Abstract
Helicobacter pylori (H. pylori) is a common human pathogen responsible for various gastric diseases. This bacterium relies on the production of urease and hydrogenase to inhabit the acidic environment of the stomach. Nickel is an essential cofactor for urease and hydrogenase. H. pylori has to uptake sufficient nickel ions for the maturation of urease, and on the other way, to prevent the toxic effects of excessive nickel ions. Therefore, H. pylori has to strike a delicate balance between the import of nickel ions, its efficient intracellular storage, and delivery to nickel-dependent metalloenzymes when required. The assembly and maturation of the urease enzyme is a complex and timely ordered process, requiring various regulatory, uptake, chaperone and accessory proteins. In this review, we focus on several nickel trafficking proteins involved in urease maturation: NikR, NixA, HypAB, UreEFGH, HspA, Hpn and Hpnl. The work will deepen our understanding of how this pathogenic bacterium adapts to severe habitant environments in the host.
Keywords: Urease, Histidine-rich protein, NikR, NixA, Helicobacter pylori
Core tip: Helicobacter pylori (H. pylori) is responsible for various gastric diseases. The nickel containing urease and hydrogenase are essential for the successful infections of H. pylori in the stomach. Nickel is an essential cofactor for urease and hydrogenase. In this review we discussed the various regulatory, uptake, chaperone and accessory proteins involved in the maturation of urease, especially the proteins NikR, NixA, HypAB, UreEFGH, HspA, Hpn and Hpnl. The work will deepen our understanding of how this pathogenic bacterium adapts to severe habitant environments in the host.
INTRODUCTION
Helicobacter pylori (H. pylori), a micro-aerophilic Gram-negative spirobacterium, infects around half of the people worldwide and is responsible for gastric diseases such as chronic gastritis, peptic ulcer and gastric cancer[1]. The bacterium is widely present in the mucus layer of the stomach, the mucus glands in the stomach cavity and the surface of gastric epithelial cells as well as within the cells. Due to the wide presence in the differential parts of the stomach, it is difficult to completely eradicate the pathogen during gastric disease therapy[2]. The commonly used treatment for H. pylori related diseases is the so-called triple therapy, which consists of two antibiotics and either a proton pump inhibitor (PPI) or one kind of bismuth-based colloidal drug[3,4]. In some countries, standard triple therapy combining one PPI, amoxicillin and clarithromycin is the best option. However, in countries where clarithromycin resistance rate is over 20%, bismuth-containing quadruple therapy, or non-bismuth sequential or concomitant therapies are the preferred option. The medical and social impact of the discovery of H. pylori was acknowledged by the award of the 2005 Noble Prize in Physiology and Medicine to Marshall and Warren.
Around 80% of H. pylori cells inhabit the moderately acidic gastric mucus. Once entry into the stomach, the first hurdle for H. pylori is to be quickly transmitted through the extremely acidic gastric lumen, exhibiting a median pH of approximately 1.4[5]. H. pylori multiplies in an environmental pH from 6.0 to 8.0[6], and cannot survive when the pH < 4.0 or > 8.2[7]. In order to live in the gastric environment, H. pylori has developed various acid-resistant mechanisms. Time-independent acid resistance depends on the high isoelectric points of the inner and outer membrane proteins to reduce proton permeability[8]. Acute acid resistance depends on the constitutive synthesis of urease that catalyzes the hydrolysis of urea to ammonia and carbamate, the latter of which is further degraded to ammonia and carbonic acid. The end products are in an equilibrium between their protonated and de-protonated forms, leading to an elevation of the surrounding pH from absolutely acidic to approximately neutral[9]. Urease is an oligomeric Ni2+-containing heterodimer of UreA and UreB subunits and is essential for H. pylori to infect in all animal models so far examined[10-12]. The substrate gastric juice urea is able to rapidly access intrabacterial urease through a pH-gating urea channel, UreI[13], when the periplasmic pH falls < 6.2.
H. pylori urease is produced in a high level, accounting for up to 10% of total cellular proteins[14]. Expression of urease protein is constitutive[15], primarily due to the housekeeping σ80-dependent promoters for the transcription of both ureAB and ureEFGHI[16,17]. Under in vitro growth conditions without additionally added Ni2+, only 2% of the active sites were filled with Ni2+[18,19]. Urease produces NH3 from gastric juice urea with maximal efficiency at millimolar concentrations[14,20], 1014 times faster than uncatalyzed reactions. The enzymatic hydrolysis of urea causes an abrupt overall pH increase, resulting in negative side effects for human and positive effects in the buffering of the periplasm and maintenance of a proton motive force adequate for ATP synthesis of the bacterium[21]. H. pylori urease was shown to be a giant 1.1 MDa complex containing 12 subunits of UreA and UreB (Figure 1), with two Ni2+ needed for enzyme activity[6,22]. The assembly of the urease enzyme is a complex, timely ordered process, and the UreEFGH accessory proteins are absolutely necessary[23,24]: UreH stabilizes the apoprotein[25]; UreF facilitates carbamylation of the Ni2+-bridging lysine residue and blocks premature Ni2+ binding to the active site[26]; UreG provides energy during urease assembly[27]; and UreE facilitates Ni2+ incorporation into the active center[28]. The hydrogenase accessory proteins HypA and HypB are also necessary to maintain the urease activity, indicating that the bacterium utilizes both maturation systems for the activation of its urease[18]. This present review intends to cover the reports and discoveries in the field of nickel trafficking system in urease maturation of H. pylori, which may deepen our understanding of how this pathogenic bacterium adapts to severe habitant environments in the host.
Figure 1.
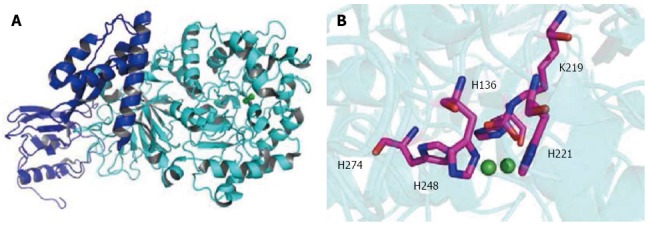
Structure of Helicobacter pylori urease. A: The urease enzyme subunit UreAB (UreA, blue; UreB, cyan; PDB code: 1E9Z); B: The active sites of H. pylori urease with the side chains of the enzyme involved in the chelation of the catalytic di-nickel center shown (Ni, green; O, red; N, blue; C, pink). H. pylori: Helicobacter pylori.
NICKEL REGULATORY PROTEIN NIKR
Bacteria have developed sophisticated mechanisms to regulate levels of intracellular nickel ions, to ensure sufficient nickel for enzyme processes in one way and to prevent excessive toxic free ions in the other way[29]. NikRs, a novel class of ribbon-helix-helix nickel regulatory proteins, are homotetrameric transcription factors that repress and/or activate specific genes in response to nickel availability. H. pylori NikR, a tetrameric protein made of two dimeric N-terminal DNA-binding domains (DBD) and C-terminal domains for tetramerization and metal binding (MBD), binds stoichiometric nickel with picomolar affinities[30,31], comparable to NikRs from other species[32-34]. The DBD and MBD are connected by a flexible linker, allowing for differential conformations (open, trans and cis) of NikR. In E. coli and Pyrococcus horikoshii, the apo-NikRs adopt an open conformation, whereas the apo-NikR shows an unusual closed trans-conformation and asymmetrical quaternary arrangement, where the DBDs are on the opposite sides of the transmembrane domain[35]. Computational and NMR studies suggest that NikR is interconverting among the open, trans and cis forms in solution and nickel binding facilitates the interconversion[36].
At non-physiologically low pH (4.6-5.6), NikR had three types of nickel-binding sites: the final high affinity site (F) with square-planar geometry, the intermediate site (I) involving residues belonging either to the F or external site, and the external sites (X) with an octahedral geometry[35,37]. Whereas in physiological conditions (pH 5.6-7.5), NikR binds four low-spin Ni2+ at the protein tetramerization interface, although differential nickel coordination modes are proposed. Michel’s group suggests that two nickels are bound at 4-coordinate square-planar sites with His3Cys ligands (i.e., 4-sites) and the other two are coordinated by His3(H2O)2-3 in square pyramidal or octahedral geometries (i.e., 5/6 sites)[37]. Ciurli’s group reports a structure with all four nickel ions bound to 4 sites[38], and the four binding sites are classified into two sets (2/2), with binding affinities differing by one order of magnitude[39]. The findings may suggest that an equilibrium exists between the two nickel-bound forms of the protein.
The biological role of NikR is to regulate the transcription of multiple genes as a function of nickel availability[40,41]: up-regulated genes in nickel metabolism (nikABCDE, nixA, ureA, ureB, hpn and hpn-like); down-regulated genes in iron uptake and storage (pfr, fur and exbB/exbD), motility (cheV, flaA and flaB), and stress responses to outer membrane proteins (omp11, omp31 and omp32)[40]. The nickel-responsive binding of NikR to target promoters pUreA, pNikR, pexbB and pFur have been characterized by the in vitro gel shift and DNase I footprinting studies. Michel’s[42] group proposed a mechanism for nickel-mediated DNA recognition by NikR. NikR prefers binding Ni at 5/6 sites. Upon addition of two Ni, the ligands are rearranged to two 4-sites. Addition of two more Ni results in mixed coordination geometry (two 4-sites and two 5/6-sites) and makes the protein binding to target DNA. The binding to DNA changes the orientation of the DBD from trans to cis, an orientation that is stabilized at the MBD/DBD interface[42].
Controversial opinions exist for the roles of NikR in urease activation as a function of pH. One opinion goes that under acidic conditions, the greater availability of Ni2+ leads to the formation of Ni2+-NikR complexes which further increase the expression of urease, Ni2+ transporter NixA and iron regulator Fur[43,44]. Whereas, Pflock et al[45] found that a two-component system ArsRS (acid responsive signaling) regulated urease expression in response to low pH, and further proposed that urease expression is mediated by two distinct mechanisms: one in response to increasing Ni2+ concentration (NikR) and one in response to decreasing pH (ArsR).
NICKEL UPTAKE
Due to the essential stasis of Ni2+-containing urease for the host colonization and infection of H. pylori, a constant supply of Ni2+ into H. pylori is required. The concentration of nickel ions in the environment is relatively low: around 30 nM in seawater and 5 nM in freshwater, a condition requiring highly specific importers of Ni2+ ions for H. pylori[46]. Thus far, two types of nickel uptake strategies have been identified in H. pylori[46]: (1) NixA[47], a member of the nickel-cobalt transporter family (NiCoT)[48]; and (2) the multiple-component ATP-binding protein cassette (ABC)-transporters, which are believed to be a four-gene operon designated as abcABCD[49].
NixA is required for effective H. pylori colonization, as disruption of the gene led to reduced colonization[50]. NixA is predicted to have eight transmembrane-spanning helices, and transports Ni2+ with a Vmax of 1750 pmol Ni2+/min per 108 cells and a Km of 11.3 nmol[51,52], thus enabling H. pylori to efficiently scavenge nickel ions in the range of 2-11 nmol from the human body[53]. NixA transcription was shown to be repressed by NikR in a nickel-dependent manner to prevent excess toxic in vivo nickel[44,54].
NixA deletion mutants still retained urease activity in some levels (up to 50% in some strains)[50,55], indicating the existence of an alternative nickel transporter. Further analysis identified the abcABCD genes, a component of the ATP-dependent nickel transport system to be potentially involved in NixA-independent nickel uptake, as mutations in abcCD decreased urease activity[49]. Another work identified FrpB4 to be a potential outer membrane nickel uptake protein as energized by the TonB/ExbB/ExbD machinery[56], indicating that the established iron uptake machinery may be involved in nickel uptake. However, further work is needed to confirm their role and mechanism in nickel transport.
CHAPERONES
Similar to other bacteria, H. pylori has to maintain a delicate balance between the import of nickel ions, its efficient intracellular storage, and delivery to nickel-dependent metalloenzymes when required. Metals, such as nickel, pose problems for the cell because they are required for the growth, whereas they inhibit growth and exhibit toxic effects when present in excess. In this section, we would like to discuss the proteins involved in metallocenter assembly in urease.
HypA and HypB
HypA and HypB are named to emphasize their roles in the maturation and activation of NiFe hydrogenase (hyp, hydrogenase pleiotropic). However HypA and HypB are also found to be accessory proteins for urease[57], as reflected by the reduced urease activity (40-200 folds) upon hypA or hypB disruption[18] and the competition between HypA and UreG for UreE (see below) recognition[58]. HypA binds nickel and zinc ions and HypB is a P-loop GTPase to provide energy during nickel insertion in hydrogenase. HypA and HypB exist as homodimers in solution and form heterodimers with each other[59,60] with a low affinity (Kd of 52.2 ± 8.8 μmol)[61]. HypA and HypB also make heterodimers with UreE[62] and SlyD[63], respectively in solution. The NMR structure of zinc-bound HypA monomer indicates that the nickel binding site is located at the N-terminus and nickel is bound to four nitrogens in a square planar geometry[64]. A thermodynamic study indicates that the zinc binding site has a much higher affinity to zinc than nickel and zinc binding induces a great change in the secondary structure of HypA to exert its structural role in the metalloprotein[65]. Further study with XAS showed that HypA dimer has a unique structural flexibility of the zinc site and has roles in sensing nickel binding and pH[66,67]: a decrease of pH from 7.2 to 6.3 induces a change of the zinc binding ligands from Cys4 to Cys2His2 and results in a change of the nickel binding stoichiometry from one Ni per monomer to one Ni per dimer[66]. Cys106 and His107 of HypB are required for nickel binding and metal-dependent dimerization[68]. Nickel binding of HypB is possibly facilitated by SlyD via its IF (insert-in-flap) domain[63]. Zinc binding significantly inhibits the GTPase activity of HypB[68]. Nickel binding is reported to either slightly[68] or highly[61] stimulate the activity of HypB, with reasons for these discrepancies yet unknown. The regulation of HypB activities by metal binding may contribute to the maturation of the hydrogenase and urease.
UreEFGH
UreEFGH is a group of accessory proteins involved in the synthesis of the urease active site[41], which has been excellently covered recently in a review by Farrugia et al[69]. This review will only briefly discuss their respective roles. The information about UreH is quite limited primarily due to its insolubility in solution, although it is believed to be the first protein to bind to apo-urease[70]. UreE is the chaperone to deliver nickel to urease and UreF activates the GTPase activity of UreG[29,41]. UreE is capable of binding Ni and Zn (Kd of 0.15 and 0.49 μmol, respectively) in a stoichiometry of one per dimer[71,72]. Apo-UreE is a dimer and the metal-bound protein is a tetramer (dimer of dimer) formed by the coordination of the metal ion by His104 from each subunit[73]. A second UreE crystal structure indicates that Ni is six-coordinate (His102 from one monomer, His102, His152, Glu4 from the other, a water molecule and one unidentified ligand)[74]. His152 is disordered in the crystal and could be replaced by UreG residues, thus leading to the transfer of nickel from UreE to UreG. In the calculated structure of UreDEFG through computational modeling, the convex surface of the UreG dimer is in direct contact only with the shallow crevice at the interface of the two UreF monomers through weak van der Waals and polar interactions[75]. UreF and UreH can form dimer of heterodimers in solution with concomitant conformational changes in two distinctive regions of UreF[76]: (1) the flexible C-terminus becomes ordered to form an extra helix α10 and a loop stabilized by hydrogen bonds involving Arg250; and (2) the first turn of helix α2 uncoils to expose a conserved residue Tyr48. Both Arg250 and Tyr48 are critical for the heterotrimeric formation of UreG-UreF-UreH and urease maturation[76]. One crystal structure of UreGFH indicates that UreFH facilitates UreG dimerization and assembles its metal binding sties by juxtaposing two Cys66-Pro67-His68 motifs at the interface to form the (UreGFH)2 complex[77].
HspA, Hpn and Hpn-like
HspA, Hpn and Hpn-like (Hpnl) proteins in H. pylori are histidine-rich in full or in part. HspA is a bacterial GroES homologue with a unique cysteine- and histidine-rich C-terminal domain[78]. HspA binds 2 Ni per monomer with a dissociation constant of 1.1 μmol in vitro[79]. The in vivo work showed that HspA is involved in intracellular nickel sequestration and detoxification, and plays a role as a specific nickel chaperone in the maturation of hydrogenase, while not for urease[80]. Hpn (Figure 2A) is a histidine rich protein (accounting for around half of its amino acids) and highly abundant in the cell cytoplasm (approximately 2% of all protein synthesized)[81]. The majority of histidines are located within the central part of the protein and include two separated stretches of 6 and 7 consecutive histidine residues. There are two internal short repeats of Glu-Glu-Gly-Cys-Cys, four sets of paired histidine residues and an X-X-His motif at the N-terminus. All these sequence features indicate that this protein would strongly bind metal ions. Mutated strains of H. pylori lacking the hpn gene are four times more sensitive to ranitidine bismuth citrate, a metal-containing drug widely used to treat H. pylori infections, than the wild type[3,82,83]. Hpn exists in solution as a range of multimeric forms with the 20-mer to be potentially physiologically relevant[84]. The protein can bind nickel in a stoichiometry of five Ni per monomer with a Kd of 7.1 μmol. Therefore it is possible that nickel may be transferred from Hpn to stronger nickel binding proteins, such as HypA (Kd of 1.3 μmol) and HspA (Kd of 1.8 μmol). Nickel can be released from Hpn by decreasing pH (pH1/2 of 6.3) or by adding nickel chelating agent EDTA[84,85], which indicates that Hpn could provide stored nickel ions to the relevant chaperone proteins for the subsequent urease maturation upon intracellular pH decrease. The nickel release from Hpn by EDTA is a two-step process consisting of a rapidly established equilibrium (formation of Hpn-Ni•EDTA, K) followed by a rate-determining step (dissociation of Hpn-Ni•EDTA to Ni-EDTA and apo-Hpn, k2)[85]. The data was fitted in Figure 2B which suggests that lower pH favors both the formation of the Hpn-Ni•EDTA intermediate and its decomposition to the Ni-exchanged products[85]. Later work by our group showed that this His-rich protein can form amyloid-like structures and exhibit some cytotoxic effects to gastric epithelia cells[86], indicating that Hpn may be involved in the pathological roles of H. pylori other than the nickel storage role in the maturation of nickel specific enzymes[87]. Hpnl is a histidine- and glutamine-rich protein in H. pylori, the N-terminus (46 residues) of which shows 56% identity to Hpn. Hpnl binds two nickel ions per monomer in the histidine-rich domain with a dissociation constant of 3.8 μmol[88]. Nickel release experiments established that Hpnl is similar to Hpn, as nickel can be release from Hpnl at acidic pH (pH1/2 of 4.6) and in the presence of EDTA. One in vivo study by Maier’s group indicated H. pylori can utilize stored nickel ions via Hpn and Hpnl to aid colonization of the host[89].
Figure 2.
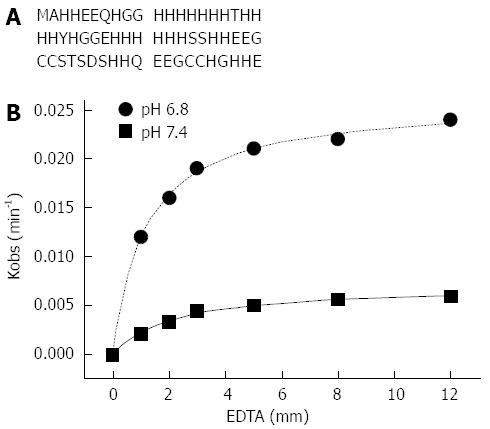
Amino acid sequence of Hpn (A) and the apparent rate of nickel release vs concentrations of ethylenediaminetetraacetic acid at pH 6.8 and 7.4 and the best nonlinear fit of the data (B). EDTA: Ethylenediaminetetraacetic acid.
CONCLUSION
H. pylori is an established agent causing various gastric diseases. The nickel containing urease and hydrogenase are essential for the successful infections of H. pylori in the stomach. Nickel is an essential cofactor for urease and hydrogenase. Various nickel-binding proteins play key roles in microbial nickel homeostasis by shuttling nickel within the cells. In this review we discussed the regulatory, uptake, chaperone and accessory proteins involved in the maturation of urease, especially the proteins NikR, NixA, HypAB, UreEFGH, HspA, Hpn and Hpnl. The proteins function in a coordinated way to maturate the urease in an efficient way for the successful inhabitation of the bacterium in the stomach (Figure 3). The work will deepen our understanding of how this pathogenic bacterium adapts to severe habitant environments in the host.
Figure 3.
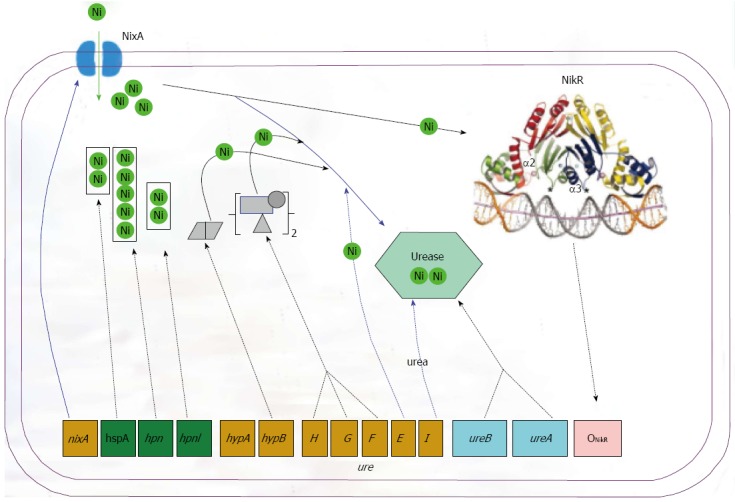
Complex network controlling urease synthesis and activity in Helicobacter pylori. The different levels of control comprise (1) expression of the UreAB structural subunits fine-tuned by acidity and the nickel-dependent transcriptional regulator NikR; (2) nickel uptake into cells via NixA importer; (3) nickel storage in histidine-rich proteins such as Hpn, Hpnl and HspA; (4) nickel incorporation into urease as mediated by accessory proteins UreEFGH and HypAB; and (5) urea substrate entry via UreI. H. pylori: Helicobacter pylori.
Footnotes
Supported by National Natural Science Foundation of China, No. 20801061, No. 31000373 and No. 21371182; The Fundamental Research Funds for the Central Universities, No. 10lgpy19 and No. 21611201; Pearl River Rising Star of Science and Technology of Guangzhou City, No. 2011J2200079 and No. 2011J2200003
P- Reviewers: McGee DJ, Roychoudhury S, Wang WH S- Editor: Cui XM L- Editor: Wang TQ E- Editor: Wang CH
References
- 1.Blaser MJ. Helicobacter pylori: its role in disease. Clin Infect Dis. 1992;15:386–391. doi: 10.1093/clind/15.3.386. [DOI] [PubMed] [Google Scholar]
- 2.Ricci V, Zarrilli R, Romano M. Voyage of Helicobacter pylori in human stomach: odyssey of a bacterium. Dig Liver Dis. 2002;34:2–8. doi: 10.1016/s1590-8658(02)80051-2. [DOI] [PubMed] [Google Scholar]
- 3.Sun H, Zhang L, Szeto KY. Bismuth in medicine. Met Ions Biol Syst. 2004;41:333–378. [PubMed] [Google Scholar]
- 4.Ge R, Chen Z, Zhou Q. The actions of bismuth in the treatment of Helicobacter pylori infections: an update. Metallomics. 2012;4:239–243. doi: 10.1039/c2mt00180b. [DOI] [PubMed] [Google Scholar]
- 5.Teyssen S, Chari ST, Scheid J, Singer MV. Effect of repeated boluses of intravenous omeprazole and primed infusions of ranitidine on 24-hour intragastric pH in healthy human subjects. Dig Dis Sci. 1995;40:247–255. doi: 10.1007/BF02065405. [DOI] [PubMed] [Google Scholar]
- 6.Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 7.Morgan DR, Freedman R, Depew CE, Kraft WG. Growth of Campylobacter pylori in liquid media. J Clin Microbiol. 1987;25:2123–2125. doi: 10.1128/jcm.25.11.2123-2125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutknecht J. Proton/hydroxide conductance through lipid bilayer membranes. J Membr Biol. 1984;82:105–112. doi: 10.1007/BF01870737. [DOI] [PubMed] [Google Scholar]
- 9.Murakami M, Yoo JK, Teramura S, Yamamoto K, Saita H, Matuo K, Asada T, Kita T. Generation of ammonia and mucosal lesion formation following hydrolysis of urea by urease in the rat stomach. J Clin Gastroenterol. 1990;12 Suppl 1:S104–S109. doi: 10.1097/00004836-199001001-00018. [DOI] [PubMed] [Google Scholar]
- 10.Eaton KA, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrutis KA, Fox JG, Schauer DB, Marini RP, Murphy JC, Yan L, Solnick JV. Inability of an isogenic urease-negative mutant stain of Helicobacter mustelae to colonize the ferret stomach. Infect Immun. 1995;63:3722–3725. doi: 10.1128/iai.63.9.3722-3725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weeks DL, Gushansky G, Scott DR, Sachs G. Mechanism of proton gating of a urea channel. J Biol Chem. 2004;279:9944–9950. doi: 10.1074/jbc.M312680200. [DOI] [PubMed] [Google Scholar]
- 14.Hu LT, Mobley HL. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobley HL, Hu LT, Foxal PA. Helicobacter pylori urease: properties and role in pathogenesis. Scand J Gastroenterol Suppl. 1991;187:39–46. [PubMed] [Google Scholar]
- 16.Davies BJ, de Vries N, Rijpkema SG, van Vliet AH, Penn CW. Transcriptional and mutational analysis of the Helicobacter pylori urease promoter. FEMS Microbiol Lett. 2002;213:27–32. doi: 10.1111/j.1574-6968.2002.tb11281.x. [DOI] [PubMed] [Google Scholar]
- 17.Shirai M, Fujinaga R, Akada JK, Nakazawa T. Activation of Helicobacter pylori ureA promoter by a hybrid Escherichia coli-H. pylori rpoD gene in E. coli. Gene. 1999;239:351–359. doi: 10.1016/s0378-1119(99)00389-3. [DOI] [PubMed] [Google Scholar]
- 18.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 19.Stingl K, De Reuse H. Staying alive overdosed: how does Helicobacter pylori control urease activity? Int J Med Microbiol. 2005;295:307–315. doi: 10.1016/j.ijmm.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Dunn BE, Campbell GP, Perez-Perez GI, Blaser MJ. Purification and characterization of urease from Helicobacter pylori. J Biol Chem. 1990;265:9464–9469. [PubMed] [Google Scholar]
- 21.Jahns T. Ammonium/urea-dependent generation of a proton electrochemical potential and synthesis of ATP in Bacillus pasteurii. J Bacteriol. 1996;178:403–409. doi: 10.1128/jb.178.2.403-409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 23.Ferrero RL, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchar J, Hausinger RP. Biosynthesis of metal sites. Chem Rev. 2004;104:509–525. doi: 10.1021/cr020613p. [DOI] [PubMed] [Google Scholar]
- 25.Park IS, Carr MB, Hausinger RP. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc Natl Acad Sci USA. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moncrief MB, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moncrief MB, Hausinger RP. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am J Physiol Gastrointest Liver Physiol. 2003;284:G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- 29.Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 30.Abraham LO, Li Y, Zamble DB. The metal- and DNA-binding activities of Helicobacter pylori NikR. J Inorg Biochem. 2006;100:1005–1014. doi: 10.1016/j.jinorgbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V. In vitro analysis of protein-operator interactions of the NikR and fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J Bacteriol. 2005;187:7703–7715. doi: 10.1128/JB.187.22.7703-7715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chivers PT, Sauer RT. NikR is a ribbon-helix-helix DNA-binding protein. Protein Sci. 1999;8:2494–2500. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chivers PT, Sauer RT. Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–19741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 34.Wang SC, Dias AV, Bloom SL, Zamble DB. Selectivity of metal binding and metal-induced stability of Escherichia coli NikR. Biochemistry. 2004;43:10018–10028. doi: 10.1021/bi049405c. [DOI] [PubMed] [Google Scholar]
- 35.Dian C, Schauer K, Kapp U, McSweeney SM, Labigne A, Terradot L. Structural basis of the nickel response in Helicobacter pylori: crystal structures of HpNikR in Apo and nickel-bound states. J Mol Biol. 2006;361:715–730. doi: 10.1016/j.jmb.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 36.Musiani F, Bertosa B, Magistrato A, Zambelli B, Turano P, Losasso V, Micheletti C, Ciurli S, Carloni P. Computational study of the DNA-binding protein Helicobacter pylori NikR: the role of Ni2. J Chem Theory Comput. 2010;6:3503–3515. doi: 10.1021/ct900635z. [DOI] [PubMed] [Google Scholar]
- 37.West AL, St John F, Lopes PE, MacKerell AD, Pozharski E, Michel SL. Holo-Ni(II)HpNikR is an asymmetric tetramer containing two different nickel-binding sites. J Am Chem Soc. 2010;132:14447–14456. doi: 10.1021/ja104118r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benini S, Cianci M, Ciurli S. Holo-Ni2+ Helicobacter pylori NikR contains four square-planar nickel-binding sites at physiological pH. Dalton Trans. 2011;40:7831–7833. doi: 10.1039/c1dt11107h. [DOI] [PubMed] [Google Scholar]
- 39.Zambelli B, Bellucci M, Danielli A, Scarlato V, Ciurli S. The Ni2+ binding properties of Helicobacter pylori NikR. Chem Commun (Camb) 2007;(35):3649–3651. doi: 10.1039/b706025d. [DOI] [PubMed] [Google Scholar]
- 40.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol Microbiol. 2003;49:947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 41.Higgins KA, Carr CE, Maroney MJ. Specific metal recognition in nickel trafficking. Biochemistry. 2012;51:7816–7832. doi: 10.1021/bi300981m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West AL, Evans SE, González JM, Carter LG, Tsuruta H, Pozharski E, Michel SL. Ni(II) coordination to mixed sites modulates DNA binding of HpNikR via a long-range effect. Proc Natl Acad Sci USA. 2012;109:5633–5638. doi: 10.1073/pnas.1120283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Vliet AH, Ernst FD, Kusters JG. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 2004;12:489–494. doi: 10.1016/j.tim.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Wolfram L, Haas E, Bauerfeind P. Nickel represses the synthesis of the nickel permease NixA of Helicobacter pylori. J Bacteriol. 2006;188:1245–1250. doi: 10.1128/JB.188.4.1245-1250.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pflock M, Kennard S, Delany I, Scarlato V, Beier D. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect Immun. 2005;73:6437–6445. doi: 10.1128/IAI.73.10.6437-6445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eitinger T, Mandrand-Berthelot MA. Nickel transport systems in microorganisms. Arch Microbiol. 2000;173:1–9. doi: 10.1007/s002030050001. [DOI] [PubMed] [Google Scholar]
- 47.Mobley HL, Garner RM, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 48.Eitinger T, Suhr J, Moore L, Smith JA. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals. 2005;18:399–405. doi: 10.1007/s10534-005-3714-x. [DOI] [PubMed] [Google Scholar]
- 49.Hendricks JK, Mobley HL. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nolan KJ, McGee DJ, Mitchell HM, Kolesnikow T, Harro JM, O’Rourke J, Wilson JE, Danon SJ, Moss ND, Mobley HL, et al. In vivo behavior of a Helicobacter pylori SS1 nixA mutant with reduced urease activity. Infect Immun. 2002;70:685–691. doi: 10.1128/iai.70.2.685-691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fulkerson JF, Garner RM, Mobley HL. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J Biol Chem. 1998;273:235–241. doi: 10.1074/jbc.273.1.235. [DOI] [PubMed] [Google Scholar]
- 52.Wolfram L, Bauerfeind P. Conserved low-affinity nickel-binding amino acids are essential for the function of the nickel permease NixA of Helicobacter pylori. J Bacteriol. 2002;184:1438–1443. doi: 10.1128/JB.184.5.1438-1443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sunderman FW. Biological monitoring of nickel in humans. Scand J Work Environ Health. 1993;19 Suppl 1:34–38. [PubMed] [Google Scholar]
- 54.Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, Kusters JG, van Vliet AH. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect Immun. 2005;73:7252–7258. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bauerfeind P, Garner RM, Mobley LT. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schauer K, Gouget B, Carrière M, Labigne A, de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol Microbiol. 2007;63:1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 57.Maier RJ, Benoit SL, Seshadri S. Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals. 2007;20:655–664. doi: 10.1007/s10534-006-9061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benoit SL, McMurry JL, Hill SA, Maier RJ. Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochim Biophys Acta. 2012;1820:1519–1525. doi: 10.1016/j.bbagen.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atanassova A, Zamble DB. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J Bacteriol. 2005;187:4689–4697. doi: 10.1128/JB.187.14.4689-4697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia W, Li H, Yang X, Wong KB, Sun H. Metallo-GTPase HypB from Helicobacter pylori and its interaction with nickel chaperone protein HypA. J Biol Chem. 2012;287:6753–6763. doi: 10.1074/jbc.M111.287581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benoit SL, Mehta N, Weinberg MV, Maier C, Maier RJ. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology. 2007;153:1474–1482. doi: 10.1099/mic.0.2006/003228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng T, Li H, Yang X, Xia W, Sun H. Interaction of SlyD with HypB of Helicobacter pylori facilitates nickel trafficking. Metallomics. 2013;5:804–807. doi: 10.1039/c3mt00014a. [DOI] [PubMed] [Google Scholar]
- 64.Xia W, Li H, Sze KH, Sun H. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J Am Chem Soc. 2009;131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 65.Rowinska-Zyrek M, Potocki S, Witkowska D, Valensin D, Kozlowski H. The zinc-binding fragment of HypA from Helicobacter pylori: a tempting site also for nickel ions. Dalton Trans. 2013;42:6012–6020. doi: 10.1039/c2dt32195e. [DOI] [PubMed] [Google Scholar]
- 66.Herbst RW, Perovic I, Martin-Diaconescu V, O’Brien K, Chivers PT, Pochapsky SS, Pochapsky TC, Maroney MJ. Communication between the zinc and nickel sites in dimeric HypA: metal recognition and pH sensing. J Am Chem Soc. 2010;132:10338–10351. doi: 10.1021/ja1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kennedy DC, Herbst RW, Iwig JS, Chivers PT, Maroney MJ. A dynamic Zn site in Helicobacter pylori HypA: a potential mechanism for metal-specific protein activity. J Am Chem Soc. 2007;129:16–17. doi: 10.1021/ja066958x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sydor AM, Liu J, Zamble DB. Effects of metal on the biochemical properties of Helicobacter pylori HypB, a maturation factor of [NiFe]-hydrogenase and urease. J Bacteriol. 2011;193:1359–1368. doi: 10.1128/JB.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the urease metallocenter. J Biol Chem. 2013;288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zambelli B, Musiani F, Benini S, Ciurli S. Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Acc Chem Res. 2011;44:520–530. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]
- 71.Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Helicobacter pylori UreE, a urease accessory protein: specific Ni(2+)- and Zn(2+)-binding properties and interaction with its cognate UreG. Biochem J. 2009;422:91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 73.Shi R, Munger C, Asinas A, Benoit SL, Miller E, Matte A, Maier RJ, Cygler M. Crystal structures of apo and metal-bound forms of the UreE protein from Helicobacter pylori: role of multiple metal binding sites. Biochemistry. 2010;49:7080–7088. doi: 10.1021/bi100372h. [DOI] [PubMed] [Google Scholar]
- 74.Banaszak K, Martin-Diaconescu V, Bellucci M, Zambelli B, Rypniewski W, Maroney MJ, Ciurli S. Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking. Biochem J. 2012;441:1017–1026. doi: 10.1042/BJ20111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biagi F, Musiani F, Ciurli S. Structure of the UreD-UreF-UreG-UreE complex in Helicobacter pylori: a model study. J Biol Inorg Chem. 2013;18:571–577. doi: 10.1007/s00775-013-1002-8. [DOI] [PubMed] [Google Scholar]
- 76.Fong YH, Wong HC, Chuck CP, Chen YW, Sun H, Wong KB. Assembly of preactivation complex for urease maturation in Helicobacter pylori: crystal structure of UreF-UreH protein complex. J Biol Chem. 2011;286:43241–43249. doi: 10.1074/jbc.M111.296830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fong YH, Wong HC, Yuen MH, Lau PH, Chen YW, Wong KB. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol. 2013;11:e1001678. doi: 10.1371/journal.pbio.1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, Bergeret M, Braimi N, Dupont C, Labigne A. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol. 1996;147:661–669. doi: 10.1016/0923-2508(96)84023-x. [DOI] [PubMed] [Google Scholar]
- 79.Cun S, Li H, Ge R, Lin MC, Sun H. A histidine-rich and cysteine-rich metal-binding domain at the C terminus of heat shock protein A from Helicobacter pylori: implication for nickel homeostasis and bismuth susceptibility. J Biol Chem. 2008;283:15142–15151. doi: 10.1074/jbc.M800591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schauer K, Muller C, Carrière M, Labigne A, Cavazza C, De Reuse H. The Helicobacter pylori GroES cochaperonin HspA functions as a specialized nickel chaperone and sequestration protein through its unique C-terminal extension. J Bacteriol. 2010;192:1231–1237. doi: 10.1128/JB.01216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gilbert JV, Ramakrishna J, Sunderman FW, Wright A, Plaut AG. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect Immun. 1995;63:2682–2688. doi: 10.1128/iai.63.7.2682-2688.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mobley HL, Garner RM, Chippendale GR, Gilbert JV, Kane AV, Plaut AG. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter. 1999;4:162–169. doi: 10.1046/j.1523-5378.1999.99286.x. [DOI] [PubMed] [Google Scholar]
- 83.Ge R, Sun H. Bioinorganic chemistry of bismuth and antimony: target sites of metallodrugs. Acc Chem Res. 2007;40:267–274. doi: 10.1021/ar600001b. [DOI] [PubMed] [Google Scholar]
- 84.Ge R, Watt RM, Sun X, Tanner JA, He QY, Huang JD, Sun H. Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochem J. 2006;393:285–293. doi: 10.1042/BJ20051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ge R, Zhang Y, Sun X, Watt RM, He QY, Huang JD, Wilcox DE, Sun H. Thermodynamic and kinetic aspects of metal binding to the histidine-rich protein, Hpn. J Am Chem Soc. 2006;128:11330–11331. doi: 10.1021/ja062589t. [DOI] [PubMed] [Google Scholar]
- 86.Ge R, Sun X, Wang D, Zhou Q, Sun H. Histidine-rich protein Hpn from Helicobacter pylori forms amyloid-like fibrils in vitro and inhibits the proliferation of gastric epithelial AGS cells. Biochim Biophys Acta. 2011;1813:1422–1427. doi: 10.1016/j.bbamcr.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 87.Ge R, Sun X. The in vivo functions of a histidine-rich protein Hpn in Helicobacter pylori: linking gastric and Alzheimer’s diseases together? Med Hypotheses. 2011;77:788–790. doi: 10.1016/j.mehy.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 88.Zeng YB, Zhang DM, Li H, Sun H. Binding of Ni2+ to a histidine- and glutamine-rich protein, Hpn-like. J Biol Inorg Chem. 2008;13:1121–1131. doi: 10.1007/s00775-008-0397-0. [DOI] [PubMed] [Google Scholar]
- 89.Benoit SL, Miller EF, Maier RJ. Helicobacter pylori stores nickel to aid its host colonization. Infect Immun. 2013;81:580–584. doi: 10.1128/IAI.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


