Summary
Progenitor cells in the cerebral cortex sequentially generate distinct classes of projection neurons. Recent work suggests the cortex may contain intrinsically fate-restricted progenitors marked by expression of Cux2. However, the heterogeneity of the neocortical ventricular zone as well as the contribution of lineage-restricted progenitors to the overall cortical neurogenic program remains unclear. Here we utilize in vivo genetic fate mapping to demonstrate that Fezf2-expressing radial glial cells (RGCs) exist throughout cortical development and sequentially generate all major projection neuron subtypes and glia. Moreover, we show that the vast majority of CUX2+ cells in the VZ and SVZ are migrating interneurons derived from the subcortical telencephalon. Examination of the embryonic cortical progenitor population demonstrates that Cux2+ RGCs generate both deep- and upper-layer projection neurons. These results identify Fezf2+ radial glial cells as a multipotent neocortical progenitor and suggest that the existence, and molecular identity, of laminar-fate-restricted RGCs awaits further investigation.
Introduction
The neocortex contains six layers of projection neurons and glia. Projection neurons in each cortical layer display similar morphologies, axonal projections and gene expression patterns (Kwan et al., 2012). During development, cortical projection neurons are generated from radial glial cells (RGCs) and basal progenitors in an inside-out pattern such that deep-layer neurons are generated first, followed by upper-layer neurons (Molyneaux et al., 2007). Three decades of work based upon transplantation experiments (Desai and McConnell, 2000; McConnell, 1985; McConnell and Kaznowski, 1991), viral lineage tracing (Luskin et al., 1988; Walsh and Cepko, 1988) and in vitro culture of single RGCs (Shen et al., 2006) suggests that cortical projection neuron subtype is sequentially determined by birthdate through progressive lineage restriction of a common RGC (Leone et al., 2008). However, the identification of early Cux2-expressing (Cux2+) RGCs, which were reported to be intrinsically specified to generate late-born, upper-layer neurons (Franco et al., 2012), calls into question this decades-old model and raises the possibility that deep-layer projection neurons are similarly generated from lineage-restricted progenitors (Franco and Muller, 2013; Marin, 2012).
The transcription factor Fezf2 (also known as Fezl and Zfp312) is expressed in early cortical progenitors and deep-layer neurons, and is critical for the fate-specification of subcerebral projection neurons (Chen et al., 2005a; Chen et al., 2005b; Molyneaux et al., 2005). In Fezf2-/- mice, subcerebral projections are absent and deep-layer neurons instead switch their identity to become corticothalamic or callosal projection neurons (Chen et al., 2005a; Chen et al., 2008; Chen et al., 2005b; Han et al., 2011; McKenna et al., 2011; Molyneaux et al., 2005). Several studies suggest that ectopic expression of Fezf2 in late cortical progenitors (Chen et al., 2008) or immature neurons (De la Rossa et al., 2013; Rouaux and Arlotta, 2012) redirects these cells to differentiate into subcerebral projection neurons. These results indicate that expression of Fezf2 in cortical progenitors may be sufficient to confer a subcerebral neuron identity, and thus Fezf2-expressing (Fezf2+) cortical progenitor cells may be lineage-restricted to generate deep-layer neurons (Franco and Muller, 2013; Woodworth et al., 2012).
To investigate the lineage potential of Fezf2+ progenitor cells we performed in vivo genetic fate mapping using the Fezf2 locus. Here we show that Fezf2+ cortical progenitor cells are RGCs that exist throughout cortical neurogenesis. Temporal fate mapping demonstrated that Fezf2+ RGCs sequentially generate projection neuron subtypes and glia based upon the birthdate of these cells. Furthermore, Fezf2+ RGCs generated upper-layer neurons without expressing detectable levels of CUX2 protein. Finally, we demonstrate that cells labeled by Cux2-Cre and Cux2-CreERT2 generate both deep- and upper-layer projection neurons. Collectively, these results indicate that Fezf2+ RGCs are a multipotent progenitor for neocortical projection neurons, astrocytes and oligodendroctytes, and suggest that laminar-fate-restricted RGCs remain to be identified.
Results
Lineage traced Fezf2-expressing progenitor cells are RGCs
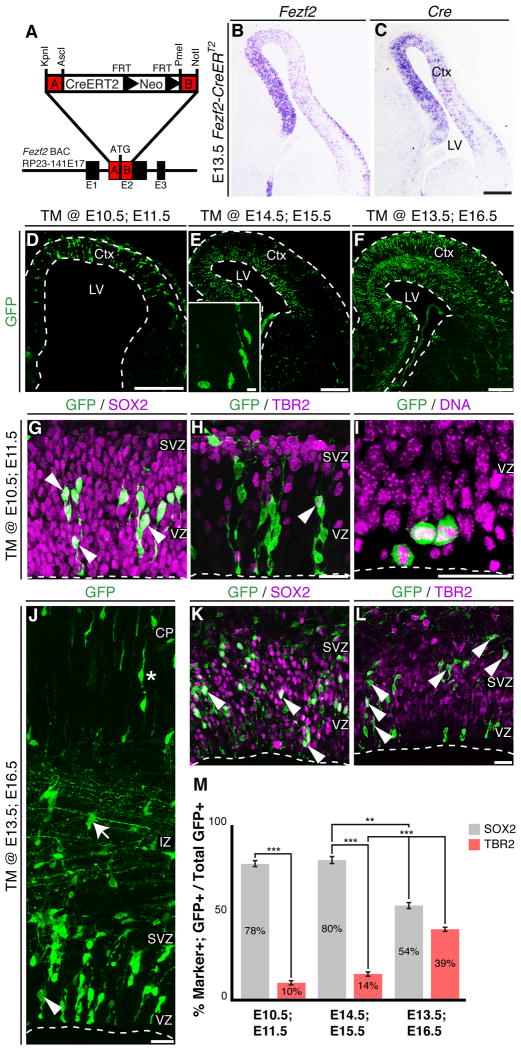
We first characterized Fezf2 expression by in situ hybridization. As previously reported (Hirata et al., 2004), we detected Fezf2 expression in early neocortical progenitors (Figure 1B). Interestingly, Fezf2 expression in the ventricular zone (VZ) persisted postnatally, long after deep-layer neuron generation has ceased (Figure S1A). This was confirmed by GFP expression in Fezf2-GFP BAC transgenic mice (Gong et al., 2003; Shim et al., 2012), which revealed GFP+ cells in the VZ during late embryonic and early postnatal stages (Figure S1B). To assess the differentiation potential of Fezf2+ progenitor cells, we generated nine independent Fezf2-CreERT2 BAC transgenic mouse lines (Figure 1A). In situ hybridization for Cre and Fezf2 showed that Cre expression was identical to that of endogenous Fezf2 (Figures 1B-C and S1C-D). Breeding these mice to three different Cre reporter lines (RCE-GFP, R26R-LacZ or TauR-mGFP) (Friedrich and Soriano, 1991; Hippenmeyer et al., 2005; Sousa et al., 2009) revealed that the fused CreERT2 protein was tightly regulated by tamoxifen (Figure S1E-I). Although Cre mRNA was expressed in deep-layer neurons (Figure 1C and S1D), we observed tamoxifen-induced recombination in these neurons with only the TauR-mGFP reporter (Figure S1H-I). No recombination was observed in postmitotic neurons upon tamoxifen administration with the Rosa26R-LacZ or RCE-GFP reporters (Figure S1G, J-J'). Critically, this allowed us to perform lineage-tracing experiments for Fezf2+ cortical progenitor cells using the RCE-GFP reporter without the ambiguity caused by Cre-mediated recombination in postmitotic neurons.
Figure 1.
Fezf2-expressing progenitors are RGCs. (A) Strategy for generation of Fezf2-CreERT2 mice. (B-C) In situ hybridization for Fezf2 (B) and Cre (C) at E13.5. (D-F) Low magnification images of GFP+ cells in the cortex of Fezf2-CreERT2; RCE-GFP mice following CRE-mediated recombination. (G-H) 24 hours after tamoxifen induction, most GFP+ cells expressed SOX2 (78 ± 3%) (G), and few cells expressed TBR2 (10 ± 2%) (H). (I) GFP+ cells dividing at the ventricular surface. (J-L) In TM @ E13.5; E16.5 brains, Fezf2+ RGCs gave rise to basal progenitors. (J) GFP was expressed in both VZ and SVZ progenitors (arrowhead), migrating neurons in the intermediate zone (arrow), and cortical neurons (asterisk). (K) GFP+ cells expressed SOX2 (54% ± 3) and showed typical RGC morphology. (L) Many GFP+ cells expressed TBR2 (39% ± 2). (M) Quantification of the percentages of GFP+SOX2+ RGCs and GFP+TBR2+ basal progenitors among all the GFP+ cells ± SEM. * P < 0.05, ** P < 0.005, *** P < 0.0001. CP, cortical plate; Ctx, cerebral cortex; IZ, intermediate zone; LV, lateral ventricle; SVZ, sub-ventricular zone. Thal, thalamus; TM; tamoxifen; VZ, ventricular zone. Scale bars: (C-F) 250 µm, (E insert) 10 µm, (H, I, J, L) 25 µm. See also Figure S1.
Examination of Fezf2-CreERT2; RCE-GFP mice after tamoxifen induction revealed that recombination specifically marked Fezf2+ RGCs (Figure 1D-M). Twenty-four hours after tamoxifen administration, approximately 80% of GFP+ cells expressed the RGC marker SOX2, while about 10% of GFP+ cells expressed the basal progenitor marker TBR2 (Figure 1D-E, G-H, M). The majority of GFP+ cells were located in the VZ; many had both apical and basal processes and divided at the ventricular surface (Figure 1G-I), all of which are characteristic of RGCs. The few TBR2+GFP+ cells were likely basal progenitors newly generated from Fezf2+ RGCs. Supporting this, three days after an E13.5 tamoxifen administration (TM @ E13.5; E16.5), the fraction of TBR2+GFP+ cells increased to 39% (Figure 1F, J-M). These results indicate that lineage-traced Fezf2+ progenitors are RGCs.
Fezf2+ RGCs sequentially generate deep-layer and upper-layer cortical projection neurons and glia
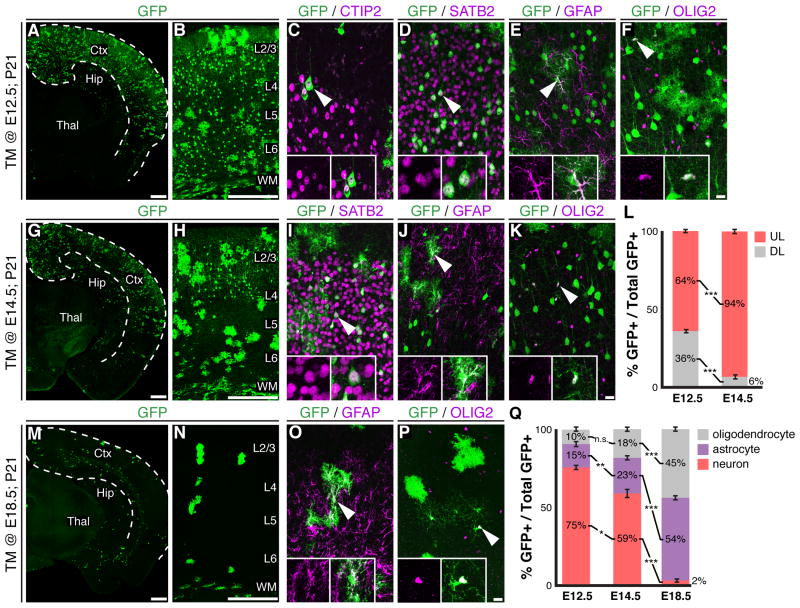
To assess the lineage potential of Fezf2+ RGCs, we administered tamoxifen to Fezf2-CreERT2; RCE-GFP mice at different embryonic stages. In TM @ E12.5; P21 brains, GFP+ cells were detected throughout the cortical plate and included cortical projection neurons (75%) and glia (25%) (Figures 2A-F, Q, S2A). GFP+ neurons were present in both deep (36%) and upper (64%) layers (Figure 2A-D, L). Those in layer 5 expressed high-levels of CTIP2 (also known as BCL11B), indicating a subcerebral neuron identity (Figure 2C). Many GFP+ cells expressed the callosal neuron marker SATB2, and were observed in both deep- and upper-layers (Figure 2D). Indeed, GFP labeled axon tracts demonstrated that Fezf2+ RGCs generated callosal, subcerebral and corticothalamic projection neurons (Figure S2D-F). In addition to projection neurons, 15% of GFP+ cells showed an astrocytic morphology and expressed GFAP (Figure 2E, Q) while 10% of GFP+ cells expressed the oligodendrocyte marker OLIG2 (Figures 2F, Q, S2A). These results demonstrate that at E12.5, Fezf2+ RGCs are multipotent.
Figure 2.
Fezf2+ RGCs sequentially generate projection neurons and glia. (A-F) Immunohistochemical analysis of TM @ E12.5; P21 brains. (A, B) GFP+ cells were present throughout the cortex. GFP+ cells expressed CTIP2 (C), SATB2 (D), GFAP (E) or OLIG2 (F). (G-K) Immunohistochemistry on brain sections from TM @ E14.5; P21 mice. (G, H) GFP+ neurons were mostly in layers 2-4. GFP+ cells expressed SATB2 (I), GFAP (J), or OLIG2 (K). (L) Percentage of GFP+ neurons in deep and upper layers ± SEM. (M-P) Immunohistochemical analysis of brains from TM @ E18.5; P21 mice. GFP+ cells included astrocytes (O) and oligodendrocytes (P). (Q) Percentages of GFP+ cells that were neurons, astrocytes or oligodendrocytes ± SEM. * P < 0.05, ** P < 0.005, *** P < 0.0001. Ctx, cerebral cortex; Hip, hippocampus; Thal, thalamus; TM, tamoxifen; WM, white matter. Scale bars: (A, G, M) 500 µm, (B, H, N) 250 µm, (F, K, P) 25 µm. See also Figure S2.
In TM @ E14.5; P21 brains, GFP+ cells included projection neurons, astrocytes and oligodendrocytes (Figures 2G-K, Q, S2B). However, 94% of GFP+ neurons were present in upper layers, and many expressed SATB2 (Figure 2G-I, L). Retrograde tracing from the contralateral cortical hemisphere showed that GFP+SATB2+ cells projected callosal axons (Figure S2G-O). In TM @ E18.5; P21 brains only 2% of GFP+ cells were projection neurons (Figure 2Q). Instead, the GFP+ cells consisted of astrocytes (53%) and oligodendrocytes (45%) (Figures 2M-Q, S2C). Collectively, these results indicate that Fezf2+ RGCs are multipotent and sequentially generate neocortical projection neurons and glia according to their birthdate.
Clonal analysis of Fezf2+ RGCs
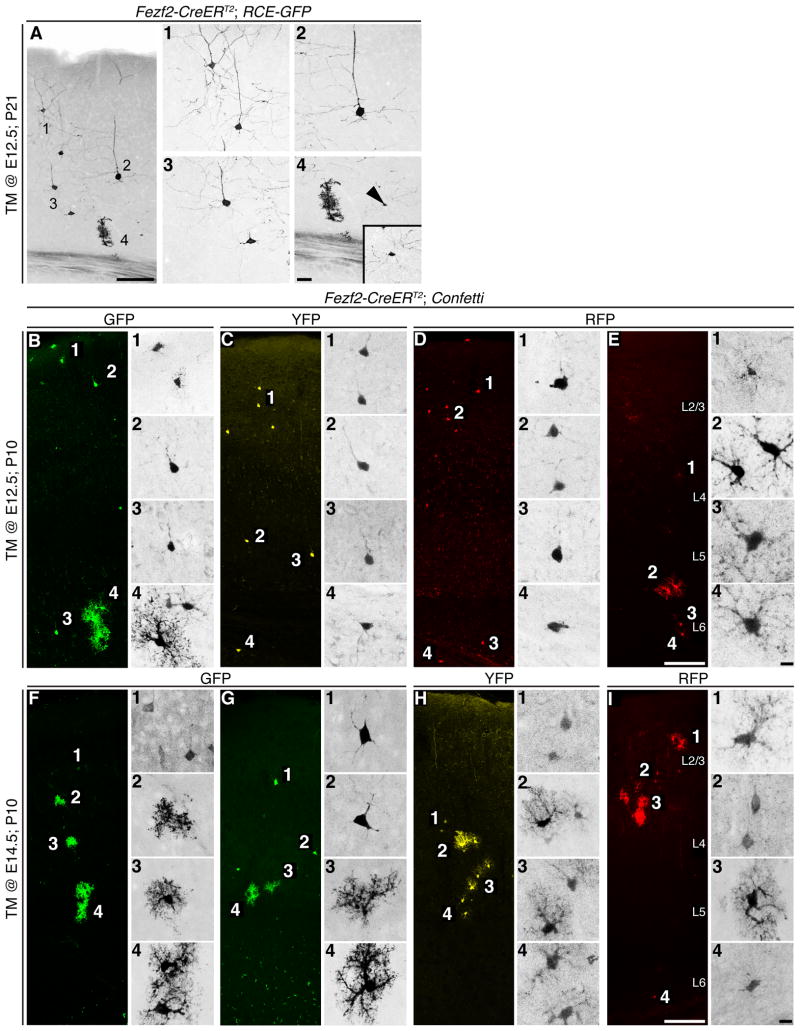
To further examine the lineage of Fezf2+ RGCs we preformed in vivo clonal analysis. Low efficiency tamoxifen induction at E12.5 using the RCE-GFP reporter labeled putative RGC clones, which consisted of neurons in both deep- and upper-layers, astrocytes and ligodendrocytes (Figure 3A). To extend our clonal analysis we crossed Fezf2-CreERT2 mice to the Confetti reporter line (Snippert et al., 2010), which enabled the identification of individual RGC clones based upon the exclusive expression of either CFP, GFP, YFP or RFP. Early (TM @ E12.5) Fezf2+ RGCs produced clones that consisted of neurons in deep- and upper-layers and glia (Figure 3B-D). In contrast, when tamoxifen was administered at E14.5, clones consisted of only upper-layer neurons and glia (Figure 3F-I). Of note, we occasionally observed clones consisting of only glia (Figure 3E), consistent with previous viral based clonal analysis of cortical progenitors (Luskin et al., 1988; Walsh and Cepko, 1988). These results support the conclusion that Fezf2+ RGCs sequentially generate cortical projection neuron subtypes and glia.
Figure 3.
Clonal analysis of Fezf2+ RGCs. (A) GFP+ clone from a P21 Fezf2-CreERT2; RCE-GFP brain that received TM at E12.5, indicating that early Fezf2+ RGCs generate deep- and upper-layer neurons, astrocytes and oligodendrocytes. (B-I) Fezf2-CreERT2; Confetti mice enabled clonal anaylys of RGCs based upon flourescent protein expression. (B-D) Eamples of TM @ E12.5; P10 brains demonstrating that clones included both deep- and upper-layer neurons and glia. (E) We occasionally observed clones that contained only glia. (F-I) TM @ E14.5; P10 brains contained clones with only upper-layer neruons and glia. TM, tamoxifen. Scale Bars: (A, E, I) 100 µm, (A4) 25 µm, (E4, I4) 10 µm.
CUX2+ cells in the VZ and SVZ are migrating interneurons derived from the ventral telencephalon
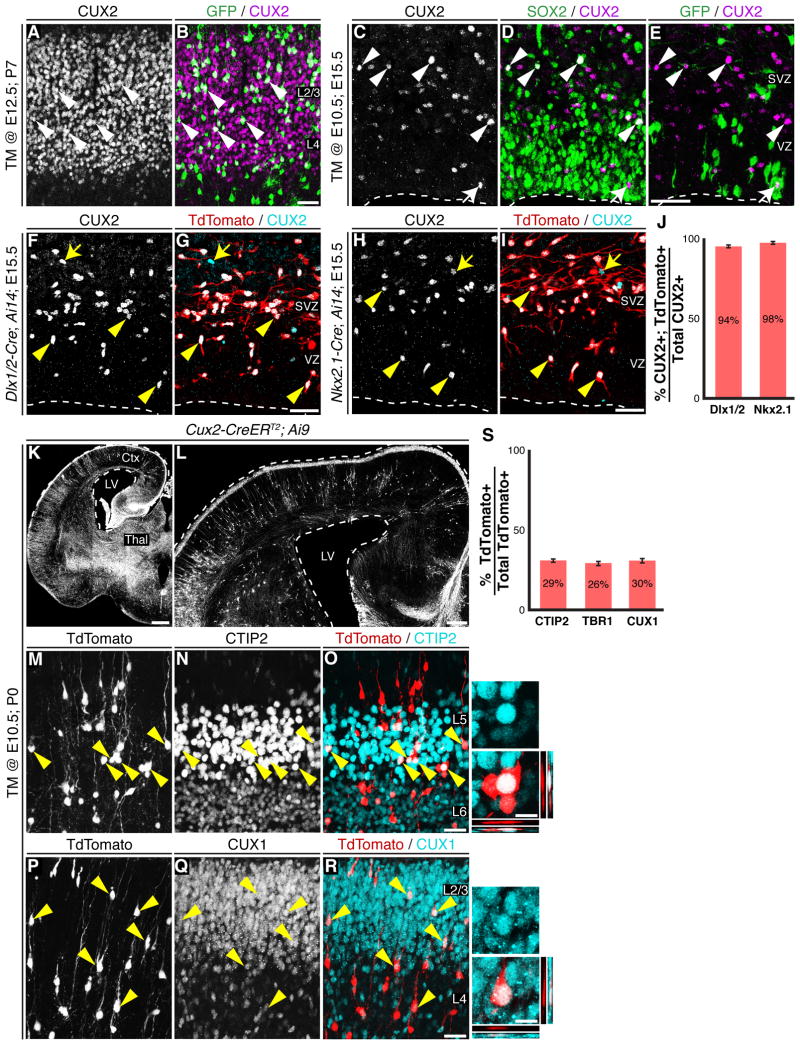
The finding that Fezf2+ RGCs contribute substantially to upper-layer neurogenesis is consistent with the classic progressive restriction model (Leone et al., 2008). However, this is in contrast to a newly proposed model that suggests all upper-layer neurons are generated from Cux2+ RGCs (Franco and Muller, 2013). To resolve this difference, we explored the relationship between Fezf2+ RGCs and CUX2+ cortical cells using two different CUX2 antibodies, which recognize different regions of the CUX2 protein (Conforto et al., 2012; Iulianella et al., 2008; Laz et al., 2007). We found that 72% of upper-layer neurons generated from Fezf2+ RGCs expressed CUX2 (Figure 4A-B). Since Cux2+ projection neurons were reported to arise from Cux2+ RGCs (Franco et al., 2012), we next investigated whether Fezf2+ RGCs transit through a CUX2+ RGC stage to generate upper-layer neurons. In agreement with previous reports (Cobos et al., 2006; Cubelos et al., 2008a; Cubelos et al., 2008b; Franco et al., 2012; Franco et al., 2011; Nieto et al., 2004; Zimmer et al., 2004), we detected robust CUX2 expression beginning at E14.5, including expression in neocortical interneurons (Figure S3). We examined the relationship between Fezf2+ RGCs and CUX2+ progenitors using TM @ E10.5; E15.5 Fezf2-CreERT2; RCE-GFP brains, since the Cux2+ RGC population was reported to peak at this age (Franco et al., 2012). We found that while some CUX2+ cells resided in the VZ, the majority were in the SVZ (Figure 4C). We rarely observed GFP+CUX2+ cells in the VZ/SVZ (Figure 4E), and these cells were SOX2- (Figure 4D), suggesting they were not RGCs, but likely basal progenitors generated from Fezf2+ RGCs. Taken together, these results indicate that Fezf2+ RGCs that generate upper-layer neurons do not express significant levels of CUX2 protein.
Figure 4.
CUX2+ cells in VZ/SVZ are migrating interneurons and Cux2-Cre/CreER T2 labeled RGCs generate both deep- and upper-layer projection neurons. (A-B) Fezf2+ RGCs generated CUX2+ upper-layer neurons. (C-E) Immunohistochemical analysis of TM @ E10.5; E15.5 Fezf2-CreERT2; RCE-GFP brains. Few CUX2+ cells in the SVZ expressed SOX2 and these cells re located at the VZ/SVZ boundary (C-D). (E) Rare CUX2+GFP+ cell in the VZ/SVZ. These CUX2+GFP+ cells did not express SOX2 (D-E). The arrowheads in C-E point to the CUX2+SOX2+ cells in VZ/SVZ, the arrows point to a rare GFP+CUX2+ cell. (F-J) Dlx1/2-Cre; Ai14 and Nkx2.1-Cre; Ai14 mice revealed that the majority of CUX2+ cells in the E15.5 neocotical VZ/SVZ were interneurons generated from Dlx1/2 (F-G) or Nkx2.1 (H-I) lineages. (J) Quantification of the percentage of CUX2+TdTomato+ cells among all CUX2+ cells in the VZ/SVZ ± SEM. (K-L) TM @ E10.5; P0 Cux2-CreERT2; Ai9 brains contained TdTomato+ cells in all cortical layers. (M-O) TdTomato+ cells in deep-layers expressed CTIP2. (P-R) Upper-layer TdTomato+ cells expressed CUX1. (S) Percentage of TdTomato+ cells in layers 2-6 that expressed CTIP2, TBR1 or CUX1 ± SEM. CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; TM, tamoxifen; VZ, ventricular zone. Scale Bars: (B, E, G, I, O, R) 25 µm, (K) 500 µm, (L) 200 µm, (close-ups from O, R) 10 µm. See also Figures S3 and S4.
The lack of GFP+CUX2+ RGCs in the VZ/SVZ of TM @ E10.5; E15.5 brains prompted us to investigate the identity of CUX2+ cells in the neocortical VZ/SVZ. Since CUX2 was previously reported to be expressed in interneurons (Zimmer et al., 2004), we determined the percentage of CUX2+ cells in the E15.5 neocortical VZ/SVZ that originated from the ventral telencephalon. To label cortical interneurons, we crossed either the Dlx1/2-Cre (Potter et al., 2009) or the Nkx2.1-Cre (Xu et al., 2008) alleles to mice harboring the Ai14 reporter (Madisen et al., 2010). In the mice carrying both Cre and Ai14 alleles, cortical interneurons were labeled by TdTomato expression (Figure 4G, I). Examination of E15.5 Dlx1/2-Cre; Ai14 or Nkx2.1-Cre; Ail 4 brains revealed that the vast majority (94% or 98% respectativly) of CUX2+ cells in the neocortical VZ/SVZ were TdTomato+ (Figure 4F-J). This result demonstrates that in the developing neocortex, nearly all CUX2+ cells in the VZ/SVZ are immature interneurons derived from the ventral telencephalon.
Cux2-Cre and Cux2-CreERT2 label RGCs that generate both deep- and upper-layer neurons
The lack of CUX2+ RGCs in the developing neocortex seemed to contradict the previous report that Cux2+ RGCs, marked by Cux2-Cre and Cux2-CreERT2 alleles, generate upper-layer projection neurons (Franco et al., 2012). To resolve this discrepency, we obtained Cux2-Cre and Cux2-CreERT2 mice and analyzed them using RCE-GFP, Ai14 and Ai9 (Madisen et al., 2010) reporter lines. In the brains of E15.5 Cux2-Cre; RCE-GFP and Cux2-Cre; Ai14 embryos, many cells were labeled by GFP or TdTomato (Figure S4E-F). Labelled cells in the neocortical VZ/SVZ expressed the RGC marker PAX6 (Figure S4G) or the basal progenitor marker TBR2 (Figure S4H), consistent with the Cux2 in situ hybridization results (Figure S4A-D, see also Franco et al., 2012). Immunohistochemistry using CUX2 and CRE antibodies revealed that most of the lineage traced cells in the VZ/SVZ did not express CUX2 or CRE protein (Figure S4J-K), indicating that the Cre-reporter system may be more sensitive than immunohistochemistry for detecting low levels of Cux2 expression. Aside from cortical progenitor cells, Cux2-Cre also labelled many neurons in the cortical plate that expressed CTIP2, a marker for early-born subcortical projection neurons (Figure S4I).
We next investigated the lineage potential of Cux2+ RGCs using Cux2-CreERT2; Ai9 mice. In TM @ E10.5; P0 brains, TdTomato+ cells were observed throughout the brain (Figure 4K) including in both deep and upper cortical layers (Figure 4L). Indeed, within layers 2-6 of the neocortex, 29% and 26% of TdTomato+ cells expressed the deep-layer makers CTIP2 and TBR1 respectativly, while 30% expressed the upper-layer marker CUX1 (Figures 4M-S and S4N). CTIP2 expression in TdTomato+ cells in P0 Cux2-CreERT2;Ai9 brains is consistent with the observed expression in labelled cells of E15 Cux2-Cre brains (Figure S4I). In addition to CTIP2 and TBR1, TdTomato+ cells also expressed NFIB and SOX5 (Figure S4L-M), both of which mark early-born subcortical projection neurons at P0 (Betancourt et al., 2013; Kwan et al., 2008; Lai et al., 2008; McKenna et al., 2011). Collectivly, these results demonstrate that early Cux2-Cre and Cux2-CreERT2-labeled neocortical RGCs are not lineage-restricted in that they generate both deep- and upper-layer projection neurons.
Discussion
Sequential generation of projection neurons and glia by multipotent RGCs
Our lineage analysis demonstrates that both early and late Fezf2+ RGCs are multipotent. Early Fezf2+ RGCs generate all major cortical projection neuron subtypes as well as astrocytes and oligodendrocytes. Late Fezf2+ RGCs generate upper-layer neurons, astrocytes and oligodendroctes. Thus, they sequentially generate projection neurons and glia in accordance with the classic model of cortical neurogenesis (Leone et al., 2008). However, our results do not exclude the existence of intrinsically fate-restriced RGCs. Rather, because Cux2-Cre and Cux2-CreERT2-labeled neocortical RGCs generate both deep- and upper-layer neurons, this suggests that laminar-fate-restricted RGCs cannot be identified by Cux2 expression alone.
A comparison between Fezf2-CreERT2 mice and the Cux2-Cre and Cux2-CreERT2 mice used by Franco et al. reveals possible reasons for the divergent conclusions reached by these two studies. Upon tamoxifen administration, the Fezf2-CreERT2 allele induces recombination of the RCE-GFP reporter only in progenitor cells and not in postmitotic neurons. This was observed when tamoxifen or 4-hydroxytamoxifen was administered from E12 to adult stages. However, the Cux2-Cre and Cux2-CreERT2 alleles induced recombination in both progenitor cells and postmitotic neurons (Franco et al., 2012; Franco et al., 2011), possibly masking the true lineage potential of Cux2+ progenitor cells. Further, Fezf2 is not expressed in neocortical interneurons or their progenitors. In contrast, Cux2 is expressed at high levels in interneurons that are born in the subpallium and migrate into the neocortex (Cobos et al., 2006).
Fezf2 expression in neocortical progenitors
Fezf2 is essential for subcerebral neuron development (Chen et al., 2005a; Chen et al., 2008; Chen et al., 2005b; Han et al., 2011; McKenna et al., 2011; Molyneaux et al., 2005). Indeed, overexpression of Fezf2 in late cortical progenitors redirects these cells to generate subcerebral projection neurons (Chen et al., 2008; Chen et al., 2005b; Molyneaux et al., 2005). Here we demonstrate that during normal brain development, Fezf2 is expressed in late RGCs as these cells are generating upper-layer projection neurons and glia. This suggests it is not simply Fezf2 expression, but rather its expression level, in RGCs that influences the differentiated cell types that are produced. Going forward it will important to understand the mechanisms that precisely regulate Fezf2 transcription levels across different expression domains in order to functionally dissect its involvement in neocortical development.
Heterogenity of embryonic neural stem cells
Previous studies suggest that neocortical RGCs are a heterogeneous cell population. Subsets of RGCs were reported to differentially express markers such as RC2, GLAST and BLBP (Hartfuss et al., 2001). In addition, in vivo lineage analysis of neocortical progenitors using retroviral vectors produced clones that consisted of only neurons or glia (Luskin et al., 1988; Walsh and Cepko, 1988), suggesting the existence of neuron- or glia-restricted progenitor cells. Further, recent work indicates that the neocortical progenitor pool contains both outer radial glia cells (Hansen et al., 2010; Wang et al., 2011) as well as short neural precursors (Gal et al., 2006; Stancik et al., 2010; Tyler and Haydar, 2013) that are molecularly and morphologically distinct from ventricular zone RGCs.
Results from our study indicate that progressive lineage restriction of multipotent RGCs is a common mechanism for generating cellular diversity during cortical development. However, the finding that Cux2-Cre and Cux2-CreERT2-labeled RGCs are not upper-layer fate-restricted progenitors does not exclude the possibility that some cortical projection neurons may originate from fate-restricted RGCs. Indeed, our clonal analysis of Fezf2+ RGCs produced clones that consisted of only glial cells (Figure 3E), suggesting the possibility of lineage-restricted subpopulations within the Fezf2+ RGC population. Moreover, it is possible that Fezf2+ RGCs are a heterogeneous population in that some Fezf2+ RGCs may express Cux2 mRNA or protein at levels below the detection threshold of in situ hybridization or immunohistochemistry. Many genes expressed in RGCs have been identified (Molyneaux et al., 2007; Woodworth et al., 2012). Genetic fate mapping using these loci will further expand our knowledge of progenitor cell heterogeneity within the neocortical progenitor pool and uncover the mechanisms that generate projection neuron diversity.
Experimental Procedures
All experiments were carried out in accordance with protocols approved by the IACUC at University of California at Santa Cruz, and were performed in accordance with institutional and federal guidelines. Fezf2-CreERT2 mice were generated according to previously established strategies (Lee et al., 2001) by modifying the RP23-141E17 BAC.
In situ hybridization and immunohistochemistry were preformed as previously described (Eckler et al., 2011). The rabbit anti-CUX2 antibody (Conforto et al., 2012; Laz et al., 2007) was a gift from Dr. David Waxman. We also confirmed CUX2 expression using a second rabbit anti-CUX2 antibody (Iulianella et al., 2008; Iulianella et al., 2009) kindly provided by Angelo Iulianella.
Supplementary Material
Highlights.
Fezf2+ RGCs exist throughout cortical neurogenesis
Fezf2+ RGCs sequentially generate cortical projection neuron subtypes and glia
Cux2-Cre/CreERT2 labeled RGCs generate deep- and upper-layer projection neurons
Acknowledgments
We thank Drs. Lindsay Hinck, Susan McConnell and Jennifer Betancourt for critical reading of the manuscript. We thank Drs. David Waxman and Tara L. Conforto (Boston University) and Dr. Angelo Iulianella (Dalhousie University) for providing CUX2 antibodies. Drs. Guo-Ping Feng, Xue-wen Cheng and Zhi-qi Xiong kindly provided the pERFN vector. We thank Drs. Ulrich Mueller, Susan McConnell, and Pushkar Joshi for providing Cux2-Cre and Cux2-CreERT2 mice, and Dr. Gord Fishell for providing the RCE-GFP mice. Dr. Ben Abrams and the UCSC Microscopy center provided support with image acquisition. This work was funded by R01MH082965 from the National Institutes of Health (to B. C.), a New Faculty Award RN1-00530-1 from the California Institute of Regenerative Medicine (CIRM) (to B. C.), a training grant TG2-01157 from CIRM (to C. G. and W. L. M.), and R01MH049428 from the National Institutes of Health (to J. L. R. R).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Betancourt J, Katzman S, Chen B. Nuclear factor one b regulates neural stem cell differentiation and axonal projection of corticofugal neurons. The Journal of comparative neurology. 2013 doi: 10.1002/cne.23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cerebral cortex. 2006;16(Suppl 1):i82–88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Conforto TL, Zhang Y, Sherman J, Waxman DJ. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Molecular and cellular biology. 2012;32:4611–4627. doi: 10.1128/MCB.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, Nieto M. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cerebral cortex. 2008a;18:1758–1770. doi: 10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Kim S, Redondo JM, Walsh C, Nieto M. Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Developmental neurobiology. 2008b;68:917–925. doi: 10.1002/dneu.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rossa A, Bellone C, Golding B, Vitali I, Moss J, Toni N, Luscher C, Jabaudon D. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nature neuroscience. 2013;16:193–200. doi: 10.1038/nn.3299. [DOI] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Eckler MJ, McKenna WL, Taghvaei S, McConnell SK, Chen B. Fezf1 and Fezf2 are required for olfactory development and sensory neuron identity. The Journal of comparative neurology. 2011;519:1829–1846. doi: 10.1002/cne.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Muller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes & development. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Developmental biology. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS biology. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Suda Y, Nakao K, Narimatsu M, Hirano T, Hibi M. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development. 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Vanden Heuvel GB, Trainor PA. Cux2 functions downstream of Notch signaling to regulate dorsal interneuron formation in the spinal cord. Development. 2009;136:2329–2334. doi: 10.1242/dev.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Laz EV, Holloway MG, Chen CS, Waxman DJ. Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver. Endocrinology. 2007;148:3327–3337. doi: 10.1210/en.2006-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Current opinion in neurobiology. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskin MB, Pearlman AL, Sanes JR. Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron. 1988;1:635–647. doi: 10.1016/0896-6273(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. Brain development: The neuron family tree remodelled. Nature. 2012;490:185–186. doi: 10.1038/490185a. [DOI] [PubMed] [Google Scholar]
- McConnell SK. Migration and differentiation of cerebral cortical neurons after transplantation into the brains of ferrets. Science. 1985;229:1268–1271. doi: 10.1126/science.4035355. [DOI] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. The Journal of comparative neurology. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Molecular and cellular neurosciences. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nature cell biology. 2012;15:214–221. doi: 10.1038/ncb2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nature neuroscience. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cerebral cortex. 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WA, Haydar TF. Multiplex Genetic Fate Mapping Reveals a Novel Route of Neocortical Neurogenesis, Which Is Altered in the Ts65Dn Mouse Model of Down Syndrome. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:5106–5119. doi: 10.1523/JNEUROSCI.5380-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C, Cepko CL. Clonally related cortical cells show several migration patterns. Science. 1988;241:1342–1345. doi: 10.1126/science.3137660. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nature neuroscience. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth MB, Custo Greig L, Kriegstein AR, Macklis JD. SnapShot: cortical development. Cell. 2012;151:918–918. e911. doi: 10.1016/j.cell.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. The Journal of comparative neurology. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cerebral cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.