Abstract
EWS (Ewing's Sarcoma) gene encodes an RNA/DNA-binding protein that is ubiquitously expressed and involved in various cellular processes. EWS deficiency leads to impaired development and early senescence through unknown mechanisms. We found that EWS regulates the expression of Drosha and microRNAs (miRNAs). EWS deficiency resulted in increased expression of Drosha, a well-known microprocessor, and increased levels of miR-29b and miR-18b. Importantly, miR-29b and miR-18b were directly involved in the post-transcriptional regulation of collagen IV alpha 1 (Col4a1) and connective tissue growth factor (CTGF) in EWS knock-out (KO) mouse embryonic fibroblast cells. The upregulation of Drosha, miR-29b and miR-18b and the sequential downregulation of Col4a1 and CTGF contributed to the deregulation of dermal development in EWS KO mice. Otherwise, knockdown of Drosha rescued miRNA-dependent downregulation of Col4a1 and CTGF proteins. Taken together, our data indicate that EWS is involved in post-transcriptional regulation of Col4a1 and CTGF via a Drosha–miRNA-dependent pathway. This finding suggests that EWS has a novel role in dermal morphogenesis through the modulation of miRNA biogenesis.
Keywords: EWS, Drosha, microRNA, CTGF, dermal development
Ewing sarcoma (EWS) gene is a member of the TET family proteins (TLS/FUS, EWS, and TAF15) and encodes a RNA and single-stranded DNA-binding protein. A role for EWS in RNA splicing has been implicated from its interactions with various splicing factors.1 EWS also interacts with subunits of transcription factor II D, CREB-binding protein (CBP) and RNA polymerase II complexes, suggesting a role in basic transcription.2 Subsequent studies have shown that EWS acts as a transcriptional activator for BRN3A, HNF3, and OCT4 in a cell-type- and promoter-specific manner.3, 4, 5 Interestingly, EWS gene is frequently rearranged by chromosomal translocations in several cancers, leading to its fusion with many transcription factors including FLI1, ATF1, and WT1. The resulting chimeric fusion proteins, such as EWS–FLI1, EWS–ATF1, and EWS–WT1, function as aberrant transcription factors that drive proliferation, survival, and transformation.6 Although much effort has been focused on the EWS fusion oncoproteins, very little effort has been invested on understanding the function of wild-type (WT) EWS. A recent study has shown that EWS has essential roles in precursor B lymphocyte development and meiosis.7 Furthermore, loss of Ews resulted in premature cellular senescence in mouse embryonic fibroblasts (MEFs) and in hematopoietic stem progenitor cells.8 Therefore, EWS is a multifunctional protein with roles in many different cellular processes.
Skin development is organized by complex and balanced mechanism of gene activation and silencing. Collagen IV is a major part of the dermal–epidermal junction and has special roles in the maintenance of basement membrane integrity in the skin.9, 10 Previous studies demonstrated that reduced or depleted Collagen IV leads to impaired basement membrane stability.11 Connective tissue growth factor (CTGF) is a member of the CCN family of a cysteine-rich protein.12 CTGF induces cell adhesion and expression of the extracellular matrix (ECM) protein collagen type I.13 Several studies have shown that the CTGF stimulates proliferation, angiogenesis, migration, ECM production, and cell attachment.14
MicroRNAs (miRNAs) are a small noncoding RNAs (∼22 nucleotides) that regulate gene expression at the post-transcriptional level. miRNA processing is initiated by RNA polymerase II as primary miRNAs (pri-miRNAs). Drosha cleaves pri-miRNA to precursor miRNAs (pre-miRNAs) in the nucleus.15 Exportin-5 facilitates the exit of the pre-miRNAs and Dicer removes the loop of the pre-miRNAs to produce the mature miRNA duplex. One strand of the duplex is combined with the RNA-induced silencing complex (RISC).16 The miRNA processing machinery (microprocessor) including Drosha and DGCR8 may participate in the processing of miRNAs for the proper execution of gene expression programs during normal development, including skin development.17 The level of cell-type-specific molecules and factors implicated in skin development are directly regulated by miRNAs. Recent studies report that miRNAs have a quite important role in skin development.18
EWS has been found in the complex containing Drosha by mass spectrometry analysis.19 However, the role of EWS in miRNA biogenesis or regulation has not been studied. To determine whether EWS has any roles in miRNA expression and/or regulation, we performed miRNA array and RNA-sequencing analyzing genome-wide data to identify which miRNAs and mRNAs are altered under EWS deficiency. We further validated targets and molecular pathways that are associated with the identified miRNAs in vivo. Our results indicate that EWS has a crucial role in the expression of genes that are essential for the normal development of skin through the regulation of specific miRNAs.
Results
EWS modulates miRNA biogenesis
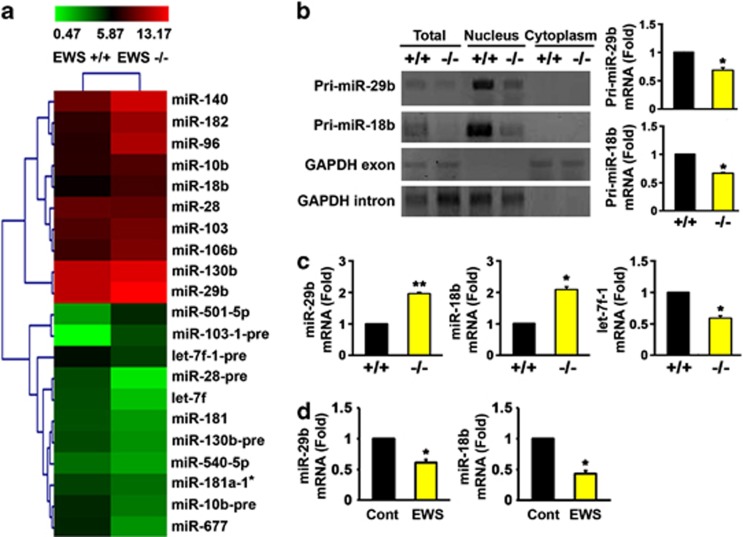
In the first series of experiments, we performed a microarray analysis of miRNA expression to determine the role of EWS in miRNA biogenesis using EWS WT (+/+) and knock-out (KO) (−/−) MEF cells (Figure 1a). We found upregulated versus downregulated miRNAs in the absence of EWS. We chose to focus our studies on the regulation and function of miR-29b and miR-18b in the context of EWS-deficient conditions because they are known to target mRNAs related to the formation of ECM, specifically collagen IV alpha 1 (Col4a1) and CTGF.7, 20 Furthermore, levels of miR-29b and miR-18b were significantly elevated in EWS KO mice, which have abnormal skin development (Supplementary Table 1 and Supplementary Figures 1a and b). We performed quantitative real-time PCR (qRT-PCR) analysis of miR-29b and miR-18b and confirmed that both miRNAs are highly and significantly induced in EWS KO cells (Figure 1c). We proposed that the increase of mature miRNAs is likely mediated through the activity of microprocessor via subsequent processing of pri-miRNA in the EWS KO cells (Figure 1b).15 In this regard, we measured the levels of pri-miR-29b and pri-miR-18b in subcellular fractions (Figure 1b). As we expected, the levels of pri-miR-29b and pri-miR-18b were significantly decreased in the nuclear factions of EWS KO cells. To further confirm whether the processing of miR-29b and miR-18b are regulated by EWS, we ectopically expressed EWS (Supplementary Figure 4d) and measured miR-29b and miR-18b levels (Figure 1d). Overexpression of EWS significantly reduced miR-29b and miR-18b levels. This result shows that EWS regulates the processing of miR-29b and miR-18b.
Figure 1.
EWS deficiency alters the level of miRNAs. (a) The heat map of miRNA array showed that relative expressions of miR-29b and miR-18b are highly increased in EWS KO MEF. The relative expression of miRNAs is displayed as colors: higher (red) or lower (green). The heat map represents the average of two samples. (b) RT-PCR analysis verified that the levels of pri-miR-29b and pri-miR-18b are decreased in subcellular fractions of EWS KO MEF compared with EWS WT MEF. The intron and exon of mouse GAPDH were amplified as markers of nuclear and cytoplasmic RNA, respectively. The graphs (right panel) represent the average±S.E.M. of three separate experiments. (c) qRT-PCR analysis verified that miR-29b and miR-18b are significantly increased in EWS KO MEF. Let-7f-1 was decreased in EWS KO MEF. The data represent the average±S.E.M. of three separate experiments. (d) qRT-PCR analysis showed that overexpression of EWS significantly reduces miR-29b and miR-18b levels in EWS KO MEFs. The data represent the average±S.E.M. of three separate experiments. Significantly different at *, P<0.05; **, P<0.005
miR-29b and miR-18b negatively regulate Col4a1 and CTGF expression
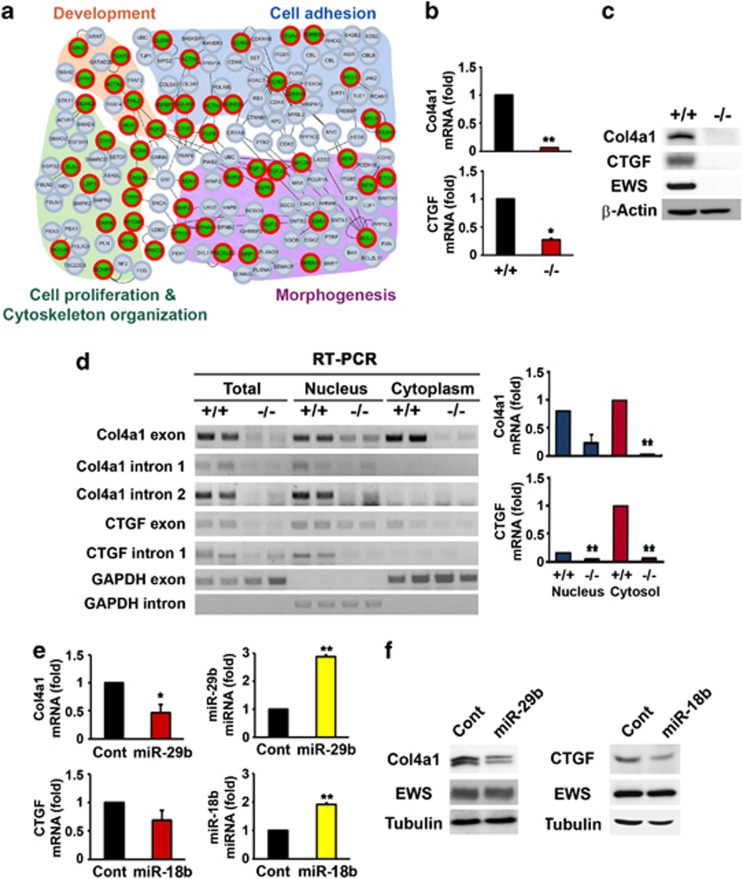
In order to examine whether proposed targets of miRNAs are altered at the transcript level and what other profiles of transcriptome (mRNA) are modulated under EWS deficiency condition, we carried out the whole transcriptome sequencing (RNA-seq) and gene analysis. We found that dermal development related gene Col4a1 and CTGF, target of miR-29b and miR18b, and other genes (FGF10, Col8a1, DMPK, FGFR2, MGP, MYOM1, ASPN, ENG, PRRX1, and Serpinb1a) were altered in EWS KO cells in comparison to EWS WT cells (Supplementary Figures 2a and b). We further performed functional enrichment analysis for the transcriptome data and found that 46 genes among the 161 genes are mainly involved in processes associated with cell adhesion, cell proliferation and cytoskeleton organization, development, and morphogenesis. Then, we generated a biological network to define relationships among 46 genes and their specified processes as shown in Figure 2a. The network showed that EWS deficiency deregulates genes that are closely linked to cell proliferation and impairs development and morphogenesis. Col4a1 and CTGF protein levels were significantly decreased in EWS KO cells (Figure 2b). qRT-PCR analysis confirmed that the mRNA levels of Col4a1 and CTGF also were dramatically decreased in EWS KO cells (Figure 2c). These results suggest that the expression of Col4a1 and CTGF might be regulated at the post-transcriptional level. To test this, we performed RT-PCR with RNAs isolated from nuclear and cytoplasm of EWS WT and KO cells. Amplification of Col4a1 (first and second) and CTGF (first) introns were markedly decreased in the nucleus and the cytoplasm of EWS KO cells, which is consistent with reduced post-transcription of Col4a1 and CTGF in EWS KO cells (Figure 2d). Amplification of Col4a1 and CTGF exons in EWS KO cells were also lower than those of EWS WT cells (Figure 2d). We also examined whether Col4a1 and CTGF transcripts are regulated post-transcriptionally by EWS deficiency (Figure 2d). qRT-PCR showed that levels of Col4a1 and CTGF mRNA are significantly reduced in the cytoplasmic fraction and only slightly decreased in the nuclear fraction of EWS KO cells. This suggests that EWS is involved in transcriptional regulation in part and more substantially in post-transcriptional regulation of Col4a1 and CTGF. As we were interested in determining how of Col4a1 and CTGF mRNA levels are reduced in the cytoplasmic fraction given that miRNA (miR-29b and miR-18b) levels are significantly expressed in EWS KO cells, we chose to study post-transcriptional regulation of Col4a1 and CTGF through miRNA in the context of EWS deficiency. To verify whether miR-29b and miR-18b target and regulate Col4a1 and CTGF mRNA, we overexpressed miR-29b and miR-18b and measured the protein levels of Col4a1 and CTGF, respectively. miR-29b and miR-18b markedly decreased the protein levels of Col4a1 and CTGF (Figure 2f). qRT-PCR results showed that the reduction of Col4a1 and CTGF mRNAs are inversely correlated with high levels of ectopically expressed miR-29b and miR-18b (Figure 2e).
Figure 2.
Col4a1 and CTGF are targeted by miR-29b and miR-18b under EWS deficiency condition. (a) A biological network delineating cellular processes associated with cell adhesion, cell proliferation and cytoskeleton organization, development and morphogenesis is represented by functionally enriched 46 genes. Node colors denote the downregulation (green with red circle) or no change (gray) of mRNA expression under EWS deficiency. Genes in the network were grouped according to their interaction and function. The lines represent activation information obtained from KEGG pathways. (b) qRT-PCR analysis verified that the mRNA levels of Col4a1 and CTGF were significantly reduced in EWS KO MEF. The data represent the average±S.E.M. of three separate experiments. (c) Western blot analysis confirmed that Col4a1 and CTGF were downregulated in EWS KO MEF. (d) RT-PCR analysis verified that mRNAs of Col4a1 (exon, intron1 and 2) and CTGF (exon and intron 1) the Col4a1 and CTGF are differently modulated in subcellular fractions of EWS WT and KO MEFs. The intron 1 and exon of mouse GAPDH were amplified as markers of nuclear and cytoplasmic RNA, respectively. The data represent the average±S.E.M. of three separate experiments. (e) Overexpression of miR-29b and miR-18b downregulated the mRNA levels of Col4a1 and CTGF in EWS WT MEFs. The mRNAs were detected by qRT-PCR. The data represent the average±S.E.M. of five separate experiments. Significantly different at *, P<0.05; **, P<0.005. (f) Overexpression of miR-29b and miR-18b reduced the protein level of Col4a1 and CTGF in EWS WT MEFs
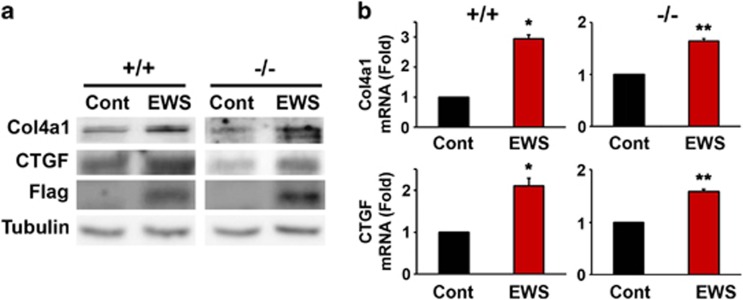
To investigate whether EWS directly affects the expression of Col4a1 and CTGF, we ectopically expressed EWS and measured the transcript and protein levels of Col4a1 and CTGF. EWS increased mRNA and protein levels of Col4a1 and CTGF in EWS WT and KO MEFs (Figures 3a and b). Although Col4a1 and CTGF protein levels were robustly reduced in EWS KO MEFs in which the levels of miR-29b and miR-18b were elevated, the ectopic overexpression of miR-29b and miR-18b only moderately downregulated Col4a1 and CTGF protein levels. The mechanism responsible for this discrepancy is unclear but in any case it seems that endogenously expressed miR-29b and miR-18b are more efficient post-transcriptional regulators than their exogenous counterparts in EWS KO MEFs. The underlying causes of this difference will require further investigation.
Figure 3.
Overexpression of EWS leads to increases of Col4a1 and CTGF expression. (a) Overexpression of EWS increased the protein levels of Col4a1 and CTGF were in EWS MEFs. (b) qRT-PCR analysis represented that overexpression of EWS increased the mRNA levels of Col4a1 and CTGF in EWS MEF cells. The data represent the average±S.E.M. of three separate experiments. Significantly different at *, P<0.05; **, P<0.005
EWS regulates Drosha expression
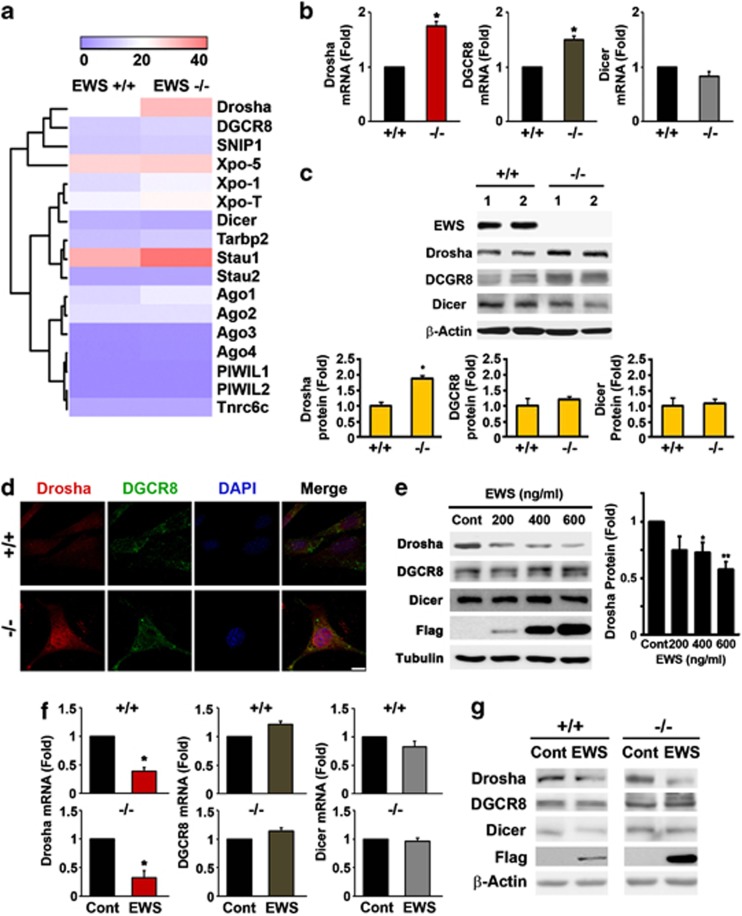
As we discovered that Col4a1 and CTGF are regulated by abnormal increases of miR-29b and miR-18b under EWS deficiency, we proposed that microprocessor might also be affected and they are consequently involved in the generation of mature miRNAs in EWS MEF cells. In this context, we performed functional enrichment analysis for the transcriptome data to find out the expression of miRNA processing-related genes including microprocessors (Drosha, DGCR8, and Dicer). The heat map showed alterations of miRNA processing-related genes (Figure 4a). Interestingly, we found that both mRNA and protein level of Drosha were highly and significantly elevated in EWS KO cells (Figures 4b and c). The protein levels of DGCR8 and Dicer did not show significant change in EWS KO cells (Figure 4c). We further performed subcellular fractionation and identified that Drosha is highly increased in the nucleus of EWS KO cells in comparison to EWS WT cells (Supplementary Figure 3a). In conformity with the fractionation data, the immunofluorescence staining and confocal microscopy showed that the strong immunoreactivity of Drosha is found in the nucleus of EWS KO cells, whereas the immunoreactivity of DGCR8 is barely increased in the nucleus (Figure 4d). To examine whether EWS directly affects the transcription of Drosha, we performed a Drosha promoter assay and Chromatin Immunoprecipitation (ChIP). Overexpression of EWS repressed the transcriptional activity of Drosha promoter in a dose-dependent manner while EWS-DNA occupancy within the region of the Drosha promoter was increased in NIH 3T3 cells (Supplementary Figures 4a and b). In addition, the protein level of Drosha was consistently decreased by EWS in a dose-dependent manner, whereas the protein levels of DGCR8 and Dicer were not changed by EWS (Figure 4e). To study whether the level of Drosha is directly regulated by EWS, we ectopically expressed EWS and measured the mRNA and protein levels of Drosha in EWS WT and KO MEFs. Both mRNA and protein levels of Drosha were downregulated by EWS, whereas DGCR8 and Dicer levels were not affected (Figures 4f and g). As we expected, the level of Drosha mRNA was inversely correlated with the level of EWS (Supplementary Figures 4c and d). These data indicate that EWS modulates the expression of Drosha.
Figure 4.
EWS regulates Drosha expression. (a) The heat map of transcriptome analysis showed relative expressions of miRNA processing-related genes in between EWS WT and KO MEFs. Drosha among many microprocessors was increased in EWS KO cells, wherease other genes were barely changed. The relative expression level of genes was displayed as colors: higher (pale red) or lower (violet). (b) qRT-PCR verified that the mRNA level of Drosha was most significantly increased in EWS KO MEF. The data represent the average±S.E.M. of three separate experiments. (c) Western blot analysis confirmed that the protein level of Drosha elevated in EWS KO MEF. Two independent experiments were performed for western blot analysis. The graphs represent the average±S.E.M. of three separate experiments. (d) The confocal microscopy presented that the immunoreactivity of Drosha is highly increased in the nucleus of EWS KO MEF, whereas the immunoreactivity of DGCR8 is not changed. Scale bar, 10 μm. (e) Overexpression of EWS reduced the protein level of Drosha in a dose-dependent manner. The graph represents the average±S.E.M. of five separate experiments. (f) Overexpression of EWS decreased the mRNA level of Drosha both in EWS WT and KO MEFs. The data represent the average±S.E.M. of five separate experiments. Significantly different at *, P<0.05; **, P<0.005. (g) Overexpression of EWS reduced the protein levels of Drosha both in EWS WT and KO MEFs
Drosha alters miR-29b and miR-18b processing and involves in Col4a1 and CTGF expression
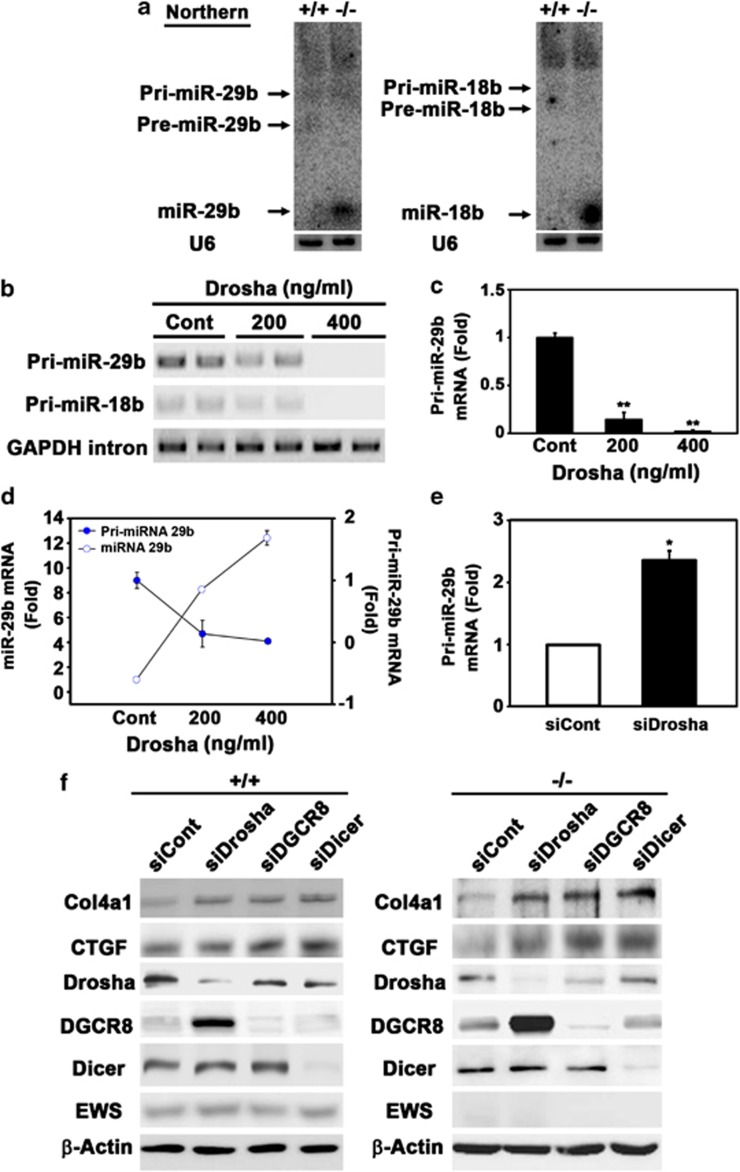
In order to identify the expression of pre- and mature forms of miR-29b and miR-18b in EWS KO MEFs, we further carried out northern blot analysis. The levels of mature miR-29b and miR-18b were markedly elevated in EWS KO MEFs (Figure 5a). To verify whether Drosha has an effecter role for the biogenesis of miR-29b and miR-18b, we determined pri- and mature forms of miR-29b and miR-18b by RT-PCR and qRT-PCR after the induction of Drosha or the knockdown of Drosha. Overexpression of Drosha significantly decreased the levels of pri-miR-29b and pri-miR-18b in a dose-dependent manner (Figures 5b and c). qRT-PCR analysis verified that Drosha reduces the level of pri-miR-29b in a dose-dependent manner. In addition, Drosha increased the level of miR-29b in a dose-dependent manner. This data indicated that Drosha directly regulates the processing of pri-miR-29b to mature forms of miR-29b (Figure 5d). In contrast, knockdown of Drosha significantly restored the level of pri-miR-29b (Figure 5e).
Figure 5.
Drosha regulates the processing of pri-miR-29b and pri-miR-18b and involves in Col4a1 and CTGF expression. (a) Northern blot analysis of miR-29b and miR-18b in EWS MEF cells. Mature forms of the miR-29b and miR-18b were highly expressed in EWS KO MEFs. U6 was used as a loading control. (b) RT-PCR analysis revealed that the levels of pri-miR-29b and pri-miR-18b are decreased by Drosha in a dose-dependent manner (c) qRT-PCR analysis verified that Drosha reduces the level of pri-miR-29b in a dose-dependent manner. The data represent the average±S.E.M. of three separate experiments. (d) Drosha increased the level of miR-29b in a dose-dependent manner. The data represent the average±S.E.M. of three separate experiments. (e) Knockdown of Drosha increased the level of pri-miR-29b. The data represent the average±S.E.M. of three separate experiments. Significantly different at *, P<0.05; **, P<0.005. (f) Knockdown of Drosha, DGCR8, and Dicer increased Col4a1 and CTGF protein levels both in EWS WT and KO MEFs
To prove a hypothetical pathway that the alteration of Drosha activity under EWS deficiency subsequently contributes to miRNA-mediated post-transcriptional regulation of Col4a1 and CTGF, we applied RNAi technique and observed the effect of microprocessor depletion on the level of Col4a1 and CTGF (Figure 5f). When we knock down DGCR8 with verified siRNAs, the protein level of Drosha of was slightly reduced. This might be caused by off-target effects.15 The protein levels of Col4a1 and CTGF were specifically increased by the depletion of microprocessors in EWS MEFs (Figure 5f). We also confirmed increases in Col4a1 and CTGF mRNA levels by knocking down of Drosha, DGCR8, and Dicer, which demonstrates that microprocessor-dependent miRNA pathway involves in the post-transcriptional regulation of Col41 and CTGF in the cytoplasm (Supplementary Figures 5a and b). Furthermore, we addressed whether the regulatory mechanism of Col4a1 and CTGF found in EWS KO cells is reproducible in an established fibroblast cell line such as NIH 3T3 cells. As expected, overexpression of EWS induced the expression of Col4a1 and CTGF (Supplementary Figure 6a). Consistent with our previous results, expression of EWS led to a decrease in Drosha protein level (Supplementary Figure 6a), and an increase of Col4a1 and CTGF mRNA levels in NIH 3T3 cells (Supplementary Figure 6b). The mRNA levels of Drosha were also decreased by EWS in NIH 3T3 cells (Supplementary Figure 6b). Otherwise, knockdown of the microprocessor increased mRNA and protein levels of Col4a1 and CTGF in NIH 3T3 as similar to the results in EWS MEF cells (Supplementary Figures 6c and d). When miR-29b and miR-18b were highly expressed, both mRNA and protein levels of Col4a1 and CTGF were decreased in NIH 3T3 cells (Supplementary Figure 7a). The exogenous induction of miR-29b and miR-18b in NIH 3T3 cells was confirmed by qRT-PCR (Supplementary Figures 7b and c). To further verify the target specificity of miR-29b and miR-18b against Col4a1 and CTGF, we performed luciferase reporter assay using 3′ UTR of Col4a1 and CTGF in NIH 3T3 cells. The post-transcriptional regulation of Col4a1 and CTGF was significantly downregulated by miR-29b and miR-18b, respectively (Supplementary Figures 7d and e). We also carried out locked nucleic acid inhibitor experiments to decrease miR-29b and miR-18b. Both miR-29b inhibitor (anti-29b) and miR-18b inhibitor (anti-18b) markedly increased Col4a1 and CTGF protein and mRNA levels and decreased miR-29b and miR-18b levels in NIH 3T3 cells (Supplementary Figures 7f and g). These findings support that the alteration of microprocessor activity by EWS contributes to miRNA-mediated post-transcriptional regulation of Col4a1 and CTGF.
EWS deficiency upregulates Drosha, miR-29b, and miR-18b and downregulates Col4a1 and CTGF in vivo
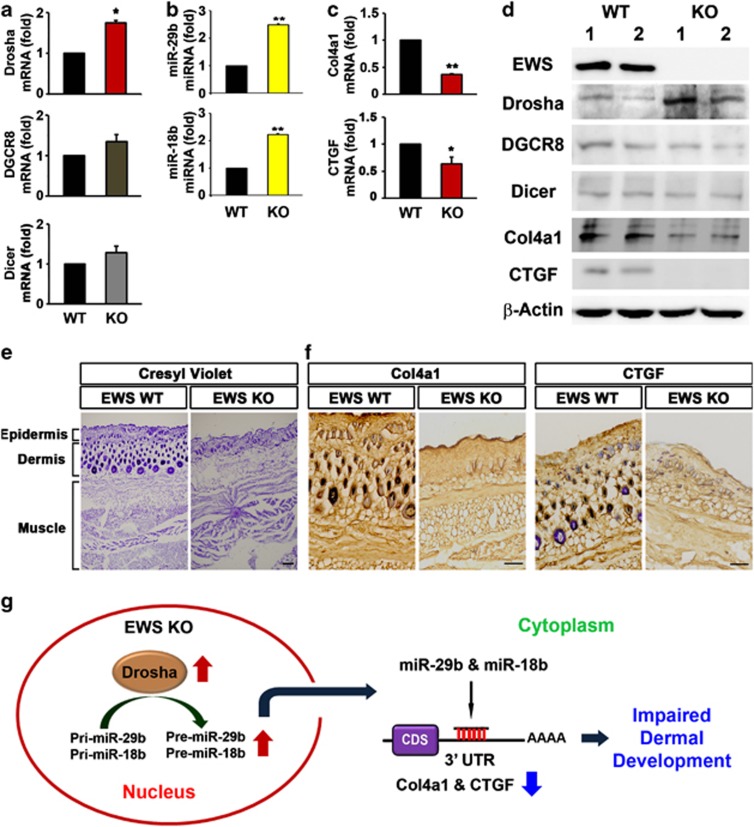
To examine whether in vitro findings on the role of EWS in the regulation of Drosha, miR-29b and miR-18b, and Col4a1 and CTGF are reproducible in vivo, we detected their mRNA and protein levels in the skin of EWS WT and KO mice. The mRNA level of Drosha was significantly increased in the skin of EWS KO mice (Figure 6a). In addition, the protein level of Drosha was significantly increased in the skin tissues of EWS KO mice, whereas proteins levels of DGCR8 and Dicer were not altered (Figure 6d). Importantly, the expression levels of miR-29b and miR-18b were significantly increased in the skin of EWS KO mice (Figure 6b). qRT-PCR analysis confirmed that the mRNA levels of Col4a1 and CTGF were significantly downregulated in the skin of EWS KO mice (Figure 6c). The protein levels of Col4a1 and CTGF were significantly lower in the skin of EWS KO mice than in EWS WT mice (Figure 6d). These results indicate that EWS deficiency deregulates the expression of Col4a1 and CTGF by altering Drosha–miRNA-dependent post-transcription. The Nissl staining represented abnormal development in the skin of EWS KO mice in comparison to EWS WT mice (Figure 6e). The immunohistochemistry showed that both immunoreactivity of Col41a and CTGF were markedly reduced in the skin of EWS KO mice (Figure 6f).
Figure 6.
EWS KO mice show the alteration of Drosha, miRNAs, and Col4a1 and CTGF. (a) Drosha mRNA levels were increased in EWS KO mice. The data represent the average±S.E.M. of three separate experiments. (b) miR-29b and miR-18b levels were markedly increased in EWS KO mice. (c) Col4a1 and CTGF mRNA levels were reduced in EWS KO mice. The data represent the average±S.E.M. of three separate experiments. Significantly different at *, P<0.05; **, P<0.005. (d) The protein levels of Drosha were increased in EWS KO mice whereas the protein levels of Col4a1 and CTGF were downregulated in the skin tissues of EWS KO mice (n=2). (e) Nissl staining showed severely altered skin structure of EWS KO mice. (f) Col4a1 and CTGF immunoreactivity were decreased in EWS KO mice skin tissues. All scale bars: 100 μm. (g) A schematic diagram illustrates that the upregulation of Drosha under EWS deficiency accelerates the processing of pri-miR-29b and pri-miR-18b in the nucleus. Increased miR-29b and miR-18b negatively regulate the mRNAs of Col4a1 and CTGF in the cytoplasm. As a result, EWS deficiency leads to impaired dermal development
Discussion
EWS is a member of the TET family of proteins and is found frequently fused to several transcription factors containing different DNA-binding domains.3, 8 EWS fusion proteins, such as FLI1,1 or EWS interaction partners, such as CBP,3 can control the transcription of mRNA of many genes. It has been previously demonstrated that EWS is part of the Drosha/DGCR complex, but the potential roles for EWS in miRNA biogenesis or regulation has not been examined. In the present study, we found that several miRNAs are altered as a result of EWS inactivation. Among them, we discovered that miR-29b and miR-18b are significantly upregulated in the absence of EWS and subsequently, expression of Col4a1 and CTGF were decreased in EWS KO cells. Interestingly, we found that Drosha, one of the most important miRNAs processing molecules, was significantly elevated in EWS KO cells as seen on RNA sequencing analysis, by showing the inverse correlation with the levels of Col4a1 and CTGF. This data led us to hypothesize that the processing of miRNA might be affected in a Drosha-dependent manner and that altered miRNAs may participate in the post-transcriptional regulation of Col41 and CTGF under EWS deficiency.
Drosha is an miRNA processor and is essential for miRNA maturation. Drosha interacts with DGCR8 to process pri-miRNA to pre-miRNA.21 Both Drosha and DGCR8 proteins are presented abundantly and ubiquitously but the expression level of these proteins depends on the cell types.22 If Drosha levels are elevated in the cell, more mature form of pre-miRNAs will be produced and, consequently, more mature form of miRNAs will be processed in a cell-specific manner. In this paradigm, our study indicates that Drosha is directly regulated by EWS as an upstream target of the miRNA processing pathway. Drosha promoter assay showed that the transcription of Drosha is downregulated by EWS in a dose-dependent manner. Our EWS ChIP data showing that EWS-DNA occupancy is highly increased within the 5′-UTR promoter region of Drosha, supports a role of EWS in the regulation of Drosha transcription. As Drosha is upregulated in EWS KO cells, it is expected that the processing of pri-miRNA to pre-miRNA could subsequently be facilitated.23 Dicer then converts the pre-miRNA into a small RNA duplex, which combines with the RISC.22
As predicted by the increased Drosha expression in EWS KO cells, pri-miRNAs for miR-29b and miR-18b were markedly decreased in EWS KO cells, whereas the two miRNAs were highly increased in EWS KO cells. The increases in the level of miR-29b and miR-18b were correlated with post-transcriptional regulation of Col4a1 and CTGF in the cytoplasm of EWS KO cells. Overexpression of miR-29b and miR-18b significantly downregulated the mRNA of Col4a1 and CTGF, respectively, in a dose-dependent manner. In contrast, anti-29b and anti-18b upregulated the protein and mRNA levels of Col4a1 and CTGF. It is apparent that miR-29b and miR-18b induce the degradation of target mRNA as a major mode of action.17 Importantly the levels of miR-29b and miR-18b were significantly elevated in the skin tissues of EWS KO mice. In addition, both the mRNA and the protein levels of Drosha were elevated in the skin tissues of EWS KO mice, whereas the protein levels of DGCR8 and Dicer were slightly changed. In contrast, the mRNA and protein levels of Col4a1 and CTGF were significantly reduced in the skin tissues of EWS KO mice. These results indicate that Col4a1 and CTGF are post-transcriptionally regulated by Drosha-dependent miRNA pathway under EWS deficiency in vivo. Previous studies have shown that Col4a1 and CTGF have a critical role in the dermal development.9, 14 An inverse correlation between the level of miR-18/19 and the level of CTGF clearly shows that small noncoding miRNAs regulate gene expression at the post-transcriptional level.20 Thus, the changes of specific miRNA and its-targeted ECM protein are implicated in the aging process.20 Otherwise, the effects of miR-29b on the expression of collagen genes that are related to the constitution of ECM and the possible protective role of miR-29b in the renal injury have been examined.17, 24 However, the direct regulatory mechanism of these miRNAs by microprocessor complex and its role in the skin development has not been determined. For the first time, our study present that Drosha-dependent miRNA processing of the miR-18b and miR-29b is directly associated with a failure of skin development in EWS KO mice. Indeed, knockdown of Drosha, DGCR8, and Dicer by siRNAs recovered the mRNA and protein levels of Col4a1 and CTGF. In this context, EWS modulation of Drosha and miRNAs may enhance the expression of Col4a1 and CTGF to improve dermal morphogenesis.18
In summary, we provide evidence that EWS modulates the expression of Col4a1 and CTGF by regulating miRNA-29b and miRNA-18b (Figure 6g). Furthermore, increased Drosha expression mediates the processing of pre-miRNAs, promoting the biogenesis of miR-29b and miR-18b, and subsequently reducing post-transcription levels of Col4a1 and CTGF under EWS deficiency. We found that EWS has a permissive role in the upregulation of Drosha and miRNA processing. The exact nature by which EWS regulates Drosha expression and the microprocessor complex needs to be further examined. Identifying the targets of other miRNAs that are altered as a result of EWS inactivation will reveal additional roles for the multifunctional EWS in different and as yet undescribed cellular processes.
Materials and Methods
EWS MEF cell culture
EWS MEF cells were grown in Dulbecco's modified Eagle's medium (Hyclone, Logan, UT, USA) supplemented with 10% FBS (Hyclone), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen Life Tech, Grand Island, NY, USA), and 10% MEM Non-essential Amino acid solution (SIGMA, St. Louis, MO, USA).
Subcellular fractionation
EWS MEF cells were grown in a 10 cm dish and they were harvested in 450 ul of ice-cold buffer A (10mM HEPES at pH 7.9, 10 mM KCl, 1 mM dithiothreitol, and 0.1 mM EDTA (ethylenediaminetetraacetic acid) at pH 8.0). EWS MEF cells were dispersed by pipetting and incubated for 25 min on ice. Then 5 ul of 10% NP-40 was added, and cells were incubated for 2 min on ice. The nuclei were precipitated by centrifugation at 5000 r.p.m. for 3 min at 4 °C. The supernatant was taken as the cytoplasmic fraction.
Quantitative real-time PCR
Total RNA was extracted from EWS MEF cells and NIH 3T3 by TRIzol reagent (MRC, Cincinnati, OH, USA). RNA was measured in a spectrophotometer at 260-nm absorbance. RNA analysis was conducted as follows. Fifty nanograms of RNA were used as a template for qRT-PCR amplification, using SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan). Primers were standardized in the linear range of cycle before the onset of the plateau. Primer sequences are given in Supplementary Table 1. Mouse GAPDH was used as an internal control. Two-step PCR thermal cycling for DNA amplification and real-time data acquisition were performed with an ABI StepOnePlus Real-Time PCR System using the following cycle conditions: 95 °C for 1 min × 1 cycle, and 95 °C for 15 s, followed by 60 °C for 1 min × 40 cycles. Fluorescence data were analyzed by the ABI StepOnePlus software and expressed as, Ct, the number of cycles needed to generate a fluorescent signal above a predefined threshold. The ABI StepOnePlus software set baseline and threshold values. Supplementary Tables 2 and 3 show primer sequences that are used for qRT-PCR and reverse transcriptase PCR (RT-PCR).
Plasmid construction
miR-29b (forward primer, 5′-CGCGGATCCAACTATTGCACGGACTTCAC-3′ and reverse primer, 5′-CCGCTCGAGACACTGGACACTTACTTCAG-3′) and miR-18b (forward primer, 5′-CGCGGATCCACCATGGTGATTTAATCAGA-3′ and reverse primer, 5′-CCGCTCGAGCCGTTCAAATCATTTCTCAA-3′) were amplified from EWS MEF cDNA by PCR. PCR product was cloned into pCDNA3 (Invitrogen Life Tech) with BamHI and XhoI (NEW ENGLAND BioLabs, Ipswich, MA, USA) restriction enzyme sites and sequenced.
RNA interference experiments and western blot analysis
30 nM of siRNA duplex were transfected in NIH 3T3 and EWS KO MEF cells with RNAiMax transfection reagent (Invitrogen Life Tech) according to the manufacturer's instructions. siRNAs and inhibitor of miR-29b and miR-18b were synthesized from COSMO GENETECH (Seoul, Korea). Target sequences of mouse siDrosha, siDGCR8, and siDicer are 5′-AGAUCACCGUCUCUAGAAA-3′, 5′-AACAAUUUGGAGCUAGAUGAA-3′, and 5′-ACACAGCAGUUGUCCUAAA-3′. Inhibitor miR-29b sequences are 5′-UAAACCACCAUAUGAAACCAGC-3′ and miR-18b sequences are 5′-CUAACAGCACUAGAUGCACCUUA-3′. EWS MEF and NIH 3T3 cells were collected at 72 h after siRNA and Flag-EWS transfection. The cells were dispersed by pipetting in lysis buffer (10 mM Tris at pH 7.4, 1 mM EDTA at pH 8.0 500 mM NaCl, and 0.5% Triton X-100) and incubated for 30 min on ice. Primary antibodies used in this study are rabbit anti-Col4a1 antibody (Upstate Biotechnologies and abCam, Cambridge, UK), rabbit anti-CTGF antibody (Upstate Biotechnologies and abCam), rabbit anti-Drosha antibody (Upstate Biotechnologies and abCam), rabbit anti-DGCR8 antibody against recombinant DGCR8 protein prepared in Escherichia coli,16 goat anti-DGCR8 antibody (Upstate Biotechnologies and abCam), rabbit anti-Dicer (Santa Cruz Biotechnology, Dallas, TX, USA), rabbit anti-Ago2 (Santa Cruz Biotechnology), mouse anti-EWS (Santa Cruz Biotechnology), mouse anti-Flag (SIGMA), mouse anti β-actin, and mouse anti-tubulin (Millipore, Billerica, MA, USA).
Confocal microscopy
Immunofluorescence staining and confocal microscopy was used to determine mouse anti-EWS (Santa Cruz Biotechnology). Images were analyzed using a spinning disk confocal microscope (Olympus DSU, Shinjuku, Japan). Deconvolution and three-dimensional construction of the confocal image was performed by AQI-X-COMBO-CWF program (Media cybernetics Inc., Rockville, MD, USA). Control experiments were performed in the absence of primary antibody or in the presence of blocking peptide.
RNA-seq and gene analysis
For the mRNA-Seq sample preparation, the Illumina standard kit was used according to the manufacturer's protocol. Briefly, 3 μg of each total RNA sample was used for polyA mRNA selection using streptavidin-coated magnetic beads, followed by thermal mRNA fragmentation. The fragmented mRNA was subjected to cDNA synthesis using reverse transcriptase (SuperScript II, Grand Island, NY, USA) and random primers. The cDNA was further converted into double-stranded cDNA and, after an end repair process (Klenow fragment, T4 polynucleotide kinase, and T4polymerase), was finally ligated to Illumina paired end adapters. Size selection was performed using a 2% agarose gel, generating cDNA libraries ranging in size from 200 to 250 bp. Finally, the libraries were enriched using 10 cycles of PCR and purified by the QIAquick PCR purification kit (Qiagen, Venlo, Netherlands). The enriched libraries were diluted with Elution Buffer to a final concentration of 10 nM. Each library was run at a concentration of 8 pM on one Genome Analyzer (GAIIx) lane using 53 bp sequencing. Reads were then processed and aligned to the mouse genome UCSC build mm9 using GSNAP.25 GSNAP uses the normalized RNA-Seq 8 fragment counts to measure the relative abundances of transcripts. The unit of measurement is RPKM (Reads Per Kilobase of exon per Million fragments mapped).26
Functional enrichment analysis and network analysis
For the functional enrichment analysis we selected genes whose RPKM levels in EWS KO (−/−) MEFs were three times smaller than in EWS WT (+/+) MEFs. Five hundred and twenty genes were selected and grouped in terms of GO biological processes (GOBPs) and KEGG pathway using DAVID.27 Four groups by GOBPs were selected: (1) development, (2) cell adhesion, (3) cell proliferation and cytoskeleton organization, and (4) morphogenesis. Then direct neighbors of the 91 genes were collected by querying each of the 91 genes to the NCBI Entrez EUtilities Web Service Client provided by Cytoscape (v. 2.8.0)28 and STRING.29 Finally, 46 genes were selected by removal of some genes without neighboring and were displayed in a network with 161 genes including neighbors by using Cytoscape(v. 2.8.0). Genes in the network were grouped according to their interaction and function.
Transcriptional profile of miRNA processing genes
To visualize heat map transcriptional profiles of miRNA processing genes, we collected miRNA processing-related genes from the NCBI Entrez E-utility Web Service using Cytoscape (v 2.8.0). Seventeen miRNA-processing-related genes with RPKM ≥1.0 were finally selected. The mRNA expression levels of miRNA-processing-related genes in EWS WT (+/+) and EWS KO (−/−) were plotted in a heat map using gplots package in R Development Core Team.
Histopathological evaluation
Serially cut skin tissue sections were immunostained for Col4a1 (Upstate Biotechnologies and abCam) and CTGF (Upstate Biotechnologies and abCam) using a previously reported conjugated secondary antibody method in skin tissue samples. Preabsorption with excess target proteins, omission of the primary antibodies, and omission of secondary antibodies were performed to determine the amount of background generated from the detection assay.
Statistical analysis
Data are presented as the mean±standard error of the mean (S.E.M.). Data analysis was performed by Student's t-test or one-way ANOVAs followed by Mann–Whitney and Kruskal–Wallis tests. Differences were considered statistically significant when P<0.05.
Acknowledgments
We thank Dr. V Narry Kim for providing Drosha, DGCR8, and Dicer plasmids and Dr. Fukamizu for providing EWS plasmid. We also thank Elizabeth (Eun Kyung) Park (Boston University School of Medicine) for the preparation of manuscript. This study was supported by Brain Science Flagship Grant (2E24320) (HR) from Korea Institute of Science and Technology and NIH Grant (NS 067283-01A1) (HR).
Glossary
- EWS
Ewing's Sarcoma
- Col4a1
collagen IV alpha 1
- CTGF
connective tissue growth factor
- KO
knock-out
- MEFs
mouse embryonic fibroblasts
- CBP
CREB-binding protein
- ECM
extracellular matrix
- miRNAs
microRNAs
- pri-miRNA
primary miRNAs
- pre-miRNAs
precursor miRNAs
- RISC
RNA-induced silencing complex
- microprocessor
miRNA processing machinery
- qRT-PCR
quantitative real-time PCR
- RNA-seq
whole transcriptome sequencing
- ChIP
Chromatin Immunoprecipitation
- LNA
locked-nucleic-acid inhibitors
- TFIID
transcription factor II D
- anti-29b
miR-29b inhibitor
- anti-18b
miR-18b inhibitor
- TET family of proteins
TLS/FUS, EWS and TAF15
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Erkizan HV, Uversky VN, Toretsky JA. Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing's sarcoma. Clin Cancer Res. 2010;16:4077–4083. doi: 10.1158/1078-0432.CCR-09-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossow KL, Janknecht R. The Ewing's sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61:2690–2695. [PubMed] [Google Scholar]
- Araya N, Hirota K, Shimamoto Y, Miyagishi M, Yoshida E, Ishida J, et al. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J Biol Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- Kovar H. Dr. Jekyll and Mr. Hyde: the two faces of the FUS/EWS/TAF15 protein family. Sarcoma. 2011;2011:1–13. doi: 10.1155/2011/837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Rhee BK, Bae GY, Han YM, Kim J. Stimulation of Oct-4 activity by Ewing's sarcoma protein. Stem Cells. 2005;23:738–751. doi: 10.1634/stemcells.2004-0375. [DOI] [PubMed] [Google Scholar]
- Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. doi: 10.1016/j.gene.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Shen H, Yu H, Li H, Cheng T, Lee SB, et al. Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence. Blood. 2011;117:1156–1166. doi: 10.1182/blood-2010-04-279349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poschl E, Schlotzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–1628. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- Abreu-Velez AM, Howard MS. Collagen IV in normal skin and in pathological processes. N Am J Med Sci. 2012;4:1–8. doi: 10.4103/1947-2714.92892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaud Guittot S, Verot A, Odet F, Chauvin MA, le Magueresse-Battistoni B. A comprehensive survey of the laminins and collagens type IV expressed in mouse Leydig cells and their regulation by LH/hCG. Reproduction. 2008;135:479–488. doi: 10.1530/REP-07-0561. [DOI] [PubMed] [Google Scholar]
- Hertel M, Tretter Y, Alzheimer C, Werner S. Connective tissue growth factor: a novel player in tissue reorganization after brain injury. Eur J Neurosci. 2000;12:376–380. doi: 10.1046/j.1460-9568.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- Blom IE, Goldschmeding R, Leask A. Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy. Matrix Biol. 2002;21:473–482. doi: 10.1016/s0945-053x(02)00055-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Abraham DJ, Shi-Wen X, Pearson JD, Black CM, Lyons KM, et al. CCN2 (connective tissue growth factor) promotes fibroblast adhesion to fibronectin. Mol Biol Cell. 2004;15:5635–5646. doi: 10.1091/mbc.E04-06-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2008;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta M, Choi YS, Okegbe T, Wong G, Tam OH, Chong MM, et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development. 2012;139:1405–1416. doi: 10.1242/dev.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.