/0147-6513(91)90070-6. [DOI] [PubMed] [Google Scholar]
- Snell TW, Fields AM, Johnston RK. Antioxidants can extend lifespan of Brachionus manjavacas (Rotifera), but only in a few combinations. Biogerontology. 2012;13:261–275. doi: 10.1007/s10522-012-9371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söti C, Csermely P. Chaperones and aging: role in neurodegeneration and in other civilizational diseases. Neurochem Int. 2002;41:383–389. doi: 10.1016/S0197-0186(02)00043-8. [DOI] [PubMed] [Google Scholar]
- Soti C, Csermely P. Aging and molecular chaperones. Exp Gerontol. 2003;38:1037–1040. doi: 10.1016/S0531-5565(03)00185-2. [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab Rev. 1998;30:225–243. doi: 10.3109/03602539808996310. [DOI] [PubMed] [Google Scholar]
- Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30–30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- Traber MG, Stevens JF. Vitamins C and E: beneficial effects from a mechanistic perspective. Free Radic Biol Med. 2011;51:1000–1013. doi: 10.1016/j.freeradbiomed.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Zhang Z, Fitzgerald GF, Van Sinderen D. Molecular characterization of hsp20, encoding a small heat shock protein of Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:4695–4703. doi: 10.1128/AEM.02496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen CJ, Loeschcke V. Longevity and the stress response in Drosophila. Exp Gerontol. 2007;42:153–159. doi: 10.1016/j.exger.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Misra LM, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. 2005;126:929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Weithoff G. Dietary restriction in two rotifer species: the effect of the length of food deprivation on life span and reproduction. Oecologia. 2007;153:303–308. doi: 10.1007/s00442-007-0739-6. [DOI] [PubMed] [Google Scholar]
- Wheelock C, Wolfe M, Olsen H, Tjeerdema R, Sowby M. Hsp60-induced tolerance in the rotifer Brachionus plicatilis exposed to multiple environmental contaminants. Arch Environ Contam Toxicol. 1999;36:281–287. doi: 10.1007/s002449900472. [DOI] [PubMed] [Google Scholar]
- Wheelock C, Baumgartner T, Newman J, Wolfe M, Tjeerdema R. Effect of nutritional state on Hsp60 levels in the rotifer Brachionus plicatilis following toxicant exposure. Aquat Toxicol. 2002;61:89–93. doi: 10.1016/S0166-445X(02)00044-9. [DOI] [PubMed] [Google Scholar]
- Winklhofer KF, Tatzelt J, Haass C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dong S, Jiang Q, Kuang T, Huang W, Yang J. Changes in expression of manganese superoxide dismutase, copper and zinc superoxide dismutase and catalase in Brachionus calyciflorus during the aging process. PLoS One. 2013;8:e57186. doi: 10.1371/journal.pone.0057186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga T, Hagiwara A, Tsukamoto K. Effect of periodical starvation on the life history of Brachionus plicatilis O.F. Müller (Rotifera): a possible strategy for population stability. J Exp Mar Biol Ecol. 2000;253:253–260. doi: 10.1016/S0022-0981(00)00268-9. [DOI] [PubMed] [Google Scholar]