Centrosomes establish a mitotic spindle geometry that facilitates correct kinetochore–microtubule attachments, contributing to the fidelity of chromosome segregation.
Abstract
Most animal cells contain a centrosome, which comprises a pair of centrioles surrounded by an ordered pericentriolar matrix (PCM). Although the role of this organelle in organizing the mitotic spindle poles is well established, its precise contribution to cell division and cell survival remains a subject of debate. By genetically ablating key components of centriole biogenesis in chicken DT40 B cells, we generated multiple cell lines that lack centrioles. PCM components accumulated in acentriolar microtubule (MT)-organizing centers but failed to adopt a higher-order structure, as shown by three-dimensional structured illumination microscopy. Cells without centrioles exhibited both a delay in bipolar spindle assembly and a high rate of chromosomal instability. Collectively, our results expose a vital role for centrosomes in establishing a mitotic spindle geometry that facilitates correct kinetochore–MT attachments. We propose that centrosomes are essential in organisms in which rapid segregation of a large number of chromosomes needs to be attained with fidelity.
Introduction
Centrosomes undergo a tightly controlled duplication process, which involves the hierarchical assembly of a small number of proteins (Gönczy, 2012). In brief, in S phase, each old centriole templates the assembly of a procentriole around a ninefold symmetrical structure, the cartwheel. The procentriole grows by elongation of triplet and doublet microtubules (MTs) comprising its wall. CEP152 and STIL are essential for centriole formation, and mutations in either genes lead to developmental disorders (Kleylein-Sohn et al., 2007; Cizmecioglu et al., 2010; Hatch et al., 2010; Klingseisen and Jackson, 2011; Mahmood et al., 2011; Tang et al., 2011; Arquint et al., 2012; Vulprecht et al., 2012).
During mitosis, the two centrosomes nucleate and organize MTs at the spindle poles. However, bipolar spindles can assemble in Drosophila melanogaster cells lacking centrioles (Bettencourt-Dias et al., 2005; Basto et al., 2006) and in mammalian cells after laser ablation of centrosomes (Khodjakov et al., 2000). The prevailing view is that chromatin-dependent spindle assembly renders centrosomes dispensable for bipolar spindle formation (Meunier and Vernos, 2012). If so, why do the majority of proliferating animal cells contain centrosomes? Studies aiming to eliminate centrosomes from vertebrate cells have so far relied on laser ablation (Khodjakov et al., 2000), microsurgical removal of centrosomes (Maniotis and Schliwa, 1991; Hinchcliffe et al., 2001; Hornick et al., 2011), or antibody injections against centriole components (Bobinnec et al., 1998). Although very informative, these methods are not suitable for tracking cell fate over many cell divisions.
Here, we report the consequences of permanent centriole loss in vertebrate cells after disruption of CEP152 and STIL. Our results collectively dispute the notion that centrosomes are dispensable for mitosis because vertebrate cells without centrioles display marked mitotic delay, chromosome instability, and aneuploidy.
Results and discussion
Disruption of CEP152 and STIL causes loss of centrioles in vertebrate cells
We disrupted CEP152 and STIL genes in the hyperrecombinogenic chicken B cell line, DT40. Protein-null CEP152 knockout (KO) cells were generated by removing exons encoding aa 1–433, a conserved domain in CEP152 that mediates binding to a key regulator of centriole biogenesis, PLK4 (Fig. S1, A–E; Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). STIL-KO alleles were created by removing exons encoding aa 785–1,130 of STIL, comprising the conserved STAN (STIL/Ana2) motif (Fig. S1, F and G; Stevens et al., 2010a). Disruption of STIL causes embryonic lethality and abnormal centrosome function in zebrafish and mouse, but the extent of centriole impairment in these models is not known (Izraeli et al., 1999; Pfaff et al., 2007; Castiel et al., 2011).
Centrosome ultrastructure was analyzed by serial section transmission EM (TEM) in two independent clones of CEP152-KO (#1 and #2) and STIL-KO cells (#1 and #2). Centrioles are normally embedded in the pericentriolar matrix (PCM), the site of MT nucleation. Clusters of electron-dense granules, called centriolar satellites, are also associated with the PCM in interphase cells (Kubo et al., 1999). Unlike wild-type (WT) cells, in which intact centrioles were frequently observed (Fig. 1 A), centriole-like electron-dense structures were rare in KO cells. When a mitotic pole or a clump of centriolar satellites was found, the region was examined by serial sectioning. In both mutants, satellite clumps were often associated with an area containing a high density of cytoplasmic MTs, termed acentriolar MT-organizing centers (aMTOCs; Hornick et al., 2011). In about half of CEP152-KO cells, aMTOCs also contained structures resembling disrupted centriolar walls together with dissociated doublet and triplet MTs (Fig. 1, B, C, and G), which were absent in STIL-KO cells (Fig. 1, D and E). The spindle poles of KO cells contained no centrioles (Fig. 1, F and G). Thus, CEP152-KO and STIL-KO cells lack intact centrioles.
Figure 1.
CEP152-KO and STIL-KO DT40 cells lack intact centrioles. (A–G) TEM of WT, CEP152-KO, and STIL-KO cells. Arrowheads indicate centriolar satellites. Bars, 100 nm. (A) A centriole pair is illustrated in WT. (B and C) CEP152-KO cells contain electron-dense structures reminiscent of partial centriole walls. Insets in A and C show a cross section of triplet MTs at high magnification. (D–F) STIL-KO cells lack centriolar structures and contain only aMTOCs. (F) Mitotic spindle pole in STIL-KO. (G) Table depicts quantification of the major phenotypes observed by TEM. Asterisks indicate electron-dense structures that can potentially correspond to centrioles.
The presence of centriolar MT triplets in CEP152-KO, but not in STIL-KO, highlights differences in their respective roles in centriole biogenesis. CEP152, STIL, and the essential cartwheel component SAS-6 have all been implicated in centrosomal targeting of CPAP, a protein thought to recruit centriolar MTs, with STIL and SAS-6 being interdependent for localization (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Stevens et al., 2010b; Tang et al., 2011; Vulprecht et al., 2012). However, CEP152-KO aMTOCs contain centriole wall remnants and triplet MTs, indicating the presence of CPAP. Thus, STIL and SAS-6 (or other components) can recruit functional CPAP in the absence of CEP152. The lack of stable cylindrical centrioles in CEP152-KO cells, however, strongly implicates CEP152 in establishing and/or stabilizing the ninefold symmetry of centrioles.
aMTOCs assemble in the absence of centrioles but are disordered
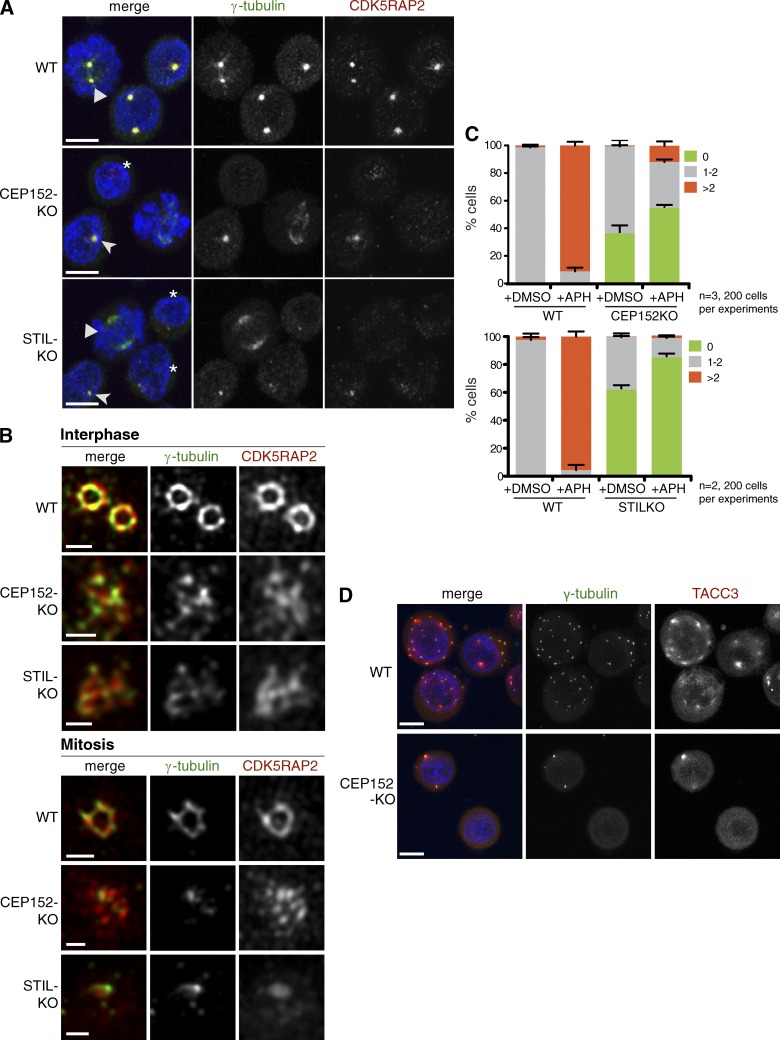
Consistent with the presence of aMTOCs in KO cells, immunofluorescence microscopy revealed cytoplasmic foci enriched in the PCM components CDK5RAP2 and γ-tubulin. The two proteins exhibited significant cell to cell and cell cycle–dependent variations in their localization: during mitosis, γ-tubulin spread along MTs, whereas CDK5RAP2 seemed more dispersed (Fig. 2 A). In interphase, the majority of cells possessed a single aMTOC (Fig. S2 A). aMTOCs in KO cells are reminiscent of structures reported upon disruption of centrosomes in Drosophila and mammalian cells (Maniotis and Schliwa, 1991; Debec et al., 1995; Khodjakov et al., 2000; Hinchcliffe et al., 2001; Basto et al., 2006; Moutinho-Pereira et al., 2009; Hornick et al., 2011; Lecland et al., 2013).
Figure 2.
aMTOCs in KO cells are disordered and unable to overduplicate. (A) CEP152-KO and STIL-KO cells contain cytoplasmic foci enriched in γ-tubulin and CDK5RAP2. Triangles indicate mitotic cells. Asterisks and arrowheads indicate cells with undetectable or weak PCM signal, respectively. DNA is shown in blue. Bars, 5 µm. (B) 3D-SIM images of interphase and mitotic centrosomes stained with antibodies against γ-tubulin and CDK5RAP2 are shown. Bars, 0.5 µm. (C) Centrosome amplification during aphidicolin (APH)-induced arrest. Cells were treated with DMSO or aphidicolin for 24 h. Graphs show the percentages of cells without centrosome (0), one to two centrosomes (1–2), or more than two centrosomes (>2). Centrosomes are defined as γ-tubulin–positive foci. Although ∼10% CEP152-KO cells show centrosome amplification, unlike WT, these contained up to three foci. Error bars are means ± SEM. (D) Representative fields of aphidicolin-treated cells corresponding to C. Cells were stained with antibodies against γ-tubulin and the spindle pole protein TACC3. DNA is shown in blue. Bars, 10 µm.
Super-resolution microscopy has revealed ordered PCM domains in centrosomes, which appear as concentric rings (Sir et al., 2011; Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). To address whether PCM proteins can self-organize in the absence of centrioles, localization of γ-tubulin and CDK5RAP2 was viewed by 3D structured illumination microscopy (SIM; Fig. 2 B). Both proteins appeared as rings in 100% of WT centrosomes (n = 46) but were disordered in all KO aMTOCs (n = 44 CEP152-KO and n = 35 STIL-KO). Maintenance of aMTOCs does not seem to require MTs (Fig. S2 A). Interphase MT networks were comparable between CEP152-KO and WT (Fig. S2 B), and some aMTOCs were able to nucleate MTs (Fig. S2 C). Prolonged treatment with the DNA replication inhibitor aphidicolin induced centrosome amplification only in WT cells, indicating that aMTOCs cannot overduplicate in KO cells (Fig. 2, C and D).
Cells without intact centrioles proliferate slower but are DNA repair proficient
We obtained several CEP152-KO and STIL-KO cell lines at the expected gene-targeting frequencies, implying that loss of centrioles does not preclude clonal growth. Although KO cells could be maintained in log phase indefinitely, they showed increased doubling time and apoptosis (Fig. 3, A and B). Even after deducting apoptotic cells, doubling times were ∼10% higher than WT, indicative of longer cell cycles. Cell cycle profiles were not markedly disrupted; although a ∼1.4-fold increase in G2/M and a decrease in S-phase populations were seen (Fig. 3 C). Regulatory components of cell cycle progression and DNA damage response localize to the PCM, implicating the centrosome in these fundamental signaling pathways (Doxsey et al., 2005; Löffler et al., 2006). We therefore asked whether cells lacking intact centrosomes are defective in their ability to deal with DNA damage. Cells were subjected to a range of genotoxic treatments, and their colony formation potential was assayed. KO cells showed no increase in sensitivity to ultraviolet-C (not depicted), double-strand break-inducing γ-irradiation, or the cross-linking agent cisplatin (Fig. 3 D). Therefore, intact centrosomes are dispensable for survival after DNA damage. Because cytoplasmic PCM foci are present in ∼60% of CEP152-KO and 40% STIL-KO (albeit much reduced in size; Fig. 2, A and C), these aMTOCs may perform a limited role in signaling.
Figure 3.
Cells lacking intact centrioles proliferate slowly but are proficient in DNA repair. (A) CEP152-KO and STIL-KO cells display growth impairment. (B) KO cells show increased apoptosis compared with WT. n = 4. Error bars indicate SEMs, analysis of variance, Tukey’s test. (C) Quantification of cell cycle profiles by FACS analysis with propidium Iodide (PI) and anti-BrdU antibodies, with >15,000 events per genotype. Percentages of cells per each cell cycle stage are indicated. (D) Exponential one phase decay graph illustrates clonogenic survival of WT, CEP152-KO, and STIL-KO cells after genotoxic treatments. The percentage of colony formation was normalized against untreated controls of the same genotype. Cells were treated with indicated doses of cisplatin for 1.5 h or irradiation (IR). Error bars indicate SDs.
Centrosomes are required for efficient bipolar spindle formation and timely anaphase onset
Mitotic KO cells displayed disorganized and unfocused MT arrays (Fig. 4, A–C), which appeared to contact kinetochores (Fig. 4 B). Time-lapse microscopy of GFP-tubulin–expressing cells revealed that WT cells entered mitosis with two MT asters and formed bipolar spindles within 3–6 min after nuclear envelope breakdown (NEBD; Fig. 4 D and Video 1). In contrast, KO cells displayed no discernible MT asters until NEBD, when a surge of MT formation was seen (Videos 2 and 3). Initially, unfocused MTs eventually assembled into bipolar spindles, indicative of spindle self-organization. KO cells also exhibited a prominent delay in anaphase onset (Fig. 4 E), a consistent phenotype of centrosome disruption in experimental model systems (Basto et al., 2006; Pfaff et al., 2007; Castiel et al., 2011; Hornick et al., 2011). About 60% of CEP152-KO and 40% of STIL-KO cells contained one aMTOC in interphase (Fig. 2 C). However, live imaging revealed no monoasters, indicating that most aMTOCs cannot form robust spindle poles. To identify sites of MT assembly during mitosis, we examined MT regrowth after depolymerization. In WT cells, MTs were predominantly produced at centrosomes, whereas in KO cells, nascent MTs concentrated near the chromatin (Fig. 4 F). Thus, chromatin-dependent MT formation is likely to drive anastral spindle assembly in KO cells, but augmin-dependent MT assembly may also contribute (Duncan and Wakefield, 2011; Meunier and Vernos, 2012). In summary, we show that centrosome-driven spindle assembly facilitates bipolar spindle formation, thereby ensuring normal mitotic timing.
Figure 4.
Bipolar spindle formation and anaphase onset are delayed in cells lacking intact centrioles. (A) CEP152-KO and STIL-KO cells contain disorganized mitotic spindles. Cells were stained with antibodies against CDK5RAP2 and α-tubulin. DNA is shown in blue. Asterisks mark cells with disorganized spindles. (B) Unfocused MTs contact kinetochores in STIL-KO cells. Cells were stained with antibodies against the kinetochore protein, CENP-O, and α-tubulin. DNA is shown in blue. A single focal plane is shown along with higher magnification of framed area. (C) Graphs show the percentage of mitotic cells with disorganized spindles in CEP152-KO and STIL-KO (n = 3, >165 cells per genotype per experiment; means ± SD). (D) Still frames from time-lapse experiments show GFP-tubulin–expressing WT, CEP152-KO, and STIL-KO cells (Videos 1, 2, and 3). NEBD marks the first frame after NEBD. Times are given in hours, minutes, and seconds. (E) Graph shows distribution of time intervals from NEBD to anaphase onset. Medians and interquartile ranges are indicated, from Kruskal–Wallis and Dunn’s test. (F) MT regrowth in mitotic cells after depolymerization. Note the absence of MTs at 0 min and enrichment of MTs near chromatin in STIL-KO cells after 3 min of regrowth. Cells were stained with antibodies against CDK5RAP2 and α-tubulin. Bars, 5 µm.
Cells lacking intact centrioles are chromosomally unstable and aneuploid
We next asked whether abnormal spindle assembly in KO cells affects mitotic fidelity. Chromosome behavior was followed in live cells expressing histone H2B-GFP. Nearly 30% of KO exhibited chromosome missegregation (Fig. 5, A and B; and Videos 4, 5, and 6). To exclude that this phenotype was caused by electroporation used in gene targeting, we filmed a cell line, which underwent gene targeting but contained normal centrosomes. Importantly, neither WT nor these so-called WT-2 cells displayed chromosome missegregation (Fig. 5 B; Sir et al., 2011). Cleavage furrow positioning appeared normal in KO cells (Fig. S3 A). Like WT, furrow ingression occurred within 3 min of anaphase onset followed by the appearance of the cytokinetic bridge 3–6 min later (Fig. S3 B). Thus, intact centrosomes are dispensable for progression through telophase and cytokinesis.
Figure 5.
CEP152-KO and STIL-KO cells exhibit chromosome instability and aneuploidy. (A) Still frames from time-lapse experiments show H2B-GFP–expressing WT, CEP152-KO, and STIL-KO cells (Videos 4, 5, and 6). Lagging chromatids (red arrowheads) or larger chromatin masses (yellow arrowheads) are evident during anaphase in KO cells. Pictures with blue asterisks are shown with increased gain setting in the framed area. Bars, 2 µm. (B) Table summarizes chromosome missegregation phenotypes from time-lapse experiments. Overlays of H2B-GFP and bright-field images were analyzed. (C) Lagging chromatids in anaphase cells show merotelic attachments. Cells were stained with antibodies against CENP-O and α-tubulin. DNA is shown in blue. Framed areas are shown at higher magnification. Bars, 5 µm. (D) Chromosome (chr) spreads of WT and CEP152-KO cells. WT cells show normal ploidy. CEP152-KO image on the left illustrates CA (trisomy of chromosome 3 but normal copy number of chromosomes 1, 2, 4, and Z) and, on the right, CIN (trisomy of chromosome 4 but normal copy number of chromosomes 1, 2, 3, and Z). Pie charts on the bottom depict the percentage of cells with indicated ploidies. Numbers of chromosomes 1–4 and Z were scored. Bar, 8 µm.
KO cells frequently displayed lagging chromatids during anaphase (Fig. S3 C). In addition, some cells exhibited three unequal chromatin masses in anaphase, but eventually underwent bipolar cytokinesis. We suspect that such segregation defects result from an extra spindle pole persisting into anaphase because spindles in KO cells often showed multipolar configurations before attaining bipolarity (Fig. 4 A). Merotelic attachments, in which one kinetochore is simultaneously attached to MTs emanating from two spindle poles, can lead to lagging chromatids (Gregan et al., 2011). Anastral spindles in KO cells permit formation of kinetochore–MT attachments before spindle pole focusing (Fig. 4 B), a potential source of merotely. Indeed, merotelic attachments were detectable on lagging chromatids in anaphase KO cells (Fig. 5 C). Such lagging chromatids may get trapped in the midbody and interfere with cleavage furrow ingression (Janssen et al., 2011), which could explain the rare cytokinesis failures in the KO. Lagging chromatids often integrate into one of the daughter nuclei, but those that fail to do so before nuclear envelope reassembly give rise to micronuclei instead (Crasta et al., 2012). KO cells exhibited examples of both fates (Fig. 5 A and Fig. S3, D and E). Lagging chromatids are expected to segregate randomly between daughter cells, although a bias for correct segregation has been noted (Cimini et al., 2004; Thompson and Compton, 2011). Therefore, although ∼25% of KO cells display lagging chromatids, these probably cause fewer than 10% missegregation events.
To assess whether chromosome missegregation precipitates aneuploidy, chromosome spreads were analyzed. Chicken has a diploid karyotype of 2n = 78 chromosomes (4 pairs of macrochromosomes, 34 pairs of microchromosomes, and 1 pair of sex chromosomes ZW). DT40 cells are trisomic for chromosome 2. Scoring macrochromosomes 1–4 and Z revealed aberrant ploidy in CEP152-KO and STIL-KO cells (Fig. 5 D). Interestingly, we noted genotype-specific alterations: ∼70% of cells from two CEP152-KO clones contained an extra copy of chromosome 3 as opposed to ∼4% of STIL-KO, whereas ∼70% of cells from two STIL-KO clones showed disomy of (the otherwise trisomic) chromosome 2 as opposed to ∼6% of CEP152-KO (Fig. 5 D). Because of their prevalence, we refer to trisomy of chromosome 3 in CEP152-KO and disomy of chromosome 2 in STIL-KO as constitutional aneuploidies (CAs). Upon disruption of CEP152 and STIL, complete loss of centrioles is attained only after two to three cell cycles, and thus, an intermediate cell population containing one to two centrioles is generated. Such cells are prone to form transient monopolar spindles (Sir et al., 2011), potent inducers of merotely (Thompson and Compton, 2008). Segregation errors early in clonal expansion may be the cause of CA in KO cells. In addition to CA, sporadic instances of numerical chromosome alterations were detected in ∼10–20% of KO cells, indicative of chromosomal instability (CIN). Single chromosome losses and gains constituted the majority of CIN. Although there is evidence to suggest that aneuploidy can drive CIN (Pfau and Amon, 2012), our result that a significant proportion of KO cells displays CIN, but not CA, argues against CA being the cause of CIN (Fig. 5 D).
These findings collectively reveal two key roles for centrosomes in vertebrate cells. First, centrosomes increase the speed of spindle assembly. Mitosis takes twice as long in cells without functional centrosomes, a delay that could diminish the proliferative capacity of untransformed cells and their progenitors (Uetake and Sluder, 2010). Second, intact centrosomes improve mitotic fidelity. Remarkably, merotelic attachments are common in mouse oocytes that normally lack centrioles and in cells that fail to separate their centrosomes before NEBD (Kitajima et al., 2011; Silkworth et al., 2012). Thus, establishment of MT–kinetochore attachments seems exquisitely sensitive to spindle geometry in early mitosis (Magidson et al., 2011). Unlike chromatin-driven self-assembly, the presence of two separated centrosomes at NEBD imparts a spindle geometry, which minimizes the incidence of erroneous attachments. Multipolar spindle configurations during anastral spindle formation in KO cells could also contribute to merotely (Thompson and Compton, 2008; Ganem et al., 2009; Silkworth et al., 2009, 2012). Although we favor the explanation that it is the altered spindle geometry, which reduces mitotic fidelity in cells lacking centrioles, CEP152 and STIL could possess yet uncharacterized roles in regulating MT dynamics and/or chromosome segregation. A relatively small increase in aneuploidy was reported upon loss of centrioles in fruit flies (3 vs. 1% in WT; Basto et al., 2006). Anastral spindle formation in centrin mutant Chlamydomonas reinhardtii produced a chromosome loss rate of 0.002 per chromosome per cell division (Zamora and Marshall, 2005). Multiplying 0.002 by 34, the number of chromosomes in C. reinhardtii, suggests that chromosome loss may occur in ∼6% of cell divisions, which is close to our estimate of ∼10% in KO cells lacking centrioles. There are only eight chromosomes in Drosophila, in contrast to 34 and 78 in C. reinhardtii and chicken, respectively. We therefore postulate that centrosome-driven spindle assembly is important for mitotic fidelity predominantly in organisms with high chromosome numbers. Collectively, our work establishes a need for centrosomes in the rapid and faithful segregation of chromosomes in vertebrates. We speculate that impaired mitotic timing and CIN might underlie developmental disorders caused by mutations in centrosomal genes.
Materials and methods
Cell culture, drug treatments, and colony formation assays
DT40 cells were cultured in suspension in RPMI 1640 medium (Invitrogen), which was supplemented with 10% FBS, 1% chicken serum, 110 U/ml penicillin, 10 mg/ml streptomycin, and 50 µM β-mercaptoethanol at 40°C with 5% CO2. Nocodazole (Sigma-Aldrich) was used at 1 µg/ml. For metaphase spreads, DT40 cells were treated with 1 µg/ml colcemid. To assay centrosome amplification, DT40 cells were incubated with 5 µg/ml aphidicolin (Sigma-Aldrich).
For colony formation assays, DT40 cells at 105 cells/ml were treated at 5, 10, 15, 20, and 25 µM clinical-grade cisplatin (gift from J. Brenton, Cancer Research UK Cambridge Institute, Cambridge, England, UK) for 1.5 h or subjected to ionizing radiation (IR) at 2, 4, 6, 8, and 10 Gy to generate DNA lesions. Cells were serially diluted by 10- and 100-fold. 100 µl of each concentration was plated in duplicates onto methylcellulose. Colony numbers were scored after 12 d. Percentage of cell survival was calculated at different concentrations of cisplatin and doses of IR by normalizing against untreated controls of the same genotype.
Homologous gene targeting in DT40 cells
For generating the targeting construct, left and right arm homology regions were PCR amplified from DT40 genomic DNA using DNA polymerase (Phusion High-Fidelity; Finnzymes) or left arm Taq DNA polymerase (Takara Bio Inc.). PCR reactions were performed according to the manufacturer’s instructions using MgCl2-supplemented PCR buffers. The primers used to amplify the left and right arm sequences are listed in Table S1. Homology arms were cloned into pBluescript II SK− (pSK) (Arakawa et al., 2001). A drug resistance cassette (neomycin, blasticidin, or puromycin) was cloned into pSK between BamHI sites. The two alleles of CEP152 and STIL were targeted sequentially: for both CEP152- and STIL-KO, the first allele was targeted with neomycin, and the second was targeted with puromycin. For C-terminal tandem affinity purification (TAP) tagging, CEP152 homology arms were cloned into a pBluescript II SK− vector carrying a GsTAP (protein G and streptavidin binding protein–containing TAP) sequence (Bürckstümmer et al., 2006). The left arm contained the C-terminal part of CEP152 lacking the STOP codon and was inserted in frame upstream of the GsTAP tag. The blasticidin-resistance cassette was cloned into this construct. All final constructs were linearized and transfected as described in Sir et al. (2011). Antibiotic-resistant clones were picked after 7–10 d. Targeted integration of the resistance cassettes was screened by PCR. Primers used for PCR reactions are listed in Table S1. WT-2 cells refer to a previously reported DT40 cell line (TAP-CEP63), which was generated by sequential gene targeting of WT cells to have an in-frame TAP tag into both alleles of CEP63 gene. TAP-CEP63 cells were shown to display normal mitotic spindle morphology (Sir et al., 2011).
Antibodies and immunostainings
Primary antibodies used in this study were CDK5RAP2 and chicken anti-TACC3 raised against aa 126–442 of Gallus gallus TACC3 (Barr et al., 2010), centrin-3 (Abnova), protein G, protein G–HRP (Abcam), α-tubulin (Dm1α; Sigma-Aldrich), γ-tubulin (GTU88; Sigma-Aldrich), BrdU (B44; BD), chicken CENP-O (Medical and Biological Laboratories, Co.), and myosin IIA (Sigma-Aldrich). DNA was stained with Hoechst 33258 (Sigma-Aldrich). Before fixation, DT40 cells were settled onto poly-l-lysine–coated coverslips for 10 min at 40°C. For visualization of mitotic spindles and centrosomal proteins, cells were fixed in cold 100% methanol for 5 min at –20°C and washed with PBS/0.1% Tween 20. For 3D-SIM experiments, DT40 cells were treated with PBS/1% Triton X-100/0.5% NP-40 for 5 min at room temperature after methanol fixation. For visualization of kinetochores, cells were fixed in 3.7% formaldehyde containing 100 mM Pipes, 1 mM MgCl2, 0.5 mM CaCl2, and 0.4% Triton X-100, pH 6.8, and then postfixed in cold methanol for 5 min. All primary antibodies were diluted at 0.25–1 µg/ml in PBS/5% BSA and incubated with coverslips for 2 h at 37°C or overnight at 4°C. Secondary antibodies conjugated to Alexa Fluor 488, 555, and 594 (Invitrogen) were used at 1 µg/ml in PBS/5% BSA and incubated with coverslips for 1 h at 37°C. Coverslips were mounted in antifade medium (ProLong Gold; Invitrogen) containing 1.5 µg/ml Hoechst 33258 (Sigma-Aldrich).
Image acquisition and processing
Imaging of fixed cells was performed on a scanning confocal microscope (Eclipse 90i; Nikon) except for kinetochore stainings, which were acquired on an OMX DeltaVision microscope (Applied Precision) in conventional mode. Cells were mounted in antifade medium (ProLong Gold) and imaged with 100×, 1.4 NA objective (Nikon). Images presented here are 3D projections of z sections taken every 0.5 µm across the cell. Images of any individual figures were acquired using the same settings and were imported into Volocity (5.0; PerkinElmer) and Photoshop (CS6; Adobe) and were adjusted to use the full range of pixel intensities. Super-resolution microscopy was performed using a Structured Illumination Microscope by OMX DeltaVision. Cells were imaged with 60×, 1.4 NA objective (Olympus). Data were reconstructed using softWoRx software (Applied Precision) and then imported into Volocity (5.0) and Photoshop (CS6) and were adjusted to use the full range of pixel intensities. Fluorochromes used in this study are Alexa Fluor fluorophores (Molecular Probes) Alexa Fluor 488, Alexa Fluor 555, and Alexa Fluor 594. DNA was visualized by Hoechst staining. For time-lapse imaging of DT40 cells expressing GFP–α-tubulin and histone H2B–GFP (Kanda et al., 1998), cells were settled onto 60-µm dishes (µ-Dish35mm,low; ibidi) and supplemented with Leibovitz’s L-15 medium (Invitrogen) containing 10% FBS. Cells were imaged at 40°C in a humidified incubation chamber (Tokai Hit) and were imaged using a spinning-disc confocal system (PerkinElmer) mounted on an inverted microscope (Eclipse TE2000-S; Nikon) and equipped with an electron-multiplying charge-coupled device digital camera (C9100-13; Hamamatsu Photonics). Imaging was performed with a frame rate of 3 min with z steps of 1.5 µm using Volocity 2D. Volume-rendered image sequences were exported as QuickTime files (Apple). Bright-field images were taken at the same interval or occasionally at a frame rate of 6 min. For still images, snapshots were taken in Volocity and processed in Photoshop.
EM
DT40 cells were pelleted and immediately fixed in prewarmed 1% glutaraldehyde in PHEM (60 mM Pipes, 25 mM Hepes, 2 mM MgCl2, and 1 mM EGTA, pH 6.9) or PBS buffer for 1 h at 37°C. Cell pellets were washed in 0.1 M Na cacodylate buffer, pH 7.2, at room temperature for 5 min each and postfixed in 1% osmium tetroxide in Na cacodylate buffer for 1 h at room temperature or 30 min at 4°C. Pellets were then washed in Na cacodylate buffer twice for 30 min. Pellets were gradually dehydrated by immersing them in a graded ethanol solution from 50, 70, to 90% and three times in 100% ethanol. Dehydrated cell pellets were embedded in Epoxy medium using Epoxy Embedding kit (Fluka) or Araldite, and serial sections were cut.
MT regrowth
For MT regrowth experiments, cells were treated with 1 µg/ml nocodazole for 1 h (Fig. 4 E) or 3 h (Fig. S3) at 40°C followed by 1 h on ice. Cells were washed three times for 5 min with cold PBS and then transferred to poly-l-lysine–coated coverslips for 30 min on ice. Cells on the coverslip were incubated for 3 min at 40°C to polymerize MTs.
Western blotting
DT40 cells were lysed in hypotonic buffer containing 10 mM Tris-HCl, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 10 µM β-mercaptoethanol, and protease inhibitor cocktail (Sigma-Aldrich) by passing them through a 23-gauge needle. Cleared cytoplasmic extracts were obtained by centrifuging cell lysates at 16,000 g for 20 min at 4°C. Lysates were separated on 3–8% Tris-acetate or 4–12% Bis-Tris SDS-PAGE gels (Invitrogen) and transferred onto nitrocellulose for Western blot analysis.
Apoptosis assay and flow cytometry
For cell cycle analysis, DT40 cells were pulsed with 20 µM BrdU for 10 min. Cells were then fixed in 70% ice-cold ethanol and incubated with 2 M HCl/0.5% Triton X-100 in PBS at 37°C. Cells were then incubated with anti-BrdU antibody at 1:35 for 1 h at 37°C with shaking. Cells were washed and incubated with FITC-conjugated anti–mouse antibody (Jackson ImmunoResearch Laboratories, Inc.). Cells were then treated with 40 µg/ml propidium iodide and 100 µg/ml RNase A for 15 min. Apoptosis assays were performed using the MitoProbe JC-1 Assay kit (Invitrogen) according to the manufacturer’s instruction. The fluorescence emission shift from green to red was measured on the cytometer (FACSCalibur; Cytek) and analyzed with the software FlowJo v10 (Tree Star, Inc.).
Statistical analysis
Statistical analysis and graphs were performed using Excel (Microsoft) or Prism (GraphPad Software). The number of experimental repeats (n values) are reported for each dataset in the figures and figure legends. Data are presented as means ± SEM unless stated otherwise. One-way analysis of variance for multiple comparisons was performed on all data followed by Tukey’s test using Prism 6. When normal distribution could not be confirmed, the nonparametric Kruskal–Wallis was used followed by Dunn’s multiple comparison posttest.
Online supplemental material
Fig. S1 summarizes gene targeting. Fig. S2 and Fig. S3 show analyses of aMTOCs and chromosome missegregation phenotypes, respectively. Table S1 shows primer sequences. Videos 1, 2, and 3 show mitosis in GFP–α-tubulin–expressing WT (Video 1), CEP152-KO (Video 2), and STIL-KO (Video 3) cells. Videos 4, 5, and 6 show mitosis in H2B-GFP–expressing WT (Video 4), CEP152-KO (Video 5), and STIL-KO (Video 6) cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201309038/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201309038.dv.
Supplementary Material
Acknowledgments
We thank the Gergely laboratory and K.J. Patel for helpful comments and the core facilities at the Cambridge Institute for support. J.V. Kilmartin thanks Matthew Freeman and Sean Munro for scientific support.
This work was made possible by Science Foundation Ireland Principal Investigator awards 08/IN.1/B1029 and 10/IN.1/B2972 to C.G. Morrison, Fanconi Anemia Research Fund grant for J.-H. Sir, core funding from Cancer Research UK to J.-H. Sir, M. Pütz, M. Dunning, and F. Gergely, and a Royal Society University Research Fellowship to F. Gergely.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- aMTOC
- acentriolar MT-organizing center
- CA
- constitutional aneuploidy
- CIN
- chromosomal instability
- IR
- ionizing radiation
- KO
- knockout
- MT
- microtubule
- NEBD
- nuclear envelope breakdown
- PCM
- pericentriolar matrix
- SIM
- structured illumination microscopy
- TAP
- tandem affinity purification
- TEM
- transmission EM
- WT
- wild type
References
- Arakawa H., Lodygin D., Buerstedde J.M. 2001. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 1:7 10.1186/1472-6750-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., Sonnen K.F., Stierhof Y.D., Nigg E.A. 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125:1342–1352 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- Barr A.R., Kilmartin J.V., Gergely F. 2010. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 189:23–39 10.1083/jcb.200912163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. 2006. Flies without centrioles. Cell. 125:1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., Bornens M. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. 2006. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 3:1013–1019 10.1038/nmeth968 [DOI] [PubMed] [Google Scholar]
- Castiel A., Danieli M.M., David A., Moshkovitz S., Aplan P.D., Kirsch I.R., Brandeis M., Krämer A., Izraeli S. 2011. The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J. Cell Sci. 124:532–539 10.1242/jcs.079731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Cameron L.A., Salmon E.D. 2004. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 14:2149–2155 10.1016/j.cub.2004.11.029 [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., Hoffmann I. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debec A., Détraves C., Montmory C., Géraud G., Wright M. 1995. Polar organization of gamma-tubulin in acentriolar mitotic spindles of Drosophila melanogaster cells. J. Cell Sci. 108:2645–2653 [DOI] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303–311 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Duncan T., Wakefield J.G. 2011. 50 ways to build a spindle: the complexity of microtubule generation during mitosis. Chromosome Res. 19:321–333 10.1007/s10577-011-9205-8 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D.M. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Fu J., Glover D.M. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Gregan J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., Cimini D. 2011. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 21:374–381 10.1016/j.tcb.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Miller F.J., Cham M., Khodjakov A., Sluder G. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 291:1547–1550 10.1126/science.1056866 [DOI] [PubMed] [Google Scholar]
- Hornick J.E., Mader C.C., Tribble E.K., Bagne C.C., Vaughan K.T., Shaw S.L., Hinchcliffe E.H. 2011. Amphiastral mitotic spindle assembly in vertebrate cells lacking centrosomes. Curr. Biol. 21:598–605 10.1016/j.cub.2011.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izraeli S., Lowe L.A., Bertness V.L., Good D.J., Dorward D.W., Kirsch I.R., Kuehn M.R. 1999. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature. 399:691–694 10.1038/21429 [DOI] [PubMed] [Google Scholar]
- Janssen A., van der Burg M., Szuhai K., Kops G.J., Medema R.H. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 333:1895–1898 10.1126/science.1210214 [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67 10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Ohsugi M., Ellenberg J. 2011. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 146:568–581 10.1016/j.cell.2011.07.031 [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., Nigg E.A. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Klingseisen A., Jackson A.P. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25:2011–2024 10.1101/gad.169037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G.D., Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Lecland N., Debec A., Delmas A., Moutinho-Pereira S., Malmanche N., Bouissou A., Dupré C., Jourdan A., Raynaud-Messina B., Maiato H., Guichet A. 2013. Establishment and mitotic characterization of new Drosophila acentriolar cell lines from DSas-4 mutant. Biol. Open. 2:314–323 10.1242/bio.20133327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler H., Lukas J., Bartek J., Krämer A. 2006. Structure meets function—centrosomes, genome maintenance and the DNA damage response. Exp. Cell Res. 312:2633–2640 10.1016/j.yexcr.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., Khodjakov A. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood S., Ahmad W., Hassan M.J. 2011. Autosomal Recessive Primary Microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J. Rare Dis. 6:39 10.1186/1750-1172-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis A., Schliwa M. 1991. Microsurgical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell. 67:495–504 10.1016/0092-8674(91)90524-3 [DOI] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., Agard D.A. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S., Vernos I. 2012. Microtubule assembly during mitosis - from distinct origins to distinct functions? J. Cell Sci. 125:2805–2814 10.1242/jcs.092429 [DOI] [PubMed] [Google Scholar]
- Moutinho-Pereira S., Debec A., Maiato H. 2009. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol. Biol. Cell. 20:2796–2808 10.1091/mbc.E09-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff K.L., Straub C.T., Chiang K., Bear D.M., Zhou Y., Zon L.I. 2007. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol. Cell. Biol. 27:5887–5897 10.1128/MCB.00175-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfau S.J., Amon A. 2012. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 13:515–527 10.1038/embor.2012.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Paul R., Mogilner A., Cimini D. 2012. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell. 23:401–411 10.1091/mbc.E11-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J.H., Barr A.R., Nicholas A.K., Carvalho O.P., Khurshid M., Sossick A., Reichelt S., D’Santos C., Woods C.G., Gergely F. 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43:1147–1153 10.1038/ng.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., Nigg E.A. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Dobbelaere J., Brunk K., Franz A., Raff J.W. 2010a. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188:313–323 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Roque H., Raff J.W. 2010b. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell. 19:913–919 10.1016/j.devcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.J., Lin S.Y., Hsu W.B., Lin Y.N., Wu C.T., Lin Y.C., Chang C.W., Wu K.S., Tang T.K. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30:4790–4804 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2011. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl. Acad. Sci. USA. 108:17974–17978 10.1073/pnas.1109720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Sluder G. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., Krämer A. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Zamora I., Marshall W.F. 2005. A mutation in the centriole-associated protein centrin causes genomic instability via increased chromosome loss in Chlamydomonas reinhardtii. BMC Biol. 3:15 10.1186/1741-7007-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.