Summary
DNA methylation and histone H3 lysine 9 trimethylation (H3K9me3) play important roles in silencing of genes and retroelements. However, a comprehensive comparison of genes and repetitive elements repressed by these pathways has not been reported. Here we show that in mouse embryonic stem cells (mESCs), the genes up-regulated following deletion of the H3K9 methyltransferase Setdb1 are distinct from those de-repressed in mESC deficient in the DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b, with the exception of a small number of primarily germline-specific genes. Numerous endogenous retroviruses (ERVs) lose H3K9me3 and are concomitantly de-repressed exclusively in SETDB1 knockout mESCs. Strikingly, ~15% of up-regulated genes are induced in association with de-repression of promoter proximal ERVs, half in the context of “chimaeric” transcripts that initiate within these retroelements and splice to genic exons. Thus, SETDB1 plays a previously unappreciated yet critical role in inhibiting aberrant gene transcription by suppressing the expression of proximal ERVs.
Introduction
Transcription in eukaryotes is influenced by a wide variety of chromatin-associated factors that affect nucleosome structure and/or positioning and in turn accessibility of RNA polymerases to DNA (Kouzarides, 2007). DNA methylation plays an important role in reinforcing the silent state of a subset of tissue-specific and imprinted genes (Fouse et al., 2008), as well as repetitive elements (Walsh et al., 1998). This epigenetic mark is established primarily by the de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B, which are most active in the germline and in early embryogenesis, and maintained by the related DNMT1 (Law and Jacobsen, 2010). While DNA methylation in promoter regions is associated with transcriptional silencing, the presence of this mark in the gene body can also influence elongation efficiency (Lorincz et al., 2004), splicing (Chodavarapu et al., 2010) and initiation from intragenic alternative promoters (Maunakea et al., 2010).
Post-translational histone modifications on the other hand, can act either to promote or inhibit transcription depending on the histone residue modified, the nature of the modification and the position of the marked nucleosome relative to the transcription start site (TSS). A subset of these covalent histone modifications influence transcription in part by promoting or inhibiting de novo DNA methylation. Methylation of histone H3 on lysine 4 (H3K4) for example, inhibits binding of the germline-specific DNMT3A2/B cofactor DNMT3L to peptides corresponding to the amino-terminus of histone H3, indicating that this mark may protect specific genomic regions against DNA methylation and associated transcriptional repression (Ooi et al., 2007). Indeed, a recent study combining chromatin immunoprecipitation (ChIP) with genome-wide DNA methylation analyses revealed that H3K4me2 and H3K4me3 are anti-correlated with DNA methylation in mESCs and differentiated cells (Mohn et al., 2008).
In contrast, in plants and filamentous fungi, the H3K9 specific lysine methyltransferases (KMTases) KYP and DIM5 respectively, are required for de novo DNA methylation, and deletion of the genes encoding these KMTases leads to reactivation of the repetitive elements marked by H3K9 and DNA methylation (Freitag and Selker, 2005). In mice, deletion of the H3K9 KMTases SUV39H1 and SUV39H2 leads to a loss of both H3K9me3 and DNA methylation at major satellite repeats (Lehnertz et al., 2003). Similarly, the H3K9 KMTase G9a, which is responsible for H3K9me2 in euchromatin, is required for DNA methylation at a subset of genes and repetitive elements in mESCs (Dong et al., 2008). Similarly, deletion of Dnmt1 or Dnmt3a and Dnmt3b has no effect on H3K9me3 of pericentromeric heterochromatin in mice (Lehnertz et al., 2003). While these observations indicate that H3K9 KMTases can act upstream of DNMTs in specific genomic contexts, DNA methylation at genic promoters is only weakly correlated with H3K9me2 or H3K9me3 in somatic cells and/or mESCs (Edwards et al., 2010; Yokochi et al., 2009), indicating that H3K9 methylation may generally act independently of DNA methylation to negatively regulate gene expression.
Unlike genic promoter regions, class I and II ERVs are both densely DNA methylated and marked by H3K9me2/3 in mESCs (Dong et al., 2008). Recently, we showed that the H3K9 KMTase SETDB1 (ESET/KMT1E), which plays an important role in stem cell maintenance (Bilodeau et al., 2009; Yuan et al., 2009), is required for H3K9me3 marking and silencing of several ERV subfamilies in mESCs (Matsui et al., 2010). Surprisingly, the overall level of DNA methylation at these ERVs was unchanged or only modestly reduced in Setdb1 conditional knockout (SETDB1 KO) cells. Conversely, H3K9me3 at these elements was not reduced in Dnmt1/Dnmt3a/Dnmt3b triple knockout (DNMT TKO) mESCs, nor was transcription comparably induced, indicating that SETDB1 functions independently of DNA methylation in these cells. However, a comprehensive genome-wide comparison of the role of SETDB1 versus DNA methylation in transcriptional silencing of genes and ERVs has not been performed.
To identify those genes and/or repetitive elements regulated by DNA methylation and/or SETDB1 genome-wide, and to determine whether SETDB1-mediated deposition of H3K9me3 and associated transcriptional silencing is perturbed in the absence of DNA methylation, we conducted RNA-seq and H3K9me3 Native-ChIP (NChIP)-seq experiments on SETDB1 KO, DNMT TKO and corresponding WT mESCs. We show that disrupting these two epigenetic pathways results in the de-repression of predominantly distinct sets of genes and repetitive elements in mESCs. Furthermore, depletion of Setdb1 leads to widespread reactivation of class I and II ERVs and unexpectedly, to the aberrant expression of numerous chimaeric RNAs that originate in such ERVs and splice to canonical genic exons.
Results
Genome-wide profiling of gene expression in SETDB1 KO and DNMT TKO mESCs
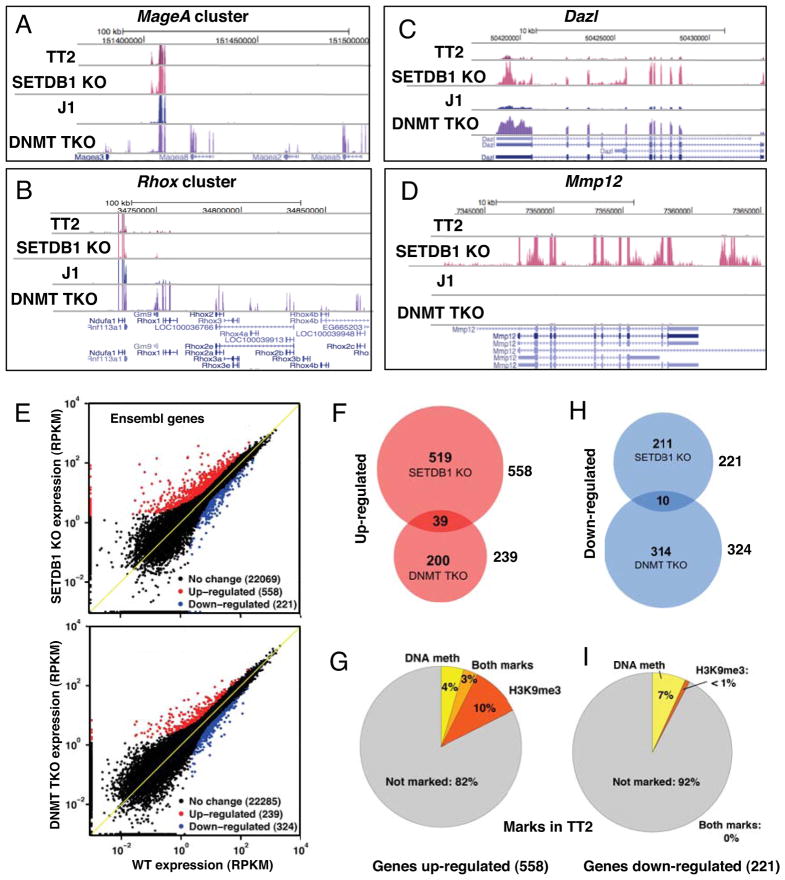
We isolated mRNA from SETDB1 KO (Matsui et al., 2010) and DNMT TKO (Tsumura et al., 2006) mESCs and their parent lines TT2 and J1, respectively and performed RNA-seq as described previously (Morin et al., 2008). Greater than 20M paired-end reads for each cell line were aligned to mouse genome and transcriptome resources (see Experimental Procedures and Figure S1A). Several genes within the MageA and Rhox gene clusters reported previously to be DNA methylated and repressed in mESCs, including MageA5, MageA8, Rhox2 and Rhox4 (Fouse et al., 2008; Oda et al., 2006), as well as the MageA4 and Rhox1 genes, were de-repressed in the DNMT TKO line (Figure 1A–B). None of these genes were de-repressed in the SETDB1 KO line. In contrast, the germline-specific gene Dazl was de-repressed in both the DNMT TKO and SETDB1 KO lines, while the macrophage-specific gene Mmp12 was de-repressed exclusively in the SETDB1 KO line (Figure 1C–D). Consistent with these observations, de-repression of Dazl and Mmp12 was reported previously in an independently derived SETDB1 deficient mESC line (Bilodeau et al., 2009). De-repression of Dazl in both KO lines was validated by qRT-PCR (data not shown), confirming that for a subset of genes, disruption of either pathway is sufficient for transcriptional activation.
Figure 1. SETDB1 and DNA methylation are required for silencing of predominantly distinct sets of genes.
RNA-seq was performed on SETDB1 KO and DNMT TKO mESCs and their parent lines TT2 and J1, respectively. A–D. UCSC genome browser (mm9) screen shots showing mRNA levels across the MageA and Rhox gene clusters, as well as the germline-specific gene Dazl and the Mmp12 gene. E. Two-dimensional plots of all protein-coding Ensembl genes (22,848 total) with non-zero read coverage in either WT or KO lines are shown. Up- and down-regulated genes showing Z-score ≥ 1.2 and fold-change ≥ 2.0 are highlighted. F. The overlap in up-regulated genes is shown, along with G. the fraction of genes up-regulated in the SETDB1 KO line that are marked in the TT2 line by DNA methylation (Myant et al., 2011) and/or H3K9me3 in the promoter region (TSS +/−500bp). H–I. Similar analyses are shown for the down-regulated genes (see Figure S1 and S2).
To characterize gene expression patterns in the mutant and WT RNA-seq datasets, we generated reads per kilobase per million mapped reads (RPKM) (Mortazavi et al., 2008) values for every annotated exon for all ENSEMBL protein-coding genes (22,848 total). In the SETDB1 KO and DNMT TKO lines, 558 (2.4%) and 239 (1.0%) genes were found to be de-repressed respectively, applying combined thresholds based on Z-score (>1.2) and fold-change (≥2) (Figure 1E and Table S1). While ~17% of genes reported previously via expression microarray to be up-regulated >2-fold in a related DNMT deficient ES line (Fouse et al., 2008) were also scored as up-regulated in our RNA-seq analysis, the majority were not. This likely reflects the greater specificity of high-throughput sequencing (Marioni et al., 2008) under the stringent threshold applied (Fig. S1B).
Strikingly, only 7.0% (39/558) of genes de-repressed in the SETDB1 KO line were also de-repressed in the DNMT TKO line (Figure 1F and Table S1), and gene ontology (GO) analysis of the genes up- or down-regulated in the DNMT TKO and SETDB1 KO lines revealed that none of the GO terms identified are common to both KO lines (Figure S1C–D and Table S2). Furthermore, analysis of a recently published DNA methylation dataset revealed that only 7% of the promoter regions of genes up-regulated in the SETDB1 KO are DNA methylated in the WT TT2 line (Myant et al., 2011) (Figure 1G), indicating that SETDB1 and the DNMTs are required for silencing of predominantly distinct sets of genes.
To determine the genome-wide distribution of H3K9me3, and whether this mark is perturbed in the absence of SETDB1 and/or DNA methylation, we conducted NChIP-seq (O’Neill and Turner, 2003) on the SETDB1 KO and DNMT TKO lines as well as their parent lines, using an antibody specific for H3K9me3 (Figure S2A–B). ~255Mb (13%) or ~215Mb (11%) of the mappable mouse genome (analyzed in 800 bp bins) is marked by H3K9me3 in the TT2 and J1 parent lines, respectively. While >50% of H3K9me3 marked regions lost this mark in the SETDB1 KO line, only ~15% did so in the DNMT TKO line (data not shown). Fewer than 1% of the 221 genes down-regulated in the SETDB1 KO line are marked by H3K9me3 in the promoter region in WT cells (Figure 1H–I), implicating SETDB1 predominantly as a transcriptional repressor, as expected. Surprisingly however, only 13% of the promoter regions of genes up-regulated in the SETDB1 KO line are marked by H3K9me3 in the WT line (Figure 1I), revealing that only a minority of induced genes are direct targets of SETDB1.
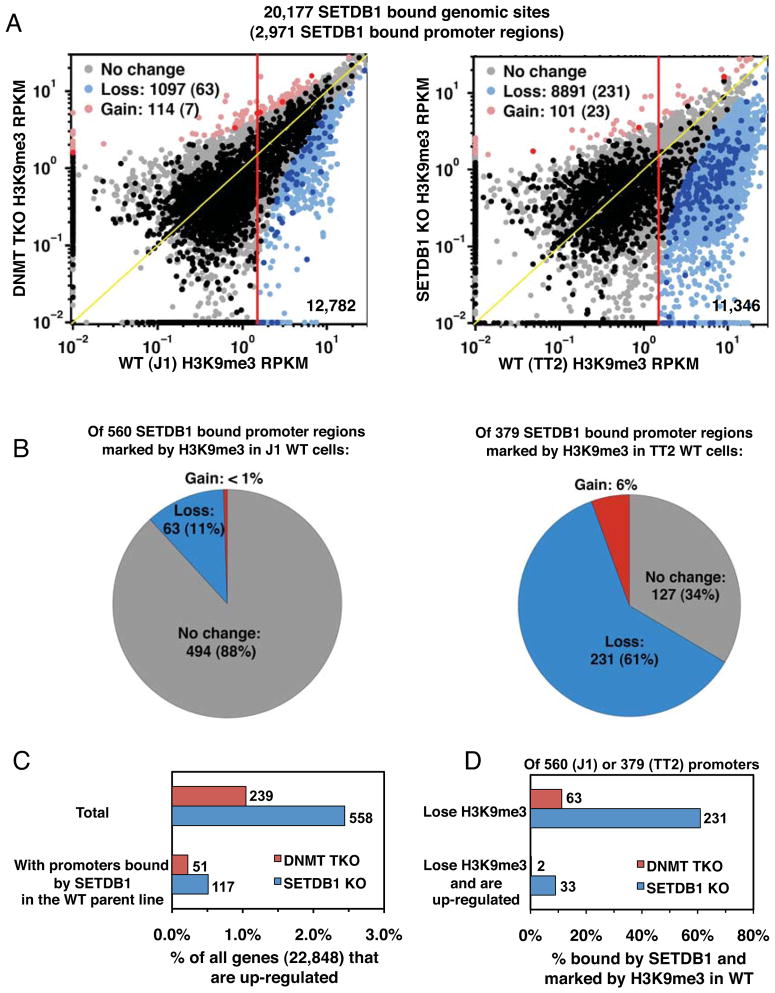
To focus specifically on direct genomic targets of this H3K9 KMTase, we realigned a previously reported mESC SETDB1 ChIP-seq dataset (Yuan et al., 2009) to the genome and identified 20,177 high confidence SETDB1 binding sites using FindPeaks (Fejes et al., 2008). Of these, 67.3% and 64.8% were marked by H3K9me3 in TT2 and J1 ES cells, respectively, comparable to the 65.3% of these sites marked by H3K9me3 in the original study (Yuan et al., 2009) (Figure S2C). A 3-way comparison between the parental mESC lines yielded 87–93% overlap (11,100 common sites) between H3K9me3 enriched regions at SETDB1 binding sites, revealing that our NChIP data are highly correlated with those generated by Yuan et al.
Genome-wide analysis of H3K9me3 enrichment at all SETDB1 bound regions, measured in terms of RPKM, revealed that while only 9% (1,097/12,782) of H3K9me3 marked sites are lost in the DNMT TKO line, 78% (8,891/11,346) of sites are lost in the SETDB1 KO (Figure 2A). Similarly, analysis of all SETDB1 bound promoter (+/−500bp of the TSS) regions marked by H3K9me3 revealed that 11% and 61% lost this mark in the DNMT and SETDB1 KO lines, respectively (Figure 2A–B). Taken together, these results indicate that SETDB1-mediated deposition of H3K9me3 is generally not dependent upon the presence of DNA methylation.
Figure 2. SETDB1 bound loci are depleted of H3K9me3 in SETDB1 KO but not DNMT TKO cells.
A. H3K9me3 RPKM values at genomic (light shading) or promoter (heavy shading) regions bound by SETDB1 are plotted for DNMT TKO vs. J1 and SETDB1 KO vs. TT2 lines and the number of genomic sites or promoter regions (in parentheses) losing or gaining H3K9me3 in the KO lines is shown. B. The number and percentage of SETDB1 bound, H3K9me3 marked promoter regions losing, gaining or showing no change in H3K9me3 in DNMT TKO and SETDB1 KO lines is shown. C. The percent and number of all genes or genes bound by SETDB1 in their promoter regions that are up-regulated are shown for each KO line. D. The percent and number of genes with SETDB1-bound promoters that lose H3K9me3 and are up-regulated in each KO line are shown (see Figure S1 and S2).
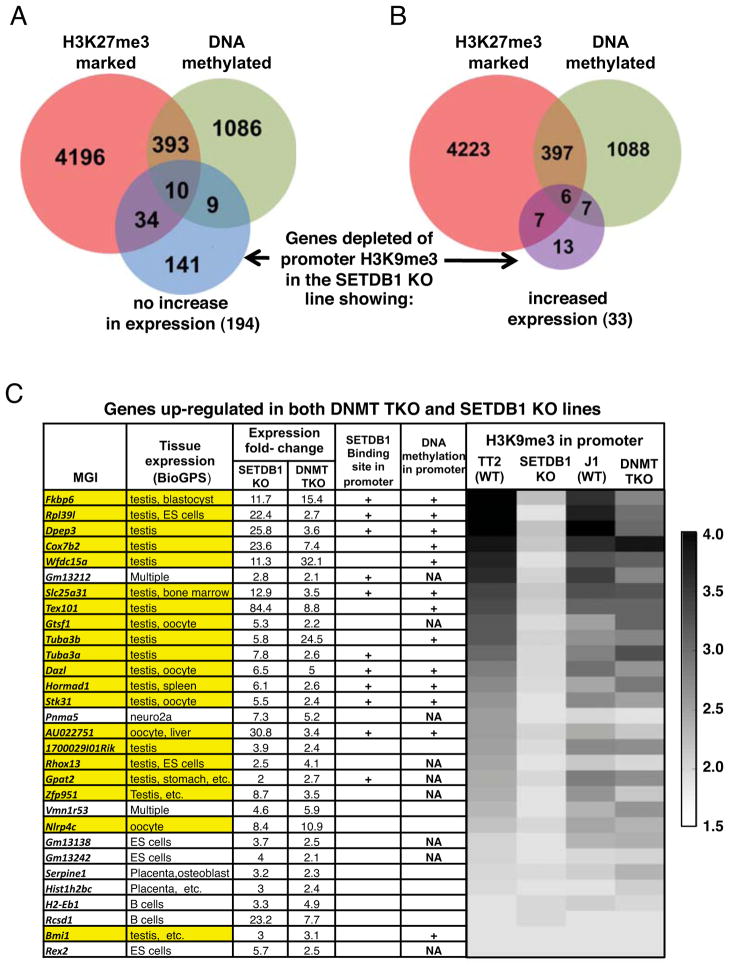
Integration of the RNA-seq and ChIP-seq datasets revealed that SETDB1 is bound to the promoter regions of only 21% (117 of 558) of up-regulated genes in wildtype (WT) cells (Figure 2C), confirming that the majority of such genes are induced as a result of downstream effects of SETDB1 loss. Surprisingly, of the 231 genes bound by SETDB1 in their promoter regions that lose H3K9me3 in the SETDB1 KO line, 86% are not up-regulated (Figure 2D). Analysis of the promoter regions of 194 of these 198 gene promoters for which DNA methylation data are available (Myant et al., 2011) reveals that only 9.8% are DNA methylated in the TT2 line (Figure 3A), similar to the 9.8% (1,498/15,252) of all ENSEMBL protein coding gene promoters that are methylated. Furthermore, analysis of previously published ChIP-seq data (Mikkelsen et al., 2007) reveals that only 22.7% of these promoter regions are marked by H3K27me3 (Figure 3A). Thus, the majority of these H3K9me3 marked genes are not marked concurrently by DNA methylation or H3K27me3 in WT mESCs.
Figure 3. Genes depleted of promoter H3K9me3 in the SETDB1 KO are generally not marked by DNA methylation or H3K27me3.
The DNA methylation (Myant et al., 2011) and H3K27me3 (Mikkelsen et al., 2007) states of genes depleted of H3K9me3 in their promoter regions (TSS +/−500bp) in the SETDB1 KO line A. showing no increase or B. increased expression, are shown. C. The tissue specificity of genes represented in the BioGPS database that are de-repressed in both the SETDB1 KO and DNMT TKO lines (30 of 39 total) is shown, along with the DNA methylation (Myant et al., 2011), H3K9me3 and SETDB1 binding (Yuan et al., 2009) states in the promoter regions of these genes (see Figure S2, Tables S1 and S2). Genes highlighted in yellow are expressed in the germline. NA, promoters of MGI gene not represented in the DNA methylation dataset.
In contrast, of the 33 genes that lose H3K9me3 and are concomitantly de-repressed in the SETDB1 KO line, 40.6% are also DNA methylated in TT2 cells (Figure 3B), prompting us to analyze the 39 genes de-repressed in both KO lines (see Fig. 1F) in greater detail. Strikingly, 20 of the 30 genes de-repressed in both KO lines for which gene expression information is available in the BioGPS database (GNF1M Gene Atlas data set) (Wu et al., 2009) are expressed in testis and/or oocytes (Figure 3C and Table S1). In TT2 cells, 18 of these germline-specific genes are marked by H3K9me3 in their promoter regions, all but one of which lose this mark in the SETDB1 KO, indicating that they are direct SETDB1 targets. Furthermore the presence of 13 SNPs in the promoter region of the Tuba3a gene allowed us to confirm that both alleles are marked by H3K9me3. Indeed, 18 and 11 reads from the TT2 H3K9me3 dataset definitively mapped to the C57BL/6 and CBA alleles, respectively. The majority of these genes are also DNA methylated in TT2 cells (Myant et al., 2011) and many show reduced H3K9me3 in DNMT TKO cells (Figure 3C). Taken together, these results indicate that DNA methylation and H3K9me3 act in cis at a specific set of germline-specific genes in mESCs and play critical non-redundant roles in silencing of these genes.
Genome-wide profiling of ERV expression and H3K9me3
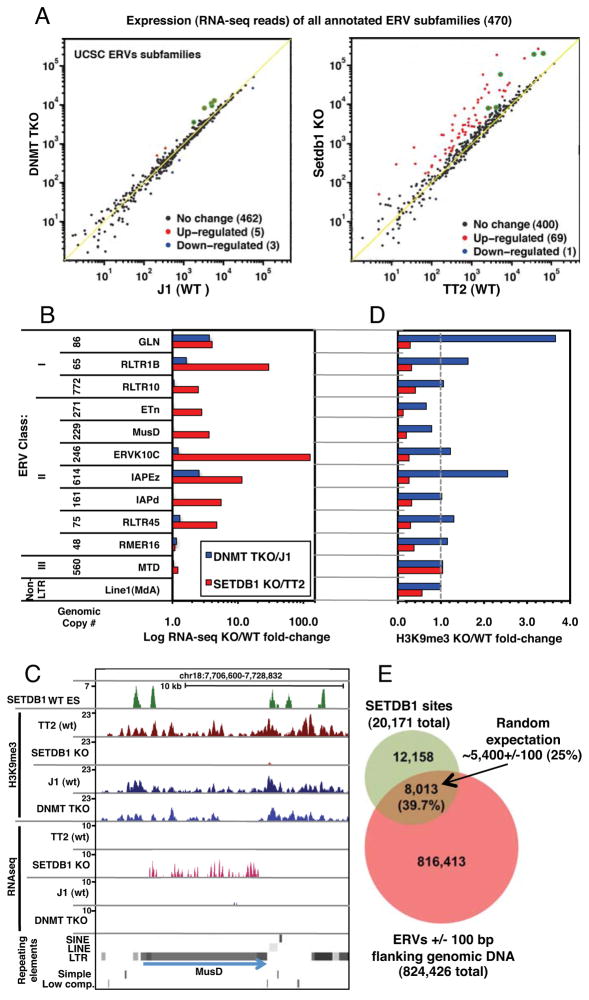
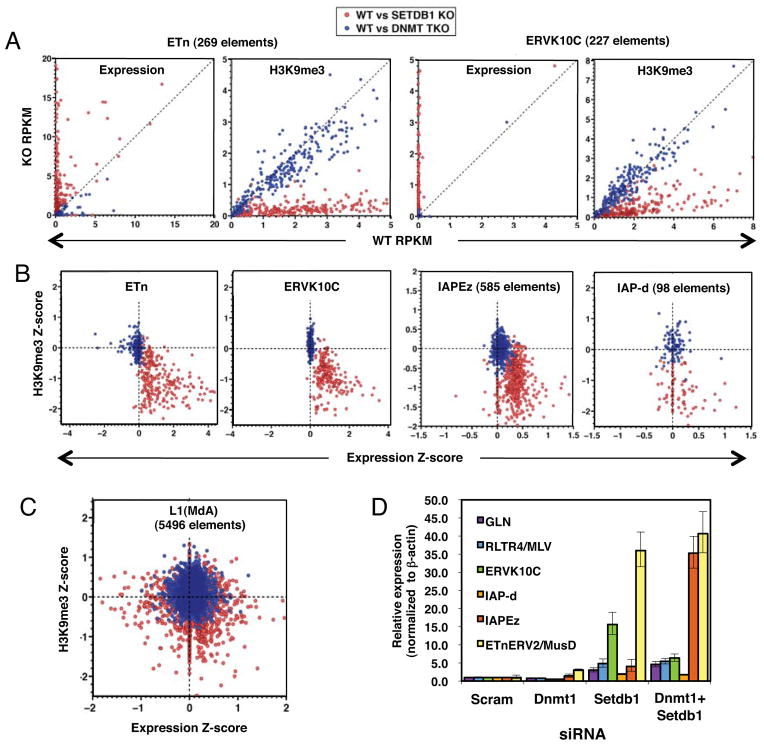
The observation that a relatively small number of genes are de-repressed as a direct result of SETDB1-deposited H3K9me3 at genic promoters indicates that this KMTase may be principally engaged in repressing non-coding and/or repetitive elements in mESCs. Previously, we showed that SETDB1 plays a more important role in silencing of several subfamilies of ERVs in mESCs than does DNA methylation (Matsui et al., 2010). However, we did not address whether increased proviral expression was the result of activation of a limited number of specific ERVs, or disseminated reactivation of multiple ERVs within each subfamily. To distinguish between these two possibilities and to expand our analysis to include all annotated subfamilies of ERVs, we determined the relative RNA levels in the KO and parental mESC lines of all Repbase (Jurka et al., 2005) annotated ERVs. Strikingly, while 69 ERV subfamilies were de-repressed in the SETDB1 KO line, only 5 were de-repressed in the DNMT TKO line (Figure 4A) and 4 of the latter were de-repressed to a greater extent in the SETDB1 KO line. Analysis of uniquely mapped reads aligning to the annotated internal regions of all ERV subfamilies present at >50 copies in the genome revealed that between 4 and 20% of all genomic copies of 10 Class I or II ERVs subfamilies, including RLTR1B, GLN, ERVK10C, ETn, ETnERV, MMTV, ETnERV2/MusD, RLTR45, IAP-d, and RLTR10, were de-repressed in the SETDB1 KO line (Table S3). Reactivation of a subset of these ERVs was confirmed by qRT-PCR (Figure S3A). In contrast, no ERV subfamily showed reactivation of ≥4% of genomic copies in DNMT TKO cells. Summing the total normalized RNA-seq coverage over “intact” ERVs (annotated internal regions flanked by their cognate LTRs) confirmed that the majority of these elements were significantly de-repressed exclusively in the SETDB1 KO line (Figure 4B and S3B), indicating that the difference observed between the two KO lines is unlikely to be due to polymorphisms in mapped ERVs between mouse strains (Zhang et al., 2008). A similar trend was observed when only uniquely aligned reads were considered (Table S3).
Figure 4. ERVs are de-repressed in SETDB1 KO but not DNMT TKO mESCs.
A. The sum of RNA-seq reads aligned to each annotated ERV subfamily was normalized to the total number of exonic reads and plotted for SETDB1 KO vs. TT2 and DNMT TKO vs. J1 lines. ERV subfamilies up or down-regulated in the KO lines are shown in red and blue, respectively. Subfamilies up-regulated in both lines are highlighted in green. B. For analysis of intact ERVs, the total normalized RNA-seq coverage for all annotated ERV internal regions flanked by their cognate LTRs was determined for representative class I, II and III ERV subfamilies, as well as LINE1MdA elements. The fold-change in expression for each pair of cells lines is shown. C. A screen shot of a representative ETnERV2/MusD element, including H3K9me3 NChIP-seq, RNA-seq and SETDB1 ChIP-seq (Yuan et al., 2009) tracks, is shown. D. The fold-change in H3K9me3 (including 1 kb of flanking genomic sequence) relative to the parent line for each subfamily presented in panel B is shown. E. The overlap between all annotated ERVs (+/−100 bp of flanking sequence) and mapped SETDB1 binding sites (threshold height >8) reveals that ~40% of all SETDB1 binding sites map within or near an annotated ERV (see Figures S3, S4 and Table S3). Random expectation of ~25% is based on 20 bootstraps (p-value <0.05).
To determine whether reactivation of these ERVs was accompanied by loss of H3K9me3 in cis, we analyzed the H3K9me3 status of all annotated ERVs. Inspection of full-length elements revealed that H3K9me3 frequently spreads at least 1 kb into flanking genomic DNA (Figure 4C and Figure S4), as previously described (Mikkelsen et al., 2007). Scoring H3K9me3 across ERVs and 1 kb into their flanks revealed a consistent and dramatic decrease in this repressive mark exclusively in the SETDB1 KO line (Figure 4D). In fact, enrichment of this mark is increased at several ERVs in the DNMT TKO line. Strikingly, analysis of the SETDB1 ChIP-seq dataset described above (Yuan et al., 2009) revealed that ~40% of the 20,171 SETDB1 binding sites in the mouse genome overlap with, or occur within 100 bp of an annotated ERV, a significantly greater number than predicted based on random expectation (Figure 4E). This likely significantly underestimates the true overlap, as ChIP-seq reads that map to sites within multi-copy ERVs that show no sequence variation are excluded from the analysis.
We next analyzed the expression and H3K9me3 states of individual full-length ERVs, considering only uniquely aligned reads (Table S4). Analysis of a subset of the class I and II ERV subfamilies de-repressed in the SETDB1 KO line revealed a significant increase in expression and loss of H3K9me3 at the majority of elements in the SETDB1 KO, but only modest changes in expression and H3K9me3 in the DNMT TKO line in all cases (Figures 5A and S5). Plotting expression vs. H3K9me3 levels of the parental and KO mESC lines for each of these ERV subfamilies revealed a strong correlation between loss of H3K9me3 and induction of ERV expression (Figure 5B and Figure S5), although not all of the ERVs depleted of H3K9me3 showed increased expression, perhaps due to the fact that a number of these elements are transcriptionally inert. In contrast, representative Class III ERVs and non-LTR LINE1 elements were generally not marked by H3K9me3 (consistent with a previous report (Mikkelsen et al., 2007)), nor de-repressed in either KO line (Figure 5C and Figure S5).
Figure 5. Class I and II ERVs are simultaneously de-repressed and lose H3K9me3 exclusively in SETDB1 KO mESCs.
Unambiguous RNA-seq and ChIP-seq reads aligning to ERVs with internal regions flanked by their cognate annotated LTRs were assembled as described in the Supplemental Information. A. RNA-seq and H3K9me3 RPKM values for ETn and ERVK10C ERVs are shown for TT2 vs. SETDB1 or J1 vs. DNMT TKO lines. B. Plotting H3K9me3 vs. RNA-seq Z-scores reveals that numerous ERVs lose H3K9me3 and are concomitantly de-repressed exclusively in the SETDB1 KO line. C. In contrast, L1 elements show no consistent changes in expression or H3K9me3 in either KO line. D. TT2 cells were transfected with siRNAs specific for Dnmt1 or Setdb1, alone or in combination and expression values relative to a scrambled siRNA control, was determined for several ERVs by qRT-PCR (technical replicates, mean +/− SD)(see also Figure S5, Table S4).
As DNA methylation may play a role in maintaining a subset of these elements in a silent state in the absence of H3K9me3, we determined whether simultaneous depletion of DNA methylation and SETDB1 leads to a higher level of ERV reactivation than depletion of SETDB1 alone. Dnmt1 and Setdb1 were targeted via RNAi either alone or in combination, and expression of several ERV subfamilies was monitored via qRT-PCR (Figure 5D). While knockdown (KD) of SETDB1 induced expression of GLN, RLTR4/MLV, ERVK10C, IAPE-z and in particular ETnERV2/MusD ERVs, KD of Dnmt1 had a relatively modest effect on expression of these proviruses. For each of these subfamilies, simultaneous KD of Setdb1 and Dnmt1 did not increase the level of expression over that observed upon KD of Setdb1 alone, with the exception of the young IAPE-z subfamily, for which the double KD behaves synergistically. Taken together, these data reveal that while SETDB1 plays a dominant role in silencing of class I and II ERVs, for a subset of these elements, DNA methylation provides an additional layer of silencing in the absence of H3K9me3.
Aberrant ERV transcription in the SETDB1 KO line leads to expression of chimaeric transcripts with downstream genes
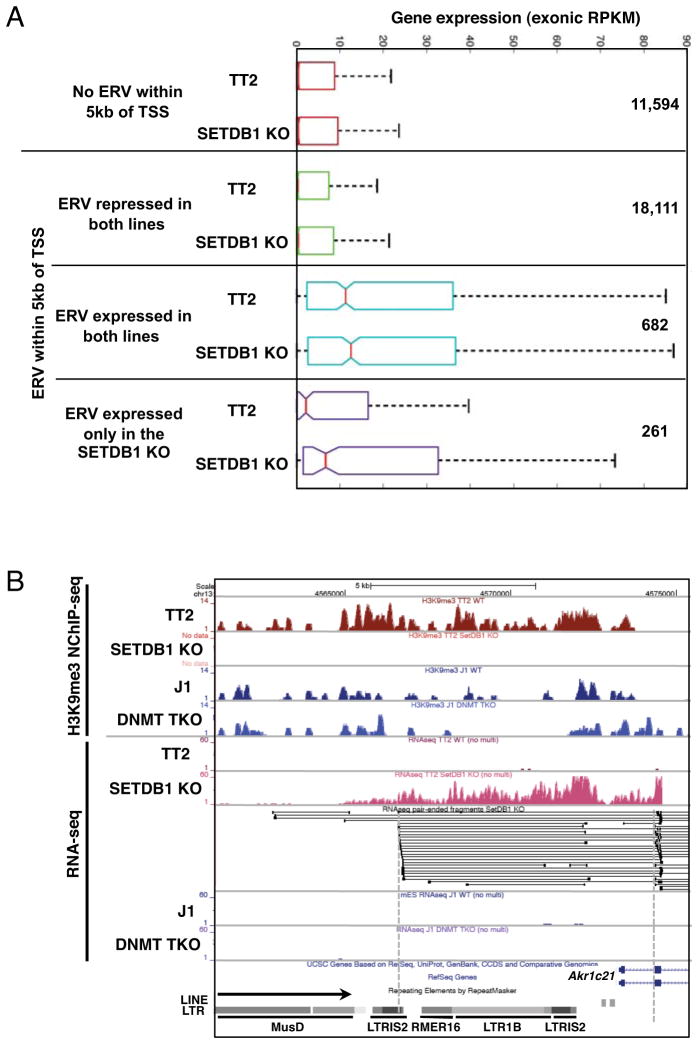
The widespread de-repression of ERVs in the SETDB1 KO line prompted us to explore the possibility that a subset of the genes showing ectopic transcription were induced as a consequence of de-repression of proximal ERVs. We classified all genes based on the absence or presence of an annotated ERV +/−5 kb from the annotated TSS and further subdivided the latter on the basis of RNA-seq coverage over these ERVs in the TT2 WT and/or SETDB1 KO lines. Intriguingly, genes 3′ of ERVs transcribed in both lines (RNA-seq RPKM>1) were generally expressed at higher levels than genes lacking an ERV within 5 kb of the TSS, or genes in which an ERV is present but not transcribed (coverage < 1 RPKM) in either line (Figure 6A). Deletion of Setdb1 had little effect on these relationships. Strikingly however, 56 of the 261 genes with a promoter proximal ERV showing a ≥10-fold increase in transcription (and a minimum expression level of RPKM >1) in the SETDB1 KO line are themselves concomitantly up-regulated, representing ~10% of the 558 up-regulated genes (shown in Figure 1F and listed in Table S1) in this line. This is significantly greater than the 2.4% of all genes showing an increase in expression in this line (P-value <10−15), indicating that constitutively expressed ERVs positively influence the expression of proximal genes, and that aberrant activation of ERVs may alter the expression of neighboring genes.
Figure 6. Increased genic expression in SETDB1 KO mESCs is associated with increased expression of promoter proximal ERVs.
A. Protein coding genes were grouped according to the presence of an annotated ERV within 5kb of the annotated TSS(s) and then classified solely on the basis of the presence or absence of RNA-seq reads over these promoter proximal ERVs in the TT2 and/or SETDB1 KO lines. The distribution of RNA-seq coverage (normalized exonic RPKM) for genes with no proximal ERV is shown, along with genes harboring promoter proximal ERVs that are: 1. repressed in both lines (RNA-seq coverage <1.0 aRPKM)(See Supplemental Experimental Procedures); 2. expressed in both lines (RNA-seq coverage ≥1.0 aRPKM and SETDB1 KO aRPKM/TT2 aRPKM between .75 and 1.3); or 3. expressed predominantly in the SETDB1 KO line (RNA-seq coverage ≥1.0 aRPKM and SETDB1 KO aRPKM/TT2 aRPKM ≥10). The number of genes in each category is also shown. B. UCSC genome-browser screen shot of the 5′ end of the Akr1c21 gene, showing H3K9me3 NChIP-seq and RNA-seq tracks, alignment of the split paired-end RNA-seq reads in the locus and ERVs 5′ of the gene (see Figure S6).
Surprisingly, inspection of paired-end read alignments at several such genes revealed the presence of numerous “chimaeric” transcripts (Peaston et al., 2004) (Van De Lagemaat et al., 2003), with one mate pair read mapping within the promoter proximal ERV and the other within an annotated genic exon. For example, 20 paired-end reads in the Akr1c21 locus show one mate-pair read mapping to an ERV within a cluster of elements upstream of the TSS and the other to the 5′ end of the 2nd annotated exon (Figure 6B). Similar observations were made for the Angptl6 and Cyp2b23 loci (Figure S6). To identify additional chimaeric mRNAs, we surveyed paired-end RNA-seq reads for the presence of individual transcripts with one of the mate-pair reads aligning to an ERV and the other to an annotated genic exon. Numerous genes with such chimaeric reads were found in all four cell lines (Figure S7A and Table S5). Analysis of the 117 genes associated with such constitutive chimaeric transcripts in the TT2 and SETDB1 KO lines revealed that the genic expression levels (RPKM over annotated exons) were similar in most cases, with only 5 of these genes showing increased expression in the SETDB1 KO line. Furthermore, the ERVs identified were generally distinct from those de-repressed in the SETDB1 KO line (compare Figures 4 and S3 with S7B).
To identify chimaeric transcripts induced as a result of Setdb1 deletion, we screened for genes showing a ≥ 4-fold increase in such reads in the SETDB1 KO (and an arbitrary minimum of 3 chimaeric reads). Strikingly, we identified 84 such genes, 63 of which show 3 or more chimaeric reads in the SETDB1 KO but none in the TT2 line (Table S5). Interestingly, none of these genes showed a ≥ 4-fold increase in chimaeric reads in the DNMT TKO line (Table S5). Furthermore, in contrast to the genes associated with constitutive chimaeric transcripts, 38 of these genes, representing 6.8% of all up-regulated genes in the SETDB1 KO, intersect with the list of genes showing increased expression (as measured by total exonic RNA-seq coverage; Table S1) in this line (Figure S7A). Thirteen of these chimaeric genes are among the 56 up-regulated genes associated with a de-repressed promoter proximal ERV (identified in Figure 6A), yielding a total of 81 genes up-regulated in association with de-repression of a nearby ERV. Strikingly, 4 of the top 10 and 17 of the top 100 genes ranked in terms of fold-increase in expression in the SETDB1 KO line are included in this list (Table S1), indicating that genes associated with ERV-initiated chimaeric transcripts can be transcribed at very high levels. The annotated ERVs associated with chimaeric transcripts are generally truncated elements, indicating that transcription is more likely to extend into flanking genomic sequence when the splice and/or polyA sites of the ERV is deleted. Taken together, these results indicate that transcription from promoter-proximal ERVs can increase mRNA levels of associated downstream genes, frequently in association with the generation of chimaeric transcripts.
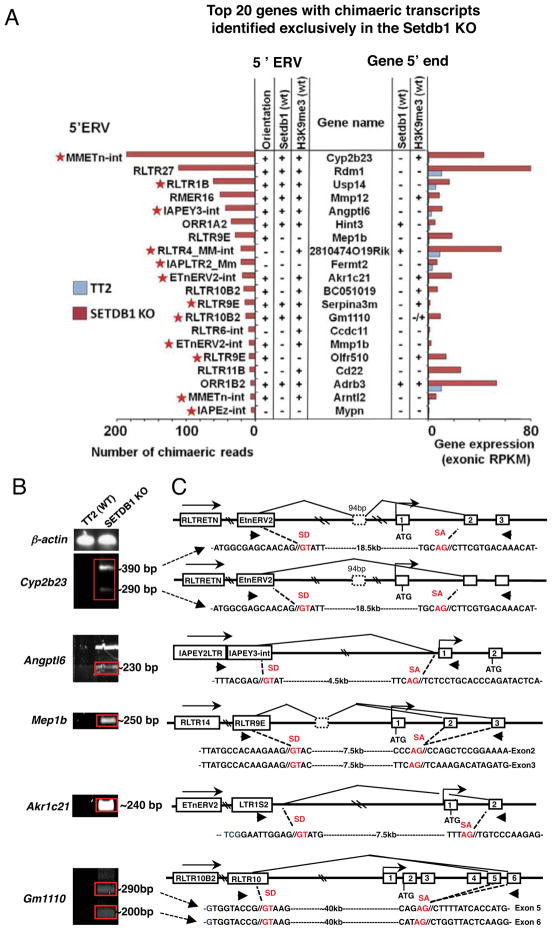
To further characterize the positive correlation between the number of chimaeric paired-end reads detected and the read coverage (across all exons) of associated genes, we analyzed the top 20 genes ordered in terms of the number of chimaeric transcripts in the SETDB1 KO line in greater detail (Figure 7A). Intriguingly, the majority of cognate genic promoters are not marked by H3K9me3 or bound by SETDB1, while 16 of the 20 ERVs in which transcription apparently initiates are marked by H3K9me3. Furthermore, many of the ERVs in which transcription of these chimaeric mRNAs initiate, such as IAP, Etn and RLTR1B elements, are in the same subfamilies of ERVs that are broadly de-repressed in SETDB1 KO cells (see Figures 4B and S3).
Figure 7. Chimaeric transcripts initiating in LTR elements 5′ of genic TSSs and splicing to canonical genic exons are detected exclusively in the SETDB1 KO line.
A. Genes with one paired-end read mapping to an annotated ERV and the other to a genic exon were identified. The top 20 genes in the SETDB1 KO, in terms of the number of chimaeric reads identified, are shown, along with RNA-seq coverage over genic exons. Annotation of the ERV in which transcription initiates, the orientation of the ERV in relation to the gene and the presence of SETDB1 or H3K9me3 in the ERV or at the 5′ end of the gene is also shown. Stars indicate the subclasses of ERVs that are broadly reactivated. B. The presence of chimaeric transcripts of the Akr1c21, Angptl6, Gm1110, Mep1b and Cyp2b23 genes was validated by RT-PCR using primers (arrows) designed within the 50 bp regions to which the chimaeric paired-end reads aligned. β-actin was used as a control. C. Amplicons were cloned and sequenced and the structure of the chimaeric RNAs, the orientation and subfamily of the ERV in which transcription initiates and the annotated genic TSS and exons (numbered) are shown for each locus. The sequence of the relevant novel splice donor (SD) and genic splice acceptor (SA) sites are also shown. For several genes, splicing to several genic exons was observed (see Figure S7).
To validate the existence of these SETDB1 KO-dependent chimaeric transcripts, RT-PCR was conducted using primers specific for the genomic regions complementary to the chimaeric paired-end reads of five of these genes (Figure 7B), including Akrc21, Angptl6, Gm1110, Mep1b and Cyp2b23. As expected, PCR products were only observed in the SETDB1 KO. The upstream sequences of these transcripts, including any cryptic splice site junctions, were subsequently determined by Sanger sequencing, confirming that they frequently splice from cryptic splice donor sites embedded in an ERV itself or in 3′ flanking genomic DNA, to 5′ genic splice acceptor sites (Figure 7C). Analysis of the coding potential of these novel transcripts reveals that the complete native ORF of only the Angptl6 gene is retained, but a cryptic upstream ORF is also encoded which likely precludes the expression of the Angptl6 protein. To establish the coding potential of the remaining chimaeric genes, we carried out ab initio transcript assembly using Cufflinks (Trapnell et al., 2010). Of the 38 induced chimaeric genes, transcript modeling revealed that 20 initiate in an ERV and extend to a genic exon, 13 of which are associated with genes up-regulated in the SETDB1 KO (Table S5). While 9 of these chimaeric transcripts encode the native genic ORF, only three, CD209c, 2810474O19Rik and 2010005H15Rik, do not also encode a cryptic upstream ORF. Thus, paradoxically, while the level of transcript over genic exons is dramatically up-regulated for a number of the chimaeric transcripts identified, translation of the native ORF is likely to be reduced for the majority of constitutively expressed genes associated with chimaeric transcripts, due to the presence of cryptic upstream ORFs.
Discussion
DNA methylation and post-translational histone modifications are highly dynamic epigenetic marks, particularly early in development, when transcriptional networks undergo reprogramming associated with differentiation. A recent microarray study revealed that the majority of genes de-repressed in the absence of DNA methylation are not marked by H3K27me3, suggesting that DNA methylation acts independent of H3K27me3 to maintain genes in a silent state (Fouse et al., 2008). Here, we show that the majority of genes de-repressed in the absence of DNA methylation are not de-repressed in the absence of SETDB1/H3K9me3, and vice-versa. Genes in the MageA and Rhox clusters that are reactivated in the DNMT TKO line for example, are not marked by H3K9me3 in WT cells, nor reactivated in the SETDB1 KO line, while genes that are reactivated in the SETDB1 KO line are not DNA methylated in WT cells, nor reactivated in the DNMT TKO line. In contrast, a relatively small number of predominantly germline-specific genes are DNA methylated and marked by H3K9me3 in their promoter regions and de-repressed in both KO lines. Why H3K9me3 and DNA methylation marks are required to repress these germline-specific genes remains unknown, but may reflect the expression of multiple transcriptional activators that can act independently to promote transcription, each of which must be inhibited by one or the other of these pathways to maintain their promoter regions in an inaccessible state.
Genome-wide reactivation of Class I and Class II ERVs in mESCs lacking SETDB1 but not DNA methylation confirms our previous qRT-PCR and northern blotting-based observations (Matsui et al., 2010) and is consistent with recent reports showing that many of the same ERV families are de-repressed in mESCs and blastocysts deficient in the SETDB1 binding partner KAP1 (Rowe et al., 2010), but not in two independently derived DNMT TKO lines (Hutnick et al., 2010; Matsui et al., 2010; Tsumura et al., 2006). Taken together, these data clearly show that while DNA methylation may be critical for silencing of these ERVs in somatic cells and at specific stages in germline development (Walsh et al., 1998), an alternative silencing pathway maintains these elements in a silent state in mESCs and early in embryonic development. The relatively high turnover of DNA methylation in primordial germ cells and in the early embryo (Morgan et al., 2005) may reduce the efficacy of this pathway at these stages. Regardless, given that newly retrotransposed ETn and IAP ERVs are responsible for a significant number of mouse germline mutations (Maksakova et al., 2006), at least some of these elements are clearly capable of evading the host silencing machinery in the germline or early in embryonic development.
Exogenous (Bushman et al., 2005; Lewinski et al., 2006) and young endogenous (Medstrand et al., 2002) viruses generally integrate within or near genes. However, given their propensity to interfere with gene expression, ERVs are generally excluded from genes and adjacent regions by natural selection (Medstrand et al., 2002). Nevertheless, perhaps due to their high transcriptional activity, a number of ERVs have been domesticated to provide new regulatory elements for tissue- or cell-specific expression of developmentally regulated genes (Van De Lagemaat et al., 2003). MT and MuERV-L class III ERVs for example, are highly expressed in oocytes and 2-cell embryos and drive expression of chimaeric transcripts that comprise 14% and 3% of all ESTs at these stages, respectively (Peaston et al., 2004). Moreover, a recent genome-wide analysis of cap-selected mouse and human transcripts from different tissues and developmental stages revealed that up to 30% of transcripts initiate within repetitive elements, many of them tissue-specific (Faulkner et al., 2009).
Our genome-wide analyses revealed a number of chimaeric mRNAs expressed predominantly in SETDB1-deficient cells that are initiated primarily by the same subclasses of class I and II ERVs that are broadly de-repressed in these cells. The majority of ERVs in which these chimaeric transcripts initiate (15 of 21) are in the sense orientation, consistent with a previous report showing that promoter proximal LTR elements are more likely to be used as gene promoters when in the sense orientation (Dunn et al., 2005). While these chimaeric transcripts are not detected in WT mESCs due to SETDB1-mediated silencing, 11 of the top 20 chimaeric transcripts identified in the SETDB1 KO line, including the Cyp2b23, Mmp12, Angptl6 and Mep1b chimaeras, are expressed in a subset of normal and/or tumor tissues, according to the AceView cDNA database (Thierry-Mieg and Thierry-Mieg, 2006) (Table S5), indicating that silencing of a number of the ERV-initiated chimaeric transcripts, while robust in mESCs, is relaxed in other tissues. Intriguingly, aberrant expression of several ERV-initiated proto-oncogenes is linked to transformation in mice (Howard et al., 2008; Lee et al., 1999) and humans (Lamprecht et al., 2010).
Further evidence for ERV-mediated perturbation of gene expression comes from studies of mouse mutants harboring novel ERV insertions (Druker et al., 2004; Duhl et al., 1994; Maksakova et al., 2006; Vasicek et al., 1997). The most well known example is the Avy epiallele, an epimutation resulting from the insertion of an IAP element in a pseudoexon upstream of the Agouti gene (Waterland and Jirtle, 2003). A cryptic promoter in the IAP element promotes constitutive ectopic expression of a chimaeric transcript consisting of a novel IAP 5′LTR-encoded exon spliced to the canonical splice acceptor site of exon 2 of the Agouti gene (Duhl et al., 1994). This chimaeric mRNA encodes a functional Agouti protein, the aberrant expression of which leads to yellow fur, obesity and tumorigenesis in the Avy mouse at non-Mendelian ratios. Another example involves a distinct IAP insertion in the Pcdaα v8 gene, which results in reduced expression of this gene in brain tissue due to DNA methylation of the IAP element. Strikingly, Pcdaα v8 expression is induced over 100-fold in neuroblastoma cell lines in association with up-regulation of this IAP element (Sugino et al., 2004).
In summary, we find that the widespread reactivation of Class I and Class II ERVs triggered exclusively by Setdb1 deletion is accompanied by the expression of novel ERV-initiated genic transcripts, many of which encode novel ORFs upstream of the canonical genic ORF that likely preclude expression of the native protein. Nevertheless, the regulatory elements within these ERVs may represent a reservoir of alternative promoters that have the potential to be domesticated, should they confer a selective advantage. Regardless, the results presented here clearly reveal that SETDB1 is not only required for silencing of a subset of genes in mESCs, but also plays a critical role in protecting the integrity of the transcriptome in these cells by inhibiting the aberrant expression of ERVs and ERV-initiated transcripts that splice to genic exons.
Experimental Procedures
Cell Culture, RNA isolation, qRT-PCR and RNA-seq
J1 and TT2 mESCs were passaged as described (Matsui et al., 2010). RNA was isolated using the GenElute Kit (Sigma-Aldridge). For RT analysis, RNA was reverse transcribed using SuperScript III (Invitrogen) as per the manufacturer’s instructions and qRT-PCR was carried out using SsoFAST EvaGreen Supermix (BioRad) on StepOne Software v2.1 (Applied Biosystems). RNA-seq libraries were constructed from mRNA as described in Morin et al. (Morin et al., 2008) from 10μg of DNAseI treated total RNA and paired-end sequencing performed on an Illumina Genome Analyzeriix, following the recommended protocol (Illumina Inc., Hayward, CA). Sequence reads were aligned to the mouse reference genome (mm9) using MAQ v0.7.1 (Li et al., 2008) with Smith-Waterman alignment disabled and annotated exon-exon junctions compiled from Ensembl (Flicek et al., 2010), RefSeq (Pruitt and Maglott, 2001) and UCSC (Rhead et al., 2010) (downloaded from http://genome.ucsc.edu on 03/17/09). Oligonucleotide sequences used in RNAi and PCR experiments are listed in Supplemental Experimental procedures.
NChIP, Data Normalization, RPKM, and Z-score
NChIP was conducted as described in the Supplementary Information. To compare expression and H3K9me3 coverage levels across samples, we calculated RPKM values in regions of interest for both RNA-seq and NChIP-seq samples (Mortazavi et al., 2008), as described in detail in the Supplemental Information. For pair wise sample comparisons, a Z-score was calculated assuming the distribution of read coverage for each sample follows a Poisson model:
, where RPKMA and RPKMB are RPKMs in the region of interest of A and B samples, respectively. Sequencing reads have been deposited in the Gene Expression Omnibus under accession number GSEXXXXX.
Supplementary Material
Acknowledgments
We thank M. Okano for Dnmt TKO mESCs and T. Jenuwein for Suv39h DKO mESCs. We are also grateful to Jim Bone, Bryeanna Villani, Nina Thiessen and to members of the GSC Functional Genomics and Sequence Production groups and Lorincz lab for technical support. We thank Keji Zhao, Ting Wang, Irina Stancheva and Dirk Schübeler for helpful discussions. This work was supported by grants from the Genome Network Project from MEXT of Japan to Y.S. and CIHR grants 77805 to M.C.L. and 92093 to M.C.L. and M.H.. M.C.L. is an MSFHR Scholar and a CIHR New Investigator.
References
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Dunn CA, Van De Lagemaat LN, Baillie GJ, Mager DL. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene. 2005;364:2–12. doi: 10.1016/j.gene.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Edwards JR, O’Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010 doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Selker EU. Controlling DNA methylation: many roads to one modification. Curr Opin Genet Dev. 2005;15:191–199. doi: 10.1016/j.gde.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16:571–579. 571. doi: 10.1038/nm.2129. following 579. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Haruna T, Ishimoto A, Honjo T, Yanagawa S. Intracisternal type A particle-mediated activation of the Notch4/int3 gene in a mouse mammary tumor: generation of truncated Notch4/int3 mRNAs by retroviral splicing events. J Virol. 1999;73:5166–5171. doi: 10.1128/jvi.73.6.5166-5171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AHFM. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL. Retroelement Distributions in the Human Genome: Variations Associated With Age and Proximity to Genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. BioTechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Myant K, Termanis A, Sundaram AY, Boe T, Li C, Merusi C, Burrage J, Heras JI, Stancheva I. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21:83–94. doi: 10.1101/gr.108498.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E, Okano M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006;20:3382–3394. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Meth. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Sugino H, Toyama T, Taguchi Y, Esumi S, Miyazaki M, Yagi T. Negative and positive effects of an IAP-LTR on nearby Pcdaalpha gene expression in the central nervous system and neuroblastoma cell lines. Gene. 2004;337:91–103. doi: 10.1016/j.gene.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12, 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Van De Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Vasicek TJ, Zeng L, Guan XJ, Zhang T, Costantini F, Tilghman SM. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147:777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge C, Haase J, Janes J, Huss J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biology. 2009;10:R130–R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci U S A. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Maksakova IA, Gagnier L, Van De Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genetics. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.