Abstract
BACKGROUND
Thymidylate synthase (TS) is important for maintenance of the intracellular thymidine pool, which is crucial for DNA synthesis and repair. TS messenger RNA and protein levels are predictive of response to 5-fluorouracil-containing therapy for patients with colorectal cancer and gastric cancer. High levels of expression of 2 other genes important in DNA synthesis and repair, RRM1 and ERCC1, are prognostic of survival in early stage nonsmall-cell lung cancer (NSCLC) patients. We hypothesized that intratumoral TS expression would be prognostic of outcome in stage I NSCLC.
METHODS
Cytoplasmic tumoral TS was determined by automated in situ protein quantification (AQUA) in 160 patients with completely resected NSCLC that had not received chemotherapy or radiation. It was also determined by real-time quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) in 85 similar patients. The 2 datasets were partially overlapping (N = 32). The optimal cut-point was determined by the maximal log-rank method with adjustment of the P-values for multiple looks.
RESULTS
TS protein expression was significantly associated with patient survival (P = .0013, adjusted P = .034). The optimal cutpoint was at the 25% percentile; the group with low expression (≤57.02) had a median overall survival (OS) of 51.7 months, and the high expression group (>57.02) had a median OS of 81.3 months. TS mRNA expression was not significantly associated with patient survival or TS protein expression. In a multivariate analysis adjusting for tumor stage, TS remained significantly prognostic of survival (P = .0013, adjusted P = .032).
CONCLUSIONS
In situ cytoplasmic TS protein expression in tumors of patients with resected stage I NSCLC is a clinically important determinant of survival.
Keywords: thymidylate synthase, lung cancer, adenocarcinoma of the lung, squamous cell carcinoma of the lung, biomarkers, immunohistochemistry
Thymidylate synthase (TS) catalyzes the conversion of deoxyuridine monophosphate (dUMP) to (deoxy)thymidine monophosphate (TMP), which requires oxidization of tetrahydrofolate to dihydrofolate. TMP is subsequently phosphorylated to TTP, which is required for DNA synthesis and repair. 5-Fluorouracil (5FU) inhibits TMP synthesis and is an effective chemotherapeutic agent.1–3 High tumoral levels of TS have been associated with resistance to 5FU-based chemotherapy, particularly in patients with colorectal carcinoma (CRC) and gastric carcinoma.4–6
Recent work has demonstrated that genes involved in nucleotide metabolism and DNA repair are important determinants of the phenotypic behavior of early stage nonsmall-cell lung cancer (NSCLC); ie, they are prognostic of patients’ survival. Specifically, RRM1, the regulatory subunit of ribonucleotide reductase, and ERCC1, a component of the 5′ nuclease involved in nucleotide excision repair, are prognostic of patient outcomes.7–10 High levels of mRNA and protein expression of these genes are associated with long survival in patients and reduced metastasis formation in animal models.11 In addition, high RRM1 levels in transgenic animals were found to be protective of carcinogen-induced lung tumor formation.12
Because TS is downstream of ribonucleotide reductase and crucial for the formation of 1 of the deoxynucleosides required for DNA synthesis and repair, we investigated if TS expression at the protein and mRNA levels is prognostic of outcome in patients with completely resected stage I NSCLC who did not receive additional chemotherapy or radiation and if in situ TS expression is correlated with RRM1 and ERCC1 expression.
MATERIALS AND METHODS
Study Populations
The 2 study populations consisted of patients that underwent a complete resection of NSCLC at the Moffitt Cancer Center from 1991 to 2001, as described elsewhere.10 In brief, patients had to have pathologic stage IA or IB disease; adenocarcinoma, squamous cell carcinoma, or large-cell carcinoma; no perioperative chemotherapy or radiation; no prior lung cancer; and no prior radiation to the chest. We identified 187 patients with sufficient tumor tissue for construction of tissue microarrays that comprised the population for assessment of gene expression at the protein level (Table 1). We identified 92 patients with fresh-frozen tumor tissue that comprised the population for assessment of gene expression at the mRNA levels (Table 2). Thirty-two patients with gene expression values at the protein and mRNA levels overlapped between the 2 populations. Follow-up was done at the Moffitt Cancer Center or by referring physicians and recommended as 3-monthly visits for 2 years, 6-monthly visits for 3 years, and then annual visits. Information from outside physicians was obtained by mail and telephone contacts. For overall survival (OS) the time from diagnosis to death was recorded and verified using vital statistics records.
TABLE I.
Patient Characteristics, TS Protein Expression, and Survival
| Patient characteristics
|
TS by AQUA
|
|||||
|---|---|---|---|---|---|---|
| No. (187) | Median overall Survival, mo | Log-rank P | Median | No. (160) | Kruskal-Wallis P | |
| Stage | ||||||
| IA | 85 | 100.5 | .019 | 99.6 | 71 | .818 |
| IB | 102 | 62.2 | 97.3 | 89 | ||
| Histology | ||||||
| Adeno | 78 | 72.0 | .061 | 98.4 | 67 | .427 |
| BAC | 18 | 87.4 | 84.6 | 17 | ||
| Squamous | 68 | 101.8 | 101.5 | 55 | ||
| Large cell | 23 | 41.8 | 93.3 | 21 | ||
| Performance status | ||||||
| 0 | 128 | 79.0 | .034 | 99.8 | 109 | .191 |
| 1 | 48 | 52.7 | 90.4 | 41 | ||
| Weight loss, >5% in 3 mo | ||||||
| Absent | 159 | 74.9 | .823 | 94.3 | 133 | .278 |
| Present | 14 | 81.3 | 102.9 | 14 | ||
| Smoking status | ||||||
| Never | 11 | NR | .077 | 101.2 | 8 | .326 |
| Quit | 113 | 77.9 | 94.4 | 98 | ||
| Active | 49 | 61.4 | 105.0 | 41 | ||
| Sex | ||||||
| Women | 86 | 81.3 | .143 | 103.6 | 73 | .284 |
| Men | 101 | 69.4 | 91.6 | 87 | ||
TS indicates thymidylate synthase; AQUA, automated quantitative analysis; NR, not reached.
TABLE 2.
Patient Characteristics, TS mRNA Expression, and Survival
| Patient characteristics
|
TS by RT-PCR
|
|||||
|---|---|---|---|---|---|---|
| No. (92) | Median overall survival, mo | Log-rank P | Median | No. (85) | Kruskal-Wallis P | |
| Stage | ||||||
| IA | 39 | NR | .671 | 3.42 | 37 | .642 |
| IB | 53 | 74.9 | 3.65 | 48 | ||
| Histology | ||||||
| Adeno | 41 | 62.2 | .642 | 2.25 | 39 | .002 |
| BAC | 7 | 87.4 | 0.50 | 6 | ||
| Squamous | 33 | NR | 4.73 | 30 | ||
| Large cell | 11 | 69.4 | 5.08 | 10 | ||
| Performance status | ||||||
| 0 | 52 | 80.4 | .771 | 3.47 | 52 | .853 |
| 1–2 | 33 | 62.1 | 3.87 | 26 | ||
| Weight loss, >5% in 3 mo | ||||||
| Absent | 75 | 80.4 | .020 | 3.52 | 69 | .703 |
| Present | 6 | 34.1 | 5.60 | 6 | ||
| Smoking status | ||||||
| Never | 6 | 31.2 | .898 | 4.53 | 4 | .057 |
| Quit | 55 | 74.9 | 2.26 | 51 | ||
| Active | 27 | 88.2 | 5.16 | 26 | ||
| Sex | ||||||
| Women | 39 | 88.2 | .605 | 4.04 | 35 | .652 |
| Men | 53 | 74.9 | 2.33 | 50 | ||
TS indicates thymidylate synthase; RT-PCR, reverse-transcriptase polymerase chain reaction; NR, not reached.
Tissue Microarray Construction
Tumor specimens were collected prospectively, fixed in neutral-buffered formalin (10% v/v), and completely embedded in paraffin wax. Whole tissue sections were hematoxylin and eosin-stained and representative tumor areas were marked. Tissue cores with a diameter of 0.6 mm were punched and arrayed into a recipient block using a tissue arrayer (Beecher Instrument, Silver Spring, Md). Sections of 5 μm thickness were cut, transferred to 4× adhesive-coated slides (Instrumedics, Hackensack, NJ), and exposed to UV light for 30 seconds to enhance adherence.
In Situ Detection and Quantification of TS Protein Expression
Immunohistochemistry (IHC) based on immunofluorescence combined with automated quantitative analysis (AQUA) was used to assess in situ expression of the target molecules.13 Antigens were retrieved by microwave oven treatment for 15 minutes in 0.01 mol/L of Na-citrate at pH 6.0. The slides were blocked for 30 minutes with 0.3% BSA and then incubated overnight at room temperature with the primary antibody (mouse clone TS-106, 1:30, #MS-471-P, Lab Vision, Fremont, Calif). For identification of carcinomatous cells, an antiserum to cytokeratin was used (rabbit antihuman pancytokeratin AE1/AE3, 1:200, #Z0622, Dako Cytomation, Carpinteria, Calif). Slides were washed and incubated with 2 different secondary antibodies for 1 hour (Envision labeled polymer-HRP antimouse, #K4007, and Alexa 555 goat antirabbit, #A21429, 1:200, Dako Cytomation). For fluorescence amplification, slides were exposed to Cy5-Tyramide (1:50) for 10 minutes at room temperature. They were mounted with Prolong Gold antifade reagent with DAPI (4′-6-diamidino-2-phenylindole) mound solution. The final tissue microarray slides were scanned with SpotGrabber and image data were analyzed with AQUA (PM-2000, HistoRx, New Haven, Conn). The lowest possible AQUA score is 0 and the highest is 255.
RNA Isolation and TS mRNA Expression Analysis
Fresh-frozen tumor specimens had been prospectively collected on 92 patients that fulfilled the same selection criteria as described above (Table 2). RNA was isolated from tumor specimens with ≥60% of cells consisting of tumor and cDNA was generated using oligo-DT and random primers with reverse transcriptase (QuantiTect Reverse Transcription Kit, Qiagen, Valencia, Calif). Approximately 5 ng of sample cDNA was used in triplicate for TS expression analysis by real-time technology (ABI 7900HT, Foster City, Calif). The relative amount of TS mRNA in a sample was determined by comparing the threshold cycle with the standard curve, and the standardized amount was then determined by dividing the TS amount (ABI, HS00426591-m1, amplicon size 87 bp) by the 18SrRNA amount (ABI, #4319413E).
Western Blot Analysis
Cytoplasmic and nuclear extracts from permanent genetically modified cultures of cell line NCI-H23 were prepared using a nuclear/cytosol fractionation kit (BioVision, Mountain View, Calif). Protein extracts (50 μg) were separated through 10% Novex Tris-glycine gels (Invitrogen, Carlsbad, Calif) and blotted onto pure nitrocellulose membranes (Bio-Rad Laboratories, Hercules, Calif). The blots were incubated with TS antibody (mouse clone TS-106, 1:200, Lab Vision), Oct-1 antiserum (#3342-100, 1:1000, BioVision), and GAPDH antiserum (#sc-20,357, 1:1000, Santa Cruz Biotechnology, Santa Cruz, Calif) at 4°C overnight. Protein bands were observed with antirabbit, antimouse, or antigoat IgG horseradish peroxidase secondary antibody (1:1000; Santa Cruz) and SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, Ill). The house-keeping gene GAPDH was used as equal loading control.
Confocal Microscopy
Lung adenocarcinoma cell lines (H23, H125, H292, H322, A549) and colon carcinoma cell lines (H498, H508, H747, SNU-C2A, SNU-C4) were grown directly on Lab-Tek chamber slides. Adherent cells were washed in phosphate-buffered saline (PBS), fixed by incubation for 20 minutes in 4% paraformaldehyde in PBS, and washed in PBS. They were permeablized for 1 hour in 0.25% Triton X-100/PBS and washed in PBS. TS (1:100) antibodies were diluted in binding buffer (1%BSA/0.1%NP40/PBS), added to the chambers, and incubated for 1 hour. After washing in PBS the slides were incubated for 45 minutes with 1:500 dilutions of Alexa Fluor 555 antimouse IgG (Molecular Probes, InVitrogen, Eugene, Ore). The slides were washed with PBS and covered using ProLong Gold antifade reagent with DAPI (Molecular Probes, InVitrogen). As negative controls the same procedure was performed without primary antibody. Samples were viewed with an inverted Zeiss LSM 510 confocal microscope with a ×63/1.20NA water immersion objective. Nuclei were observed with DAPI. Images were produced with dual photomultiplier detectors and the LSM 5 v. 3.2.0.115 software suite.
Statistical Methods
The average values for AQUA scores from 2 replicate readings were calculated and treated as independent continuous variables. The average of TS mRNA expression was calculated from triplicate readings and was treated as an independent continuous variable. The primary objective was to assess the association between in situ TS protein expression and OS. For this we did not classify TS expression into high and low categories a priori. Instead, we used the maximal log-rank method for optimal cutpoint determination and adjusted the P-values for multiple looks.14,15 Cut-points within the central 80% of ordered TS protein expression were considered. The association between survival and low and high TS expression was then summarized in a Kaplan-Meier survival curve using the cutpoint percentile(s) determined by TS protein expression. A multivariate Cox regression analysis using backward elimination with a significance level-to-stay of 0.15 was performed to assess the impact of TS protein expression on survival while adjusting for the potential covariates tumor stage, performance status, sex, RRM1 protein expression, and ERCC1 protein expression. RRM1 and ERCC1 protein expression were included as continuous variables. Because the dichotomized split for TS protein expression was obtained by optimal selection, its Cox regression P-value was adjusted. The maximal log-rank method for optimal cutpoint determination was also used to examine the association of TS mRNA expression and OS.
Associations between continuous variables were analyzed using the Spearman correlation coefficient. Associations between continuous and discrete variables were analyzed using the Wilcoxon rank sum test or the Kruskal-Wallis test depending on the number of discrete groups. The log-rank test was used to assess survival differences between demographic and clinical groups.
RESULTS
In Situ TS Protein Expression
The monoclonal TS antibody used for the study is commercially available through LabVision (order #MS-471-P, lot #471P504B). It was generated in mice using recombinant human TS as the antigen. The antibody detected a dominant band of approximately 38 kD, the expected molecular mass of TS, with cytoplasmic localization on Western blots of lysates of lung cancer cell line H23 (Fig. 1). By using confocal microscopy we confirmed the predominant cytoplasmic localization of TS in 5 lung cancer cell lines (Fig. 2A); however, nuclear expression was observed in all, and it was lowest in H23 and highest in A549. In contrast, in 5 cell lines derived from colon carcinoma TS expression was predominantly nuclear (data not shown).
FIGURE 1.
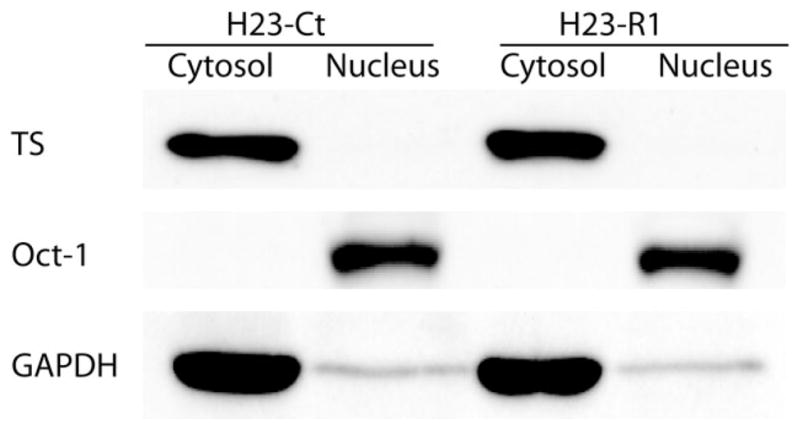
Western blot of cell lines for thymidylate synthase (TS). Cytosolic and nuclear extracts were prepared from cell lines H23-Ct and H23-R1. They were separated in polyacrylamide gels, transferred to membranes, and probed with antibodies directed to TS, Oct-1 (a nuclear protein), and GAPDH (a cytosolic protein). A band of 38 kD representing TS was found in the cytoplasm.
FIGURE 2.
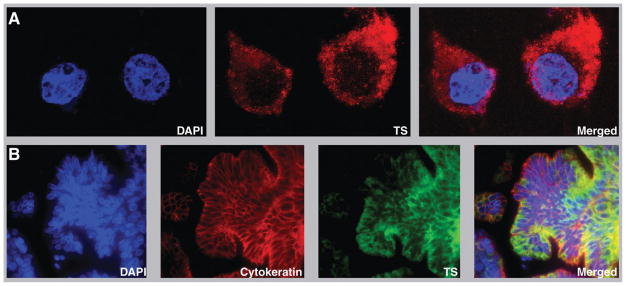
Confocal microscopy and automated quantitative analysis (AQUA) for thymidylate synthase (TS). (A) A predominantly cytoplasmic localization for TS in a lung cancer cell line (H23-Ct). (B) TS staining in a lung cancer specimen by immunofluorescence. Nuclei are stained in blue by DAPI, cytokeratin is stained in red, and TS is stained in green. The amount of cytoplasmic in situ TS expression was determined by AQUA.
In situ TS protein expression was determined by AQUA in the tumor cytoplasm in 2 replicate tissue microarrays that encompassed a total of 187 patients with completely resected NSCLC that had not received perioperative chemotherapy or radiation (Fig. 2B). TS expression data were obtained on 160/187 (86%) patients. Duplicate expression data were not available for 27 specimens because the spots were absent on the section or had washed off during the processing. Average values ranged from 5.8 to 238.6 (on a scale from 0–255) with a median of 98.4 and a mean of 98.3 (standard deviation, 53.0). The data were near normally distributed and a data transformation for normalization was not performed. There was a significant correlation in TS expression between replicate arrays (r = .599, P <.001).
There was no statistically significant association between TS protein expression and tumor stage, histology, performance status, absence or presence of weight loss, smoking status, or sex (Table 1). The unidimensional tumor diameters in this patient cohort ranged from 0.5 to 10.8 cm and did not correlate significantly with TS expression (r = .051, P = .52). We also did not find a significant correlation with the previously reported RRM1 and ERCC1 protein levels (r = .13 and −.07, respectively, P >.1).10
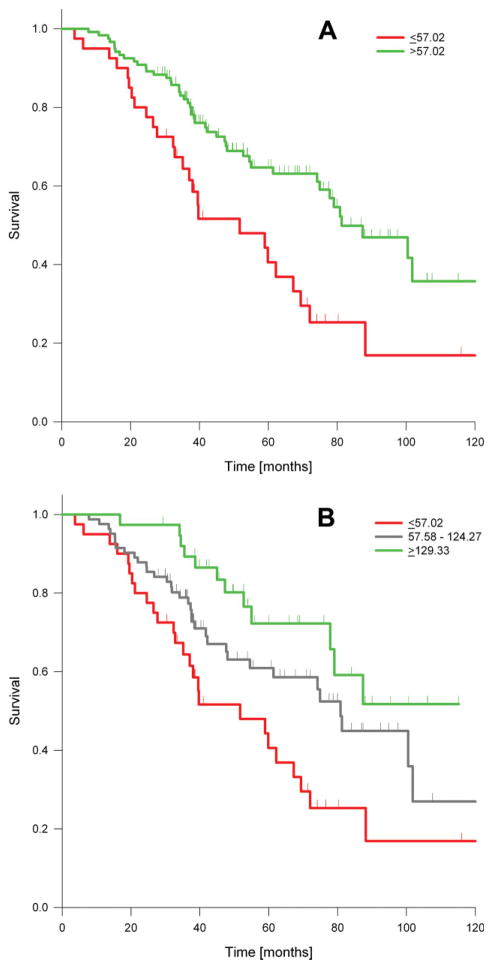
By using the maximal log-rank method for optimal cutpoint determination, we found that the most significant OS difference was seen if patients with levels of ≤57.02 were separated from those with levels >57.02, which was the 25th percentile (40/160 patients) of TS expression (Fig. 3A). The median survival in the low TS group was 51.7 months and was 81.3 months for those in the high TS group (P =.0013). This survival difference remained significant after adjusting the P-value for multiple looks (P =.034).
FIGURE 3.
Kaplan-Meier overall survival estimates by thymidylate synthase (TS) expression. (A) Survival of 160 patients by in situ protein expression. Green lines depict patients with marker expression >57.02 (N = 120; median = 81.3 months, 95% confidence interval [CI], 62.9–99.6), and red lines depict expression ≤57.02 (N = 40; median = 51.7 months, 95% CI, 21.5–81.9). The unadjusted P-value was .0013. (B) Survival of the same group of patients incorporating the second-best cutpoint. Green lines depict patients with marker expression ≥129.33 (N = 18; median not reached), blue depict patients with marker expression from 57.58 to 124.27 (N = 82; median, 80.8 months), and red lines depict expression ≤57.02 (N = 40; median = 51.7 months). The unadjusted P-value was .002.
In the final multivariate model, which included TS protein expression and tumor stage (Table 3), TS remained significantly associated with OS (P =.0013, adjusted P = .032). The hazard ratio for death was 0.45 for high versus low TS protein expression.
TABLE 3.
Cox Regression Model
| Variable | No. | Univariate hazard ratio (95% CI*) | Univariate P | Multivariate hazard ratio† (95% CI) | Multivariate P |
|---|---|---|---|---|---|
| TS group (low=reference) | 160 | .46 (.28–.75) | .040** | .45 (.28–.73) | .032‡ |
| Stage (IA=reference) | 160 | 1.72 (1.06–2.78) | .029 | 1.76 (1.08–2.87) | .022 |
| Sex (Women=reference) | 160 | 1.23 (.77–1.96) | .393 | ||
| ERCC1 expression | 160 | .994 (.986–1.002) | .130 | ||
| RRM1 expression | 160 | .988 (.975–1.002) | .095 | ||
| Performance status (0=reference) | 150 | 1.46 (.87–2.44) | .156 |
CI indicates confidence interval.
Based on 160 patients because performance status was eliminated from the final multivariate analysis.
Adjusted P to account for optimal cutpoint selection.
In addition to the planned survival analysis, exploratory examinations of the association of TS expression and survival were conducted. TS expression was included in a Cox regression analysis as a continuous variable and yielded low P-values both unadjusted (P = .006) and adjusted for tumor stage (P = .002). We also identified a second cutpoint for TS protein expression with an AQUA score of 129.33 (89th percentile). Inclusion of this cutpoint in the survival analysis yielded a classification of patients into 3 survival groups (Fig. 3B). The median survival in the highest TS group was >120 months; it was 80.9 months in the medium TS group and 51.7 months in the lowest TS group (P =.002).
A dichotomization of the TS expression data using the sample median of 98.4 as a cutpoint revealed a median OS of 81.3 months for patients with high TS expression and 62.2 months for those with low TS expression (P =.071).
TS mRNA Expression
TS mRNA expression was determined in fresh-frozen tumor specimens from 92 patients with completely resected stage I NSCLC. Data were obtained from 85 patients (92%), and expression ranged from 0.2 to 104.3 with a median of 3.5 and a mean of 6.7 (standard deviation, 12.9). mRNA levels were not significantly associated with tumor stage, performance status, absence or presence of weight loss, smoking status, or sex (Table 2). They were, however, significantly different among the histologic subtypes; the levels were lower in adenocarcinomas compared with squamous and large cell carcinomas (Table 2). The unidimensional tumor diameters in this patient cohort ranged from 1.4 to 10.8 cm and they did not correlate with TS expression (r = .135, P =.218).
We used the maximal log-rank method for optimal cutpoint determination, and no statistically significant cutpoint for TS mRNA expression was found after adjusting for multiple looks (adjusted P = 1.0).
Thirty-two patients were common to both data-sets and thus had TS expression values at both the protein and mRNA levels. There was a near zero correlation between the levels (r = .028, P =.877).
DISCUSSION
The level of expression of 2 genes involved in nucleotide synthesis and DNA damage repair, RRM1 and ERCC1, have recently been reported as prognostic of survival in patients with surgically resected NSCLC.9,10 Patients whose tumors express high levels have a better outcome that those with low levels. Several potential mechanisms may contribute to this. One of these is the reported suppression of meta-static propensity in tumors genetically engineered to overexpress RRM1 in vitro and in experimental animal models.11,12,16 Another, and possibly complementary mechanism, may be the increased ability of overexpressing cells to repair DNA damage.12
Among the genes involved in nucleotide synthesis and DNA damage repair, TS is a particularly attractive candidate, which may have similar properties because of its gatekeeper function for TTP production. Our investigations have shown that, similar to RRM1 and ERCC1, high levels of TS protein expression are prognostic of long survival in patients with surgically resected NSCLC that did not receive perioperative chemotherapy or radiation. In addition, we found no association between the levels of TS and those of RRM1 and ERCC1, which suggests that TS is an independent marker of survival. This possibility was corroborated by the multivariate analysis that included clinical variables as well as all 3 proteins. These data together support the notion that tumors with efficient nucleotide metabolism and DNA repair pathways have a less aggressive clinical behavior than those with inefficient pathways.
We were unable to obtain TS values in 14% (26/186) of the cases. This lack of TS values is a result of loss of individual spots from TMA sections during processing. We have experienced spot evaluation rates ranging from 49%, ie, loss of 51% of spots, to 100%. With the use of adhesive-coated slides our average spot evaluation rates are 83%, which is consistent with missing TS values in 14% of cases. However, an evaluation of TS expression in NSCLC cases on full section specimens should be possible in all cases that contain well-preserved viable tumor cells.
Although our study determined gene expression levels and not gene function, there is presently no evidence for biologically significant posttranslational modification of TS, which suggests there is a direct relationship between TS protein levels and function. Notably, we did not observe a correlation (r = 0.028) between mRNA and protein levels of TS. This is in contrast to a reported correlation (r = 0.6) between these levels in colorectal and gastric cancers.4 The authors assessed TS protein levels by Western blotting and mRNA levels by reverse-transcriptase polymerase chain reaction (RT-PCR) in 9 colorectal and 12 gastric cancers collected specifically for gene expression analysis. The entire specimen, collected by core needle biopsy, was processed, and the correlation coefficient between mRNA and protein expression was 0.6 in these 21 specimens.4 Thus, the methodology used for determination of TS protein expression is substantially different from the quantitative in situ method used in our study, which may account for the discrepancy. Our result, which failed to detect an association between TS protein and mRNA expression using methods specifically developed for quantitative analysis, is consistent with a more recent report demonstrating a lack of correlation between protein and mRNA levels in lung cancers for the majority of genes.17 However, we were only able to obtain simultaneous mRNA and protein levels on 32 cases, which raises concern with regard to general applicability of the result. We did not perform TS mRNA evaluation on FFPE specimens because of the limited amount of specimen availability.
Of interest are the seemingly lower TS mRNA levels in adenocarcinomas compared with squamous cell carcinomas. Although we did not observe this at the protein level, this has been previously described by Ceppi et al.18 in a cohort of 56 patients with resected NSCLC.
By using a rigorous statistical approach to account for multiple testing, we found that increased cytoplasmic TS expression was associated with prolonged survival. Two exploratory analyses supported this finding. First, had we chosen a priori to look for a decrease in the risk of death with increasing TS levels by Cox regression analysis, we would have obtained a statistically significant result (P = .006). Second, we found that the risk of death shifts dramatically at 2 distinct levels of TS expression, 57.02 and 129.33. This survival model divided our patients into a lower 25% group, middle 64% group, and upper 11% group. It is desirable to have these results confirmed in a future study.
Our result, demonstrating improved survival for NSCLC patients with high cytoplasmic TS protein expression, appears to contrast a report of improved survival for patients with low tumoral TS expression in colorectal cancer.19 The authors studied TS expression by standard IHC in formalin-fixed and paraffin-embedded specimens from 70 patients with colorectal cancer. The cohort included patients with all stages of colorectal cancer, and many received perioperative or palliative 5FU-based chemotherapy and radiation. TS expression was scored by staining intensity as high versus low. Notably, 76% of specimens had high intensity and 24% had low intensity, a dichotomization that is similar to our results. In a subgroup analysis that included the 61 patients with Dukes’ stages A-C, the authors found no significant association between TS expression and overall survival. Only if the 9 patients with metastatic carcinoma were included in the analysis was low TS intensity significantly associated with improved survival. Prior work had demonstrated that low TS expression at the mRNA level is associated with improved survival in patients with Dukes’ stage D colorectal carcinoma receiving 5FU-based chemotherapy.5,6 Thus, the results by Edler et al.19 may be explained by the increased efficacy of chemotherapy given to the patients with Dukes’ stage D disease and low TS expression. Alternatively, the impact of TS expression on patient outcomes may be associated with the subcellular compartment of its predominant localization. It is noteworthy that our confocal analyses in lung and colon cancer cell lines indicate that TS is expressed mostly cytoplasmic in lung and nuclear in colon cancers.
Finally, standard IHC with visual scoring in an attempt to quantify protein expression has significant technical limitations. These include the nonquantitative chemistry of routine immunoperoxidase stains and the subjective light intensity perception of the human eye.20 As a result, the impact of the level of expression of a target protein, such as TS, which has a continual, unimodal spectrum of expression on patient outcome may remain unrecognized. Thus, we conclude that the use of an objective and quantitative in situ protein expression methodology, such as AQUA, has allowed us to uncover the prognostic impact of TS on the survival of patients with completely resected NSCLC.
Although 5FU, a chemotherapeutic agent with efficacy in gastrointestinal and breast carcinomas, is not currently recommended for treatment of NSCLC (NCCN clinical practice guidelines in oncology, non-small-cell lung cancer, V.I.2007, http://www.nccn.org), it is important to recall that UFT, an oral agent consisting of tegafur and uracil (1:4), showed a small (3%) but significant (P = .04) improvement in 5-year OS in a randomized adjuvant phase III trial that included 979 Japanese patients with completely resected stage IA and IB adenocarcinoma of the lung.21 Given our results indicating that patients with high TS levels have a better survival than those with low levels, that TS mRNA levels are lower in adenocarcinomas than other NSCLC subtypes, and the prior work indicating better benefit from 5FU for colorectal patients with low compared with high TS expression, it is intriguing to speculate that patients with completely resected stage I adenocarcinoma and low TS expression are likely to derive maximal benefit from agents such as UFT.
Acknowledgments
Supported in part by Grant No. R01 CA102726 from the National Cancer Institute.
References
- 1.Washtien WL. Increased levels of thymidylate synthetase in cells exposed to 5-fluorouracil. Mol Pharmacol. 1984;25:171–177. [PubMed] [Google Scholar]
- 2.Moertel CG, Fleming TR, McDonald JS, et al. Levamisole and fluorouracil for adjuvant therapy of resected colon carcinoma. N Engl J Med. 1990;322:352–358. doi: 10.1056/NEJM199002083220602. [DOI] [PubMed] [Google Scholar]
- 3.Heidelberger C, Chaudhuri NK, Danneberg P, et al. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–666. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 4.Johnston PG, Lenz HJ, Leichman CG, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human color-ectal and gastric tumors. Cancer Res. 1995;55:1407–1412. [PubMed] [Google Scholar]
- 5.Leichman CG, Lenz HJ, Leichman L, et al. Quantification of intratumoral thymidylate synthase expression predicts for disseminated colorectal cancer response and resistance to protracted-infusion fluorouracil and weekly leucovorin. J Clin Oncol. 1997;15:3223–3229. doi: 10.1200/JCO.1997.15.10.3223. [DOI] [PubMed] [Google Scholar]
- 6.Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001;19:4298–4304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 7.Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Simon GR, Sharma S, Cantor A, Smith P, Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127:978–983. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 9.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. The DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 11.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 12.Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 13.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 14.Miller R, Siegmund D. Maximally selected chi-square statistics. Biometrics. 1982;38:1011–1016. [Google Scholar]
- 15.Hilsenbeck SG, Clark GM. Practical p-value adjustment for optimally selected cutpoints. Stat Med. 1996;15:103–112. doi: 10.1002/(SICI)1097-0258(19960115)15:1<103::AID-SIM156>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Zhou BS, Hsu NY, Pan BC, Doroshow JH, Yen Y. Overexpression of ribonucleotide reductase in transfected human KB cells increases their resistance to hydroxyurea: M2 but not M1 is sufficient to increase resistance to hydroxyurea in transfected cells. Cancer Res. 1995;55:1328–1333. [PubMed] [Google Scholar]
- 17.Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Ceppi P, Volante M, Saviozzi S, et al. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase. Cancer. 2006;107:1589–1596. doi: 10.1002/cncr.22208. [DOI] [PubMed] [Google Scholar]
- 19.Edler D, Kressner U, Ragnhammar P, et al. Immunohistochemically detected thymidylate synthase in colorectal cancer: an independent prognostic factor of survival. Clin Cancer Res. 2000;6:488–492. [PubMed] [Google Scholar]
- 20.Rimm DL. What brown cannot do for you. Nat Biotechnol. 2006;24:914–916. doi: 10.1038/nbt0806-914. [DOI] [PubMed] [Google Scholar]
- 21.Kato H, Ichinose Y, Ohta M, et al. A randomized trial of adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. N Engl J Med. 2004;350:1713–1721. doi: 10.1056/NEJMoa032792. [DOI] [PubMed] [Google Scholar]