Abstract
PTEN is one of the most frequently mutated or deleted tumor suppressors in human cancers. NEDD4-1 was recently identified as the E3 ubiquitin ligase for PTEN; however, a number of important questions remain regarding the role of ubiquitination in regulating PTEN function and the mechanisms by which PTEN ubiquitination is regulated. In the present study, we demonstrated that p34, which was identified as a binding partner of NEDD4-1, controls PTEN ubiquitination by regulating NEDD4-1 protein stability. p34 interacts with the WW1 domain of NEDD4-1, an interaction that enhances NEDD4-1 stability. Expression of p34 promotes PTEN poly-ubiquitination, leading to PTEN protein degradation, whereas p34 knockdown results in PTEN mono-ubiquitination. Notably, an inverse correlation between PTEN and p34/NEDD4-1 levels was confirmed in tumor samples from colon cancer patients. Thus, p34 acts as a key regulator of the oncogenic behavior of NEDD4-1 and PTEN.
Keywords: p34, NEDD4-1, PTEN, ubiquitination
The PTEN (phosphatase and tensin homolog) tumor suppressor has crucial roles in cell growth, proliferation, migration, survival, and death.1 The gene encoding PTEN, which is located on chromosome 10q23.31, is frequently mutated and deleted in a number of human cancers, including prostate, breast, glioblastoma, thyroid, melanoma, and endometrial cancers.2 In the cytoplasm, PTEN functions as a lipid phosphatase for the second-messenger phosphatidylinositol 3,4,5-triphosphate (PIP3), negatively regulating the activation of the phosphatidylinositol 3-kinase (PI3K)/AKT (also known as protein kinase B) signaling pathway via PIP3 dephosphorylation and thereby modulating downstream cellular survival and other responses, including cell growth, death, metabolism, migration, and protein translation.3
Although PTEN was initially identified as a cytoplasmic protein,4, 5 PTEN nuclear localization has since been reported in many normal and tumor cell types.6 Interestingly, nuclear PTEN, like cytoplasmic PTEN, may also exert tumor suppressor activity. Numerous studies have reported that many components of the PI3K signaling pathway, including phosphatidylinositol 4,5-bisphosphate (PIP2), PIP3, PI3K, and AKT2, are present in nuclei.7 However, recent studies have demonstrated that nuclear PTEN does not dephosphorylate nuclear PIP3,8 suggesting that the roles of nuclear PTEN are unrelated to its phosphatase activity. Functionally, nuclear PTEN has been shown to control multiple cellular events, including genomic stability, cell-cycle progression, cell death and differentiation, and gene expression.9
The tumor-suppressor functions of PTEN are controlled by a variety of modifications,10 including phosphorylation, oxidation, S-nitrosylation, acetylation, and ubiquitination. Notably, ubiquitination has been shown to regulate PTEN stability and nuclear localization.11 Recent studies have shown that increases in PTEN in nuclei are associated with increased PTEN ubiquitination, particularly monoubiquitination.7 Mono-ubiquitination enhances PTEN protein stability, whereas polyubiquitination results in proteasome-mediated PTEN degradation.12 Recently published work has revealed that NEDD4-1 (neural precursor cell expressed developmentally downregulated protein 4-1), which was identified as an E3 ubiquitin ligase for PTEN, mediates PTEN polyubiquitination4 and thereby promotes its degradation. Interestingly, PTEN mono-ubiquitination is also mediated by NEDD4-1 and results in nuclear translocation, which may protect PTEN from degradation.13, 14 However, the mechanisms underlying NEDD4-1-mediated PTEN ubiquitination remain unclear.
Although NEDD4-1 directly interacts with PTEN, resulting in the poly-ubiquitination and proteasomal degradation of endogenous PTEN, it does not completely deplete PTEN levels in all contexts. The oncogenic activity of NEDD4-1 is PTEN-dependent, as indicated by the absence of NEDD4-1 oncogenic activity in PTEN-deficient cells.15 These results prompted us to critically investigate how NEDD4-1 is regulated in the context of PTEN ubiquitination. In the present study, we identified p34 as a NEDD4-1-binding partner and established its role in regulating NEDD4-1-mediated PTEN ubiquitination and tumorigenesis.
Results
p34 interacts with NEDD4-1 and regulates its stability
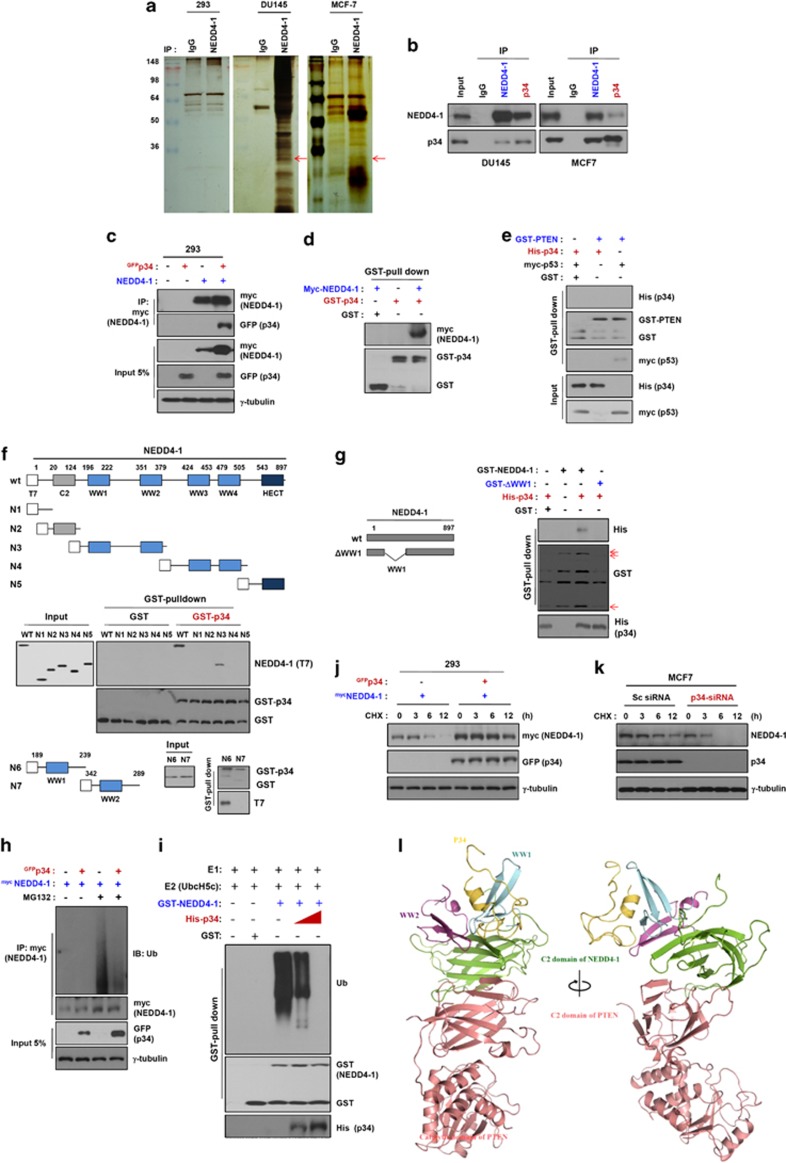
It has been previously reported that NEDD4-1 requires PTEN for its oncogenic activity.16, 17 Consistent with this observation, NEDD4-1 potentiated oncogenic Ras-induced cell transformation in a PTEN-dependent manner, suggesting that aberrant upregulation of NEDD4-1 can suppress PTEN in human cancers.12, 16, 17 On the basis of these observations, we sought to determine how NEDD4-1 is upregulated, focusing first on identifying proteins that interact with NEDD4-1. We immunoprecipitated NEDD4-1 from HEK293 (293), DU145, MCF7, and LS1034 cell lysates using an anti-NEDD4-1 antibody, followed by tandem affinity purification and mass spectrometric analyses. p34 was among the various proteins that interact with NEDD4-1 that were identified in this analysis (Figure 1a). p34 is known to bind to the cyclin-dependent kinase 4 (CDK4) and X-linked inhibitor of apoptosis protein (XIAP). This interaction was detected in the DU145 and MCF7 cancer cell lines but not in 293 cells, consistent with the results of the expression analyses, which revealed that p34 is endogenously expressed in both cancer cell lines but not in 293 cells (Supplementary Figure S1a). We first examined the interaction between endogenous p34 and NEDD4-1 in DU145, MCF7, LS1034, and MDA-MB-231 cells. Immunoprecipitation and western blot analyses detected p34 and NEDD4-1 in both p34 immunoprecipitates and NEDD4-1 immunoprecipitates, confirming that NEDD4-1 and p34 interact with one another in both cancer cell lines (Figure 1b and Supplementary Figure S2). In an effort to extend this analysis beyond DU145 and MCF7 cells, we first analyzed the protein expression levels of p34 and NEDD4-1 in a variety of cancer cell lines (Supplementary Figure S1b). Interestingly, the PTEN protein was either not expressed or only weakly expressed in cells that co-expressed p34 and NEDD4-1.
Figure 1.
p34 interacts with NEDD4-1 and enhances NEDD4-1 stability. (a) p34 is expressed in 293, DU145, MCF7, and LS1034 cancer cells (red arrow). Whole-cell lysates were immunoprecipitated using an anti-NEDD4-1 antibody, resolved using SDS-PAGE and analyzed with silver staining. Rabbit IgG was used as a negative immunoprecipitation control. (b) NEDD4-1 interacts with endogenous p34 in intact cells. DU145, MCF7, LS1034, and MDA-MB-231 cell lysates were immunoprecipitated using anti-NEDD4-1 or anti-p34 antibodies and analyzed using western blotting. (c) Exogenously expressed p34 and NEDD4-1 interact. Whole-cell lysates of 293 cells that had been transfected with GFP-p34 and/or NEDD4-1-Myc were analyzed using immunoblotting. (d) NEDD4-1 binds to p34 in vitro. GST-p34 recombinant proteins were incubated with Myc-NEDD4-1-transfected 293 cell lysates, followed by the addition of GST-Sepharose beads. The bead-bound proteins were probed using immunoblotting with antibodies that had been raised against p34 and NEDD4-1. (e) p34 does not directly bind to PTEN. p34 and PTEN binding was analyzed by incubating purified recombinant GST-PTEN with Myc-NEDD4-1. The interaction of p34 with NEDD4-1 was confirmed using His-p34-expressing cells. GST was used as a negative control for the GST pull-down binding assays. p53 was used as a positive control for the PTEN-binding assays. (f) p34 interacts with the WW1 domain of NEDD4-1. The cells were transfected with various NEDD4-1 deletion mutants were the lacking C2, WWs, and the homologous to the E6-AP carboxyl terminus (HECT) domains (NEDD4-1-N1); WWs and the HECT domains (NEDD4-1-N2); C2, WW3, WW4, and the HECT domains (NEDD4-1-N3); C2, WW1, WW2, and the HECT domains (NEDD4-1-N4); or C2 and all WW domains (NEDD4-1-N5). Twenty-four hours later, the cell lysates were incubated with GST-p34 or GST proteins, and the p34-binding domain of NEDD4-1 was analyzed using an in vitro GST pull-down assay. (g) p34 does not bind to a NEDD4-1 WW1 deletion mutant. NEDD4-1-WT or WW1 deletion mutant proteins were incubated with lysates from His-p34-expressing cells, and the binding regions of NEDD4-1 and p34 were analyzed using a GST pull-down assay. The arrows indicate GST, GST-NEDD4-1-WT, and the GST-WW1 deletion mutant. (h) NEDD4-1 auto-ubiquitination is decreased in intact cells in the presence of p34. 293 cells were transfected with NEDD4-1 and/or p34 and then treated with MG132 (25 μM) for 6 h. After harvesting the cells, anti-Myc immunoprecipitates of the cell lysates were analyzed for NEDD4-1 auto-ubiquitination. (i) NEDD4-1 auto-ubiquitination is decreased in a cell-free system in the presence of p34. GST-NEDD4-1 proteins were incubated with ATP, E1, E2, and His-p34, and auto-ubiquitination was measured. (j and k) p34 regulates NEDD4-1 protein levels. 293 and MCF7 cells were transfected with either a p34 expression plasmid for 24 h (j) or with siRNA against p34 for 48 h (k). The cells were treated with cycloheximide (50 μg/ml) at the indicated time points, followed using western blot analyses to determine the NEDD4-1 protein levels. p34 overexpression increases NEDD4-1 levels, whereas siRNA-mediated p34 knockdown decreases NEDD4-1 levels. (l) Ribbon diagram of the docking model structure of the p34/NEDD4-1/PTEN complex. The predicted mode of interactions between p34/NEDD4-1 and PTEN was based on protein–protein docking. p34 is depicted by a yellow ribbon diagram, NEDD4-1 is shown as a green ribbon diagram, and PTEN is depicted as a light red ribbon diagram. Two views from a 90-degree rotation about the vertical axis are shown
We next investigated the interaction between exogenously expressed p34 and NEDD4-1. GFP-tagged p34 was readily detected in Myc-NEDD4-1 immunoprecipitates of 293 cells co-transfected with GFP-tagged p34 and Myc-tagged NEDD4-1 expression constructs (Figure 1c). The interaction between p34 and NEDD4-1 was further examined using GST pull-down assays. Western blot analyses of purified GST-p34 proteins that had been incubated with protein extracts from 293 cells expressing Myc-tagged NEDD4-1 were used to detect p34-bound NEDD4-1 proteins (Figure 1d). GST pull-down assays followed by western blotting did not detect PTEN-bound p34 protein, indicating that p34, unlike NEDD4-1, does not bind to PTEN (Figure 1e). These results strongly suggest that p34 directly interacts with NEDD4-1 but not with PTEN.
We next analyzed which NEDD4-1 domain was responsible for the interaction with p34. We constructed expression plasmids for T7-tagged wild-type NEDD4-1 and a series of NEDD4-1 deletion mutants. Purified GST-p34 protein was then incubated with protein extracts from 293 cells expressing T7-tagged wild-type NEDD4-1 (NEDD4-1-WT) or NEDD4-1 mutants. GST-p34 was readily detected using western blot analysis following incubation with extracts of cells expressing NEDD4-1-WT or NEDD4-1-N3 (Figure 1f), indicating that p34 interacts with domains containing the WW1 and WW2 domains of NEDD4-1. We attempted to further map the domain of NEDD4-1 that was involved in p34 binding to the WW1 or WW2 domain. We created two mutant constructs of wild-type NEDD4-1, one containing only the WW1 domain and one containing only the WW2 domain. GST-p34 was clearly detected by western blotting following incubation with extracts of cells expressing the NEDD4-1 WW1 domain construct (Figure 1f). To confirm these data, we constructed an expression plasmid for a GST-tagged mutant NEDD4-1 lacking the WW1 domain (GST-delWW1). The p34 protein interacted with the wild-type NEDD4-1 protein but not with GST-delWW1 (Figure 1g), indicating that p34 directly binds to the WW1 domain of NEDD4-1.
In addition, we examined whether p34 affects the binding of PTEN to NEDD4-1. PTEN interacted with NEDD4-1 in the presence or absence of p34 (Supplementary Figure S3). However, the interaction was stronger in the presence of p34, implying that p34 may affect the binding affinity of NEDD4-1 and PTEN. Taken together, these results provide evidence that p34 interacts with the WW1 domain of NEDD4-1 but not with PTEN and enhances rather than interferes with the interaction between NEDD4-1 and PTEN. In support of the proposed effects of p34 on NEDD4-1, p34 was observed to interact with NEDD4-1 but not with PTEN.
NEDD4-1 was previously reported to be auto-ubiquitinated in cells.16 To determine whether p34 modulates this process, we overexpressed p34 and examined NEDD4-1 auto-ubiquitination. In cells that had been treated with the proteasome inhibitor MG132, p34 expression significantly decreased NEDD4-1 auto-ubiquitination (Figure 1h). To further confirm this, we evaluated NEDD4-1 ubiquitination in a cell-free system using purified proteins. Purified GST-NEDD4-1 protein was incubated with E1, UbcH5c, and extracts from 293 cells expressing His-tagged p34 from a p34 cDNA expression plasmid. p34 expression caused gradual decreases in GST-NEDD4-1 ubiquitination (Figure 1i), indicating that p34 suppresses NEDD4-1 auto-ubiquitination in both intact cells and a cell-free system. Consistent with this, NEDD4-1 protein levels were greater and more sustained in cells that had been co-transfected with GFP-tagged p34 and Myc-tagged NEDD4-1 than in cells expressing Myc-tagged NEDD4-1 alone (Figure 1j). Conversely, p34 knockdown by small interfering RNA (siRNA) significantly decreased NEDD4-1 levels compared with the treatment with scrambled siRNA (Figure 1k), indicating that p34 stabilizes the NEDD4-1 protein.
On the basis of these data, we predicted the binding mode of C2-WW1-WW2 for NEDD4-1/P34 domain/PTEN complexes (Figure 1l, Supplementary Figure S4). The WW1 domain was docked to the C-β sheets of the C2 domain of NEDD4-1 (notation of secondary structure, see reference in detail).18 The C-β sheets of the C2 domain are the interaction interfaces between the NEDD4 C2 domain and the GRB10 SH2 domain.18 The WW2 domain also docked to the C-β sheets of the C2 domain and the WW1 domain of NEDD4-1. Two salt bridge interactions were observed between the WW1 and WW2 domains. A partial homology model structure of p34 could be built (residues 71–113). The p34 model domain bound to the WW1 domain with few hydrogen bonds and hydrophobic interactions. The C-terminal 170 amino acids folded into a β sandwich structure that consisted of two antiparallel β sheets with two short a helices. The PTEN β sandwich had a topology that was identical to the type II topology of the C2 fold, and its structure was similar to that of the C2 domains of NEDD4-1. The C2 domain of NEDD4-1 interacted with the C2 domain of PTEN but not the phosphatase domain. Taken together, our findings strongly suggest that p34 stabilizes NEDD4-1 by inhibiting NEDD4-1 auto-ubiquitination and its subsequent proteasome-mediated degradation.
p34 controls NEDD4-1-mediated PTEN poly-ubiquitination
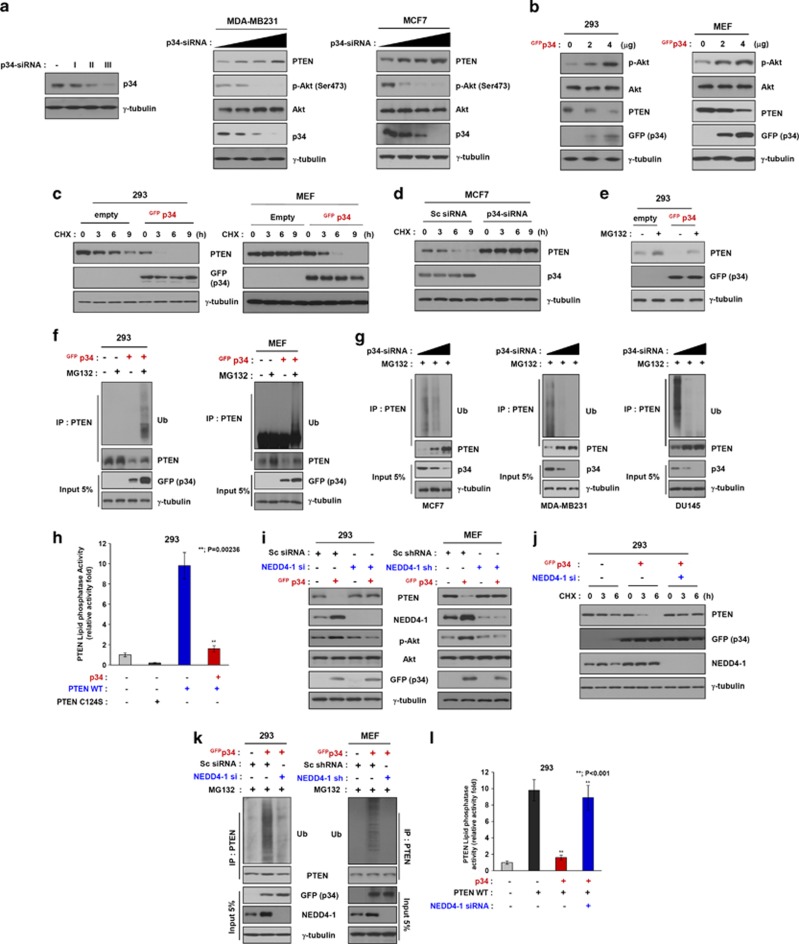
Next, we evaluated whether p34 binding to NEDD4-1 affects PTEN protein expression. We first examined the effects of p34 silencing using siRNA. p34-siRNA induced a dose-dependent increase in endogenous PTEN protein levels in MDA-MB-231 and MCF7 cells (Figure 2a). Conversely, overexpression of GFP-tagged p34 resulted in decreased levels of PTEN protein in 293 cells and mouse embryonic fibroblasts (MEFs), which do not express endogenous p34 (Figure 2b).8 In addition, the levels of phospho-AKT were inversely correlated with PTEN expression levels, which were in turn dependent upon p34 expression levels. Collectively, these results indicate that p34 may affect PTEN protein levels and PI3K signaling by binding to NEDD4-1. On the basis of these results, we further explored the role of p34 in regulating PTEN turnover by treating cells with the protein synthesis inhibitor cycloheximide and measuring changes in exogenously expressed p34 protein levels over time. In a cycloheximide-chase experiment, ectopic expression of GFP-tagged p34 led to diminished PTEN levels in 293 cells and MEFs. No such changes were observed in cells that had been transfected with the control vector (Figure 2c). Conversely, PTEN protein levels remained virtually unchanged in p34-siRNA-treated MCF7 cells, which express endogenous p34, but were decreased in cells that had been transfected with control siRNA (Figure 2d). These results suggest that p34 positively regulates the PI3K signaling pathway by decreasing PTEN protein stability. To determine whether p34 regulates PTEN protein stability via the proteasomal pathway, we transfected 293 cells with GFP-tagged p34, followed by treatment with the proteasome inhibitor MG132. The levels of the PTEN protein were significantly decreased in cells exogenously expressing p34 and MG132 compared with cells that had been transfected with the control vector (Figure 2e).
Figure 2.
p34 regulates NEDD4-1-mediated PTEN poly-ubiquitination. (a) Western blot analysis of MDA-MB-231 and MCF7 cell lines that had been transfected with p34-siRNA revealed that p34-siRNA induces a dose-dependent increase in PTEN levels and a decrease in phospho-AKT (p-AKT). (b) A Western blot analysis of 293 and MEF cells that had been transfected with GFP-tagged p34 revealed that PTEN levels are decreased and p-AKT is increased with increasing doses of exogenously expressed p34. (c and d) The half-life of the PTEN protein is decreased (in 293 cells and MEFs) by p34 overexpression and increased (in MCF7 cells) by siRNA-mediated p34 knockdown. (e) Western blot analysis of PTEN in 293 cells that had been transfected with p34 and incubated with or without MG132 revealed that PTEN is decreased in MG132-treated cells exogenously expressing p34. (f) Ubiquitination assays performed in intact 293 cells (left panel) and MEFs (right panel) that had been transfected with GFP-p34 and incubated with or without MG132 revealed that the poly-ubiquitinated form of PTEN appears following the exogenous expression of p34. (g) Ubiquitination assays performed in intact MCF7, MDA-MB-231, and DU145 cells that were transfected with p34-siRNA for 24 h and incubated with or without MG132 revealed that p34-siRNA induces a dose-dependent decrease in PTEN poly-ubiquitination in these cancer cell lines. (h) p34-mediated poly-ubiquitination of PTEN inhibits PTEN phosphatase activity. The lipid phosphatase activity of PTEN in cells that had been transfected with wild-type PTEN and/or p34 for 24 h was determined by measuring the dephosphorylating activity of immunoprecipitated PTEN. The data represent the means±S.D.'s of at least three independent experiments (**P<0.001). (i) Western blot analysis of PTEN in wild-type and NEDD4-1-knockdown cells that had been transfected with a p34 expression plasmid revealed that PTEN expression is increased by p34 expression in a wild-type NEDD4-1 background but not in NEDD4-1-knockdown cells. (j) An analysis of the PTEN protein half-life in 293 cells that had been transfected with a p34 expression plasmid and NEDD4-1 siRNA revealed that PTEN stability is enhanced by NEDD4-1 knockdown in the context of p34 expression. (k) Intact-cell ubiquitination assays in wild-type and NEDD4-1-knockdown cells that were transfected with a p34 expression plasmid revealed that PTEN poly-ubiquitination is detectable in a wild-type NEDD4-1 background but not in NEDD4-1-knockdown cells. (l) The p34-mediated reductions in the lipid phosphatase activity of PTEN were attenuated in NEDD4-1 siRNA-transfected cells compared with control cells. The data represent the means±S.D.'s of at least three independent experiments (**P<0.001)
To further determine whether p34 affects PTEN poly-ubiquitination, we performed intracellular ubiquitination assays. After ectopically expressing p34 and treating the cells with MG132, the poly-ubiquitinated form of PTEN was detected in 293 cells and MEFs (Figure 2f). Conversely, siRNA-mediated knockdown of endogenous p34 in MCF7, MDA-MB-231, and DU145 cells resulted in gradual decreases in poly-ubiquitinated PTEN (Figure 2g). In addition, the lipid phosphatase activity of PTEN was significantly decreased in cells ectopically expressing p34 compared with the lipid phosphatase-deficient PTEN C124S mutant, which was used as a negative control (Figure 2h). These results indicate that p34 mediates the poly-ubiquitination and proteasomal degradation of PTEN.
PTEN ubiquitination and proteasomal degradation were recently reported to be controlled in part by NEDD4-1, which functions as an E3 ubiquitin ligase.16 As we found that p34 directly interacts with NEDD4-1 (Figure 1), we investigated whether the effects of NEDD4-1 on PTEN ubiquitination and proteasomal degradation are dependent upon p34 expression levels. First, 293 cells and MEFs, which express endogenous NEDD4-1 but not p34, were co-transfected with a GFP-tagged p34 expression plasmid and NEDD4-1-siRNA/shRNA or scrambled siRNA. The levels of the PTEN protein remained virtually unchanged in cells that had been co-transfected with NEDD4-1-siRNA/shRNA and GFP-tagged p34 but were significantly decreased in cells that had been transfected with GFP-tagged p34 alone (Figure 2i). We further explored the effects of the ectopic expression of p34 and/or NEDD4-1-knockdown on PTEN turnover using the protein synthesis inhibitor cycloheximide. PTEN levels were remarkably decreased in NEDD4-1-knockdown 293 cells upon exogenous p34 expression but gradually increased following cycloheximide treatment (Figure 2j).
To determine whether p34 mediates NEDD4-1-dependent PTEN poly-ubiquitination, we co-transfected the cells (Figure 2i) and then treated the cells with MG132. Surprisingly, poly-ubiquitinated PTEN was clearly detected in each cell type that had been transfected with GFP-tagged p34 but not in the cells that had been co-transfected with NEDD4-1-siRNA/shRNA and GFP-tagged p34 (Figure 2k). Consistent with these observations, the lipid phosphatase activity of PTEN was lost in cells that had been treated with scrambled siRNA, although it was retained in NEDD4-1-knockdown cells ectopically expressing p34 (Figure 2l). Collectively, these results indicate that NEDD4-1-induced PTEN poly-ubiquitination is dependent upon p34.
XIAP was recently reported to induce PTEN poly-ubiquitination.19 To evaluate whether p34 affects XIAP-induced PTEN poly-ubiquitination, we performed poly-ubiquitination experiments in XIAP-knockout MEFs (Supplementary Figure S5a). Transfection of XIAP-knockout MEFs with GFP-tagged p34 led to decreases in PTEN protein levels (Supplementary Figure S5b). Consistent with this observation, PTEN in XIAP-knockout MEFs that had been transfected with GFP-tagged p34 was strongly poly-ubiquitinated (Supplementary Figure S5c). Taken together, these results suggest that p34 regulates the NEDD4-1-mediated poly-ubiquitination of PTEN in an XIAP-independent manner.
Knockdown of p34 results in PTEN mono-ubiquitination
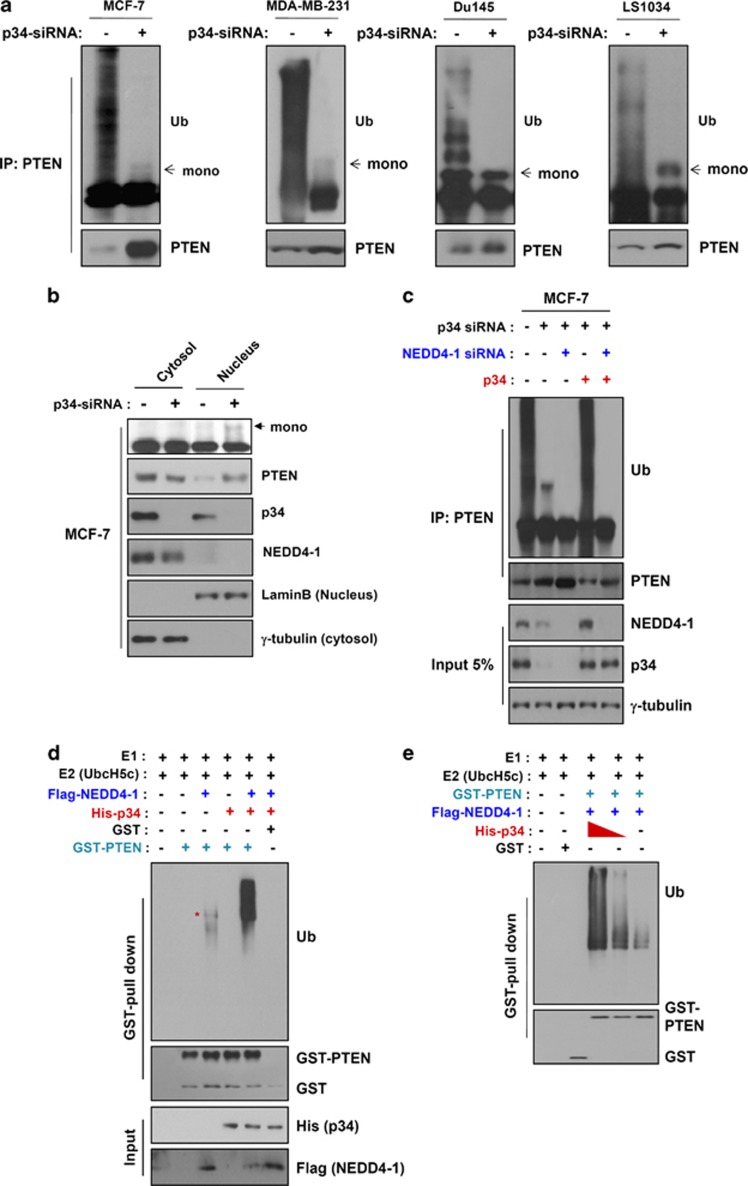
As noted above, p34 controls NEDD4-1-mediated PTEN poly-ubiquitination (Figure 2). To extend these results, we analyzed whether p34 affects PTEN mono-ubiquitination. We first transfected four cancer cell lines that endogenously express p34 and NEDD4-1 (MCF7, MDA-MB-231, DU145, and LS1034) with p34-siRNA and examined their PTEN ubiquitination statuses. Knockdown of p34 resulted in a decrease in poly-ubiquitinated PTEN (Figure 3a and Supplementary Figure S6). Consistent with this observation, PTEN proteins were strongly detected in the nuclear fraction of p34-knockdown MCF7 cells; however, virtually no cells that had been treated with scrambled siRNA displayed PTEN nuclear localization (Figure 3b). Our immunostaining experiments also revealed that the PTEN protein was mainly localized in the nuclei of cells expressing p34-siRNA but not those expressing scrambled siRNA (Supplementary Figure S7). Furthermore, mono-ubiquitinated PTEN was detected only in the nuclear fractions, indicating that the expression of p34 affects PTEN mono-ubiquitination and nuclear localization. To determine whether NEDD4-1 affects PTEN mono-ubiquitination upon p34 expression, we transfected MCF7 cells with NEDD4-1-siRNA, p34-siRNA, and/or GFP-tagged p34. Mono-ubiquitinated PTEN, which was detected in p34-knockdown cells, was not observed in cells that had been co-transfected with NEDD4-1-siRNA and p34-siRNA (Figure 3c). To further confirm this observation, we incubated purified GST-PTEN proteins with or without protein extracts from 293 cells expressing Flag-tagged NEDD4-1 and/or His-tagged p34 in the presence of E1 and E2 enzymes. PTEN was mono-ubiquitinated in the extracts that contained NEDD4-1 alone, whereas PTEN was strongly poly-ubiquitinated in the extracts containing both NEDD4-1 and p34 (Figure 3d). Furthermore, PTEN poly-ubiquitination was gradually decreased in a p34 dose-dependent manner (Figure 3e), implying that NEDD4-1-mediated PTEN mono- and poly-ubiquitination are dependent upon the expression of p34. On the basis of these results, we further investigated whether p34 acts as an E2-like enzyme in NEDD4-1-mediated PTEN ubiquitination. Purified GST-PTEN was ubiquitinated in extracts obtained from 293 cells expressing Flag-tagged NEDD4-1 and His-tagged p34 only when the E2 enzyme was present (Supplementary Figure S8), indicating that p34 does not function as an E2-like enzyme. Collectively, these results suggest that p34 may control switching between NEDD4-1-mediated PTEN mono- and poly-ubiquitination.
Figure 3.
p34 promotes switching between NEDD4-1-mediated PTEN mono- and poly-ubiquitination. (a) Intact-cell ubiquitination assays revealed that PTEN is mono-ubiquitinated in p34-knockdown cells (MCF7, MDA-MB-231, DU145, and LS1034). (b) PTEN is localized to the nuclei of p34-knockdown cells. MCF7 cells were transfected with p34-siRNA for 24 h, fractionated into nuclear and cytosolic compartments, and analyzed for PTEN expression using western blot analyses. (c) p34 expression levels dictate the switch from PTEN poly-ubiquitination to mono-ubiquitination. PTEN ubiquitination in intact cells is significantly decreased by p34 and NEDD4-1 knockdown. The cells were transfected with p34-siRNA, NEDD4-1-siRNA, and/or p34 for 24 h. PTEN ubiquitination was determined using western blot analyses of immunoprecipitated PTEN-Ub adducts. (d) Cell-free ubiquitination assays of GST-PTEN that had been incubated with NEDD4-1 in the presence or absence of p34 revealed that PTEN is mono-ubiquitinated in the presence of NEDD4-1 alone and poly-ubiquitinated in the presence of both NEDD4-1 and p34. (e) PTEN poly-ubiquitination in cell-free assays decreases with decreasing doses of p34
p34 knockdown suppresses cancer cell proliferation
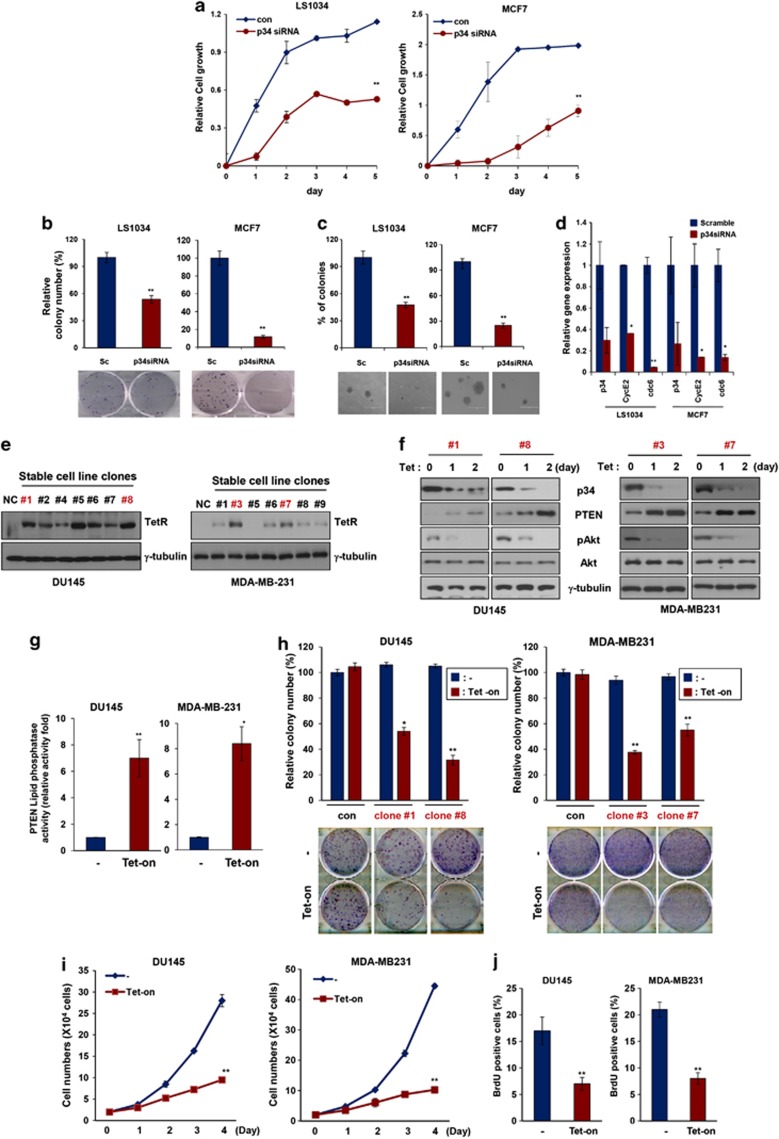
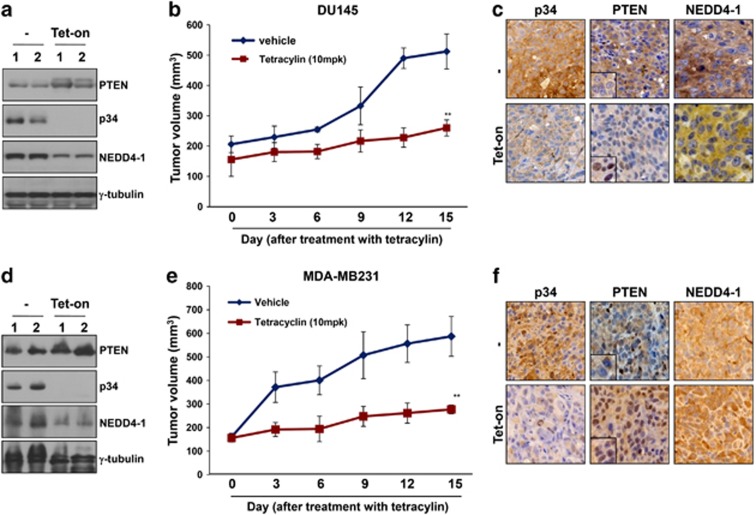
As indicated above, we postulated that p34 might function as a potent oncogene in the NEDD4-1/PTEN pathway. To test this possibility, we first examined the proliferation of cells ectopically expressing p34-siRNA. The growth of MCF7 and LS1034 cells expressing p34-siRNA was markedly decreased compared with that of cells expressing scrambled siRNA (Figure 4a). Furthermore, the frequency of colony formation by p34-siRNA-treated cells was dramatically decreased compared with control cells (Figure 4b), and the sizes of the colonies that were formed by p34-siRNA-treated cells was also decreased (Figure 4c). Consistent with these observations, PTEN was localized in the nuclei of p34-knockdown cells, and the expression levels of the cyclin E2 and Cdc6 genes, which are the targets of PTEN, were subsequently reduced (Figure 4d). Collectively, these results indicate that p34 knockdown suppresses cancer cell growth and likely does so, at least in part, by downregulating cyclin E2 and Cdc6. To further examine the inhibitory effects of p34 knockdown on cancer cell proliferation, we first established derivatives of the DU145 and MDA-MB-231 cell lines that stably express tetracycline-inducible p34-shRNA vectors (Figure 4e). Treatment with tetracycline gradually decreased p34 expression and increased PTEN expression in each clone. The levels of phosphorylated AKT were also inversely correlated with PTEN levels (Figure 4f) and lipid phosphatase activity (Figure 4g) in these clones. Tests of two clones of DU145 and MDA-MB-231 derivatives revealed that colony formation was significantly decreased after exposure to tetracycline but remained largely unchanged in untreated cells (Figure 4h). In addition, colony formation was significantly increased in Hs578T cells stably expressing p34 compared with cells that had been stably transfected with the control vector (Supplementary Figure S9). Furthermore, the growth of cells from each tetracycline-treated clone was dramatically decreased compared with that of untreated cells (Figure 4i). Consistent with these observations, BrdU incorporation was clearly reduced after exposure to tetracycline (Figure 4j). Collectively, these results indicate that p34 negatively regulates the PTEN-mediated inhibition of cancer cell proliferation.
Figure 4.
p34 regulates cancer cell proliferation. (a) Knockdown of p34 inhibits cell proliferation in the LS1034 (left panel) and MCF7 (right panel) cell lines. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01). (b) Colony-forming assays confirmed that cell proliferation is inhibited in p34-siRNA-transfected LS1034 and MCF7 cell lines. Representative photos are shown in the bottom panels. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01). (c) PTEN suppresses anchorage-independent growth in p34-siRNA-transfected LS1034 and MCF7 cell lines. All soft-agar assays were performed in triplicate. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01). (d) PTEN expression levels were determined using quantitative RT-PCR in cells that had been transfected with p34-siRNA or scrambled siRNA. The data represent the means±S.D.'s of at least three independent experiments (*P<0.05, **P<0.01). (e and f) Cells that had been stably transfected with Tet-On inducible p34-shRNA were treated with tetracycline (Tet; 1 μg/ml) to inhibit p34 expression. PTEN, p34, p-Akt, and NEDD4-1 expressions were analyzed using western blot analysis. γ-tubulin was used as an internal loading control. (g) PTEN lipid phosphatase activity was increased in tetracycline-inducible, stable p34-knockdown cell lines. The data represent the means±S.D.'s of at least three independent experiments (*P<0.05, **P<0.01). (h) Colony-forming assays confirmed that cell proliferation is inhibited in cell lines that have been stably transfected with tetracycline-inducible p34-shRNA. The data represent the means±S.D.'s of at least three independent experiments (*P<0.05, **P<0.01). (i) The growth of MDA-MB-231 and DU145 cells is inhibited in media containing tetracycline to inhibit p34. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01). (j) BrdU incorporation assays revealed that cell growth is inhibited after tetracycline treatment. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01)
p34 potentiates tumorigenicity
As p34 controls the stability of the NEDD4-1 protein and promotes switching between NEDD4-1-mediated PTEN mono- and poly-ubiquitination, we hypothesized that p34 expression would affect the oncogenic potential of NEDD4-1. To test this possibility, we established a DU145 derivative that stably expresses tetracycline-inducible shRNA against p34. These cells were subcutaneously injected into mice and were allowed to form xenograft tumors; the mice were then treated with tetracycline or vehicle. Tumors derived from two independent tetracycline-inducible stable clones of these DU145 derivatives displayed increased PTEN levels after tetracycline injection, whereas NEDD4-1 levels were decreased in association with a virtual elimination of p34 expression (Figure 5a). Furthermore, tumor growth in tetracycline-treated mice was significantly decreased compared with that in vehicle-treated control mice (Figure 5b). We next analyzed PTEN localization in tumors using immunohistochemistry (IHC). Surprisingly, PTEN displayed strong nuclear localization after tetracycline exposure (Figure 5c). In addition, p34 expression after tetracycline treatment was correlated with NEDD4-1 expression. To further confirm the inhibitory effects of p34 on tumor growth, we also established MDA-MB-231 derivatives that stably expressed shRNA against p34. Consistent with the results outlined above, p34 knockdown in these cells resulted in increased PTEN levels (Figure 5d) and decreased tumor growth (Figure 5e). PTEN was also localized to the nuclei of these tumor cells following exposure to tetracycline (Figure 5f). These results suggest that p34 potentiates the oncogenic activity of NEDD4-1.
Figure 5.
p34-shRNA inhibits xenograft tumor growth in a PTEN-dependent manner. (a) Western blot analyses of tumors derived from inducible p34-shRNA-transfected DU145 cells revealed that PTEN expression is increased after tetracycline treatment. (b) The growth of tumors derived from inducible p34-shRNA-transfected DU145 cells is reduced by the inhibition of p34 expression by tetracycline (10 mg/kg). The data represent the means±S.E.M. (**P<0.01). (c) IHC analyses demonstrated that PTEN is localized to the nucleus of DU145 tumor cells after tetracycline treatment. (d) Western blot analyses revealed that PTEN expression in tumors derived from inducible p34-shRNA-transfected MDA-MB-231 cells is increased after tetracycline treatment. (e) The growth of tumors derived from inducible p34-shRNA-transfected MDA-MB-231 cells is reduced by the inhibition of p34 expression with tetracycline (10 mg/kg). The data represent the means±S.E.M. (**P<0.01). (f) IHC analyses demonstrated that PTEN is localized to the nuclei of MDA-MB-231 tumor cells after tetracycline treatment. Representative images are shown
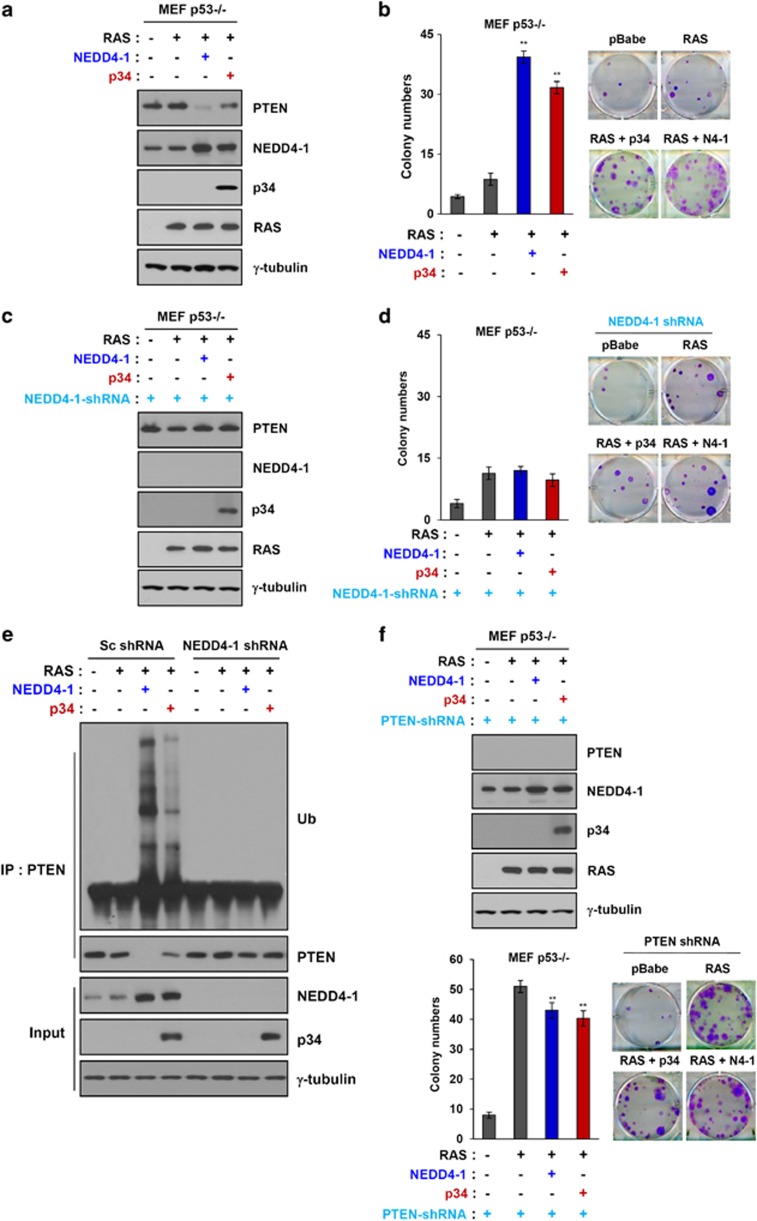
To further examine the effect of p34 on the oncogenic activity of NEDD4-1, we utilized p53-null MEFs, which do not express endogenous p34. NEDD4-1 alone is not oncogenic; however, its overexpression enhances the Ras-mediated transformation of p53-deficient MEFs.17 On the basis of this report, we first investigated the effects of p34 expression on the Ras-mediated transformation of p53-null MEFs. Consistent with the results for NEDD4-1 overexpression, p34 expression caused a decrease in PTEN levels and an increase in NEDD4-1 levels in these cells (Figure 6a). Furthermore, p34 expression resulted in an increase in colony numbers compared with that of Ras-expressing cells in p53-null MEFs (Figure 6b), implying that p34 may contribute to oncogenesis by suppressing PTEN expression, as shown in Figure 6a. Next, we examined the oncogenic activity of p34 in NEDD4-1-knockdown – p53-null MEFs using retroviral-based shRNA vectors. Importantly, p34 expression did not induce a decrease in PTEN levels in NEDD4-1KDp53−/− MEFs (Figure 6c), indicating that p34 can suppress PTEN expression in a NEDD4-1-dependent manner. Consistently, colony formation in NEDD4-1KDp53−/− MEFs was decreased to a greater extent than in the results shown in Figure 6b despite p34 expression (Figure 6d), suggesting that the oncogenic activity of p34 is NEDD4-1- and PTEN-dependent. Importantly, p34 expression resulted in PTEN poly-ubiquitination in p53-null MEFs expressing control shRNA but not in p53-null MEFs expressing NEDD4-1 shRNA (Figure 6e), indicating that p34 enhances the Ras-mediated transformation of p53-null MEFs in a NEDD4-1-dependent manner. Finally, to determine whether the effects of p34 on the oncogenic function of NEDD4-1 are PTEN-dependent, we performed colony-forming assays using p53-null MEFs expressing PTEN-shRNA. In these MEFs, p34 expression failed to enhance the transforming activity of Ras (Figure 6f), indicating that p34 enhances the oncogenic activity of NEDD4-1 in a PTEN-dependent manner. Collectively, these results suggest that p34 potentiates the PTEN-dependent oncogenic activity of NEDD4-1.
Figure 6.
PTEN-controlled tumorigenesis is dependent upon p34 and NEDD4-1. (a) Western blot analyses of PTEN, NEDD4-1, p34, and Ras expression in p53−/− MEFs that were infected with PTEN, NEDD4-1, or p34 revealed that p34 expression decreases PTEN expression and increases NEDD4-1 expression. (b) Colony-forming assays revealed that the overexpression of NEDD4-1 increases cellular transformation by Ras in p53−/− MEFs. Representative photos are shown in the right-hand panel. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01) (c) Western blot analysis of PTEN, NEDD4-1, p34, and Ras expression in NEDD4-1-knockdown p53−/− MEFs infected with Ras and PTEN or NEDD4-1 revealed that p34 does not enhance PTEN levels in the absence of NEDD4-1. (d) Colony-forming assays confirmed that cell transformation is inhibited in NEDD4-1-knockdown p53−/− MEFs. Representative photos are shown in the right-hand panel. The data represent the means±S.D.'s of at least three independent experiments. (e) Ubiquitination assays in intact cells revealed that p34 does not enhance NEDD4-1-mediated PTEN poly-ubiquitination in NEDD4-1-knockdown p53−/− MEFs. (f) Colony-forming assays (lower panel) and western blot analyses (upper panel) revealed that PTEN knockdown potentiates cellular transformation in Ras-infected p53−/− MEFs. The data represent the means±S.D.'s of at least three independent experiments (**P<0.01)
We then explored the clinical relevance of p34, NEDD4-1, and/or PTEN in human cancer tissues. We assessed the expression of p34, NEDD4-1, and PTEN in paraffin-embedded tissue microarrays (TMAs) of 191 colon tumors (Supplementary Figure S10). Of the 191 colon samples, PTEN expression was not detected in 87.2% of the 117 NEDD4-1/p34 double-positive samples. In contrast, PTEN was expressed in 15 (12.8%) of the NEDD4-1/p34 double-positive tumor samples (Supplementary Figure S10a). Therefore, our results indicate a significant inverse correlation between PTEN and p34/NEDD4-1 levels in colon cancer samples.
Discussion
PTEN is subjected to both poly- and mono-ubiquitination mediated by NEDD4-1, an E3 ubiquitin ligase, leading to PTEN degradation and nuclear localization.11, 16 However, whereas PTEN deletion induces the transformation of p53-deficient MEFs, NEDD4-1 overexpression does not, indicating that NEDD4-1 is not oncogenic alone. Moreover, using NEDD4-1-knockout MEFs,17 Fouladkou et al.14 recently demonstrated that NEDD4-1 is dispensable for the regulation of PTEN stability and localization. Thus, although the regulation of PTEN protein stability has been widely studied, the detailed mechanisms controlling its stability remain poorly understood. In the present study, we found that p34, which we identified as a binding partner for NEDD4-1, may promote switching between PTEN mono- and poly-ubiquitination via the regulation of NEDD4-1 protein stability.
p34 is a known CDK4-binding partner and has been reported to positively regulate the cell-division cycle.20, 21 p34 amplification has been observed in various human carcinomas, including pancreatic,22 lung,23 and ovarian cancers.24 p34 was recently reported to be strongly expressed in human breast cancer tissues but was absent or weakly expressed in normal tissues,25 implying that p34 may be an oncoprotein. We found that p34 (SEI-1) directly interacts with the WW1 domain of NEDD4-1, whereas its close homologs, SEI-2 and SEI-3, do not (data not shown). The mechanism by which these highly homologous proteins display distinct specificity for binding partners is of interest for future study.
We further demonstrated that the co-expression of p34 and NEDD4-1 is strongly correlated with the protein levels of PTEN in colon cancer tissues and that NEDD4-1 positively regulates tumorigenesis via the p34-dependent, proteasome-mediated degradation of PTEN. NEDD4-1 is not oncogenic alone, and its oncogenic activity is PTEN-dependent. Consistent with this observation, NEDD4-1 potentiates oncogenic Ras-induced cell transformation in a PTEN-dependent manner, suggesting that aberrant upregulation of NEDD4-1 can suppress PTEN in human cancers.12, 16, 25
The finding that NEDD4-1 is an E3 ubiquitin ligase for PTEN raises a very important question: How is NEDD4-1 regulated? Moreover, what factors are intimately involved in the NEDD4-1-mediated degradation of PTEN? The current study focused on identifying a binding partner for NEDD4-1 that does not interact with PTEN. Importantly, our study revealed that p34, a NEDD4-1-interacting protein, potentiates tumorigenesis by stabilizing the NEDD4-1 protein and inducing PTEN poly-ubiquitination. Auto-ubiquitination of NEDD4-1 has been reported to have similar effects.16 Interestingly, the relationship between p34 and NEDD4-1 is similar to that between p34 and XIAP, which functions as an E3 ligase.25 p34 directly interacts with the BIR2 domain of XIAP, promoting an increase in XIAP stability. The increased stability of XIAP, in turn, enhances its antiapoptotic activity. Notably, stabilization of the E3 ligase, NEDD4-1, via p34 binding significantly decreased PTEN protein stability, indicating that by stabilizing its E3 ligase-binding partner, p34 can affect various cellular events. In particular, the stabilization of NEDD4-1 by p34 potentiated cancer cell survival, proliferation, tumorigenesis, and resistance to anticancer agents.
High-resolution methods such as X-ray crystallography and NMR spectroscopy provide atomic-level information but encounter difficulties at the molecular-complex level. In addition to elucidating the binding mode governing the interactions of p34, NEDD4-1, and PTEN (as shown in Figure 1l), we were able to build a partial homology model structure of p34 (residues 71–113). Our model of this complex suggests that p34 does not interfere with the interaction between NEDD4-1 and PTEN. PTEN has been previously reported to bind to the C2 domain of NEDD4-1,12 an interaction that involves the C2 domain of PTEN, not the phosphatase domain. Protein–protein docking was performed using the rigid body docking method to predict protein–protein docking, as outlined in the Materials and Methods section. The disadvantage of the rigid docking method is that conformational changes will not be observed in the predicted protein–protein docking. However, the PTEN β sandwich has a topology that is identical to the type II topology of the C2 fold,26 and its structure is similar to that of the C2 domains of NEDD4-1 (http://www.rcsb.org), suggesting that the C2 domain of NEDD4-1 interacts with the C2 domain of PTEN and not with the phosphatase domain.
In the present study, we demonstrated that p34 directly interacts with the WW1 domain of NEDD4-1, forming the lowest-energy module in our model of the complex. Furthermore, p34 did not bind to purified PTEN in 293 cell extracts in vitro, suggesting that p34 indirectly modulates PTEN protein stability by binding to NEDD4-1. The observed differences in the binding affinity of NEDD4-1 for PTEN in the presence or absence of p34 warrant further study.
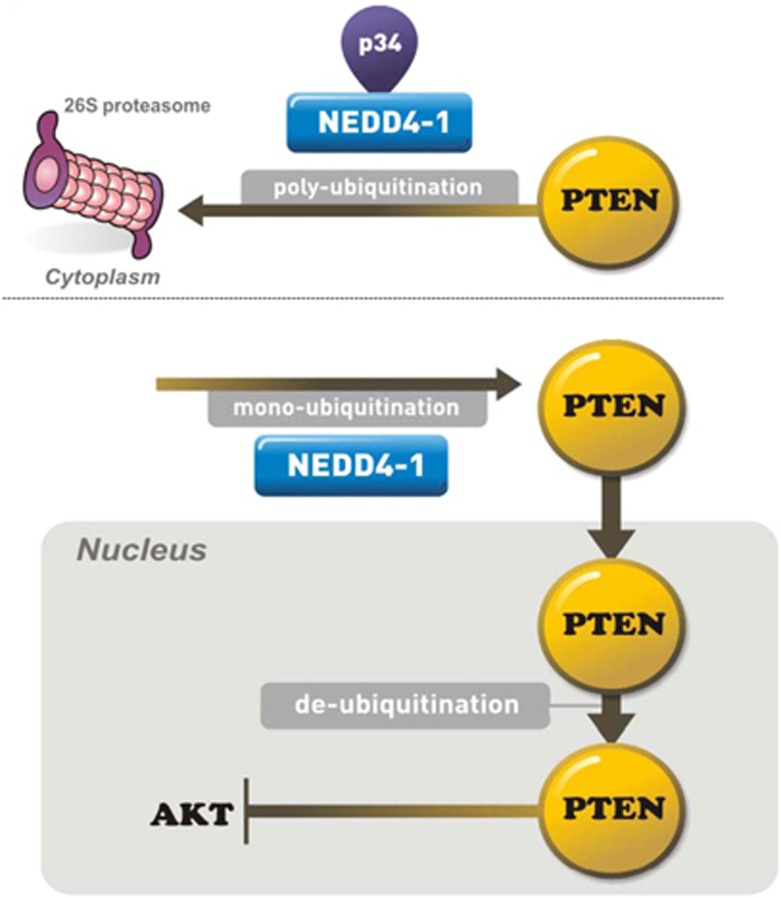
Another issue is the identity of the factors that are intimately involved in the nuclear localization of PTEN. As PTEN, unlike p53, has no transferable nuclear localization signal-like sequences, it has been suggested that PTEN nuclear localization may be predominantly dependent upon the ubiquitination system.11, 27, 28 NEDD4-1 has also been shown to dictate PTEN localization by inducing its mono-ubiquitination. Notably, the effects of NEDD4-1 on the ubiquitination activity of purified PTEN were potentiated in extracts from 293 cells that ectopically expressed p34, whereas the enzymatic activity of NEDD4-1 toward purified PTEN was decreased in the absence of p34, suggesting that p34 may modulate NEDD4-1-mediated PTEN ubiquitination. Furthermore, the absence of p34 resulted in PTEN nuclear localization in cells that endogenously express NEDD4-1. Indeed, positive regulation of PTEN nuclear transport by NEDD4-1 has been reported.11 Therefore, the presence and absence of p34 may affect the switch between the NEDD4-1-mediated mono- and poly-ubiquitination of PTEN (Figure 7).
Figure 7.
p34 interacts with NEDD4-1 and enhances its stability. NEDD4-1 associates with the enzymatic machinery that is required for PTEN ubiquitination. PTEN poly-ubiquitination is promoted by the interaction of NEDD4-1 with p34
Importantly, we also examined the clinical relevance of p34, NEDD4-1, and/or PTEN in human cancer tissues. As p34 functions as a positive regulator of NEDD4-1-mediated PTEN ubiquitination, p34 expression would be predicted to contribute to NEDD4-1-induced decreases in the levels of PTEN in human cancer samples. We confirmed this hypothesis by assessing the expression of p34, NEDD4-1, and PTEN in the TMAs of colon tissue cores. There was a significant inverse correlation between p34, NEDD4-1, and PTEN levels in colon tumors. Collectively, these results imply that the expression of p34 contributes to the NEDD4-1-mediated downregulation of PTEN in human cancers. This conclusion was further supported by the observation that PTEN levels were not affected in p34 negative and NEDD4-1 positive colon tumors (Supplementary Figure S10b). Therefore, we conclude that p34 expression contributes to the PTEN-dependent oncogenic activity of NEDD4-1 in human colon cancers.
Materials and Methods
Cell culture
Human colon cancer, breast cancer, and embryonic kidney 293 cell lines were purchased from ATCC. The cells were cultured in 5% CO2 at 37 °C in either Dulbecco's modified Eagle's medium (DMEM) or RPMI-1640 (GIBCO BRL, Grand Island, NY, USA) containing high glucose that had been supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) and penicillin/streptomycin (100 μg/ml). Primary wild-type and p53-null (p53−/−) MEFs were obtained from Millipore (Billerica, MA, USA). XIAP−/− MEFs were kindly provided by Dr. Duckett (University of Michigan Medical School). Wild-type MEFs, p53−/− MEFs, and XIAP−/− MEFs were cultured in DMEM that had been supplemented with 10% FBS and penicillin/streptomycin.
Plasmids, siRNA, shRNA, and transfection
p34, NEDD4-1, and PTEN cDNAs were purchased from Origene (Rockville, MD, USA). The NEDD4-1 deletion mutants (N1: 1–20; N2: 10–189; N3: 189–389; N4: 389–514; N5: 514–897) were subcloned into T7-tagged pCMV plasmids. The PTEN C124S mutation was introduced using a polymerase chain reaction (PCR)-based Quick change site-directed mutagenesis kit (Intron Biotechnology, Seongnam-si, Republic of Korea). All DNA and siRNA transfection studies were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's recommended protocol. The following siRNA oligonucleotides were used in the transfection studies: p34-siRNA (I), 5′-GCAAGGGUCUGAAGCGGAA-3′ p34-siRNA (II), 5′-GGAAACGGGAGGAGGAGGA-3′ p34-siRNA (III), 5′-CCGAAUUGGACUACCUCAU-3′); and NEDD4-1 siRNA, (5′-TGGCGATTTGTAAACCGAA-3′). shRNAs against NEDD4-1, p34, or PTEN were acquired from Sigma (St. Louis, MO, USA).
In vitro proliferation assays
The cells were harvested, counted, and plated in 12-well plates (2 × 104 cells per well). The cells were fixed with 10% formalin and stained with a 0.01% crystal violet solution. After staining, the cells were lysed in 10% acetic acid and analyzed using spectrophotometry.
Colony-forming assays
The clonogenic capacity was determined by plating the cells at a density of 300 cells per six-well plate. Ten to fourteen days after plating, the cells were fixed and stained with a solution of 0.01% crystal violet.
Western blot and immunoprecipitation analyses
For the western blotting analyses, the proteins were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PolyScreen membranes (New England Nuclear, Boston, MA, USA). The membranes were blocked by incubating them with 5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween-20 (TBS-T) and were then probed with antibodies. For the immunoprecipitation analyses, the cell lysates were incubated with antibodies for 12 h at 4 °C with mixing. Then, 20 μl of protein A/G Plus-Sepharose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was added, and the mixtures were incubated for an additional 2 h at 4 °C. The immunoprecipitates were washed five times with Nonidet P-40 lysis buffer and boiled in 20 μl of 2 × SDS sample buffer.
The following antibodies were used in this study: anti-PTEN, anti-phospho-AKT, anti-Myc, and anti-Ras (Cell Signaling, Beverly, CA, USA); anti-p34 (Enzo Life Sciences, Beverly, MA, USA); anti-Flag (Sigma); anti-AKT, anti-NEDD4-1, anti-GFP, anti-GST, anti-His, anti-Ub, anti-LaminB, and γ-tubulin (Santa Cruz Biotechnology); anti-TetR (Clontech, Mountain View, CA, USA); and anti-T7 (Abcam, Cambridge, MA, USA). The primary antibodies were detected using horseradish peroxidase-conjugated goat anti-mouse, goat anti-rabbit, or donkey anti-goat secondary antibodies, as appropriate, and an enhanced chemiluminescence detection system (Amersham, Buckinghamshire, UK).
Computational methods
Protein–protein docking of PTEN with the N-terminal C2, WW1, and WW2 domains of NEDD4-1 and p34 was sequentially performed using ZDOCK29 and RDOCK29, 30, 31 (Accelrys Software) (Discovery studio, version 2.1., 2007). In the docking studies, the structure of PTEN was taken from PDB 1D5R (http://www.rcsb.org), in which the structure was resolved in a complex that was bound to L-tartrate. The N-terminal C2 domain of NEDD4-1 was taken from 3B7Y. Discovery Studio 2.1 (Accelrys Software) with MODELER was used to construct a homology model of the WW1 and WW2 domains of NEDD4-1 and the p34 domain. The crystal structure of the mimivirus NDK (3DDI) was used as the template for the p34 domain (Supplementary Figure S4, www.rcsb.org). ZDOCK is a fast, rigid body, initial-stage protein–protein docking algorithm. RDOCK is used for the further refinement of complexes that are generated suing ZDOCK via CHARMm.32 In the initial stage, the protein receptor and protein ligand are treated as rigid bodies, and all six rotational and translational degrees of freedom are fully explored using scoring functions that are tolerant to conformational changes, also known as soft docking functions. An angular step of 15 degrees was used. ZDOCK uses a shape complementarity method for scoring known as Pairwise Shape Complementarity.33 For the rotational search, evenly distributed Euler angles were used. During the refinement stage of RDOCK, the 200 best poses of near native structures that were obtained during the initial stage were refined and re-ranked using a more detailed energy function that took conformational changes into account, as well as a solvation term.30
Soft-agar assays
Soft-agar plates were prepared by adding a bottom layer of 1.6% agarose in 2 × DMEM or 2 × RPMI-1640 that had been supplemented with 10% FBS to the wells of six-well plates. The cells (1 × 104) were resuspended in 0.7% agarose (Invitrogen) and 2 × DMEM or RPMI-1640 that had been supplemented with 10% FBS and seeded onto the bottom layer. The plates were then incubated for 2–3 weeks in a 37 °C incubator. The colonies were counted under a dissecting microscope.
In vitro GST pull-down binding assays
For the GST pull-down assays, 500 ng of GST-p34 or GST control were expressed in the Escherichia coli BL21(DE3) strain, as previously described,11 mixed with the lysates of NEDD4-1 deletion mutant-expressing cells in reaction buffer (20 mM Tris-HCl (pH 7.5) and 120 mM NaCl), and incubated at 30 °C for 1 h. GST pull-down buffer (20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 1% Triton X-100, 0.02% bovine serum albumen, and 5 mM 2-mercaptoethanol) was added to terminate the reaction, after which point, GST and its associated proteins were incubated for 1 h at 4 °C with glutathione-Sepharose beads (Cell Signaling, Danvers, MA, USA) and pulled down. The complexes were washed six times using GST pull-down buffer, and 2 × SDS sample buffer was added. The presence of p34 and NEDD4-1 in the immunoprecipitates, which indicates that these two proteins interact, was analyzed using immunoblotting with anti-GST and anti-T7 antibodies, respectively.
In vitro and in vivo ubiquitination assays
For the ubiquitin-binding assays, 5 μg of purified GST-PTEN, His-p34, and Flag-NEDD4-1 proteins were added to ubiquitin-binding mixtures containing 8 ng E1 (Calbiochem, San Diego, CA, USA), 500 ng E2 (GST-UbcH5c; Calbiochem), and 5 μg of ubiquitin (Sigma) in ubiquitination buffer (50 mM Tris-HCl (pH 7.4), 2 mM ATP, 5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 1 mM creatine phosphate, and 15 units/ml of creatine phosphokinase).
For the ubiquitination assays in a cell-free system, an anti-GST antibody was used to obtain PTEN protein by immunoprecipitating the protein lysates from that cells that had been transfected with PTEN-GST using an anti-GST antibody. The immunoprecipitates were extensively washed using wash buffer and then added to ubiquitination reaction mixtures containing 5 μg GST-PTEN. The reactions were incubated for 90 min at 37 °C, terminated by adding SDS sample buffer, and resolved using SDS-PAGE.
For the ubiquitination assays in intact cells, 293 cells or MEFs were transiently transfected with p34-GFP and NEDD4-1-Myc. Forty-eight hours after transfection, the cells were harvested, the insoluble fractions were removed using high-speed centrifugation (15 000 × g for 15 min at 4 °C), and 300 μg of total cellular protein from the clarified supernatants was immunoprecipitated using an anti-Myc antibody. Endogenous PTEN was detected in cells that had been transfected with p34-GFP. After 48 h, the cell lysates were prepared and immunoprecipitated with an anti-PTEN antibody. Ubiquitin adducts were detected via western blotting using an anti-ubiquitin antibody. The levels of transfected and endogenous proteins after immunoprecipitation were analyzed via western blotting using anti-GFP or anti-PTEN antibodies.
Tet-On inducible and Hs578T stable cell lines
Tet-On inducible stable cell lines were established by transfecting DU145 and MDA-MB-231 cells with the TetR-expressing plasmid, pcDNA6.0/TR (Invitrogen), followed by cell selection with blasticidin (10 μg/ml) for 3 weeks. TetR expression in the stable cell clones was determined via immunoblot analysis with an anti-TetR antibody. Two TetR-overexpressing clones were transfected with the H1-p34-shRNA vector (5′-CCCTCTTTGACCTCTCAGT-3′), and these doubly transfected cells were selected for treatment with zeocin (300 μg/ml) for 3 weeks. p34 expression in the selected, stably transfected clones was inhibited by treating the cells with tetracycline (1 μg/ml), and p34 downregulation was confirmed by immunoblotting for p34. p34 was stably overexpressed in Hs578T cells, which do not express detectable p34, by transfecting the cells with the retroviral vector pBabe-puro-p34 and selecting for the cells using puromycin (2 μg/ml) for 2 weeks. Control cells that had been stably transfected with empty vector (pBabe-puro) were prepared using the same approach. p34 expression in the stable cell lines was confirmed via western blot analyses using an anti-p34 antibody.
PTEN lipid phosphatase assays
The cells were lysed in 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, and 1% Triton X-100 buffer containing protease inhibitors. PTEN was immunoprecipitated using an anti-PTEN antibody, and the immunoprecipitates were washed and incubated with water-soluble diC8-phosphatidylinositol 3,4,5-trisphosphate in 100 mM Tris-HCl (pH 8.0) and 10 mM DTT at 37 °C for 40 min. Thereafter, the supernatants were collected and incubated with Biomol Green Reagent (Enzo Life Sciences) at room temperature for 30 min. PTEN activity was determined by measuring the release of phosphate via colorimetric detection at an optical density of 650 nm (OD650).
Quantitative reverse transcription-PCR (RT-qPCR) analyses
Total RNA was extracted from the cells using TRIzol (Invitrogen) according to the manufacturer's recommended instructions. cDNA was synthesized using RT premix (Bioneer, Daejeon, Korea) and oligo dT16-18 primers, according to the manufacturer's recommended protocol. Real-time RT-PCR was conducted using a SYBR Green PCR Master mix and a LightCycler 480 system (Roche Molecular Systems, Pleasanton, CA, USA). A 1-μl portion of the RT reaction was used to amplify p34, Cdc6, and cyclin E2 fragments using the following primer pairs: p34, 5′-TGGCCTCTAGCTCCCTCTTT-3′ (Fwd) and 5′-GCCAGTAAGTTGTCAGCCACA-3′ (Rev); Cdc6, 5′-CAGGTTCTGGACAATGCTGC-3′ (Fwd) and 5′-CTGCTGAAGAGGGAAGGAATC-3′ (Rev); and cyclin E2, 5′-TGCTGCCTTGTGCCATTT-3′ (Fwd) and 5′-GTGCTCTTCGGTGGTGTCATA-3′ (Rev). As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were quantified in parallel with the target genes using the following primer pair: 5′-AGAAGGCTGGGGCTCATTTG-3′ (Fwd) and 5′-AGGGGCCATCCACAGTCTTC-3′ (Rev). The expression levels of the targets were normalized to those of GAPDH, and fold changes in the normalized expression were calculated using the ΔΔCt method.
In vivo tumor xenografts
Xenograft tumor growth in Balb/c nude mice was used as a model in the in vivo experiments. All of the animal studies complied with animal protocols that had been approved by the Asan Medical Center Institutional Animal Care and Use Committee. Inducible p34-shRNA stable cell lines (DU145 and MDA-MB-231) were implanted subcutaneously. The tumors were measured every 3 days for 2 weeks. When the tumor sizes reached ∼180 mm3, the mice were administered 10 mg/kg of tetracycline (day 0) via the drinking water, and the tumor volumes were monitored for a 2-week period.
Immunohistochemistry
PTEN, p34, and NEDD4-1 in tumor tissues and TMAs were detected using IHC. The colon cancer TMAs were purchased from US Biomax, Inc. (Rockville, MD, USA). Antigen retrieval was performed by heating the samples in citrate buffer (pH 6.0) in a microwave. The reaction products were developed with diaminobenzidine (Dako, Hamburg, Germany), followed by counterstaining with hematoxylin. A negative control in which non-immune rabbit serum was used instead of the primary antisera did not produce any staining (data not shown). Hematoxylin was used to counterstain the IHC sections. Three pathologists independently reviewed the results.
Statistical analyses
The data were statistically analyzed using a two-tailed Student's t-test. The level of significance that was stated in the text was based on the P-values, and P-values <0.05 were considered significant. In the patient TMAs, the associations between p34(+/−), NEDD4-1(+), and negative/positive PTEN were measured using a logistic regression with a binary covariate (1: p34(+) and NEDD4-1(−); 0: others). All of the P-values were two-sided, and P-values <0.05 were considered to indicate statistical significance. R software version 2.15.1 was used for all of the statistical analyses (http://www.R-project.org).
Acknowledgments
This work was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF), which was funded by the Ministry of Education, Science and Technology (2012009160), and a grant from the Korea Health 21 R&D Project, Ministry of Health and Welfare and Family Affairs, Republic of Korea (A062254). This work also was supported by Grant 2012-231 from the Asan Institute for Life Sciences, Seoul, Republic of Korea. We thank Dr. Colin S Duckett (University of Michigan Medical School) for providing the XIAP−/− MEFs. We would like to thank Professor Kyoung Tai No for allowing to access the computational resources in his group.
Glossary
- PTEN
phosphatase and tensin homolog
- NEDD4-1
neural precursor cell expressed developmentally downregulated protein 4-1
- HECT
homologous to the E6-AP carboxyl terminus
- PI3K
phosphatidylinositol 3-kinase
- XIAP
X-linked inhibitor of apoptosis protein
- MEF
mouse embryonic fibroblast
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by J Silke
Supplementary Material
References
- Tamura M, Gu J, Tran H, Yamada KM. PTEN gene and integrin signaling in cancer. J Natl Cancer Inst. 1999;91:1820–1828. doi: 10.1093/jnci/91.21.1820. [DOI] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Aparicio C, Renner O, Leal JF, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis. 2007;28:1379–1386. doi: 10.1093/carcin/bgm052. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci USA. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamguney T, Stokoe D. New insights into PTEN. J Cell Sci. 2007;120 (Pt 23:4071–4079. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
- Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. J Cell Sci. 2008;121 (Pt 3:249–253. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata JE, Debnath S, Jenkins EC, Jr, Fournier MV. Nongenomic mechanisms of PTEN Regulation. Int J Cell Biol. 2012;2012:379685. doi: 10.1155/2012/379685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Chan AM. Post-translational modifications of PTEN and their potential therapeutic implications. Curr Cancer Drug Targets. 2011;11:536–547. doi: 10.2174/156800911795655930. [DOI] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shi Y, Wang J, Huang G, Jiang X. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem J. 2008;414:221–229. doi: 10.1042/BJ20080674. [DOI] [PubMed] [Google Scholar]
- Zhang S, Yu D. PI(3)king apart PTEN's role in cancer. Clin Cancer Res. 2010;16:4325–4330. doi: 10.1158/1078-0432.CCR-09-2990. [DOI] [PubMed] [Google Scholar]
- Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, et al. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci USA. 2008;105:8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010;285:42130–42139. doi: 10.1074/jbc.M110.143412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Themsche C, Leblanc V, Parent S, Asselin E. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J Biol Chem. 2009;284:20462–20466. doi: 10.1074/jbc.C109.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SI, Yang CM, Sim KG, Hentschel DM, O'Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J. 2001;20:2273–2285. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Nakamura T, Ohtani N, Hampson L, Hampson IN, Shimamoto A, et al. Regulation of CDK4 activity by a novel CDK4-binding protein, p34(SEI-1) Genes Dev. 1999;13:3027–3033. doi: 10.1101/gad.13.22.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund M, Gorunova L, Andren-Sandberg A, Dawiskiba S, Mitelman F, Johansson B. Cytogenetic and fluorescence in situ hybridization analyses of chromosome 19 aberrations in pancreatic carcinomas: frequent loss of 19p13.3 and gain of 19q13.1-13.2. Genes Chromosomes Cancer. 1998;21:8–16. [PubMed] [Google Scholar]
- Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, et al. Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res. 1994;54:1801–1806. [PubMed] [Google Scholar]
- Tang TC, Sham JS, Xie D, Fang Y, Huo KK, Wu QL, et al. Identification of a candidate oncogene SEI-1 within a minimal amplified region at 19q13.1 in ovarian cancer cell lines. Cancer Res. 2002;62:7157–7161. [PubMed] [Google Scholar]
- Hong SW, Kim CJ, Park WS, Shin JS, Lee SD, Ko SG, et al. p34SEI-1 inhibits apoptosis through the stabilization of the X-linked inhibitor of apoptosis protein: p34SEI-1 as a novel target for anti-breast cancer strategies. Cancer Res. 2009;69:741–746. doi: 10.1158/0008-5472.CAN-08-1189. [DOI] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Pei J, Xia H, Ke H, Wang H, Tao W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- Zhang C, Vasmatzis G, Cornette JL, DeLisi C. Determination of atomic desolvation energies from the structures of crystallized proteins. J Mol Biol. 1997;267:707–726. doi: 10.1006/jmbi.1996.0859. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Weng Z. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.