Abstract
Reproductive tract infection is a major initiator of preterm birth (PTB). The objective of this prospective cohort study of 88 participants was to determine whether PTB correlates with the vaginal microbiome during pregnancy. Total DNA was purified from posterior vaginal fornix swabs during gestation. The 16S ribosomal RNA gene was amplified using polymerase chain reaction primers, followed by chain-termination sequencing. Bacteria were identified by comparing contig consensus sequences with the Ribosomal Database Project. Dichotomous responses were summarized via proportions and continuous variables via means ± standard deviation. Mean Shannon Diversity index differed by Welch t test (P = .00016) between caucasians with PTB and term gestation. Species diversity was greatest among African Americans (P = .0045). Change in microbiome/Lactobacillus content and presence of putative novel/noxious bacteria did not correlate with PTB. We conclude that uncultured vaginal bacteria play an important role in PTB and race/ethnicity and sampling location are important determinants of the vaginal microbiome.
Keywords: PTB, vaginal microbiome, race/ethnicity
Introduction
Preterm birth (PTB; delivery < 37 weeks gestation) accounts for about 12% of United States births,1 with marked disparities of racial/ethnic PTB rates: African Americans 17.8%, caucasians 11.5%, and Asian/Pacific Islanders 10.5%.2,3 The 2012 World Health Organization global action report on PTB revealed that 15 million babies are “born too soon,” and prematurity is the world’s leading cause of neonatal death.3 It is also a major cause of neurodevelopmental/behavior disorders and other chronic morbidities.2,3 In 2005 in the United States, the cost of PTB was estimated at $26.2 billion.2 Thus, understanding the causes of PTB and attributes predisposing to PTB is a high priority in the United States and globally.2,3
Risk of PTB is multifactorial, with infection of the reproductive tract a major initiating factor.2 Intrauterine infection by known microbes is estimated in 25% of all the PTBs.4 Polymerase chain reaction (PCR) analysis of amniotic fluid from women in premature labor detected bacteria in 11% of the culture-negative samples.5 The PCR also revealed bacteria, including many uncultivated taxa, in 45% of the culture-negative amniotic fluid of participants with preterm premature rupture of membranes.6 Taken together, these studies suggest that bacteria in the vagina that do not grow in culture play an important role in the causation of PTB. Thus, we conducted a prospective cohort study of pregnant women at high and low risk of PTB and determined each participant’s vaginal microbiome and its association with PTB. We pursued a metagenomics approach, employing only a molecular technology and not culture, because it overcomes the limitations of unculturability and genomic diversity.
Methods
Participants
Participants were recruited after written informed consent under a protocol approved by the Committee on Human Research at the University of California, San Francisco (UCSF) and the Committee on the Use of Human Participants in Research at Stanford University. All participants were at UCSF. Of 113 women enrolled, 13 withdrew from participation and 88 had samples of sufficient quantity and quality for analysis. In total, 143 swabs were sequenced.
Inclusion criteria included pregnant women who had experienced ≥1 spontaneous PTBs, pregnant women who had one previous term pregnancy, nulliparious women, and women in the index pregnancy without significant comorbidities (see exclusion criteria in the subsequent paragraph); entry into the study during any trimester, 18 years of age or older, able to provide informed consent, willing to undergo serial speculum examinations and vaginal specimen collection with a swab, and willing to abstain 3 days from intercourse prior to specimen collection.
Exclusion criteria included pregnant women with symptoms or clinical evidence of vaginal, cervical, or urinary infection at first examination; significant vaginal bleeding and/or placenta previa or comorbidities of clinical evidence of urogenital infection, significant vaginal bleeding, chronic hypertension, diabetes, chronic renal disease, human immunodeficiency virus infection or other sexually transmitted infections; pregnant women with uterine abnormalities, including Müllerian anomalies and uterine septae; current multiple gestation; cervical insufficiency or previous PTB attributable to cervical insufficiency; antibiotics within 48 hours of sampling; <18 years old; or unable to provide informed consent.
Race/ethnicity was by self-identity as African American, Asian, Non-Hispanic white (caucasian), Hispanic (Latina), and Other (a heterogeneous group that included women of self-identified mixed ethnicity, Pacific Islander, native Hawaiian, Native American, and Alaska native).
Using the UCSF perinatal database, we estimated that 25% of the pregnant women with a history of unexplained spontaneous PTB (ie, not related to uterine anomalies or PTB induced for maternal morbidities) would experience another unexplained PTB. At the start of follow-up, we classified study participants with a history of unexplained PTB as “high-risk” of PTB and all other enrolled participants as “low-risk” of PTB (control). At the end of the follow-up, we classified births as preterm (<37 weeks of gestation), term, or spontaneous miscarriage (SAB).
Sampling and Sample Processing
Swabs of the posterior fornix of the vagina were obtained in each trimester (when possible) and flash frozen as described,7 deidentified, and transferred in a blinded fashion to the Stanford Genome Technology Center wheretotal DNA was purified employing a DNeasy Tissue and Blood Kit (Qiagen, Valencia, California). The final step was dialysis and concentration with Amicon Ultra Centrifugal Filters (EMD Millipore, Billerica, Massachusetts). To minimize bias in DNA extraction efficiency between gram-positive and gram-negative bacteria, the bacteria were first treated with lysozyme per the manufacturer’s instructions. If there were other (unknown) biases in the DNA extraction procedure, those biases would carry through our experiments. Each total DNA preparation for each swab was frozen in aliquots until use. One scientist (RWH) prepared all total DNAs. The swabs/DNAs were designated by their deidentified number followed by a hyphen and the trimester (-1, -2, and 3).
Chain-Terminator (Sanger) Sequencing of the 16S Ribosomal RNA Gene
An aliquot of total DNA from each swab was used as template in individual PCR reactions. The sequencing of the 16S ribosomal RNA Gene (rDNA) amplification primers were 8f and 1492r and employed as described.7,8 The 1.7-Kb product (nearly the complete gene) was purified by gel electrophoresis and cloned into a plasmid vector using a TA cloning kit from Invitrogen (Carlsbad, California). Chain-terminator (BigDye-terminator) chemistry was used for sequencing (Applied Biosystems [ABI], Foster City, California), as described.7,8 Sequencing reactions were run on an Applied Biosystems 3730xl DNA Analyzer. Average good-quality read length was >700 bases with some sequence reads >800 bases. From each vaginal swab, we achieved four 96-well plates of individual sequences (called “reads” or “sequence reads”). Individual sequence reads were trimmed for quality and vector sequences, assembled into contigs, and the contigs were hand edited as described7,8 except that the ABI BaseCaller software was used to identify bases instead of phred. 9 The software Bellerophon,10 as part of the Greengenes package,11 was used to identify and remove chimerasPlease provide supplier details for ABI BaseCaller, phred, Quantitative Insights into Microbial Ecology, and Bellerophon.. To minimize variation in these procedures, 1 scientist (MF) conducted all sequencing and processing of the sequence reads.
From Contig Consensus Sequence to Microbe
To identify bacteria, each contig consensus sequence was compared to the Ribosomal Database Project data.12–14 When the sequence match was ≥97% identity, the contig was given the name of the bacterium with the best sequence match, even when that name was “Uncultured bacterium.” When the best sequence match was <97% that bacterium was deemed “novel” and was given the name “Novel” plus the name of its closest sequence.15–17
Analyses
Dichotomous responses were summarized via proportions, and continuous variables were summarized via means and standard deviations (SDs). The primary outcome, PTB rate, was displayed as a function of important predictors to describe their relationships. In particular, the Shannon Diversity Index (SDI)18; was calculated for the microbiome of each vaginal swab (http://math.hws.edu/javamath/ryan/DiversityTest.html), and each participant’s SDI value was calculated by averaging the SDI values of all the participants’ available swabs. We report the mean (± SD) value of SDI per race/ethnicity category. We examined statistical differences among the mean SDI levels using Welch t test, which allows unequal variances.19 Chao120,21 Principal Coordinate Analysis and SDI analysis employed the “Quantitative Insights into Microbial Ecology” software (QIIME)22 ,23; with UniFrac distances.24
GenBank Accession Numbers
The putative novel rDNA sequences reported in this manuscript have been deposited in GenBank. Their accession numbers are JX871219 to JX871316.
Results
Participants and Pregnancy Outcomes
There were 46 high-risk and 42 low-risk (control) participants. Table 1 shows that of the low-risk participants, 55% were Caucasian, 12% were Asian, 12% were Hispanic, and 2% were African American, compared with 37%, 24%, 17%, and 15% of high-risk participants, respectively. Characterization of participant risk, ethnicity, pregnancy outcomes, and vaginal swabs obtained and sequenced in each trimester are in Supplemental Table S1. The number of participants in each self-identified racial/ethnic group (African American, Asian, Caucasian, Hispanic, and Other) is given in Supplemental Table S2.
Table 1.
Birth Outcomes in Relation to PTB Risk and Race/Ethnicity.a
| Risk | Outcome | Race/Ethnicity | Total No. (%) | ||||
|---|---|---|---|---|---|---|---|
| African American | Asian | Caucasian | Hispanic | Other | |||
| Low | PTB | 0 | 2 | 2 | 2 | 1 | 7 (17) |
| Term | 1 | 3 | 19 | 2 | 7 | 32 (76) | |
| SAB | 0 | 0 | 2 | 1 | 0 | 3 (7%) | |
| Total No. (%) | 1 (2%) | 5 (12%) | 23 (55%) | 5 (12%) | 8 (19%) | 42 (100%) | |
| High | PTB | 2 | 2 | 5 | 0 | 1 | 10 (22%) |
| Term | 4 | 9 | 11 | 8 | 2 | 34 (74%) | |
| SAB | 1 | 0 | 1 | 0 | 0 | 2 (4%) | |
| Total No. (%) | 7 (15%) | 11 (24%) | 17 (37%) | 8 (17%) | 3 (7%) | 46 (100%) | |
| All | PTB | 2 (25%) | 4 (25%) | 7 (18%) | 2 (15%) | 2 (18%) | 17 (19%) |
| Term | 5 | 12 | 30 | 10 | 9 | 66 (75%) | |
| SAB | 1 | 0 | 3 | 1 | 0 | 5 (6%) | |
| Total No. (%) | 8 (9%) | 16 (18%) | 40 (45%) | 13 (15%) | 11 (13%) | 88 (100%) | |
Abbreviations: PTB, preterm birth; SAB, spontaneous miscarriage.
a Risk factor for PTB: low, high; outcomes: term (= full term), PTB, and SAB (= miscarriage); race/ethnicity: Other = mixed ethnicity, Pacific Islander, native Hawaiian, Native American, and Alaska native.
The overall frequency of PTB was 19% (Table 1), higher than the national average of 12%, likely because many high-risk participants were included. The frequency of PTB was higher among participants classified as high versus low risk of PTB (22% vs 17%). The 95% confidence interval for the PTB frequency of low-risk participants is 7% to 31%. The frequency of PTB among low-risk participants, (7 of 42 = 17%), was not statistically significantly different from the national average of 12%. The PTB frequency also varied by race/ethnicity from a high of 25% among African American participants to a low of 15% among Hispanic participants (Table 1). These frequencies are in accord with the UCSF Perinatal Database and published data.1
The Vaginal Microbiome
Time and cost limit the amount of sequencing accomplished. Therefore, statistical methods (rarefaction analyses) have been devised to determine closeness of the data to saturation. Chao1 is one such method.20,21 Figure 1 presents the Chao1 analysis for each race/ethnicity. These curves suggest that saturation has not been reached for any race/ethnicity, and that, therefore, more taxa are present than accounted for within our depth of sequencing. The vaginal microbiomes for each trimester for all the participants are presented in Supplemental Table S3A. We have swabs in each of the 3 trimesters for 14 participants: 8 Caucasians, 2 Asians, 2 Hispanics, 1 African American, and 1 Other (Supplemental Table S3B). These include 2 PTBs, both in Caucasians. The vaginal microbiomes of both the PTB participants were dominated by Lactobacillus crispatus. For the Caucasian participants who carried to term, 3 vaginal microbiomes were dominated by L crispatus and 3 were dominated by Lactobacillus gasseri (except Atopobium dominated 1 third-trimester swab). The vaginal microbiomes of the 2 Hispanics and 1 Other were dominated by Lactobacillus iners.
Figure 1.
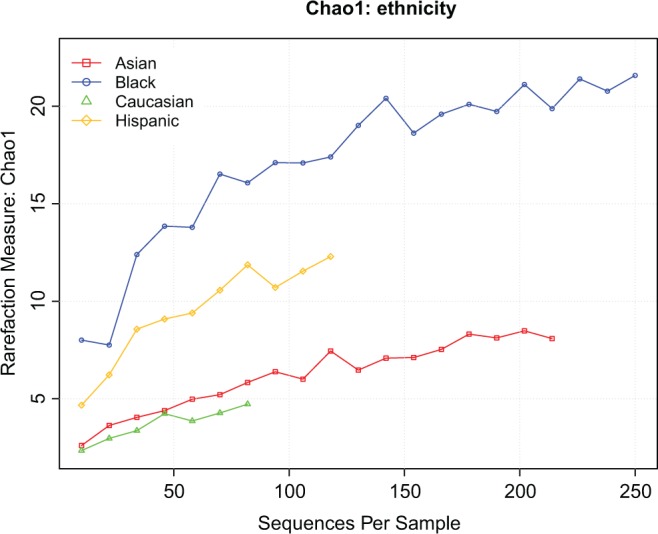
Chao1 analysis by race/ethnicity.
Lactobacillus Content as a Function of PTB Risk and Outcome
Lactobacillus is the most abundant bacterial genera in the healthy, adult vagina.25 For all bacteria except the genus Lactobacillus, the data were grouped by genera. For Lactobacillus, the bacteria were identified as species. The average percentage of Lactobacillus content of the vaginal swabs as a function of risk group and outcome is shown in Table 2. Lactobacillus content was higher among participants at low versus high risk of PTB (91.4% vs 80.7%) and did not distinguish PTB from term birth (86.2% vs 84.9%).
Table 2.
Lactobacillus Content (>1%) as a Function of PTB Risk and Outcome.a
| Risk | Outcome | Number of Participants | Average % | SD |
|---|---|---|---|---|
| Low | PTB | 7 | 80.6 | 29.7 |
| Term | 32 | 93.0 | 17.0 | |
| SAB | 3 | 100 | 0 | |
| Total | 42 | 91.2 | 14.9 | |
| High | PTB | 10 | 87.9 | 31.1 |
| Term | 34 | 79.8 | 31.6 | |
| SAB | 2 | 61.0 | 53.7 | |
| Total | 46 | 76.2 | 38.8 | |
| All | PTB | 17 | 84.9 | 29.8 |
| Term | 66 | 84.5 | 28.2 | |
| SAB | 5 | 84.4 | 34.3 | |
| Total | 88 | 84.6 | 28.5 |
Abbreviations: PTB, preterm birth; SAB, spontaneous miscarriage; SD, standard deviation.
a Risk factor for PTB: low, high; outcomes: term (= full term), PTB, and SAB (= miscarriage). Each participant’s Lactobacillus content was calculated by averaging the Lactobacillus content of all of the participant’s available swabs.
Race/ethnicity and Lactobacillus Content and Species Domination
The genus Lactobacillus was devolved into 19 species plus “uncultured Lactobacillus” and “novel Lactobacillus” (Table 3). Several species of Lactobacillus (eg, L crispatus, L iners, and L jensenii) were common in women of most or all races/ethnicities, although the vaginal microbiomes of pregnant African American and Hispanic participants were higher in L iners content. Caucasians had relatively higher L crispatus and L gasseri content, compared to other ethnicities. Putative novel species were identified in 9% and 5% of the swabs from Asian and Caucasian participants, respectively, but not from other ethnicities.
Table 3.
Lactobacillus Species Abundance as a Function of Race/Ethnicity.a
| Race/Ethnicity | |||||
|---|---|---|---|---|---|
| Species | African American (13 swabs) | Asian (23 swabs) | Caucasian (66 swabs) | Hispanic (19 swabs) | Other (17 swabs) |
| L acidophilus | 21.7 | 1.5 | |||
| L coleohominis | 4.3 | 1.5 | |||
| L crispatus | 30.8 | 26.1 | 41.5 | 15.8 | 41.2 |
| L delbrueckii | 4.3 | 5.9 | |||
| L fermentum | 1.5 | ||||
| L fornicalis | 6.2 | 5.3 | |||
| L gallinarum | 4.6 | ||||
| L gasseri | 7.7 | 17.4 | 41.5 | 10.5 | 17.6 |
| L helveticus | 3.1 | ||||
| L iners | 76.9 | 21.7 | 23.1 | 68.4 | 64.7 |
| L jensenii | 7.7 | 8.7 | 27.7 | 31.6 | 35.3 |
| L johnsonii | 5.3 | ||||
| L kefiranofaciens | 1.5 | 5.9 | |||
| L kitasatonis | 1.5 | ||||
| L reuteri | 5.9 | ||||
| L rhamnosus | 4.3 | ||||
| L sp | 1.5 | 10.5 | 5.9 | ||
| L ultunensis | 7.7 | ||||
| L vaginalis | 17.4 | 12.3 | 5.3 | 10.5 | |
| Novel | 8.7 | 4.6 | |||
| Unculturedb | 6 | 3 | 11 | 7 | 4 |
| Zero | 8 | 22 | 2 | 0 | 6 |
| Number of Participants | 8 | 16 | 40 | 13 | 11 |
Abbreviations: RDP, Ribosomal Database Project; Zero, no Lactobacillus.
a Percentage of swabs that contain, at least, 1% of the sequence reads supporting the named Lactobacillus species.
b Uncultured designated as “uncultured” in the Ribosomal Database Project. Novel matches nothing in the RDP at 97% or greater. However, the nearest named bacterium is Lactobacillus.
Some swabs contained only 1 species of Lactobacillus, more than 1 species of Lactobacillus, or no Lactobacillus (Table 4). From swabs of Asian participants, 22% were negative for all Lactobacillus species, compared to 0% to 8% for other race/ethnicities. A most striking difference was the vaginal microbiomes of pregnant African American participants, which contained <50% Lactobacillus on 46 + 8 = 54% of their swabs; whereas, from swabs of participants of other races/ethnicities, one species at more than 50% abundance was most often identified (63%-91%). Although the vaginal microbiome of Caucasian participants contained only 1 species of Lactobacillus on 93% of the swabs, the African American, Asian, and Hispanic vaginal microbiomes contained only 1 species of Lactobacillus in 69%, 70%, and 74% of the swabs, respectively.
Table 4.
Lactobacillus Species Domination by Race/Ethnicity.
| Race/Ethnicity | ||||||
|---|---|---|---|---|---|---|
| Species | African American (13 swabs) | Asian (23 swabs) | Caucasian (66 swabs) | Hispanic (19 swabs) | Other (17 swabs) | All (138 swabs) |
| One species, <50% | 46% | 0% | 2% | 11% | 0% | 7% |
| One species, >50% | 23% | 70% | 91% | 63% | 82% | 76% |
| >1 species, totals <50% | 0% | 4% | 0 | 0% | 0% | 1% |
| >1 species, totals >50% | 23% | 4% | 6% | 26% | 12% | 11% |
| Zero | 8% | 22% | 2% | 0% | 6% | 5% |
Absence or Minimal Lactobacillus and PTB
Since Lactobacillus is the most common bacterium in the healthy adult vagina, we determined whether there was a correlation between little or no Lactobacillus in the vagina and PTB. A total of 10 participants and 14 vaginal swabs evidenced little or no Lactobacillus (Supplemental Table S4). There was no correlation between the vaginal microbiome with little or no Lactobacillus and PTB.
Microbiome Changes Between Trimesters and PTB
We examined changes in microbiome content as a function of trimester (Supplemental Table S3). The 5 participants who had SABs did not progress past the first trimester and were excluded from the analysis. There were data for 7 transitions among participants with PTB, and none evidenced a change. There were data for 41 transitions among term participants, and 11 evidenced a change. Thus, there was no correlation between a change in vaginal microbiome content and PTB.
Putatively Novel and Noxious Bacteria and PTB
The presence of putatively novel bacteria in the vagina did not correlate with PTB, because they were found only in participants who carried to term (Supplemental Table S5). One participant (074) who had an SAB evidenced 33% of the sequence reads supporting a putatively novel bacterium closely related to Prevotella, and another had 100% of a putatively novel bacterium most closely related to Lactobacillus. We also examined the vaginal microbiomes for noxious bacteria, including Atopobium (Supplemental Table S6) and Prevotella (Supplemental Table S7). No participant who had a PTB harbored Atopobium (Supplemental Table S6). Other noxious bacteria were found in the vagina (Supplemental Table S8). Provocatively, 1 participant (103) with PTB had a vaginal microbiome with 97% Bifidobacterium, only 1 of 88 participants. Another with PTB (100) harbored a vaginal microbiome dominated by Ureaplasma and was the only one with this result. The data taken together suggest no correlation between noxious bacteria in the vagina and PTB, although larger studies focused on Bifidobacterium and Ureaplasma may be warranted relevant to PTB risk.
The Shannon Diversity Index and PTB
To measure the diversity of species within each vaginal microbiome, we employed the Shannon Diversity Index (SDI),18 a commonly used weighted measure of the different types of bacteria in an ecological niche. For example, when there is only one type of bacterium present, the SDI is zero and there is no diversity. The SDI was calculated from the data for each swab (Supplemental Table S9). The SDI value is given as a function of race/ethnicity in Table 5 and Supplemental Figure S1. The SDI indicated that species diversity was greatest among African American participants, followed by Hispanic and Other and was less diverse among Asians and Caucasians. The SDI for each racial/ethnic group is statistically significantly different from some of the others: for example Asian versus African American, P = .0045, by Welch t test (Table 5). Supplemental Figures S2 and S3 present principal coordinate analyses (PCoA) of race (Figure S2) and outcome (Figure S3), derived from the QIIME software pipeline.
Table 5.
Shannon Diversity Index (SDI) as a Function of Race/Ethnicity.a
| Number of Participants | SDI Mean (SD) | Pairwise P Values | |||||
|---|---|---|---|---|---|---|---|
| Race/Ethnicity | African American | Hispanic | Other | Asian | Caucasian | ||
| African American | 8 | 2.05 (1.03) | X | .11 | .023 | .0045 | .0030 |
| Hispanic | 13 | 1.28 (0.92) | X | .31 | .028 | .0082 | |
| Other | 11 | 0.94 (0.70) | X | .18 | .055 | ||
| Asian | 16 | 0.58 (0.56) | X | .46 | |||
| Caucasian | 40 | 0.46 (0.57) | X | ||||
Abbreviation: Other, mixed ethnicity, Pacific Islander, native Hawaiian, Native American, and Alaska native.; SD, standard deviation; SDI, Shannon Diversity Index.
a Each participant’s SDI value was calculated by averaging the SDI values of all of the participant’s available swabs. Statistical comparison: P value from Welch t test.
Because event rates were small, diminished by 6% of pregnancies ending in miscarriage as well as by the race/ethnicity diversity, it was not possible to disentangle the effects of PTB risk level and race/ethnicity on the birth outcome. Only among Caucasians were there sufficient participants for statistical analysis. Among these, 7 had a PTB, 3 had a miscarriage, and 30 carried to term, resulting in a PTB frequency of 7 of 40 = 17.5%. The SDI were 0.088 ± 0.082 for the 7 with PTB, and for the 30 controls who carried to term, SDI = 0.578 ± 0.608. Importantly, there was a statistically significant difference (P = .00016, by Welch t test) in the average SDI between Caucasian participants who had a PTB and those who carried to term. Figure 2 presents the QIIME 22,23 analysis of SDI for Caucasian participants by birth outcome (term birth or PTB).
Figure 2.
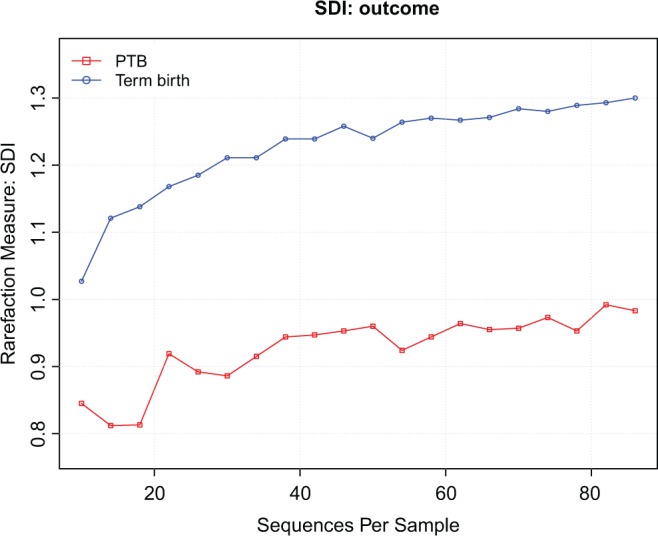
Shannon Diversity Index (SDI) for caucasians, separating preterm birth (PTB) from term birth.
To test the possibility that vaginal microbiome diversity may predict whether a pregnant woman will have a PTB, we performed Receiver–Operating Characteristic (ROC) curve analysis,26 based on a logistic regression model,27 for the prediction of participant outcome using SDI, race/ethnicity, and PTB history as predictors. The Area Under Curve (AUC) statistic 26 for this analysis was 0.70, which was significantly higher than when race/ethnicity was only used (AUC = 0.56), when only PTB history was used (AUC = 0.52), or when both ethnicity and PTB history were used (AUC = 0.59). Including Lactobacillus content as an additional predictor in the model had little effect (AUC = 0.71).
Discussion
The Vaginal Microbiome in Nonpregnant and Pregnant Women
Solely employing molecular technology, the vaginal microbiomes of healthy, nonpregnant women have been identified previously. Vaginal bacterial communities were placed into groups dominated by L crispatus, L gasseri, L iners, L jensenii, and a fifth group characterized by a much reduced Lactobacillus content.28 Importantly, the vaginal microbiomes of racial/ethnic groups could be distinguished from each other. In this context and in our results, recently published data from the Human Microbiome Project (HMP)29 are highly relevant. The HMP enrolled 113 healthy nonpregnant women and identified the microbiome at 18 sites, 3 in the vagina, including the posterior fornix. The HMP also found that the bacterial community composition of the posterior fornix was dominated by Lactobacillus, and that the vaginal microbiome correlated with race/ethnicity. Likewise, herein, the vaginal microbiome in pregnancy correlated with race/ethnicity.
The Vaginal Microbiome in Pregnancy
The vaginal microbiome during pregnancy has been investigated by several groups sampling from the posterior vaginal fornix, vaginal wall epithelium, or vaginal fluid at various times in gestation. For example, a preponderance of L acidophilus and L iners, but not L crispatus or L jensenii, were in the posterior fornix of pregnant women in Mexico City,30 and Lactobacillus ranged from 94% to 2% at delivery of 5 Mestizos and 2 Amerindian women.31 Lactobacillus species in the vaginal fluid of pregnant Japanese women with low Nugent scores detected using species-specific PCR included L crispatus 61%; L jensenii 30%; L gasseri 34%; and L iners 40%.32 Using Lactobacillus-specific culturing, L crispatus 25%; L gasseri 23%; L jensenii 15%; and L rhamnosus 11% were found in vaginal secretions of pregnant Viennese women.33 Lactobacillus species on the vaginal walls of pregnant Belgian women, detected by a combination of culturing and terminal restriction fragment length polymorphism patterns, revealed L crispatus only, 23%; L jensenii only 4%; and L gasseri/L iners (indistinguishable) only, 40%.34 In addition, more than 1 species of Lactobacillus (eg, L crispatus + L jensenii) was found in some women.34 Our results differ from some of these reports, likely due to different vaginal site sampling, race/ethnicity differences, patient personal habits (eg, douching), and other variables. Nontheless, we also detected a relatively high percentage of (separated) L gasseri and L iners in our Caucasian population.
Recently, Aagaard et al defined a “pregnant vaginal microbiome” at 3 sites, including the posterior fornix.35 They enrolled 24 pregnant participants, including 15 “non-Hispanic” women, 4 Hispanic, 3 African American, 2 Others, and no Asians and amplified 454 pyrosequenced a short, hypervariable sequence region of the 16 S ribosomal RNA gene. Two (8%)unidentified participants had a PTB. The data were not analyzed by race/ethnicity and the vaginal microbiomes of 60 control, nonpregnant women were drawn from the HMP. The depth of the sequencing was on average ∼ 6800 reads/sample that is ∼ 20-fold higher than that we achieved. Therefore, Aagaard et al35 identified the bacteria that we found and more, which is consistent with our Chao1 analysis (Figure 1), including Lactobacillales, Clostridiales, Bacteroidales, and Actinomycetales.
To compare the Aagaard data35 to ours, we consider only their identified most abundant bacteria (because of the large difference in coverage) for the posterior fornix of pregnant participants, assessed by randomForest script with Boruta selection (QIIME 1.3 Denoised; Supplemental Figure S2 in35). Lactobacillus crispatus and L iners were the most abundant bacteria, with lesser amounts of L reuteri, L jensenii, and Finegoldia magna. We found abundant L crispatus in the vaginal microbiome of our pregnant Caucasian participants and L iners abundant in the African-American and Hispanic participants. With our much lower depth of sequence, we found small amounts of L reuteri only in the heterogeneous category of Other (Table 3) and very little Fascioloides magna, for example, 2% in one vaginal swab and 3% in another (Supplemental Table S3A).
Strengths and Weaknesses of the Study
Strengths of this study include prospective cohort design, addressing whether the vaginal microbiome in pregnancy correlates with PTB, using a metagenomics approach, demonstrating that diversity of the vaginal microbiome correlates with PTB, that little or no Lactobacillus, a change in microbiome/Lactobacillus content, presence of putatively novel bacteria and noxious bacteria did not correlate with PTB, and race/ethnicity and location of sampling36 are important determinants of the vaginal microbiome in pregnancy. A major limitation of our study was its small sample size.
As with any technology, metagenomics has strengths and weaknesses. The principal strengths are that metagenomics overcomes the serious limitations of bacterial unculturability and genomic diversity. The weaknesses are related to the (1) the choice of PCR primers, (2) the depth of sequencing, (3)quantitation, and (4) the focus on the 16S rDNA sequencing, which precludes identification of other pathogens.
(1) The PCR primers employed to amplify the rDNAs are exactly matched to most such bacterial genes.37,38 However, there are known mismatches to some rDNAs, including some relevant to the vaginal microbiome, for example, Gardnerella vaginalis.39 The presence of mismatch will reduce priming efficiency, resulting in fewer sequence reads.
(2)Chain-terminator (Sanger) sequencing has great accuracy and long sequence reads (>700 bases per read). However, its slowness and expense restricted us to four 96-well plates of sequence reads per swab. We acknowledge that this sequencing method limited the number of samples sequenced due to cost. However, we chose chain-termination chemistry because it yields long, accurate reads that are very helpful in identifying putatively novel bacteria. Recently, Fettweis et al40 published a method for species identification from short 454 pyrosequencing reads of a small portion (V1-V3) of the 16S rDNA. This is a valuable approach and comparison with the nearly full-length 16S rDNA sequencing approach warrants further analysis.
As technologies have evolved, next-generation sequencing (NGS) would be pursued because the NGS read lengths have obtained significantly longer and greater than 100-fold. More sequence reads that can be achieved at a fraction of the cost with public algorithms are now available to identify and correct sequencing errors. In addition, with NGS, for eample, it is likely that more virulence genes would be identified (eg, as in G vaginalis and Sneathia amnii), and thus more bacteria, beyond those we detected, which confer pathogenicity may exist.
(3) Different bacteria have different copy numbers of rDNAs.41,42 The number of sequence reads per template is a function of the concentration of that template (eg, a bacterial genome with 20 copies of rDNA produces more sequences than a bacterial genome with 10 copies), which makes even a relative quantitation of the bacteria in a mixture tendentious.
Conclusions
This study demonstrates that the diversity of the vaginal microbiome during human pregnancy correlates with PTB, and that race/ethnicity and sampling site are important variables. The contributions of microbial communities to initiation of preterm labor and birth await further validation and hold promise for predicting PTB and early intervention to mitigate PTB and its sequelae.
Supplementary Material
Acknowledgements
We thank the study participants and M. Trebo for posting the data files on the SGTC website.
Footnotes
Authors’ Note: Supplemental tables and figures are available at http://rsx.sagepub.com/supplemental.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: March of Dimes Foundation (LCG ) and the Burroughs Wellcome Fund (RWD).
References
- 1. Martin JA, Hamilton BE, Ventura SJ, Osterman MJK, Wilson EC, Mathews TJ,. Births: Final Data for 2010. National Vital Statistics Reports, vol 61. www.cdc.gov/nchs/births.htm. Accessed August 1, 2012. [PubMed]
- 2. Behrman RE, Butler AS. Preterm Birth: Causes, Consequences, and Prevention. Washington, DC: National Academies Press; 2007. [PubMed] [Google Scholar]
- 3. World Health Organization, March of Dimes, The partnership for maternal, Newborn & Child Health, Save the Children. Born too soon: the global action report on preterm birth. 2012. whqlibdoc.who.int/publications/2012/9789241503433. Accessed February 25, 2012.
- 4. Gonçalves LF, Chaiworapongsa T, Romero R. Intrauterine infection and prematurity. Ment Retard Dev Disabil Res Rev. 2002;8(1):3–13. [DOI] [PubMed] [Google Scholar]
- 5. Gardella C, Riley DE, Hitti J, Agnew K, Krieger JN, Eschenbach D. Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor. Am J Perinatol. 2004;21(6):319–323. [DOI] [PubMed] [Google Scholar]
- 6. DiGiulio DB, Romero R, Kusanovic JP, et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol. 2010;64(1):38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci U S A. 2005;102(22):7952–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hyman RW, Herndon CN, Jiang H, et al. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet. 2012;29(2):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hyman RW, Jiang H, Fukushima M, Davis RW. A direct comparison of the KB Basecaller and phred for identifying the bases from DNA sequencing using chain termination chemistry. BMC Res Notes. 2010;3:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20(14):2317–2319. [DOI] [PubMed] [Google Scholar]
- 11. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7(6):e39315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wayne LG, Brenner DJ, Colwell RR, et al. Report of the ad-hoc-committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37(4):463–464. [Google Scholar]
- 16. Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276(5313):734–740. [DOI] [PubMed] [Google Scholar]
- 17. Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52(pt 3):1043–1047. [DOI] [PubMed] [Google Scholar]
- 18. Shannon CE. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- 19. Welch BL. The generalization of “Student’s” problem when several different population variances are involved. Biometrika. 1947;34(1-2):28–35. [DOI] [PubMed] [Google Scholar]
- 20. Chao A. Non-parametric estimation of the number of classes in a population. Scand J Statist. 1984;11:265–270. [Google Scholar]
- 21. Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Micro. 2001;67(10):4399–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caporaso JG, Kucynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011;Chap.10: Unit 10.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. White BA, Creedon DJ, Nelson KE, Wilson BA. The vaginal microbiome in health and disease. Trends Endocriinol Metab. 2011;22(10):389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(8):561–577. [PubMed] [Google Scholar]
- 27. McCullagh P, Nelder J. Generalized Linear Models. 2nd ed Boca Raton, FL: Chapman and Hall/CRC; 1989. [Google Scholar]
- 28. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hernández-Rodríguez C, Romero-González R, Albani-Campanario M, Figueroa-Damián R, Meraz-Cruz N, Hernández-Guerrero C. Vaginal microbiota of healthy pregnant Mexican women is constituted by four Lactobacillus species and several vaginosis-associated bacteria. Infect Dis Obstet Gynecol. 2011;2011:851485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamrakar R, Yamada T, Furuta I, et al. Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis. 2007;7:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kiss H, Kögler B, Petricevic L, et al. Vaginal Lactobacillus microbiota of healthy women in the late first trimester of pregnancy. BJOG. 2007;114(11):1402–1407. [DOI] [PubMed] [Google Scholar]
- 34. Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim TK, Thomas SM, Ho M, et al. Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol. 2009; 47(4):1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74(8):2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soergel DA, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16 S rRNA gene sequences. ISME J. 2012;6(7):1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Verhelst R, Verstraelen H, Claeys G, et al. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fettweis JM, Serrano MG, Sheth NU, et al. ; Vaginal Microbiome Consortium (additional members). Species-level classification of the vaginal microbiome. BMC Genomics. 2012;13(suppl 8):S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61(7):2798–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66(4):1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


