SUMMARY
The light-sensing organelle of the vertebrate rod photoreceptor, the outer segment (OS), is a modified cilium containing ~1,000 stacked disc membranes that are densely packed with visual pigment rhodopsin. The mammalian OS is renewed every ten days; new discs are assembled at the base of the OS by a poorly understood mechanism. Our results suggest that discs are formed and matured in a process that involves specific phospholipid-directed vesicular membrane targeting. Rhodopsin-laden vesicles in the OS axonemal cytoplasm fuse with nascent discs that are highly specialized with abundant phosphatidylinositol 3-phosphate (PI3P). This membrane coupling is regulated by the FYVE domain-containing protein, SARA, through its direct interaction with PI3P, rhodopsin, and SNARE protein syntaxin 3. Our model, in contrast to the previously proposed evagination model, suggests that the vesicular delivery of rhodopsin in the OS concentrates rhodopsin into discs, and this process directly participates in disc biogenesis.
INTRODUCTION
The vertebrate rod photoreceptor has a specialized light sensing organelle called the outer segment (OS). The OS consists of a plasma membrane that encloses a stack of ~1,000 closely spaced membranous discs. The OS is connected to the cell body (i.e., inner segment [IS]) by a connecting axonemal segment (i.e., connecting cilium) that serves as a bridge (Figures 1A and 1B). The OS discs are densely packed with rhodopsin for high-sensitivity light detection. OSs are continuously renewed throughout the lifetime of the animal (Young, 1967). New discs are assembled at the base of the OS by incorporating proteins and lipids that are synthesized and transported from the IS. Discs mature along their distal migration; aged discs shed at the distal tip and are engulfed by the neighboring retinal pigment epithelial cells for degradation.
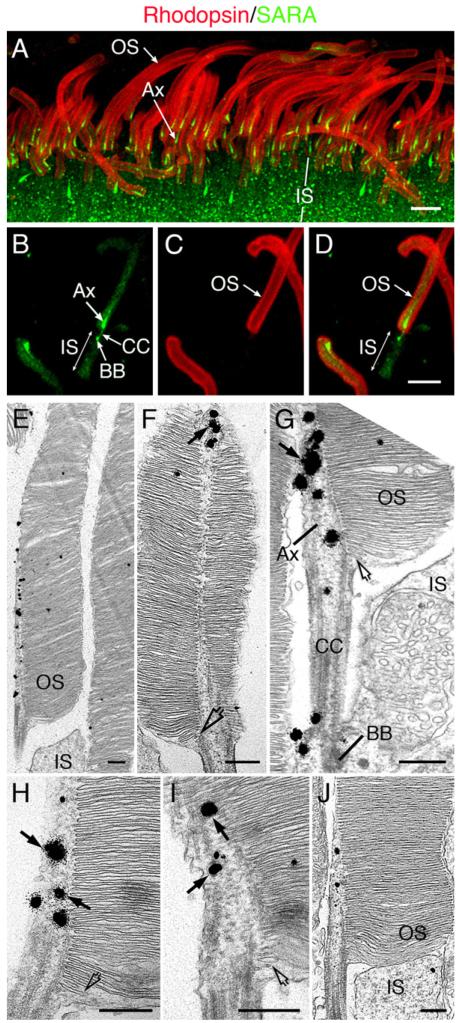
Figure 1. Organization of the Vertebrate Photoreceptor and the SARA-Rhodopsin Interaction.
(A) A schematic drawing of a mammalian rod photoreceptor, showing the OS, IS, nucleus (N), and synapse (S).
(B) A magnified view of the junction between the distal IS and the proximal OS of mouse rods. Mouse rod OS is ~30 μm in length and ~1.5 μm in diameter; connecting cilium (CC) is ~1.2 μm in length and ~0.2 μm in diameter. The microtubule-based axoneme (Ax) begins at the basal body (BB) in the distal IS and continues through the CC and into the proximal OS axoneme for ~10 μm.
(C) X-gal filter assay results of yeast transformants expressing SARA and Rho39Tr or the vector.
(D) Schematic representation of SARA.
(E) Pull-down assays were carried out by incubating purified His-SARA with either GST- or GST-Rho39-conjugated glutathione Sepharose. Glutathione eluates and input were immunoblotted with anti-SARA and anti-GST Abs, respectively.
(F) Mouse retinal lysates were immunoprecipitated with anti-rhodopsin mAb B6-30 or control mAb. The input (2 μg total protein) and the immunoprecipitates (from 5 μg total protein) were detected by immunoblotting using anti-SARA and anti-rhodopsin Abs. Bound momomeric rhodopsin was shown.
(G) Confocal images showing the triple labeling of endogenous EEA1 (red) and ectopically expressed rhodopsin (green) and Flag-SARA (cyan) in HEK cells. Arrows point to the colocalization of rhodopsin and SARA on early endosomes, which were also EEA1 labeled. SARA+/rhodopsin+/EEA1–vesicles were also occasionally observed (arrowheads). Bar = 20 μm.
Although the fascinating structure and renewal process of the OS have been described for nearly 50 years, few molecular insights have been provided to explain how discs are formed and how rhodopsin is targeted to the disc membranes. Interestingly, recent evidence suggests that rhodopsin-deficient mice fail to develop OSs (Humphries et al., 1997; Lem et al., 1999). This finding suggests that rhodopsin itself has a role in OS biogenesis, in addition to its role as a phototransduction receptor. While the molecular basis underlying rhodopsin’s participation in OS development is unknown, emerging evidence suggests that rhodopsin’s cytoplasmic C-terminal tail bears an “address signal” for its transport from its site of synthesis in the rod cell body to the OS (Tai et al., 1999; Deretic et al., 2005).
The protein-lipid interactions that regulate intracellular membrane trafficking have gained growing attention. For example, phosphatidylinositol 3-phosphate (PI3P), a product of PI3-kinase, plays a pivotal role in regulating endocytic pathway trafficking through its interaction with EEA1 (early endosomal antigen 1) on early endosomal membranes (Gaullier et al., 1998; Gillooly et al., 2000; Stenmark and Aasland, 1999). EEA1 contains a FYVE (Fab1p, YOTB, Vac1p, EEA1) domain, a highly conserved double zinc-finger motif that binds to PI3P with high specificity. The ability of EEA1 to tether vesicles and regulate assembly of the SNARE (soluble NSF attachment receptor) complex promotes endocytic membrane fusion (Christoforidis et al., 1999; Simonsen et al., 1999).
Smad anchor for receptor activation (SARA), initially isolated for its role in TGF-β signal transduction (Tsukazaki et al., 1998), is also a FYVE domain protein located on early endosomes (Hu et al., 2002; Seet and Hong, 2001). SARA also seems to play important roles in regulating endosomal membrane dynamics; overexpression of SARA induces aberrant endosomal expansion in cultured cells, likely through dysregulated fusion (Hu et al., 2002; Itoh et al., 2002; Seet and Hong, 2001). However, the molecular basis underlying SARA-mediated vesicular trafficking and its physiological relevance remains largely unknown.
In this study, we combined biochemical, morphological, and reverse genetic approaches in mammalian photoreceptors to demonstrate that the rhodopsin C-terminal tail functionally interacts with SARA. Our data suggest that the protein-protein and protein-lipid interactions organized by SARA regulate the vesicular targeting of rhodopsin-bearing axonemal vesicles to nascent discs at the base of the OS. The incorporation of rhodopsin vesicles into discs completes the OS targeting of rhodopsin and directly participates in disc biogenesis.
RESULTS
SARA Is a Novel Rhodopsin C-Terminus-Interacting Protein
Our two-hybrid screens using 39 residues of the rhodopsin cytoplasmic C terminus as a bait repeatedly isolated a bovine cDNA clone encoding an open reading frame with 97% identity to the C-terminal half (C627-V1323) of human SARA (Figure 1C). Full-length human SARA was subsequently isolated for use in interaction studies (Hu et al., 2002; Figure 1 D). We confirmed that the rhodopsin-SARA interaction is direct by using an in vitro binding assay in which purified His-SARA was pulled down by an immobilized glutathione S-transferase (GST) fusion protein containing the rhodopsin C-terminal 39 residues (GST-Rho39), but not by immobilized GST protein (Figure 1E). Furthermore, anti-rhodopsin antibody (Ab), but not control Ab, was able to coimmunoprecipitate endogenous SARA from mouse retinal extracts (Figure 1F), suggesting that SARA interacts with rhodopsin in vivo. Finally, we employed the heterologous expression system of human embryonic kidney (HEK) cells to study the spatial relationship between rhodopsin and SARA in detail. Consistent with previous reports (Hu et al., 2002; Seet and Hong, 2001), both endogenous SARA (data not shown) and transfected Flag-SARA (Figure 1G) extensively colocalized with EEA1 on early endosomes in HEK cells. Cotransfected rhodopsin was also frequently found on EEA1+/SARA+ early endosomes (Figure 1G) in addition to its plasma membrane localization (Figure 1G). These results collectively suggested that rhodopsin physically interacts with SARA on the intracellular vesicular compartments.
SARA Is Enriched in Axonemal Vesicular Structures Closely Associated with Nascent Discs
To unravel the physiological relevance of the SARA-rhodopsin interaction in vivo, we determined the subcellular distribution of SARA in rod photoreceptors. Immunofluorescent labeling of mouse retinal sections revealed that, in photoreceptors, SARA was distributed on punctate vesicular structures, presumably early endosomes, in the apical cytoplasm of the IS (Figure 2A) as well as synaptic terminals (data not shown). In the OS, SARA is particularly concentrated in the most basal portion of the axoneme (Figure 2A) and displays a comet-like labeling pattern. This unique labeling pattern is likely to be attributed to a gradually decreasing SARA level and the gradually increasing diameters of discs from the base toward the distal tips within this part of the OS. However, the colabeled rhodopsin, as predicted, was distributed throughout the entire OS (Figure 2A). SARA immunolabeling appeared to be very specific: two affinity-purified anti-SARA Abs that were generated against two different epitopes produced the same staining pattern, and this labeling was missing when the primary Ab was preabsorbed to SARA (data not shown). Unlike SARA, EEA1 was undetectable in the OS, and only weak immunoreactivity was detected in the IS (data not shown).
Figure 2. SARA Distribution in the Rod Photoreceptor.
(A–D) Confocal images of the outer part of retinal sections (A) or isolated OS/axoneme (B–D) from mouse retinas that were double immunostained with anti-SARA (green) and anti-rhodopsin Ab (red). Although rhodospin was enriched on OS plasma membrane and disc membranes, the predominant plasma membrane labeling pattern of rhodopsin was likely due to Ab sequestration. Bars = 5 μm.
(E–J) Electron micrographs of six longitudinally sectioned mouse rods demonstrating that SARA-derived silver-gold particles (arrows in [F]–[I]) were associated with tubulo-vesicular structures in the proximal portion of the OS axoneme. Only the proximal OS portions are shown in these images. At the base of the OS, profiles of small vesicles and dilated cisternae were enclosed by the plasma membrane (open arrows in [F]–[I]). Bars = 300 nm.
Immunolabeling of isolated rod OS/axonemes was subsequently performed to improve the signal detection in the ciliary region. The highest SARA signals were detected in the connecting cilium, basal body, and proximal OS axonemes (Figures 2B-2D). The estimated length of the SARA signal in the OS axoneme is ~2.5 μm. Because mouse OSs turn over every 10 days (Young, 1967) and they are 24–30 μm long, SARA was thus likely to be concentrated in the axonemal region near where discs are newly assembled and less than ~1 day old. The OS axonemal localization of SARA is distinctive and has not been reported for any other photoreceptor proteins.
The immunocytochemical studies (Figure 1G; Hu et al., 2002) in cultured cells suggested that SARA was associated with vesicular membranes. However, the limited spatial resolution provided by the OS axoneme prohibited the visualization of vesicular patterns of SARA under light microscopy. We thus set out to determine SARA’s distribution by performing immuno-electron microscopy (EM) in mouse eyes using an improved fixation and pre-embedding labeling procedure, which is excellent in preserving both fine membrane structures and antigenicity. Consistent with the light microscopic results, SARA-derived silver-intensified immunogold particles were primarily detected as 2–3 μm long arrays aligned in an axial direction in the proximal OS axoneme (Figures 2E-2J). The gold particles decorated the tubulo-vesicular structures, and these labeled structures were often near the edge of disc membranes facing the axoneme (arrows, Figures 2F-2I). SARA immunoreactivity was also detected in the connecting cilium, basal body, and IS, but to a lesser extent than in the OS (Figure 2G). Finally, our electron micrographs of rods showed that the plasma membrane of the OS appeared to enclose the entire stack of closed discs, including arrays of vesicles and/or dilated cisternae at the very base of the OS (open arrows, Figures 2F-2I and S1). The expression patterns of SARA in rods and its previously predicted function in membrane tethering and/or fusion (Hu et al., 2002) prompted us to hypothesize that SARA may regulate the membrane targeting and/or fusion between axoneme-localized rhodopsin-laden vesicles and nascent discs and, hence, disc incorporation of rhodopsin and disc assembly. We thus designed the series of experiments described below to test this hypothesis.
SARA-PI3P Interaction Is Essential for OS Delivery of Rhodopsin and Disc Biogenesis
SARA contains a characteristic FYVE domain (FYVESARA) that is known to bind to PI3P with high specificity and high avidity (Itoh et al., 2002). Consistently, recombinant SARA also specifically interacts with PI3P, but not with other modified phospholipids (Figure S2). To assess the physiological relevance of SARA, PI3P, and their interactions, we set out to determine whether and where PI3P is synthesized and distributed in rods. To this end, we first employed an assay described by Gillooly et al. (2000) to localize PI3P. In this assay, cells or tissues are incubated with biotinylated GST fusion containing a double FYVE finger of Hrs (i.e., in situ PI3P probe), and bound proteins are detected by Alexa 488-strapatvidin. The PI3P probe was first validated by its presence on EEA1-labeled early endosomes in cultured cells (Figure S3A). In rods, the PI3P probe predominantly labeled the OS, with a slight enrichment at the proximal portion (Figure S3B). Comparable results were obtained when FYVESARA was used as a probe (data not shown). In contrast, probing with control GST protein did not give rise to any specific signal in retina (data not shown).
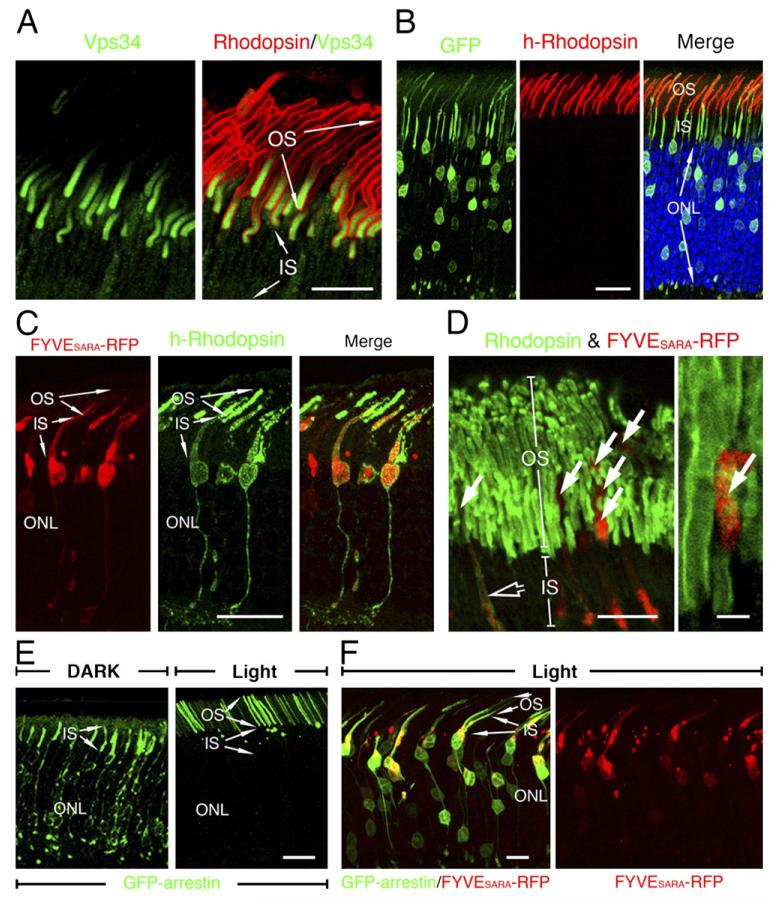
We then performed immunolabeling of the sole class III PI3-kinase, Vps34, which is the primary enzyme specifically and exclusively catalyzing the production of PI3P from phosphoinositides (Foster et al., 2003) in mouse retinas. While rhodopsin labeling marked the entire OS (Figure 3A), Vps34 immunofluorescence was displayed as an ~8 μm long gradient at the proximal portion of the OS, with the strongest signal toward the OS base (Figure 3A). No significant Vps34 signal was detected in the IS. The unique Vps34 labeling disappeared when antigen-preabsorbed Ab was used (data not shown). Note that the Vps34 labeling appeared to be present across the entire width of the proximal OS, suggesting that Vps34 was distributed on the discs. Ultrastructural localization studies also support the idea that Vps34 is present on discs (data not shown). The expression pattern of Vps34 suggested that PI3P is locally synthesized on the nascent discs at the proximal portion of the OS.
Figure 3. Localization of Vps34 and Phenotypic Examination of FYVESARA-Overexpressing Rods.
(A) Confocal images of the mouse retinas colabeled with Vps34 (green) and rhodopsin (red) Abs.
(B) Rat photoreceptors cotransfected with GFP and h-rhodopsin (red). Blue: DAPI-labeled nuclei.
(C) Confocal images of rat photoreceptors cotransfected with FYVESARA-RFP (red) and h-rhodopsin (green). While RFP signals were detected in both the IS and the proximal portion of the OS, significantly less RFP was detected in the distal portion of OS.
(D) Confocal images show the distribution of endogenous rhodopsin (green) in FYVESARA-RFP transfected photoreceptors.
(E) Confocal images of rat rods transfected with GFP-arrestin and harvested under either dark or light conditions.
(F) Confocal images of rat photoreceptors, transfected GFP-arrestin, and FYVESARA-RFP (red) and harvested under light conditions. Bars = 10 μm ([A], left panel in [D], and [F]), 20 μm (B, C, and E), and 2 μm (right panel in [D]).
The expression of SARA on axonemal vesicles and PI3P on nascent discs indicates that the FYVE domain-PI3P interaction may be used for the docking between these two opposing membrane domains. We thus predicted that overexpression of FYVESARA would block the access of PI3P to SARA and thus affect the disc biogenesis and OS targeting of rhodopsin in mammalian rods. Rhodopsin that fails to be incorporated into discs would backflow into the cell body/synapse. To demonstrate that the overexpressed FYVESARA may sequester PI3P, we showed that endogenous SARA was almost completely depleted from early endosomes in HEK cells expressing a high level of the GFP fusion of FYVESARA (Figure S4) or the red fluorescent protein (RFP) fusion of FYVESARA (data not shown). However, as expected (Itoh et al., 2002), FYVESARA-GFP fusion expressed at low level was predominantly distributed on early endosomes (Figure S4).
To study rhodopsin targeting in vivo, we employed a novel gene delivery method, retinal transfection, that permits both gain- and loss-of-function studies in rodent retina in vivo and in a cell-type-specific manner. Retinal transfection involves subretinal injection of plasmid DNA and electroporation in neonatal animal eyes. A previous study used this method to show that the chicken actin (CAG) promoter can drive gene expression that persists specifically in mature photoreceptors (Matsuda and Cepko, 2004). We first confirmed that a protein transfected in photoreceptors was specifically targeted to the proper subcellular compartment. Specifically, we examined the distribution pattern of ectopically expressed human rhodopsin (h-rhodopsin) by immunostaining transfected mouse retinas with the monoclonal Ab 3A6, which specifically recognizes human, but not rodent, rhodopsin (Li et al., 1995). Our results showed that the transfected h-rhodopsin, like endogenous rhodopsin, was specifically localized in the OS (Figure 3B). By contrast, cotransfected GFP (Figure 3B) or RFP (data not shown) was cytosolic and diffused throughout the rod cells. These results suggested that the coexpression of GFP or RFP did not interfere with the OS targeting of rhodopsin. We then examined the distribution of h-rhodopsin in rods cotransfected with FYVESARA-RFP. As shown in Figure 3C, almost all RFP-positive transfected rods had h-rhodopsin mislocalized in the cell bodies/synapses. Furthermore, it was noted that the FYVESARA-RFP ectopically expressed in the OS was predominantly distributed at the proximal region (Figure 3C), consistent with the PI3P localization studies described above. Transfected FYVESARA-GFP behaved similarly to FYVESARA-RFP (data not shown). Finally, we showed that ectopic expression of FYVE domain derived from EEA1 by transfecting 3XFYVEEEA1-GFP in rods also led to the mislocalization of cotransfected h-rhodopsin (Figure S5). Taken together, these results argue the importance of the PI3P-FYVE domain interaction in the vesicular trafficking of rhodopsin to the OS.
The mislocalization of endogenous rhodopsin was subsequently confirmed in rods singly transfected with FYVESARA-RFP. As shown in Figure 3D, FYVESARA-RFP transfected rods were often associated with reduced rhodopsin labeling in the OS (arrows) and increased rhodopsin labeling in the IS (open arrow). Furthermore, our hypothesis predicted that SARA perturbation would not have a major impact on the distributions of proteins exclusively targeted to the OS plasma membrane, such as cGMP-gated channel (Cook et al., 1989). Indeed, we found that the OS localization of cGMP-gated channel was not significantly affected by FYVESARA-RFP overexpression (Figure S6).
We noted that the rhodopsin signal that delocalized in the IS was not robust; we suspected that this might be due to Ab sequestration by the extremely high level of rhodopsin in the untransfected OS and other immunolabeling-associated technical issues (e.g., nonlinear signal and Ab penetration). To independently investigate the subcellular distribution of endogenous rhodopsin, we employed the distribution of transfected GFP-arrestin as a surrogate indicator. The rationale for this strategy is based on several previous reports that showed the following: (1) Arrestin undergoes light-dependent translocation (Whelan and McGinnis, 1988). While arrestin is primarily located in the cell bodies/synapses of dark adapted rods, it travels to the OS upon light stimulus due to its high-affinity interaction with photoactivated rhodopsin (Mendez et al., 2003; Nair et al., 2005). (2) Arrestin is almost always colocalized with mislocalized rhodopsin in the cell bodies/synapses of diseased rods, presumably because the rhodopsin molecules in the rod cell bodies are inadvertently activated, thus acting as a “sink” for arrestin to bind (Chuang et al., 2004). (3) GFP-arrestin is an intrinsically fluorescent probe that is exempt from many of the technical limitations of immunocytochemistry, and the light-regulated subcellular translocation of GFP-arrestin has been demonstrated in transgenic rods (Peterson et al., 2003). To establish our assay, we first confirmed that transfected GFP-arrestin in rods, like endogenous arrestin, traveled in and out of the OS in response to light. GFP-arrestin, detected by its green fluorescence, was primarily found in the cell bodies/synapses of dark-adapted rods, whereas GFP-arrestin was predominantly located in the OS in retinas harvested in light (Figure 3E). Cotransfection of control RFP protein had no effect on GFP-arrestin distribution (see below, Figure 5C). By contrast, cotransfected FYVESARA-RFP led to a substantial amount of GFP-arrestin accumulated in the cell body/synapse in rods that were harvested in light (Figure 3F). These results consistently suggested that the OS targeting of endogenous rhodopsin was impaired in the FYVESARA overexpressing rods.
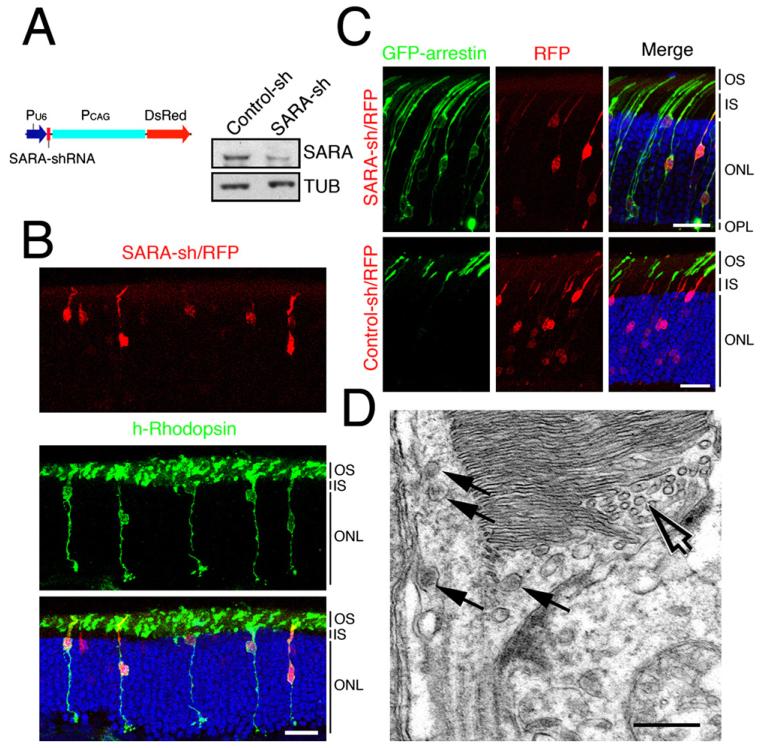
Figure 5. SARA Suppression Impairs the OS Targeting of Rhodopsin and Disc Organization.
(A) A schematic diagram of SARA-sh/RFP construct. Immunoblots of SARA and tubulin (TUB) from lysates of HEK cells transfected with control-sh or SARA-sh plasmid.
(B) Confocal images of rat retinas containing photoreceptors transfected with SARA-sh/RFP and h-rhodopsin (green). Blue: DAPI-labeled nuclei. Bar = 20 μm.
(C) Confocal images of light-adapted rat retinas cotransfected with GFP-arrestin and SARA-sh/RFP (top) or control-sh/RFP (bottom). Bars = 20 μm.
(D) Electron micrograph of a SARA-sh/RFP, GFP-arrestin cotransfected rod exhibiting a disorganized OS containing both abundant small vesicles (open arrows) and multivesicular body-like structures (arrows). Bar = 300 nm.
Finally, we examined the disc organization of FYVESARA-RFP transfected rods at the ultrastructural level. In these studies, FYVESARA-RFP transfected rods were identified by the GFP immunogold particles that were derived from the cotransfected GFP-arrestin (Figures 4A-4G). In contrast to the neighboring untransfected rods having normal disc organization (data not shown), the FYVESARA-RFP transfected rods had highly disorganized OSs. Instead of flattened, tightly packed discs, various sizes of tubules/vesicles were found at the basal OS (Figures 4A-4D). These vesicles were either clear (Figures 4A and 4B) or electron dense (arrows, Figure 4D). Large membranous sacs containing small vesicles, resembling multivesicular bodies, were also seen (Figure 4E). The abnormal accumulation of membrane vesicles can also be found underneath the swollen OS axoneme (arrows, Figure 4B) and ciliary membrane (open arrow, Figure 4B). We also frequently observed erupted OS plasma membranes associated with the accumulation of small vesicles at the extracellular space between the IS and OS (Figure 4F) and transfected rods with a completely unrecognizable OS (Figure 4G). Interestingly, no drastic morphological change was observed in the IS except for an apparent increase in the number of vesicular profiles in the transfected rods (Figure 4B). The results above collectively suggested that the SARA-PI3P interaction plays a physiologically relevant role in the vesicular trafficking of rhodopsin to OS discs as well as in disc formation.
Figure 4. Ultrastructural Analysis of the Disc Organization of FYVESARA-Overexpressing Rods.
(A–G) ImmunoEM analysis of GFP was carried out to detect rat rods transfected with FYVESARA-RFP and GFP-arrestin. GFP-derived immunogold particles are mostly distributed in the cell bodies and are largely omitted from the presented images. Representative electron micrographs covering the IS-OS junction regions of transfected rods displaying various profiles of disorganized discs. Arrows point to vesicles (A and B), tubules (C), and sacs of vesicles (D and E) accumulated beneath the basal discs and/or swollen axonemal cytoplasm. The open arrow in (B) points to vesicles aberrantly accumulated beneath the connecting cilium plasma membranes. The arrow in (F) points to the small vesicles seen in the extracellular space near the rod with OS plasma membrane erupted. (G) Electron micrograph of the severely disrupted OS in a transfected rod. Bars = 300 nm.
SARA Suppression Impairs OS Targeting of Rhodopsin and Disc Biogenesis
To corroborate the dominant-negative approach described above, the RNA interference approach was subsequently employed to analyze the specific involvement of SARA in rhodopsin’s OS targeting and disc assembly. To this end, a plasmid (SARA-sh/RFP) containing both a short-hairpin RNA (sh) against SARA and an RFP expression cassette was generated (Figure 5A). The specificity and effectiveness of SARA-sh/RFP in silencing SARA expression was validated by immunoblotting assays (Figure 5A) and immunostaining in cell cultures (Figure S7).
We analyzed how SARA suppression affects rhodopsin targeting to the OS by examining photoreceptors cotransfected with SARA-sh/RFP and h-rhodopsin (Figure 5B). We found that mislocalization of h-rhodopsin was closely associated with the RFP-positive rods that were cotransfected with SARA-sh/RFP. By contrast, h-rhodopsin was primarily restricted in the OSs of RFP-negative rods. Control experiments showed that coexpression of control-sh/RFP had no effect on the OS distribution of h-rhodopsin (data not shown). To further demonstrate the phenotypic specificity, a rescue experiment was carried out by using a plasmid that carried SARA-sh, sh-resistant SARA, and GFP (Figure S8). h-rhodopsin cotransfected with this rescue plasmid was found to be properly targeted to the OS. Finally, we showed that cotransfection of SARA-sh/RFP, but not control-sh/RFP, led to the mislocalization of GFP-arrestin in light-adapted rods (Figure 5D), suggesting that the OS targeting of endogenous rhodopsin was defective in SARA-silenced rods.
Aberrant disc organization was also seen in SARA-silenced rods by EM. SARA-sh/RFP transfected rods had an accumulation of tubulo-vesicles (open arrow;Figure 5D) and a structure resembling a multivesicular body (arrows; Figure 5D) at the most basal OS and in the swollen axoneme. These morphological changes closely resemble those seen in the FYVESARA overexpressing rods, especially those with milder phenotypes.
SNARE Protein Syntaxin 3 Binds SARA and Participates in OS Vesicular Trafficking
We predicted that SNARE protein complexes are required to render the fusion of axonemal vesicles to the nascent discs and thus are aimed to identify the specific members of SNARE fusion proteins involved in this process. Our immunolabeling survey discovered that syntaxin 3 was particularly enriched in the basal OS axonemal region (green, Figure 6A) in addition to its IS and synaptic terminal locations (data not shown). The distribution of syntaxin 3, an integral membrane protein, in OS strikingly resembled that of SARA (Figure 2A), indicating that syntaxin 3 is also distributed on axonemal vesicles at the basal OS.
Figure 6. Syntaxin 3 Distribution and Interaction with SARA and Phenotypic Examination of Syntaxin-3-Suppressed Rods.
(A) Immunolocalization of syntaxin 3 (green) and rhodopsin (red) in mouse rods. Arrows point to the proximal OS axonemal labeling of syntaxin 3. Bar = 5 μm.
(B) Distribution of myc-syntaxin 3 (green) and FLAG-SARA (red) in transfected HEK cells.
(C) Pull-down assays were carried out by incubating purified His-SARA with either GST- or GST-syntaxin 3-conjugated glutathione Sepharose. Glutathione eluates and inputs were immunoblotted with anti-SARA and anti-GST Abs, respectively.
(D) Immunoblots of input and FLAG immunoprecipitates (equivalent to three times the input of total proteins) obtained from cells transfected with FLAG-SARA and/or myc-syntaxin 3 are shown.
(E) Extracts of HEK cells coexpressing FLAG-SARA, GFP-VAMP2, myc-syntaxin 3, and SNAP25 were immunoprecipitated with anti-FLAG or anti-HA (control) Ab. Immunoblots show immunoprecipitates and input (~10% of proteins subjected to immunoprecipitation) detected by each indicated molecule. (F) Confocal images of rat photoreceptors transfected with syntaxin3-sh/GFP (green) and h-rhodopsin (red).
The subcellular distribution of syntaxin 3 was further examined in transfected HEK cells. Myc-syntaxin 3 immunofluorescence was found on both plasma membranes and SARA-positive early endosomes (Figure S9). The intracellular vesicular myc-syntaxin 3 also colocalized with FLAG-SARA in double transfected HEK cells (Figure 6B). The coincidental distributions of syntaxin 3 and SARA in both rods and cultured cells prompted us to investigate the possible interaction between SARA and syntaxin 3. Pull-down assays were first conducted. These experiments showed that purified His-SARA specifically bound to immobilized GST-syntaxin 3 (Figure 6C), but not GST-syntaxin 7 (Figure S10) nor GST, suggesting that SARA bound directlyto syntaxin 3. We then performed coimmunoprecipitation experiments using HEK cells transfected with FLAG-SARA and/or myc-syntaxin 3. Our results showed that myc-syntaxin 3 was specifically coprecipitated with FLAG-SARA by anti-FLAG Ab (Figure 6D). In HEK cells, all other components of the SNARE core complex (e.g., SNAP25 and VAMP2) were also detected in the FLAG-SARA immunoprecipitates from HEK cell lysates containing these overexpressed proteins (Figure 6E). Both the overlapping subcellular distributions of SARA and syntaxin 3 and their direct interaction indicated that these two molecules may act coordinately in the same trafficking pathway.
Finally we knocked down syntaxin 3 by transfecting photoreceptors with syntaxin3-sh/GFP (Figure S11) and examined the rhodopsin distribution by visualizing the immunoreactivity of cotransfected h-rhodopsin. As shown in Figure 6F, a strong signal from mislocalized h-rhodopsin was detected in the cell bodies/synapses of the syntaxin 3-sh/GFP transfected rods, suggesting that the fusion activity of syntaxin 3/SNARE is involved in the OS targeting of rhodopsin.
DISCUSSION
Vesicular Trafficking and Membrane Fusion Involve OS Disc Formation and Maturation
Until now, the single predominant hypothesis for disc biogenesis, the evagination/disc rim formation model proposed by Steinberg and his colleagues (Steinberg et al., 1980) was based solely upon their morphological studies of adult monkey rods. These authors observed a few outfolded plasma membranes, which were referred to as open discs, at the base of OS and thus hypothesized that discs are formed by a series of evaginations of the basal OS plasma membranes followed by a “disc rim formation” process that pinches off discs (Figure 7C). Basal open discs have also been described for rodent rods (Carter-Dawson and LaVail, 1979). Although this model has been commonly cited in the literature, experimental data supporting this model have been indirect and based only on in vitro studies (Matsumoto and Besharse, 1985; Williams et al., 1988). In addition, the evagination model is difficult to explain the distinct protein expression profile between the disc membranes and OS plasma membranes. Using modified procedures for better membrane preservation (see Supplemental Discussion), our electron micrographs showed that, at the base of rodent OS, the plasma membrane enwraps the entire disc stacks along with vesicles/cisternae (Figures 2E-2J). Structures resembling an “open disc” were rarely seen in these well-preserved samples. It is probable that the basal OS plasma membranes are more vulnerable to damage from histological processing procedures due to their close proximity to the apical IS plasma membrane; the resulting damaged membranes resemble open-disc-like structures (curved arrow in Figure S1F).
Figure 7. Molecular Mechanism Underlying OS Vesicular Trafficking and Models for OS Disc Morphogenesis in Mammalian Rods.
(A) The “vesicular targeting model” suggests new discs are assembled and “grow” via SARA-, PI3P-, and SNARE-mediated vesicular trafficking and membrane fusion events. This set of events is likely to continue until a disc membrane reaches its mature size. The axonemal vesicles and the primitive disc sacs could be derived from internalized OS basal membranes, although it is also probable that some of the axonemal vesicles are directly shipped from the IS through the connecting cilium.
(B) Our data suggest that at the basal OS, the axonemally localized vesicles recruit SARA via rhodopsin’s cytoplasmic tail. SARA tethers axonemal vesicles onto nascent discs through the interaction between its FYVE domain and PI3P. In addition, SARA may regulate SNARE fusion complex assembly and/or activity via its direct interaction with syntaxin 3.
(C) The “evagination/disc rim formation” model suggests that discs are formed by a series of evagination events of the basal OS plasma membrane, depicted in the diagram in a longitudinal view. In this model, the most proximal open disc and most basal OS plasma membrane are fused by bilaterally growing ciliary membranes (i.e., disc rims). The two growing ciliary membranes meet at the opposite side of axoneme, which results in a closed disc.
Several results presented in this report complementarily support a novel model that we refer to as the “vesicular targeting model,” in which SARA-, PI3P-, and SNARE-regulated vesicular trafficking underlies the disc incorporation of rhodopsin as well as disc assembly (Figures 7A and 7B). First, our ultrastructural analysis of adult rodent rods revealed abundant tubulo-vesicles in the proximal OS axonemal cytoplasmic space. These vesicular tubules resemble structures described for transport cargoes (Presley et al., 1997) and are likely to be the membrane carriers of lipids and proteins for the development of disc membranes. We found that these vesicular structures, often in clusters, are especially concentrated near the junctions between the distal cilium and the OS base (Figures 2E-2J; arrows in Figure S1). This is consistent with the idea that these sites are where the primitive discs are first formed. Fusion activity is thus likely to be most robust at these sites. Second, the Vps34 expression pattern suggests that a high level of PI3P is synthesized at the basal nascent discs and that the locally produced PI3P in the disc membranes is likely to be used to regulate the membrane targeting specificity of rhodopsin vesicles through the SARA-PI3P interaction. Indeed, FYVE domain overexpression rendered targeting inefficient and led to disorganized OSs filled with vesicles and tubular cisternae. Unfused vesicles backed up into the OS axonemes, which are often abnormally enlarged, and into the connecting cilium and IS as well. The build-up of vesicular profiles in these rods strongly argues that the precursors of the disc membranes are vesicles. In some cases, these vesicles are released to the extracellular space upon plasma membrane rupture. This is reminiscent of a phenotype seen in a number of retinal degenerative mouse models (Blanks et al., 1982; Hagstrom et al., 1999; Li et al., 1996). Finally, defective OS targeting of rhodopsin was consistently observed in FYVESARA-overexpressed, SARA-suppressed, and syntaxin 3-suppressed rods, in agreement with our model in which rhodopsin that cannot be incorporated into discs would back up into the cell body/synapses.
SARA was concentrated on early endosomal membranes in cultured cells (Hu et al., 2002; Itoh et al., 2002; Seet and Hong, 2001). Thus, it is tempting to speculate that SARA-positive vesicular tubular structures in the OS axoneme may have shared characteristics of early endosomes. This suggestion would be in line with a previously proposed model, based on the morphological studies using rapid-freeze, deep-etch, and rapid freezing/substitution techniques in rodents. These studies suggested that tubulo-vesicles are derived from the internalized distal ciliary membrane and/or the very basal OS plasma membrane (Miyaguchi and Hashimoto, 1992; Obata and Usukura, 1992). The occurrence of endocytosis at the base of rodent OS is further supported by a tracer experiment in which horseradish peroxidase was used to briefly pulse-label isolated adult rat retinas; only the tubules/cisternae below the stacked discs had peroxidase activity when they were examined by EM (Miyaguchi and Hashimoto, 1992). In fact, long tubule extensions connected to the basal OS plasma membrane, with profiles resembling the endocytosed membrane, were frequently detected in our survey (arrowheads, Figures S1B and S1C). It is probable that endocytosis gave rise to the vesicles or even primitive membranous sacs seen in the OS axoneme. Endocytosis has a long-recognized role in protein sorting. Such a process occurring at the OS basal membrane may provide an efficient mechanism to sort rhodopsin into disc membranes and segregate it from other proteins primarily targeted for the plasma membrane.
At present, however, we cannot rule out the possibility that some of the axonemal vesicles were directly shipped from the IS through the connecting cilium. SARA was also detected in the connecting cilium and basal body. Thus, the possibility that SARA is recruited by rhodopsin at the distal IS and serves as an adaptor protein participating in rhodopsin’s translocation through the connecting cilium remains open and warrants further investigation.
Molecular Machineries and Pathways Involved in Disc Renewal
Intracellular membrane trafficking involves budding, transport of vesicles, and fusion of vesicles from the donor membrane to their respective target membranes via the pairing between specific members of v-SNAREs (on the vesicles) and t-SNAREs (on the target organelles). Membrane targeting specificity is, in part, controlled by the spatially restricted expression patterns of specific SNARE members (Sollner et al., 1993). The t-SNAREs, such as syntaxin 3, would be generally predicted to be located on the targeting membranes. However, syntaxin 3 has been detected on both plasma membranes (Low et al., 1996; Sharma et al., 2006) as well as intracellular vesicles/organelles (Gaisano et al., 1996; Peng et al., 1997) in various cell types. Our immunostaining showed that in transfected cells, syntaxin 3 was distributed on both plasma membranes and early endosomes, indicating that syntaxin 3 could belong to a growing list of endosomal t-SNAREs (Prekeris et al., 1999; Wong et al., 1998) and may participate in the endocytic trafficking pathway. The expression of syntaxin 3 in photoreceptors appears to be tightly and spatially regulated: syntaxin 3 in the IS was found on both intracellular vesicles and plasma membranes, whereas it was absent from the plasma membrane of OSs but rather concentrated in the axonemal vesicles. While the molecular details of syntaxin 3’s action in the OS require future identification of its cognate partners, it is tempting to suggest that the strategic topological restriction of syntaxin 3 in the OS likely plays a central role in determining the sites where the membrane fusions occur. We further speculate that the syntaxin 3-mediated SNARE activity is employed for the “heterotypic fusion” between axonemal vesicles and nascent discs, the “homotypic fusion” of axonemal vesicles, or both (Figure 7A).
Our results suggested that SARA is a novel vesicle-tethering molecule that interacts with the membrane proteins rhodopsin and syntaxin 3 on axonemal vesicles. SARA may even be a regulator that pairs and acts coordinately with synatxin 3 in the mammalian OS. Almost all the vesicle-tethering proteins reported so far are either long coiled-coil proteins or scaffolds that form large protein complexes. No coiled-coil structure is found in SARA. However, ectopically expressed SARA appeared to drive its associated proteins (i.e., SMAD2/3) to form high-molecular-weight complexes in cultured cells (Jayaraman and Massague, 2000). Finally, many reported membrane tethers are downstream to the RabGTPase proteins, and SARA has been reported to act as an effector downstream to Rab5 in the endosomal trafficking of cultured cells (Hu et al., 2002). The specific Rab protein(s), if any, that are involved in disc biogenesis, however, await identification.
The high-affinity interaction between PI3P and the FYVE domain has predicted that PI3P is involved in the early endosomal recruitment of FYVE domain-containing proteins. Curiously, a significant fraction of the endogenous Vps34 proteins were also found on multivesicular endosome-like vesicles distinct from early endosomes (Stein et al., 2003). High levels of PI3P, detected by the GST-FYVEHrs, were also detectable on multivesicular endosomes in addition to its early endosomal location (Gillooly et al., 2000). In this regard, localization studies in the highly compartmentalized mammalian OSs were particularly intriguing. Using three independent methods (i.e., Vps34 localization, PI3P probe, and transfected FYVE probes), our data collectively suggested that PI3P is primarily distributed on the targeting membranes (i.e., nascent discs). In contrast, SARA was primarily enriched on cargo carriers (or axonemal vesicles). A mechanism may exist to regulate and recycle SARA back onto axonemal vesicles after fusion. Taken together with the functional analysis of FVYESARA-overexpressing rods, our data suggest that the high-affinity interaction between the FYVE domain of SARA and PI3P may provide a mechanism for the selective priming between the two heterotypic membranes. This concept is novel and distinct from the current prevailing view, based primarily on studies in vitro and in cultured cells, suggesting that the PI3P-FYVE domain interaction is used to recruit FYVE domain proteins to the PI3P-containing membrane compartments as effectors for homotypic membrane tethering (McBride et al., 1999).
Finally, our finding that the concentration of Vps34 and PI3P at the basal young discs, but not mature/aged discs, underscores the heterogeneity of disc membranes, both in their protein and lipid contents. The spatial and/or temporal control of PI3P generation may constitute, at least in part, a programmed timer for the maturation of discs.
Polarized OS Targeting of Rhodopsin and Retinal Diseases
Rhodopsin’s cytoplasmic C terminus has been shown to play an active role in guiding the postGolgi vesicles traveling through the IS via specific and distinct protein-protein interactions (Deretic et al., 2005; Tai et al., 1999). We now show that the rhodopsin C terminus is also utilized to recruit protein complexes for its delivery to the final destination, the disc membrane. The concept that rhodopsin vesicles provide “building blocks” of OS discs agrees well with the recently recognized structural role of rhodopsin (Humphries et al., 1997; Lem et al., 1999).
Previous studies showed that rhodopsin’s C terminus is a hot spot for mutations associated with autosomal dominant retinitis pigmentosa (RP), a progressive retinal degenerative disease. Several C-terminal RP rhodopsin mutants tested so far were mislocalized in transgenic rods (Li et al., 1996, 1998; Sung et al., 1994). Future studies would be of interest to test whether the targeting defect of any C-terminal RP mutant rhodopsin is a consequence of impaired binding to SARA.
A plethora of inherited retinal degenerative diseases have their manifestations in disc organization (Hagstrom et al., 1999; Hong et al., 2000). The molecular insight into OS development provided herein paves the way to begin revealing the basis of retinal degenerative diseases that involve defects in OS morphogenesis.
EXPERIMENTAL PROCEDURES
Protein-Protein Interaction Assays
Two-hybrid, immunoprecipitation, immunoblotting, and pull-down assays were carried out as described (Tai et al., 1999).
In Vivo Retinal Transfection
In vivo retinal transfection was carried out as described (Matsuda and Cepko, 2004). Briefly, plasmids were injected into the subretinal space of neonatal rats; electroporation using a pair of tweezer-type electrodes (BTX, Hawthorne, NY) that were placed across the eyes (cathode faces the sclera and the anode faces the cornea) immediately followed. Animals were housed in 12 hr light-dark cycle and harvested at postnatal day 21 under either the light- or dark-adapted conditions (Mendez et al., 2003). For the light microscopic studies of transfected retinas, eyecups were fixed overnight in 4% PFA/0.1% glutaraldehyde in 0.15 M cacodylate buffer (pH 7.4). Positive transfected areas were identified under fluorescent microscopy, dissected, and embedded in 5% low-melting agarose. Forty micrometer thick vibratome sections were subjected to immunolabeling and confocal microscopic analysis (Supplemental Data). At least three animals were examined for each experiment.
For reagents and animals, light microscopic and EM analyses of retina, and PI3P probe/PI3P localization study, see Supplemental Data.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Jonathan Backer, Connie Cepko, Thierry Galli, Paul Hargrave, Robert Molday, Paul Neilsen, Tom Sollner, Harald Stenmark, Thomas Weimbs, and Michael Yaffe for reagents and Drs. Francis Lee, Hui Sun, Tim McGraw, Anne Musch, and Tim Ryan for commenting on the manuscript. This work was supported by Foundation Fighting Blindness, Research to Prevent Blindness, The Irma T. Hirsch Trust, The Ruth and Milton Steinbach Fund, and NIH EY11307 to C.-H.S.
Footnotes
Supplemental Data
Supplemental Data include Experimental Procedures, 11 figures, Discussion, and References and can be found with this article online at http://www.cell.com/cgi/content/full/130/3/535/DC1/.
REFERENCES
- Blanks JC, Mullen RJ, LaVail MM. Retinal degeneration in the pcd cerebellar mutant mouse. II. Electron microscopic analysis. J. Comp. Neurol. 1982;212:231–246. doi: 10.1002/cne.902120303. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J. Comp. Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Chuang JZ, Vega C, Jun W, Sung CH. Structural and functional impairment of endocytic pathways by retinitis pigmentosa mutant rhodopsin-arrestin complexes. J. Clin. Invest. 2004;114:131–140. doi: 10.1172/JCI21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook NJ, Molday LL, Reid D, Kaupp UB, Molday RS. The cGMP-gated channel of bovine rod photoreceptors is localized exclusively in the plasma membrane. J. Biol. Chem. 1989;264:6996–6999. [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc. Natl. Acad. Sci. USA. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster FM, Traer CJ, Abraham SM, Fry MJ. The phosphoinositide (PI) 3-kinase family. J. Cell Sci. 2003;116:3037–3040. doi: 10.1242/jcs.00609. [DOI] [PubMed] [Google Scholar]
- Gaisano HY, Ghai M, Malkus PN, Sheu L, Bouquillon A, Bennett MK, Trimble WS. Distinct cellular locations of the syntaxin family of proteins in rat pancreatic acinar cells. Mol. Biol. Cell. 1996;7:2019–2027. doi: 10.1091/mbc.7.12.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom SA, Duyao M, North MA, Li T. Retinal degeneration in tulp1 −/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- Hong DH, Pawlyk BS, Shang J, Sandberg MA, Berson EL, Li T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3) Proc. Natl. Acad. Sci. USA. 2000;97:3649–3654. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chuang JZ, Xu K, McGraw TE, Sung CH. SARA, a FYVE domain protein, affects Rab5-mediated endocytosis. J. Cell Sci. 2002;115:4755–4763. doi: 10.1242/jcs.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, Sieving PA, Sheils DM, McNally N, Creighton P, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- Itoh F, Divecha N, Brocks L, Oomen L, Janssen H, Calafat J, Itoh S, Dijke PP. The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF-beta/Smad signalling. Genes Cells. 2002;7:321–331. doi: 10.1046/j.1365-2443.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Massague J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J. Biol. Chem. 2000;275:40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl. Acad. Sci. USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Franson WK, Gordon JW, Berson EL, Dryja TP. Constitutive activation of phototransduction by K296E opsin is not a cause of photoreceptor degeneration. Proc. Natl. Acad. Sci. USA. 1995;92:3551–3555. doi: 10.1073/pnas.92.8.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Snyder WK, Olsson JE, Dryja TP. Transgenic mice carrying the dominant rhodopsin mutation P347S: Evidence for defective vectorial transport of rhodopsin to the outer segments. Proc. Natl. Acad. Sci. USA. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wong F, Chang JH, Possin DE, Hao Y, Petters RM, Milam AH. Rhodopsin transgenic pigs as a model for human retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 1998;39:808–819. [PubMed] [Google Scholar]
- Low SH, Chapin SJ, Weimbs T, Komuves LG, Bennett MK, Mostov KE. Differential localization of syntaxin isoforms in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 1996;7:2007–2018. doi: 10.1091/mbc.7.12.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto B, Besharse JC. Light and temperature modulated staining of the rod outer segment distal tips with Lucifer yellow. Invest. Ophthalmol. Vis. Sci. 1985;26:628–635. [PubMed] [Google Scholar]
- McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- Mendez A, Lem J, Simon M, Chen J. Light-dependent translocation of arrestin in the absence of rhodopsin phosphorylation and transducin signaling. J. Neurosci. 2003;23:3124–3129. doi: 10.1523/JNEUROSCI.23-08-03124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaguchi K, Hashimoto PH. Evidence for the transport of opsin in the connecting cilium and basal rod outer segment in rat retina: rapid-freeze, deep-etch and horseradish peroxidase labelling studies. J. Neurocytol. 1992;21:449–457. doi: 10.1007/BF01191508. [DOI] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata S, Usukura J. Morphogenesis of the photoreceptor outer segment during postnatal development in the mouse (BALB/C) retina. Cell Tissue Res. 1992;269:39–48. doi: 10.1007/BF00384724. [DOI] [PubMed] [Google Scholar]
- Peng XR, Yao X, Chow DC, Forte JG, Bennett MK. Association of syntaxin 3 and vesicle-associated membrane protein (VAMP) with H+/K(+)-ATPase-containing tubulovesicles in gastric parietal cells. Mol. Biol. Cell. 1997;8:399–407. doi: 10.1091/mbc.8.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JJ, Tam BM, Moritz OL, Shelamer CL, Dugger DR, McDowell JH, Hargrave PA, Papermaster DS, Smith WC. Arrestin migrates in photoreceptors in response to light: a study of arrestin localization using an arrestin-GFP fusion protein in transgenic frogs. Exp. Eye Res. 2003;76:553–563. doi: 10.1016/s0014-4835(03)00032-0. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Yang B, Oorschot V, Klumperman J, Scheller RH. Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol. Biol. Cell. 1999;10:3891–3908. doi: 10.1091/mbc.10.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Seet LF, Hong W. Endofin, an endosomal FYVE domain protein. J. Biol. Chem. 2001;276:42445–42454. doi: 10.1074/jbc.M105917200. [DOI] [PubMed] [Google Scholar]
- Sharma N, Low SH, Misra S, Pallavi B, Weimbs T. Apical targeting of syntaxin 3 is essential for epithelial cell polarity. J. Cell Biol. 2006;173:937–948. doi: 10.1083/jcb.200603132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Gaullier JM, D’Arrigo A, Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stein MP, Feng Y, Cooper KL, Welford AM, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- Steinberg RH, Fisher SK, Anderson DH. Disc morphogenesis in vertebrate photoreceptors. J. Comp. Neurol. 1980;190:501–508. doi: 10.1002/cne.901900307. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R. FYVE-finger proteins-effectors of an inositol lipid. J. Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- Sung C-H, Makino C, Baylor D, Nathans J. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J. Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Chuang J-Z, Bode C, Wolfrum U, Sung C-H. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Whelan JP, McGinnis JF. Light-dependent subcellular movement of photoreceptor proteins. J. Neurosci. Res. 1988;20:263–270. doi: 10.1002/jnr.490200216. [DOI] [PubMed] [Google Scholar]
- Williams DS, Linberg KA, Vaughan DK, Fariss RN, Fisher SK. Disruption of microfilament organization and deregulation of disk membrane morphogenesis by cytochalasin D in rod and cone photoreceptors. J. Comp. Neurol. 1988;272:161–176. doi: 10.1002/cne.902720202. [DOI] [PubMed] [Google Scholar]
- Wong SH, Xu Y, Zhang T, Hong W. Syntaxin 7, a novel syntaxin member associated with the early endosomal compartment. J. Biol. Chem. 1998;273:375–380. doi: 10.1074/jbc.273.1.375. [DOI] [PubMed] [Google Scholar]
- Young RW. The renewal of photoreceptor cell outer segments. J. Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.