Abstract
In animals, females deposit gene products into developing oocytes, which drive early cellular events in embryos immediately after fertilization. As maternal gene products are present before fertilization, the functional manipulation of maternal genes is often challenging to implement, requiring gene expression or targeting during oogenesis. Maternal expression can be achieved through transgenesis, but transgenic approaches are time consuming and subject to undesired epigenetic effects. Here, we have implemented in vitro culturing of experimentally manipulated immature oocytes to study maternal gene contribution to early embryonic development in the zebrafish. We demonstrate phenotypic rescue of a maternal-effect mutation by expressing wild-type product in cultured oocytes. We also generate loss-of-function phenotypes in embryos through either the expression of a dominant-negative transcript or injection of translation-blocking morpholino oligonucleotides. Finally, we demonstrate subcellular localization during the early cell divisions immediately after fertilization of an exogenously provided maternal product fused to a fluorescent protein. These manipulations extend the potential to carry out genetic and imaging studies of zebrafish maternal genes during the egg-to-embryo transition.
Keywords: maternal gene, maternal-effect, reverse genetics, zebrafish, oocyte culture, oogenesis, cellular island, futile cycle, mission impossible, Sas-6, centrioles
Introduction
In animal embryos, development immediately after fertilization is driven solely by maternal mRNAs and proteins present in the egg and activated during fertilization at the egg-to-embryo transition (Lindeman and Pelegri, 2010). Perduring maternal factors also function together with zygotic genes to pattern the embryo (Lyman-Gingerich et al., 2005; Kotani and Kawakami, 2008; White and Heasman, 2008; Holloway et al., 2009; Putiri and Pelegri, 2011). Reverse genetics using conditional knock-out strategies targeting the germ line and expression of RNAi constructs during oogenesis have been successful in the study of maternal genes in mouse and X. laevis (Stein et al., 2003; Lykke-Andersen, 2006; Roy and Matzuk, 2006; Mir and Heasman, 2008; Matzuk and Burns, 2012). However, successful targeting of zebrafish maternal genes by reverse genetics remains to be demonstrated.
To observe a maternal effect (phenotype due to defects in the maternal genotype) by reverse genetic targeting of maternal genes, one must interfere with gene products deposited into oocytes as they develop into eggs, before fertilization. During oogenesis, with the exception of yolk precursor proteins such as vitellogenin, which is produced by the liver and acquired by the oocyte through endocytosis (Wallace, 1985), most RNAs and proteins are produced cell-autonomously by the oocyte. Such preloaded maternal products in the mature egg pose a problem for their functional manipulation, as standard reverse genetics methods to introduce exogenous products or reagents by injection into one-cell stage embryos or mature oocytes are ineffective for early developmental stages. For example, injection of mRNAs at these stages may not allow sufficient time to express products to have an effect on early embryonic processes, while morpholino oligonucleotides injected at these stages are unable to reduce already present maternal protein.
Currently, maternal products can be exogenously expressed by transgenic approaches (Knaut et al., 2002; Kotani and Kawakami, 2008; Yabe et al., 2009). However, the generation and evaluation of transgenic animals is labor intensive and requires fish husbandry for the months corresponding to a generation time. In addition, transgenes may be subject to position effects and silencing (Choo et al., 2006; Goll et al., 2009; Akitake et al., 2011), complicating both maintenance of stocks and analysis of the consequences of transgenic expression. Methods for the systematic down-regulation of maternal genes could also be achievable in principle through transgenic expression, however, RNA interference (RNAi) during oogenesis, which has been shown to be effective in the mouse (Roy and Matzuk, 2006), has not yet been reported in the zebrafish. Given the challenges of transgenic-based approaches for the functional manipulation of maternal factors in the zebrafish, an effective in vitro culture-based method that would allow both the up- and down-regulation of maternal gene product function using currently available tools would be a welcome alternative.
During oogenesis in zebrafish, stage I oocytes are arrested at meiosis I at the diplotene phase of prophase (Selman et al., 1993). Meiosis I resumes and completes only during stage IV, at the end of which the oocyte matures into stage V oocytes and re-arrests at metaphase of meiosis II (Selman et al., 1993). After defolliculation, exposure of eggs to water during natural matings or in vitro fertilization results in egg activation, which includes the expansion of the chorion membrane (Becker and Hart, 1996, 1999) and completion of meiosis II (Streisinger al., 1981; Selman et al., 1993; Dekens et al., 2003). Protocols for in vitro maturation of zebrafish oocytes have been previously reported (Selman et al., 1993, 1994; Seki et al., 2008). In this approach, ovaries that contain oocytes at various stages of oogenesis are removed from gravid females and cultured in maturation medium containing the hormone 17α-20β-Dihydroxy-4 Pregnen-3-one (DHP). DHP treatment allows oocytes to mature from stage III (undergoing vitellogenesis, arrested at diplotene of meiosis I with the oocyte nucleus in the center of the oocyte) through stage IV (meiosis I completed, oocyte nucleus closer to the surface of the egg) until the final stage of oogenesis at stage V (translucent, germinal vesicle breaks down, oocytes arrested in metaphase of meiosis II).
Although in vitro culture conditions for zebrafish oocyte maturation have been known for almost two decades (Selman et al., 1993, 1994), oocytes matured through such in vitro protocols could not be successfully in vitro fertilized. Recent modifications to the original culture conditions, such as a maturation medium pH closer to that of the ovarian fluid, has demonstrated that such in vitro matured oocytes can also be successfully fertilized in vitro to yield viable embryos (Seki et al., 2008). However, an in vitro approach for functional analysis of zebrafish maternal genes has not been documented.
Here, we demonstrate functional manipulations of zebrafish maternal genes in immature stage IV oocytes through a modified version of the in vitro culture and fertilization method by Seki et al. We show that injection of mRNA encoding a wild-type maternal protein rescues the corresponding maternal-effect phenotype. Conversely, a loss-of-function maternal-effect phenotype is mimicked by the injection of mRNA encoding a dominant-negative protein or translation-blocking morpholinos. We find that a morpholino targeting splice junctions is ineffective, possibly due to the presence of mature mRNAs in the developing oocyte. Finally, we show that mRNAs encoding fluorescent fusions of maternal proteins allow the visualization of subcellularly localized maternal protein immediately after fertilization. These results demonstrate an in vitro approach for functional manipulation and visualization of zebrafish maternal genes, extending the range of reverse genetics to encompass the egg-to-embryo transition.
Materials and Methods
Animal Husbandry
AB and mutant fish were maintained under standard conditions at 28.8°C. Before oocyte culture, AB and homozygous mutant females were paired for natural matings to purge the females of all mature eggs. Pairs that laid fertilized eggs were placed in smaller tanks in groups of up to 10 fish and fed twice a day. Day of egg-laying was day 0; ovaries were harvested on days 1–11 postpurging for oogenesis staging series and days 8–10 postpurging to isolate stage IV oocytes for all other experiments.
In Vitro Oocyte Culture
The general method is adapted from Seki et al. (2008) from which specific reagents described were used as follows: Leibovitz's L-15 medium with L-glutamine, pH 7.0 (Gibco), gentamycin 10 mg/ml (Sigma-Aldrich), 10% bovine serum albumin in distilled water (BSA, Sigma-Aldrich) and 17α-20β-dihydroxy-4 pregnen-3-one (DHP, Sigma-Aldrich and Santa Cruz Biotechnology). Sorting medium was 90% Leibovitz's pH 9.0 (adjusted with 10N NaOH), 0.5% BSA and 100 μg/ml gentamycin. Maturation medium was 90% Leibovitz's pH 9.0, 0.5% BSA, and 1 μg/ml DHP. For each experiment, ovaries from 1–2 females were harvested into sorting medium in a 35 × 10 mm plastic culture dish. Oocytes were dissociated by gentle pipetting with a plastic Pasteur pipette and a pair of forceps. Sorted/injected oocytes were transferred into maturation medium and cultured at 26°C for 4 hr. The duration between harvesting oocytes into sorting medium and transfer into maturation medium with or without injections can be up to 2 hours and was typically approximately 1.5 hr for all experiments described. Unmanipulated oocytes left in the sorting medium were found to spontaneously undergo chorion expansion 3–4 hr postharvesting.
In Vitro Fertilization
For in vitro matured translucent stage IV and mature stage V oocytes, the follicle membrane was removed manually using a pair of forceps by holding a part of the follicle membrane and gently rolling the oocyte out of it. Defolliculated oocytes were transferred into a clean dish in groups of 2–5 in 10–30 μl of maturation medium. 5–10 μl of sperm solution was added directly on top of the oocytes, followed by a drop of embryo medium ∼10 s later. The plate was flooded gently with embryo medium after 1 min. Sperm solution was made by shearing the testes from two AB males per experiment in 300 μl of Hank's Final solution kept on ice and used within 1 hr of preparation (Pelegri and Schulte-Merker, 1999). Fertilized embryos were observed to confirm normal chorion expansion, (indicative of successful defolliculation), which occurred maximally at 5–10 min postfertilization. In vitro fertilizations using sperm solution prepared as described resulted in ∼25% polyspermy in embryos in a given experiment. These embryos could be identified live at 40 minutes postfertilization (mpf) as described in the supplementary materials and were excluded from further analysis.
Oocyte Microinjections
The working stocks of all injected reagents were diluted in 0.2 M KCl as dilution in distilled water resulted in toxicity leading to inviable oocytes. Injections were performed by manually holding the oocytes directly on the culture dish with a pair of forceps. Oocytes were injected while in sorting medium and transferred to maturation medium immediately afterward.
Stage I–IV oocytes were injected with 100 pg of GFP mRNA. Stage IV oocytes were injected with 100 pg of wild-type aurB mRNA or 200 pg of aurB-KD mRNA (Yabe et al., 2009), 200 pg of sass6-Long-mCherry mRNA (Yabe et al., 2007), ∼2 ng of Lrmp-MO (GeneTools, tggaccgggacaagagagaaagcct), ∼2.5 ng of Dhx16-MO (Putiri and Pelegri, 2011) or 2 ng of sass6-spliceMO (GeneTools, gcctacacatgattacctcagagc). Also see the Reverse Transcriptase-Polymerase Chain Reaction section.
Immunolabeling and Imaging
Embryos were allowed to develop until the two- or eight-cell stage after which they were immunolabeled as described previously (Pelegri et al., 1999; Yabe et al., 2007). Confocal images of immunostainings were obtained on a Zeiss LSM510 and processed using ImageJ. To observe effects of the Dhx16-MO, live embryos were imaged on a Zeiss Axioscope using OpenLab at ∼9 hours postfertilization (hpf).
Reverse Transcriptase-Polymerase Chain Reaction
Total RNA was isolated from whole ovaries at day 1 postpurging and from stage IV oocytes using Trizol (Invitrogen) and DNAase treated. cDNA was synthesized using oligodT and random hexamer primers (Invitrogen) and standard polymerase chain reactions (PCRs) were performed to amplify ∼300 bps of α-tubulin, β-actin, or sass6 products. Exon primers were used on oligodT primed and intron primers on random hexamer primed cDNAs. sass6-SpliceMO efficacy was confirmed by reverse transcriptase-PCR (RT-PCR) on oligodT primed 24 hpf cDNA from embryos injected with 2ng of MO at the one-cell stage, using exon primers flanking the targeted intron. Gene-specific primers used to amplify cDNA sequences were as follows: sass6-Exon-F: cgtcatcttcatcagcgttc; sass6-Exon-R: tctgctggacagagaggttg; sass6-Intron-F: tt tccagaacaactgaccacag; sass6-Intron-R: ggcaatttcaaatctgtcacc; β-actin-Exon-F: gcccatctatgagggttacg; β-actin-Exon-R: ag gaaggaaggctggaagag; β-actin-Intron-F: ccacattaaactctcatttgtgc; β-actin-Intron-R: accgagaaatgaggaactgc. Primers to corroborate the effectiveness of the sass6 splice MO were SpliceMO-sass6-F: gaggaaaag cagcagttgcag; SpliceMO-sass6-R: gctcaga gactttggtctggagctg.
Results and Discussion
Oogenesis Occurs Synchronously in Zebrafish During Which Immature Oocytes Are Translation Competent
For the in vitro culture procedure described by Seki et al., random selection of gravid females required 4–5 females for a clutch of 40–60 in vitro matured oocytes, as we could not predict whether a female would carry immature stage IV oocytes or mature stage V oocytes. We sought to overcome this unpredictability and reduce the number of euthanized females, especially homozygous maternal-effect mutant females, whose numbers are generally limiting.
To maximize the probability of isolating immature stage IV oocytes on a given day, females were paired in natural matings to “purge” mature eggs and used to establish an oogenesis time-line. Ovaries were harvested 1–11 days postpurging (dpp) and oocytes were staged as described previously (Fig. 1; Selman et al., 1993). Oogenesis resumed predictably: stage I and II oocytes (IA: 7–20 μm; IB: 20–140 μm, both translucent; II: 140–340 μm, translucent with beginning accumulation of cortical alveoli) were first observed at 1–2 dpp (Fig. 1A), early stage III oocytes (lower range of 340– 690 μm, opaque, oocytes becoming opaque due to initiation of vitellogenesis, germinal vesicle in central position) at 3–4 dpp (Fig. 1E), late stage III oocytes (nearing 690 μm, opaque, germinal vesicle in central position) at 4–7 dpp (Fig. 1I), stage IV oocytes (690–730 μm, opaque, germinal vesicle asymmetrically located, near the cortex) at 8–10 dpp (Fig. 1M) and mature stage V oocytes (730–750 μm, translucent, germinal vesicle fully disassembled) at 11 dpp and beyond. Thus, under standard husbandry conditions, purged female zebrafish undergo oogenesis to reliably yield immature stage IV oocytes between 8 and 10 dpp. Oogenesis occurred fairly synchronously, as on a given day postpurging, in addition to oogonia, most of the immature oocytes were predominantly of one stage (this study; Selman et al., 1993).
Fig. 1.
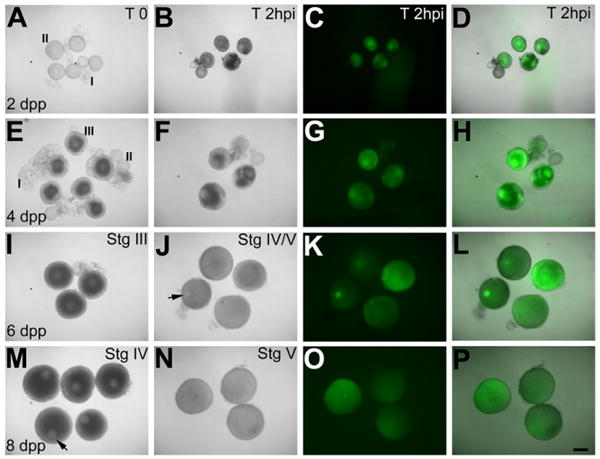
Oogenesis occurs predictably after purging and oocytes of all stages are translation competent. Live images of oocytes. A–P: Oocytes immediately after isolation from purged females on the indicated dpp at time 0 (A,E,I,M) and 2 hr after GFP mRNA injection (B,F,J,N), expressing green fluorescent protein (GFP) protein (C,D,G,H,K,L,O,P). B,F,I,J: Injected Stage I and II (B) and early III (F) oocytes degenerate, while late stage III oocytes (I) mature into stage IV transluscent oocytes (J) with germinal vesicle (arrow). Injected Stage IV oocytes (M) containing germinal vesicle (arrow) mature into stage V oocytes (N, note germinal vesicle breakdown). Scale bar = 300 μm in A–P.
We next injected GFP mRNA to assay the translation competency of oocytes during in vitro culturing (Fig. 1). Consistent with previous reports for stage I and II oocytes (Csenki et al., 2010; Bontems et al., 2009), immature zebrafish oocytes of all stages (I–IV) translated the exogenously injected GFP mRNA and accumulated detectable levels of green fluorescent protein (GFP) protein within 1.5–2 hr postinjection (Fig. 1B–D,F–H,J–L,N–P).
GFP-expressing oocytes were further assayed for their competence to mature into stage V oocytes. Stage I and II oocytes failed to mature and degenerated (Fig. 1A–H), while stage III oocytes matured into stage IV oocytes with germinal vesicles (Fig. 1I,J), which disassembled indicating their further maturation into stage V oocytes. However, unlike freshly isolated stage IV oocytes, which characteristically contain a germinal vesicle and are opaque, becoming translucent as they mature into stage V oocytes, stage III oocytes that matured in vitro into stage IV oocytes with germinal vesicles were translucent before germinal vesicle disassembly (Fig. 1J, compare with M). As germinal vesicle disassembly did occur in these stage IV oocytes and a previous study reported successful fertilization of such eggs (Li et al., 1993), we attempted to in vitro fertilize such eggs. However, we were unsuccessful in fertilizing eggs obtained from in vitro culture of stage III oocytes. Thus, although stage III oocytes mature in response to exogenous hormone, they may be limited in their potential to develop in vitro into fully mature eggs. For such stage I–III oocytes that are amenable to experimental manipulations but fail to mature in vitro, transplantation into foster female fish for in vivo maturation into eggs may be a viable strategy (Csenki et al., 2010).
Unlike stage I–III oocytes, stage IV oocytes matured into translucent stage V oocytes within 3.5–4 hr of in vitro culturing, a duration that is ideal for successful in vitro fertilization (Fig. 1N; Selman et al., 1993, 1994; Seki et al., 2008). We, therefore, restricted our in vitro oocyte culture duration to 4 hr postinjection, which based on our GFP expression results allows a maximum period of 2–2.5 hr for exogenous maternal protein expression before fertilization.
Wild-type mRNAs Injected Into Stage IV Oocytes Rescue Maternal-effect Phenotypes in Embryos
Translation of exogenously injected transcripts occurs between 2 and 3 hr postinjection, a temporal delay that precludes rescue of early maternal-effect phenotypes when the wild-type transcript is injected at the one-cell stage. Hence, phenotypic rescue of maternal-effect mutations by mRNA injections into one-cell embryos have been observed only at or after midcleavage stages (Kelly et al., 2000; Yabe et al., 2007; Holloway et al., 2009; Fukazawa et al., 2010; Putiri and Pelegri, 2011; Abrams et al., 2012). To circumvent this temporal translation lag, maternal gene expression during oogenesis has been achieved using transgenic fish (Knaut et al., 2002; Yabe et al., 2009). However, despite increases in transgenic generation efficiency (Kawakami, 2005; Kwan et al., 2007), this approach requires at least one generation-time, possibly more if transgene expression is required in a specific mutant background. Moreover, transgenes can be subject to epigenetic modifications, complicating analysis. Direct assessment of maternal gene function by injection into developing oocytes would avoid such drawbacks and simultaneously expedite the time-frame of the experiment.
Embryos from females homozygous for the maternal-effect mutation cellular island (cei) fail to undergo early cell divisions and the mutation was positionally cloned as a missense mutation in the zebrafish Aurora Kinase B (Stka/AurB; Dosch et al., 2004; Yabe et al., 2009). Phenotypic rescue of the cei mutation was previously achieved through transgenesis as injection of wild-type aurB mRNA at the one-cell stage failed to rescue the early maternal-effect phenotype in cei/aurB mutant embryos (Yabe et al., 2009). We attempted to directly rescue the cei/aurB maternal-effect phenotype by injecting wild-type aurB mRNA into stage IV oocytes from homozygous cei/aurB females. In cei/aurB mutant embryos from such injections, cleavage furrows ingressed and were robustly maintained, reflecting rescue of the mutant phenotype, while sibling cei/aurB mutant embryos from an uninjected subset of in vitro mature eggs showed characteristic rudimentary furrows (Supp. Table S1, which is available online). We confirmed rescue of the cei/aurB maternal-effect phenotype by assaying for β-catenin, which accumulates at mature cleavage furrows in wild-type (Fig. 2A–D), but not cei/aurB mutant embryos (Fig. 2E–H; Yabe et al., 2009). Confirming our observations in live embryos, β-catenin protein was found at the furrows in cei/aurB mutant embryos that were injected with wild-type aurB mRNA as stage IV oocytes (Fig. 2I–L; Supp. Table S1). These results recapitulate the rescue of the cei/aurB maternal-effect phenotype obtained through transgenic wild-type AurB copies (Yabe et al., 2009).
Fig. 2.
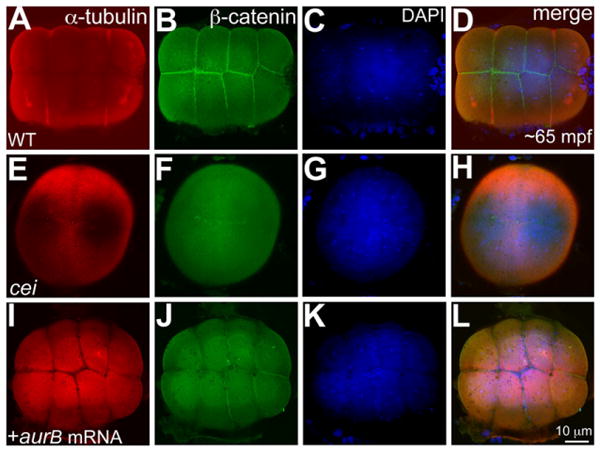
Wild-type aurB mRNA injected into stage IV cei/aurB oocytes rescues the maternal-effect phenotype in cei/aurB embryos. A–L: Animal views of 65 mpf blastodiscs immunolabeled to detect α-tubulin (A,E,I) and β-catenin (B,F,J), with DAPI (4′,6-diamidine-2-phenylidole-dihydrochloride) stainings (C,G,K) and panel merges (D,H,L). A,B,E,F: Wild-type embryos show robust furrows between cells and β-catenin accumulation at the furrows (A,B), while cei/aurB embryo from uninjected cei/aurB stage IV oocyte shows partial, rudimentary furrows (E), which do not accumulate β-catenin (F). I,J: Rescued cei/aurB embryo from cei/aurB oocyte injected with wild-type aurB mRNA labeled with α-tubulin (I) show robust furrows which accumulate β-catenin (J), indicating furrow maturation. C,G,K: DAPI images show that nuclear division proceeds normally in all cases.
In addition to cei/aurB, we have shown that early maternal-effect phenotypes in fertilized embryos of futile cycle (Lindeman and Pelegri., 2012) and motley (Nair et al., in revision) can also be rescued by injecting mRNAs of the corresponding wild-type genes into stage IV mutant oocytes. In motley, phenotypes associated with oogenesis in mature eggs are additionally rescued (Nair et al., in revision). Taken together, our results demonstrate that early maternal-effect phenotypes can be effectively rescued by injecting wild-type mRNAs into stage IV oocytes cultured to maturity in vitro.
Mutant Transcripts Predicted to Interfere With Endogenous Protein Function Cause a Maternal Dominant-Negative Effect When Injected Into Stage IV Oocytes
We next tested whether we could interfere with endogenous maternal function by injecting transcripts encoding mutant versions of a protein. A mutation in the kinase domain of AurB functions as a dominant-negative in mammalian cell lines (Terada et al., 1998). Injections of an mRNA with the same mutation engineered in the zebrafish AurB kinase domain (aurB-KD) at the one-cell stage failed to mimic the maternal-effect cei/aurB mutation (i.e., it did not affect early cell divisions), resulting instead in brain necrosis later (Yabe et al., 2009). We injected wild-type stage IV oocytes with the aurB-KD mRNA to test whether we could obtain a maternal dominant-negative effect resulting from reduced wild-type AurB function during early cleavage stages. In embryos derived from stage IV oocytes injected with the aurB-KD mRNA, we observed rudimentary cleavage furrows that failed to fully ingress and mature (Supp. Table S1). Compared with wild-type embryos (Fig. 3A–D), β-catenin also failed to accumulate at the cleavage furrows in the aurB-KD mRNA injected embryos, confirming that cytokinesis had indeed failed (Fig. 3I–L; Supp. Table S1). This phenotype resembles the maternal-effect cell division phenotype seen in cei/aurB mutant embryos from natural matings of cei/aurB homozygous females (Yabe et al., 2009) and in vitro matured cei/aurB stage IV oocytes (Figs. 2E–H, 3E–H). Thus, these results suggest that the aurB-KD mRNA injected into stage IV oocytes successfully interfered with the function of the endogenous maternal wild-type AurB protein, resulting in a maternal dominant-negative effect in embryos observed immediately after fertilization.
Fig. 3.
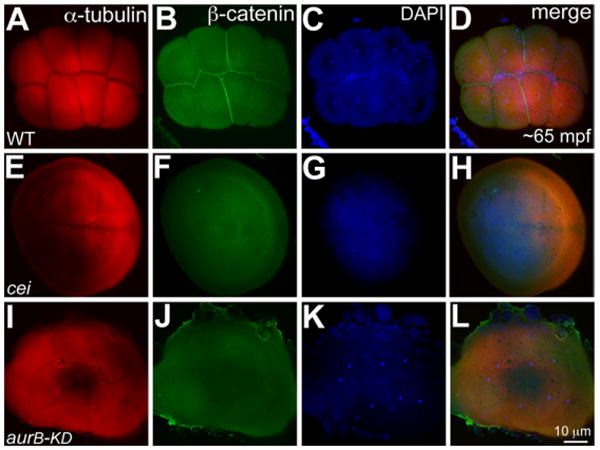
Stage IV oocyte injection of aurB-KD mRNA phenocopies the cei/aurB maternal-effect phenotype. A–L: Animal views of 65 mpf blastodiscs immunolabeled to detect α-tubulin (A,E,I) and β-catenin (B,F,J), with DAPI (4′,6-diamidine-2-phenylidole-dihydrochloride) stainings (C,G,K) and panel merges (D,H,L). A,B: Robust furrows in wild-type embryos labeled with α-tubulin (A), which accumulate β-catenin (B). E,F: α-Tubulin labeling of control cei/aurB embryos from mutant stage IV oocytes show partial rudimentary furrows (E) with no β-catenin labeling (F). I,J: α-tubulin labeled partial, immature furrows in embryos from wild-type stage IV oocytes injected with aurB-KD mRNA (I), which also fail to accumulate β-catenin (J), recapitulating cei/aurB maternal effect phenotype. C,G,K: As in Figure 2, DAPI images show that nuclear division proceeds normally in all cases.
Translation Blocking Morpholino Oligonucleotides Phenocopy Known Maternal-Effect Mutations
For maternal genes, the standard reverse-genetics approach of injecting morpholino oligonucleotides (MOs) into one-cell embryos, which successfully target zygotic transcripts (Eisen and Smith, 2008), is ineffective. This is because the wild-type maternal protein (for translation MOs), and mature mRNAs (for splice MOs), present in the egg before fertilization override any effect of the injected MO. Thus, injecting MOs into one-cell embryos for genes expressed both maternally and zygotically and for which there are maternal-effect mutations, cause zygotic phenotypes but fail to phenocopy the known maternal-effect phenotype at earlier stages (Yabe et al, 2009; Putiri and Pelegri, 2011).
To test the potential of MO injections into in vitro cultured oocytes to study maternal gene function, we targeted two maternal genes, for both of which maternal-effect mutants have been isolated by traditional forward genetics. The maternal-effect mutant futile cycle(fue) encodes a vertebrate-specific Lymphoid restricted membrane protein (Lrmp), which is required for nuclear-centrosome attachments (Lindeman and Pelegri, 2012). Translation blocking morpholinos targeting Lrmp (Lrmp-MO) when injected into one-cell stage embryos failed to phenocopy the maternal-effect phenotypes seen in fue mutant embryos from homozygous fue females: morphant embryos obtained in this manner gastrulated normally and did not manifest any nuclear-centrosome detachments typical of the maternal-effect fue mutants (our unpublished observations). We injected the same translation blocking Lrmp-MO into wild-type stage IV oocytes, which were in vitro matured into stage V oocytes, defolliculated and fertilized. In embryos derived from uninjected wild-type stage IV oocytes, at the eight-cell stage, nuclei always associated with γ-tubulin labeled centrioles and centrosomes (Fig. 4A–C). In fue mutants obtained by in vitro maturation of stage IV oocytes from homozygous fue females, karyokinesis does not occur and the nuclear patches do not associate with centrosomes (Fig. 4D–F; Lindeman and Pelegri, 2012). In Lrmp morphants obtained by injecting the translation blocking Lrmp-MO into wild-type stage IV oocytes, we observed nuclear-centrosome detachments characteristic of the maternal-effect fue mutation: failed karyokinesis, with nuclear patches that failed to associate with centrosomes (Fig. 4G–I; Supp. Table S1). Thus, injecting the Lrmp-MO into wild-type stage IV oocytes recapitulated the maternal-effect fue mutation in the resulting Lrmp morphants.
Fig. 4.
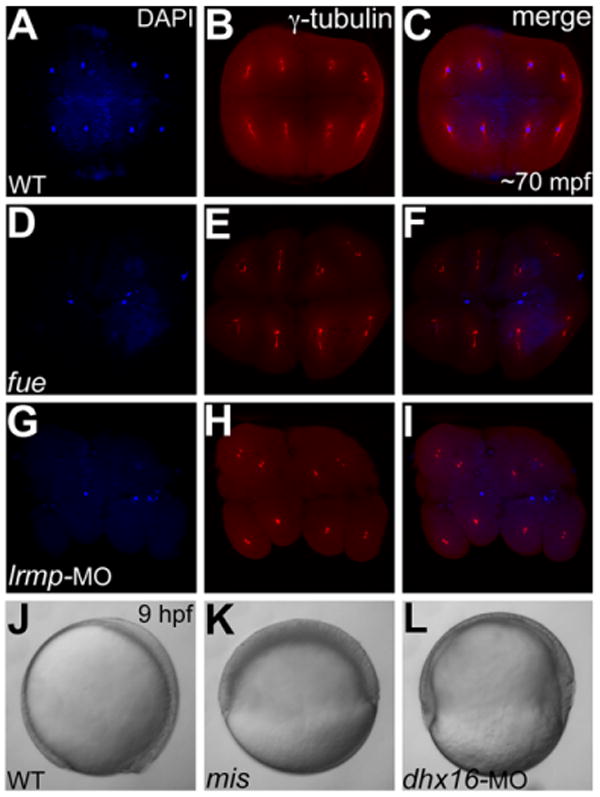
Stage IV oocyte injections of Lrmp and Dhx16 morpholinos phenocopy fue and mis maternal-effect phenotypes, respectively. A–I: Animal views of immunolabeled 70 minutes postfertilization (mpf) blastodiscs stained for DAPI (A,D,G) immunolabeled to detect γ-tubulin (B,E,H) and panel merges (C,F,I). A–C: In wild-type embryos, each nucleus associates with centrosomal γ-tubulin staining. D–F: In maternal-effect fue mutants, nuclei fail to divide, resulting in two to three patches of nuclei stainings corresponding to unfused parental pronuclei and the polar body for meiosis II (D,F), which fail to associate with γ-tubulin (E,F). G–I: In Lrmp morphants where maternal Lrmp function was inhibited, the nuclei similarly fail to divide (G) and also fail to associate with γ-tubulin (H,I). J–L: Lateral views of live embryos at 9 hours postfertilization (hpf). J,K: Wild-type embryos at 9 hpf where axis formation is evident (anterior to the top and dorsal to the right) (J), while in maternal-effect mis mutants, epiboly fails to occur (K). L: In Dhx16 morphants where maternal function was inhibited, epiboly similarly arrests, resulting in gastrulation failure.
We further validated this approach by targeting a second maternal-effect mutation mission impossible(mis), which encodes a maternal RNA heli-case, Dhx16 (Putiri and Pelegri, 2011). mis mutant embryos from homozygous mis females fail to undergo epiboly and thus fail to complete gastrulation (Putiri and Pelegri, 2011). A translation blocking morpholino targeting Dhx16 (Dhx16-MO), when injected into one-cell stage embryos, results in brain necrosis (Putiri and Pelegri, 2011), a phenotype common in zygotic knockdowns of essential genes with both maternal and zygotic contributions (Putiri and Pelegri, 2011; Yabe et al., 2011). Even though the gastrulation phenotype characteristic of the maternal-effect mis mutation occurs 4 to 9 hours after fertilization and MO injection, Dhx16 morphants obtained by injecting one-cell embryos do not manifest any gastrulation defects (Putiri and Pelegri, 2011), presumably due to the presence of functional Dhx16 protein preloaded in the egg. To test whether MO injection during oogenesis could phenocopy the maternal-effect mis mutant phenotype, we injected wild-type stage IV oocytes with translation blocking Dhx16-MO, which were in vitro matured and fertilized. In wild-type embryos derived from uninjected wild-type stage IV oocytes, epiboly and gastrulation occurred normally (Fig. 4J), while in control mis mutants derived from stage IV oocytes from homozygous mis females, epiboly fails to occur normally resulting in defective gastrulation and embryonic lysis (Fig. 4K; Putiri and Pelegri, 2011). In Dhx16 morphants derived from wild-type stage IV oocytes injected with Dhx16-MO, epiboly fails and such morphants fail to complete gastrulation and lyse (Fig. 4L; Supp. Table S1), demonstrating phenocopy of the maternal-effect mis phenotype.
Together with the aurB-KD mRNA injections, the Lrmp and Dhx16 translation blocking morpholino experiments in stage IV oocytes reaffirm the feasibility of the in vitro oocyte manipulation strategy as a reverse genetics methodology for functional studies of maternal genes. In addition to facilitating phenocopying of maternal-effect mutations obtained by forward genetics, this methodology can also aid in the dissecting of maternal vs. zygotic functions of genes, and testing the functional contribution of maternal genes with as yet unknown functions.
We further tested if splice-blocking morpholinos targeting another maternal-effect gene cellular atoll(cea), encoding the centriolar component Sass6, could recapitulate its maternal-effect cell division phenotype (Yabe et al., 2007). The sass6-spliceMO effectively caused mis-splicing of endogenous sass6 transcripts when injected at the one-cell stage, as we detected the expected alternatively spliced transcript containing the retained intron (data not shown). However, embryos derived from wild-type stage IV oocytes injected with the sass6-spliceMO appeared wild-type with no early cell divisions defects, and gastrulated normally (data not shown).
The lack of a maternal-effect phenotype with the sass6-spliceMO prompted us to investigate the splice status of cea/sass6 together with α-tubulin (data not shown) and β-actin, as control maternally expressed genes. Wild-type whole ovaries harvested at day 1 (stage I oocytes) and day 9 (stage IV oocytes) postpurging both contained exonic and intronic RT-PCR products for all three genes (Fig. S1A,B). Thus, despite ongoing splicing in stage IV oocytes, the presence of mature transcripts in stage I oocytes indicates that protein function would remain unaffected by the injection of splice MOs at stage IV, explaining the lack of a maternal-effect using sass6-spliceMO. However, whole ovaries isolated at day 1 postpurging also contain cell types such as follicle cells and immature oogonia, precluding definitive conclusion that the mature transcripts are exclusively from stage I oocytes. Regardless, our data caution that splice MOs may not be effective for the down-regulation of maternal genes, as mature transcripts at early stages of oogenesis may obscure effects of splice MOs injected into stage IV oocytes.
Transcript Encoding a Fluorescent Fusion of Maternal Protein Injected Into Stage IV Oocytes Allows Visualization of Subcellular Protein Localization in Early Embryos
Many maternal proteins are required for fundamental processes during egg activation, such as meiosis, and embryonic development postfertilization, such as male and female pronuclei fusion, karyokinesis, and cytokinesis, all of which require correct subcellular localization for biological function (Yabe et al., 2007; Bontems et al., 2009; Yabe et al., 2009; Lindeman and Pelegri, 2012; Nair et al., in revision). Expression of transcripts encoding maternal proteins fused to fluorescent reporters during oogenesis would provide a read-out of subcellular localization during early cleavage stages, especially in the absence of species-specific antibodies. We tested this rationale for zebrafish Sass6, an essential component of centrioles. It had been shown previously that fusion protein from transcripts encoding sass6∷mCherry injected at the one-cell stage retained biological function and localized to centrioles (Yabe et al., 2007). However, in such injections the fusion protein did not become visible until early gastrulation, precluding its use to visualize centrioles during the early cleavage stages. We injected mRNA coding for the same fusion construct into stage IV oocytes and assayed for localization of Sass6∷mCherry to centrioles in fertilized embryos (Fig. 5). We found that within 40 mpf, Sass6∷mCherry protein localized to foci on either side of the nuclei during prophase, presumably the centrioles (Fig. 5A–J). These Sass6∷mCherry foci colocalized with intense staining of centrosomal γ-tubulin, a known component of the peri-centriolar material associated with centrioles (Fig. 5C–E,H–J). Thus, exogenous Sass6∷mCherry translated during oogenesis localizes to centrioles in embryos immediately upon fertilization. Such expression of maternal proteins as fluorescent fusions in stage IV oocytes provides an alternative to transgenesis (Knaut et al., 2002) for analyzing subcellular localization of maternal products in the early embryo. Combined with the optical clarity of zebrafish embryos, this method should also allow live imaging of dynamic changes in maternal product subcellular localization patterns during the early cleavage stages.
Fig. 5.
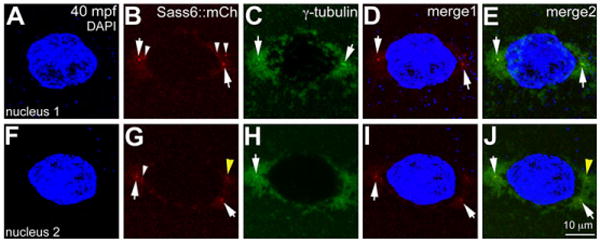
Sass6-mCherry protein from exogenous mRNA injected into stage IV oocytes localizes to centrioles during early cleavage stages. Immunolabeling for γ-tubulin and DAPI staining for nuclei in a Sass6-mCherry expressing blastodisc during prophase (40 mpf), which contains two nuclei, both of which are shown. C,E,H,J: γ-tubulin labels pericentriolar material around the nuclei, with intense foci in presumptive centrioles flanking each nuclei (arrows). B,D,E,G,I,J: Ectopically expressed Sass6-mCherry protein also localizes to foci flanking the nuclei (arrows in B,D,G,I) and colocalizes with γ-tubulin foci (arrows in E,J). B,G: Ectopic Sass6-mCherry is also observed as additional foci near the nucleus periphery (white arrowheads in B,G) and in the cytoplasm (not shown), consistent with the production of supernumerary centrioles after overexpression of maternally-derived Dsas-6 in Drosophila embryos (Peel et al., 2007). G,J: The observed putative supernumerary foci can also be associated with extra γ-tubulin foci (yellow arrowhead). Merge1: DAPI and Sass6∷mCherry; merge2: DAPI, Sass6∷mCherry and γ-tubulin.
The methods and reagents described in this study are a necessary complement to traditional forward genetics studies of maternal gene function. Furthermore, the approaches described herein could potentially facilitate functional assessment of maternal genes for which mutants are yet to be isolated or that have overlapping maternal and zygotic functions. In conclusion, our results provide proof-of-principle for effective functional manipulation and visualization of maternal gene products in zebrafish.
Supplementary Material
Key findings.
In vitro maturation method coupled to in vitro fertilization for the functional manipulation of maternal genes in zebrafish.
Injection of wild-type mRNAs into oocytes rescues maternal-effect mutations.
Functional knock down/reverse genetics can be achieved by expression of dominant-negative products or injection of translation-blocking morpholino oligonucleotides.
Expression of mRNAs coding for fluorescent fusion products allows visualizing the subcellular localization of maternal factors immediately after fertilization.
Acknowledgments
We thank Pelegri Lab animal husbandry staff for fish facility care and C. Eno for assaying viability to adulthood of embryos from the in vitro oocyte culture experiments.
Grant sponsor: NIH; Grant number: RO1 GM065303.
Footnotes
Additional Supporting information may be found in the online version of this article.
References
- Abrams EW, Zhang H, Marlow FL, Kapp L, Lu S, Mullins MC. Dynamic assembly of Brambleberry mediates nuclear envelope fusion during early development. Cell. 2012;150:521–532. doi: 10.1016/j.cell.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akitake CM, Macurak M, Halpern ME, Goll MG. Transgenerational analysis of transcriptional silencing in zebrafish. Dev Biol. 2011;352:191–201. doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Hart NH. The cortical actin sytoskeleton of unactivated zebrafish eggs: spatial organization and distribuion of filamentous actin, nonfilamentous actin, and myosin-II. Mol Reprod Dev. 1996;43:546–547. doi: 10.1002/(SICI)1098-2795(199604)43:4<536::AID-MRD17>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Becker KA, Hart NH. Reorganization of filamentous actin and myosin-II in zebrafish eggs correlates temporally and spatially with cortical granule exocytosis. J Cell Sci. 1999;112:97–110. doi: 10.1242/jcs.112.1.97. [DOI] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Choo BG, Kondrichin I, Parino S, Emeyanov A, Go W, Toh WC, Korzh V. Zebrafish transgenic Enhancer TRAP line database (ZETRAP) BMC Dev Biol. 2006;6:5. doi: 10.1186/1471-213X-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csenki Z, Zaucker A, Kovacs B, Hadzhiev Y, Hegyi A, Lefler KK, Muller T, Kovacs R, Urbanyi B, Varadi L, Muller F. Intraovarian transplantation of stage I-II follicles results in viable zebrafish embryos. Int J Dev Biol. 2010;54:585–589. doi: 10.1387/ijdb.082786zc. [DOI] [PubMed] [Google Scholar]
- Dekens MPS, Pelegri FJ, Maischein HM, Nusslein-Volhard C. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development. 2003;130:3907–3916. doi: 10.1242/dev.00606. [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 2004;6:771–780. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Fukazawa C, Santiago C, Park KM, Deery WJ, Canny SG, Holterhoff CK, Wagner DS. poky/chuk/ikk1 is required for differentiation of the zebrafish embryonic epidermis. Dev Biol. 2010;346:272–283. doi: 10.1016/j.ydbio.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Anderson R, Stainier DYR, Spradling AC, Halpern ME. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics. 2009;182:747–755. doi: 10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BA, Gomez de la Torre Canny S, Ye Y, Slusarski DC, Freisinger CM, Dosch R, Chou MM, Wagner DS, Mullins MC. A novel role for MAPKAPK2 in morphogenesis during zebrafish development. PLoS Genet. 2009;5:e1000413. doi: 10.1371/journal.pgen.1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- Kelly C, Chin AJ, Leatherman JL, Kozlowski DJ, Weinberg ES. Maternally controlled β-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000;127:3899–3911. doi: 10.1242/dev.127.18.3899. [DOI] [PubMed] [Google Scholar]
- Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C. An evolutionary conserved region in the vasa 3′UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol. 2002;12:454–466. doi: 10.1016/s0960-9822(02)00723-6. [DOI] [PubMed] [Google Scholar]
- Kotani T, Kawakami K. Misty somites, a maternal effect gene identified by transposon-mediated insertional mutagenesis in zebrafish that is essential for the somite boundary maintenance. Dev Biol. 2008;316:383–396. doi: 10.1016/j.ydbio.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Li S, Mao Z, Han W, Sun Z, Yan W, Chen H, Yan S. In vitro oocyte maturation in the zebra fish, Brachydanio rerio, and the fertilization and development of the mature egg. Chin J Biotechnol. 1993;9:247–255. [PubMed] [Google Scholar]
- Lindeman RE, Pelegri F. Vertebrate maternal-effect genes: insights into fertilization, early cleavage divisions, and germ cell determinant localization from studies in the zebrafish. Mol Reprod Dev. 2010;77:299–313. doi: 10.1002/mrd.21128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman RE, Pelegri F. Localized products of futile cycle/lrmp promote centrosome-nucleus attachment in the Zebrafish zygote. Curr Biol. 2012;22:843–851. doi: 10.1016/j.cub.2012.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K. Regulation of gene expression in mouse embryos and its embryonic cells through RNAi. Mol Biotechnol. 2006;34:271–278. doi: 10.1385/MB:34:2:271. [DOI] [PubMed] [Google Scholar]
- Lyman-Gingerich J, Westfall TA, Slusarski DC, Pelegri F. hecate, a zebrafish maternal effect gene, affects dorsal organizer induction and intracellular calcium transient frequency. Dev Biol. 2005;286:427–439. doi: 10.1016/j.ydbio.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Burns KH. Genetics of mammalian reproduction: modeling the end of the germline. Annu Rev Physiol. 2012;74:503–528. doi: 10.1146/annurev-physiol-020911-153248. [DOI] [PubMed] [Google Scholar]
- Mir A, Heasman J. How the mother can help: studying maternal Wnt signaling by anti-sense-mediated depletion of maternal mRNAs and the host transfer technique. Methods Mol Biol. 2008;469:417–429. doi: 10.1007/978-1-60327-469-2_26. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri F, Schulte-Merker S. A gynogenesis-based screen for maternal-effect genes in the zebrafish, Danio rerio. Methods Cell Biol. 1999;60:1–20. doi: 10.1016/s0091-679x(08)61891-9. [DOI] [PubMed] [Google Scholar]
- Pelegri F, Knaut H, Maischein HM, Schulte-Merker S, Nusslein-Volhard C. A mutation in the zebrafish maternal-effect gene nebel affects furrow formation and vasa RNA localization. Curr Biol. 1999;9:1431–1440. doi: 10.1016/s0960-9822(00)80112-8. [DOI] [PubMed] [Google Scholar]
- Putiri E, Pelegri F. The zebrafish maternal-effect gene mission impossible encodes the DEAH-box helicase Dhx16 and is essential for the expression of downstream endodermal genes. Dev Biol. 2011;353:275–289. doi: 10.1016/j.ydbio.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Matzuk MM. Deconstructing mammalian reproduction: using knockouts to define fertility pathways. Reproduction. 2006;131:207–219. doi: 10.1530/rep.1.00530. [DOI] [PubMed] [Google Scholar]
- Seki S, Kouya T, Tsuchiya R, Valdez DM, Jr, Jin B, Hara T, Saida N, Kasai M, Edashige K. Development of a reliable in vitro maturation system for zebrafish oocytes. Reproduction. 2008;135:285–292. doi: 10.1530/REP-07-0416. [DOI] [PubMed] [Google Scholar]
- Selman K, Wallace RA, Sarka A, Qi X. Stages of oocyte development in the Zebrafish, Brachydanio rerio. J Morphol. 1993;218:203–224. doi: 10.1002/jmor.1052180209. [DOI] [PubMed] [Google Scholar]
- Selman K, Petrino TR, Wallace RA. Experimental conditions for oocyte maturation in the Zebrafish, Brachydanio rerio. J Exp Zool. 1994;269:538–550. [Google Scholar]
- Stein P, Svoboda P, Schultz RM. Transgenic RNAi in mouse oocytes: a simple and fast approach to study gene function. Dev Biol. 2003;256:187–193. doi: 10.1016/s0012-1606(02)00122-7. [DOI] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebrafish (Brachydanio rerio) Nature. 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- Terada Y, Tatsuka M, Suzuki F, Yasuda Y, Fujita S, Otsu M. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RA. Vitellogenesis and oocyte growth in nonmammalian vertebrates. In: Browder LW, editor. Developmental biology. New York: Plenum Press; 1985. pp. 127–177. [DOI] [PubMed] [Google Scholar]
- White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zool B Mol Dev Evol. 2008;310:73–84. doi: 10.1002/jez.b.21153. [DOI] [PubMed] [Google Scholar]
- Yabe T, Ge X, Pelegri F. The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev Biol. 2007;312:44–60. doi: 10.1016/j.ydbio.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Ge X, Lindeman R, Nair S, Runke G, Mullins MC, Pelegri F. The maternal-effect gene cellular island encodes aurora B kinase and is essential for furrow formation in the early zebrafish embryo. PLoS Genet. 2009;5:e1000518. doi: 10.1371/journal.pgen.1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


