Abstract
Staphylococcal Cassette Chromosome mec (SCCmec)typing is a very important molecular tool for understanding the epidemiology and clonal strain relatedness of methicillin-resistant Staphylococcus aureus (MRSA), particularly with the emerging outbreaks of community-associated MRSA (CA-MRSA) occurring on a worldwide basis. Traditional PCR typing schemes classify SCCmec by targeting and identifying the individual mec and ccr gene complex types, but require the use of many primer sets and multiple individual PCR experiments. We designed and published a simple multiplex PCR assay for quick-screening of major SCCmec types and subtypes I to V, and later updated it as new sequence information became available. This simple assay targets individual SCCmec types in a single reaction, is easy to interpret and has been extensively used worldwide. However, due to the sophisticated nature of the assay and the large number of primers present in the reaction, there is the potential for difficulties while adapting this assay to individual laboratories. To facilitate the process of establishing a MRSA SCCmec assay, here we demonstrate how to set up our multiplex PCR assay, and discuss some of the vital steps and procedural nuances that make it successful.
Keywords: Infection, Issue 79, Microbiology, Genetics, Medicine, Cellular Biology, Molecular Biology, Biomedical Engineering, Bacteria, Bacterial Infections and Mycoses, Life Sciences (General), Methicillin-resistant Staphylococcus aureus (MRSA), Staphylococcal cassette chromosome mec (SCCmec), SCCmec typing, Multiplex PCR, PCR, sequencing
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has acquired and integrated into its genome a 21-67-kb mobile genetic element, termed the staphylococcal cassette chromosome mec (SCCmec) that harbors the methicillin resistance (mecA) gene and other antibiotic resistance determinants 1. SCCmec is characterized by the presence of terminal direct and inverted repeats, two essential genetic components (the mec gene complex and the ccr gene complex), and the J-regions (Figure 1). The mec gene complex is composed of IS431mec, mecA, and intact or truncated sets of regulatory genes, mecR1 and mecI, while the ccr gene complex encodes the recombinases (ccr/ccrAB) that mediate integration of SCCmec into and its excision from the recipient chromosome 1. Recently two novel mec resistance genes (mecB and mecC) were described, which show significant sequence divergence from the mecA gene 2. The rest of the SCCmec element is comprised of J regions (J1, J2, and J3) that are located between and around the mec and ccr complexes and contain various genes or pseudogenes. There are several classes of mec complex (such as A, B, C, D and E) and allotypes of ccr complex (such as 1-8) described. Different combinations of these complex classes and allotypes generate various SCCmec types. According to the guidelines of SCCmec nomenclature and classification by the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC), SCCmec elements are classified into types based on the nature of the mec and ccr gene complexes, and are further classified into subtypes according to differences in their J region DNA 1. MRSA clones are commonly defined by incorporation of SCCmec typing information in conjunction with that provided by (gene/genomic) sequence-based typing methods, such as multilocus sequence typing (ST) and/or staphylococcal protein A gene (spa) typing (e.g. ST8-t008-MRSA-IVa). Therefore, SCCmec typing is a very important molecular tool for understanding the epidemiology and clonal strain relatedness of MRSA, particularly with the emerging outbreaks of community-associated MRSA (CA-MRSA) occurring on a worldwide basis.
Traditional PCR typing schemes classify SCCmec by targeting and identifying the individual mec and ccr gene complex types, but require the use of many (20 to 30) primer sets and multiple individual PCR experiments. Okuma et al., for example, employed 21 primers and many single target PCR reactions in detecting SCCmec I-IVb 3. Kondo et al. subsequently developed a more complete assay for identification of ccr, mec, and major differences in the J-regions, but it also required a large number of primers and multiple PCR reactions 4. To simplify SCCmec typing, multiplex PCR (M-PCR) assays were developed allowing samples to be processed in a single PCR reaction. De Lencastre and colleagues developed an M-PCR for quick screening of SCCmec I-IV, and later updated for SCCmec I-V, with a separate M-PCR for identifying SCCmec IV subtypes 5,6,7. However, these assays are more difficult to interpret, require 2 reactions in order to classify SCCmec IV subtypes, and result in several non-typeable subtypes. In an attempt to improve SCCmec typing, we designed and published a simple multiplex PCR assay for quick-screening of major SCCmec types and subtypes I to V 8, and later updated it as new sequence information became available 9. This simple assay targets individual SCCmec types in a single reaction, is easy to interpret and has been extensively used worldwide (337 from SCOPUS; updated Jan. 30, 2013). Since the original publication, we have received many requests for assistance from groups setting up this assay as part of their in-house MRSA SCCmec typing. Due to the sophisticated nature of the assay and the large number of primers present in the reaction, there is the potential for difficulties while adapting this assay to individual laboratories. To facilitate the process of establishing a MRSA SCCmec assay, we are going to demonstrate how to set up our M-PCR for SCCmec typing, as well as discuss some of the vital steps and procedural nuances that make it successful.
Protocol
These procedures should be conducted in a certified biosafety level 2 laboratory. Appropriate personal protective equipment such as gloves and lab coats should be used at all times.
1. Sample Preparation
Fresh overnight plate cultures of MRSA are required. On the day before PCR is to be done, select a single colony of MRSA with a sterile culture stick and prepare a heavy streak of the bacteria on a Tryptic Soy Agar (TSA) plate. Multiple samples can be streaked onto a single plate. Incubate overnight at 37 °C.
Add 75 μl of sterile distilled water to a 1.5 ml microcentrifuge tube. Using a sterile culture stick pick up a small amount of bacteria from the heavy overnight streak. Swirl the bacteria in the sterile water to prepare a turbid solution (Figure 2). Repeat for all samples.
Incubate the tubes in a dry heat block set at 95 °C for 10 min to lyse the bacteria and release the DNA.
Note: If the bacterial solution is too weak or too concentrated PCR efficiency is reduced. Likewise, if the temperature is above 95 °C or the incubation is longer than 10 min the PCR efficiency is reduced.
Remove the samples from the heat block and let stand at room temperature for 5 min.
Centrifuge the samples at 13,000 rpm for 1 min, creating a cellular debris pellet and clear supernatant containing the DNA. Samples can be stored at -20 °C for future use.
Note: If using a previously prepared and frozen sample, remove the microcentrifuge tube from the freezer and let stand at room temperature to fully thaw. If the pellet has been disturbed re-centrifuge the sample.
Note: gDNA prepared by any other method, including chelex extraction10, or column extraction (QIAmp DNA Mini Kit, Qiagen) is also acceptable.
2. PCR Preparation
Note: Control samples should be included in each PCR run and contain a minimum of types I, II, IIA, III, IVa, IVb, IVc, IVd, IVE and V. Ten extra samples should, therefore, be added to the count when preparing the master mix. DNA for the controls can be prepared ahead of time and stored for extended periods of time in the freezer.
- Remove PCR reagents, including 10x PCR buffer (without MgCl2), 50 mM MgCl2, dNTPs and primers, from the -20 °C freezer and allow to thaw on the clean PCR bench (no bacteria or DNA permitted) at room temperature. 0.2 ml PCR tubes can be prepared and labeled at this time. If new stocks of primers or dNTPs are required they can be prepared as follows:
- Prepare primers by diluting concentrated stocks to 10 μM working solutions with sterile distilled water. The primer stocks and working solutions can be kept frozen at -20 °C for use at future dates.
- Prepare dNTPs by diluting commercial stocks to a working solution containing 2 μM of each dNTP. In order to minimize the number of freeze/thaw cycles that the dNTPs are subjected to, stocks of 10 μM of each dNTP can be prepared ahead of time, 50 μl aliquots placed in 0.5 ml microcentrifuge tubes and stored at -20 °C. As needed, a tube of the 10 μM is removed and diluted to 2 μM of each dNTP by adding 200 μl of sterile distilled water.
Once thawed, vortex briefly to mix all reagents, then combine as per Table 1. It is important to use platinum taq DNA polymerase (as opposed to regular taq) because this has been found to influence the success of the reaction. Generally a master mix is prepared, vortexed briefly to mix and aliquoted into the individual PCR tubes.
Close all PCR tubes and move to the general work bench. Add the template to the PCR tubes by pipetting 2 μl of the clear supernatant from the previously prepared bacterial samples.
Move to the dedicated amplification location and place the tubes into the thermal cycler under the following protocol: 94 °C for 5 min, followed by 10 cycles of 94 °C 45 sec, 65 °C 45 sec, 72 °C 1.5 min. A further 25 cycles of 94 °C 45 sec, 52 °C 45 sec, 72 °C 2 min were followed by a 10 min incubation at 72 °C and a hold at 4 °C.
3. Agarose Gel Electrophoresis
(see Lee, P.Y. et al., J. Vis. Exp. (62), e3923, doi:10.3791/3923 (2012) for more details) 11.
A dedicated gel electrophoresis location is preferred. PCR products are run on a 2.5% agarose gel in 0.5x TBE buffer (110 mM Tris; 90 mM Borate; 2.5 mM EDTA; pH 8.3).
Weigh 2.5 g of agarose and add to 100 ml of 0.5x TBE buffer in a glass Erlenmeyer flask. Cover the flask with plastic wrap and boil the solution in a microwave until fully dissolved. A 2.5% gel will boil over quickly, therefore stop the microwave and swirl the solution regularly.
Cool the agarose until comfortable to touch by swirling under cold water, or by resting on the bench for 5 min. Pour the agarose into the casting tray.
Remove tubes from the PCR thermal cycler and prepare the samples by add 2 μl of 6x DNA loading dye (40% glycerol/0.25% bromophenol blue) to each sample. Load 5-10 μl of sample per well on the gel. A 1 Kb+ or 100 bp DNA ladder is appropriate for the expected band sizes.
Ethidium bromide (EtBr) can be added to the gel prior to pouring it, or alternatively the gel can be stained in an EtBr bath prior to being visualized. Place the finished gel in a solution of 0.5 μg/ml EtBr in distilled water and mix gently on shaker for 10 min. Transfer the gel to a clean container and destain in distilled water for a further 10 min.
Representative Results
This updated M-PCR is able to determine (classify) SCCmec types I-V, as well major subtypes of SCCmec IV. The assay targets unique and specific loci of SCCmec I, II, III, IVa, IVb, IVc, IVd, IVE and V, with concomitant mecA gene detection. Tentative identification of SCCmec VI and VIII is also possible, but would require further testing to confirm. Figure 3 illustrates representative SCCmec I-VI and VIII and the locations of each target within the element.
The type I specific primers (red) target orf E008 (strain NCTC10442; AB033763) in the J1 region of SCCmec I, while the type II primers (dark green) are specific to orf NO43 (strain N315; D86934) present in the J2 of all SCCmec II and SCCmec VIII described thus far. Type III (purple), IVa (dark blue), IVb (light green) and IVd (peach) are all specific targets located in the J1 regions of the respective SCCmec types. Type III primers target orf CZ003 (strain 85/2082; AB037671), type IVa target orf CQ002 (strain CA05; AB063172), type IVb target orf CM001 (strain 8/6-3P; AB063173), and type IVd primers target orf CG001 (strain JCSC4469; AB097677). The locus for type IVc primers (burgundy) is located in orf CR008 (strain MR108; AB096217) of the J1 region of SCCmec IVc, but also present in SCCmec IVE which has the similar J1 region. The locus for type IVE primers (light blue) is located in orf IVE01 (strain AR43/3330.1; AJ810121) of the J3 region of SCCmec IVE, as well as present in SCCmec IVF which shares the same J3 region. Finally, the specific target for SCCmec V is orf V011 (strain JCSC3624; AB12121) found in the J2 region of the SCCmec element. Additional PCR targets present in the assay include the mecA gene (brown) found in all SCCmec elements and serving as an internal control, the kdp gene (yellow) located in the J1 region of the prototypical SCCmec II of strain N315, and one specific to orf CZ049 of SCCHg (orange), found adjacent to SCCmec III in the prototypical type III of strain 85/2082. Because SCCmec VIII contains the same J2 region as SCCmec II (and therefore positive for the type II target) we included a target to ccrAB4 (pink) to distinguish between them. ccrAB4 is also present in multiple SCC types like IIA, IVE and IVF as an adjoining SCC element, in addition to being the main ccr complex for types VI and VIII. Of note, SCCmec IIA shares the same J1 region as type IVb and, therefore, is also positive for the IVb target.
Figure 4 is a representative gel demonstrating the expected PCR products for all representative strains of SCCmec types I-VI and VIII, and SCCmec IV subtypes a-F. The M-PCR produced distinct bands corresponding to their respective molecular sizes, easily recognizable in an agarose gel stained with ethidium bromide. For a typical SCCmec I two bands are expected; the type I specific band at 613 bp, as well as the mecA control band at 147 bp (Figure 5). Three representative SCCmec II strains are shown, including the prototypical type II of strain N315, and the less prevalent type IIA, and type IIb. To classify a strain as SCCmec II two bands should be present, including the type II locus target at 128 bp and the mecA control (Figure 6). Strain N315 carries the dominant SCCmec II element, which is identifiable based on of the additional presence of a kdp product at 398 bp (which is carried in its longer J1 region). The less prevalent SCCmec IIA, seen in strain AR14/0298, is positive for the ccrAB4 target at 106 bp (which it carries on an accessory element), as well as the type IVb target at 193 bp (which is a consequence of it sharing the same J1 region as type IVb). SCCmec IIb is also a less prevalent type and only carries the type II and mecA targets. Two representative SCCmec III are shown on the gel, including the prototypical SCCmec III of strain 85/2082, and the less dominant type IIIA of strain JCSC290. SCCmec III are identified based on the presence of two PCR products, including the type III specific target at 257 bp, and the mecA control target (Figure 7). The dominant SCCmec III will also be positive for the mercury target at 280 bp, which is carried on an adjacent SCC element and serves to distinguish it from the less common SCCmec IIIA. Five representative SCCmec IV elements are shown, including Types IVa, IVb, IVc, IVd, IVE and IVF. Unlike with SCCmec I-III, there is no single target present in all of the SCCmec IV subtypes, apart from mecA (Figure 8). The representative SCCmec IVa strain is positive for the type IVa specific product at 776 bp, the representative SCCmec IVb strain is positive for the IVb specific product at 193 bp, the representative SCCmec IVc strain is positive for the IVc product at 200 bp and the representative SCCmec IVd strain is positive for the IVd specific product at 881 bp. The representative SCCmec IVE strain is identified based on the presence of the type IVE band, as well as the ccrAB4 product (present on an adjacent accessory element) and type IVc product (present because SCCmec IVE shares the same J1 region as SCCmec IVc). SCCmec IVF is similar to SCCmec IVE in that it shares the same J3 region and is consequently positive for the IVE target, as well as is positive for the ccrAB4 target due to an adjacent accessory element. SCCmec IVF can, however, be distinguished from SCCmec IVE in that lacks the type IVc target. A typical SCCmec V would have two PCR products present on the gel; the type V specific band at 325 bp and the mecA control at 147 bp (Figure 9). Finally, while the goal of this assay was not to provide definitive identification of either SCCmecVI or VIII, representative strains were included for both since a specific PCR pattern provides suggestive evidence for the presence of these types. Further investigation using traditional typing methods (ccrand mecgene complex typing) for confirmation of both SCCmecVI and VIII 3,4, or using a specific SCCmecVIII typing assay for confirmation of SCCmecVIII 12 would be required. In addition to the mecA target, the representative SCCmec VI would be expected to produce a positive ccrAB4 product at 106 bp as this is its main ccr complex type (Figure 10). The representative SCCmec VIII would also be positive for mecA at 147 bp and ccrAB4 at 106 bp, and would be distinguished from SCCmec VI on the basis of a positive type II specific product in SCCmec VIII (Figure 10), which results from it having the same J2 region as SCCmec II 13.
 Figure 1. The essential structure of SCCmec elements demonstrating the mec and ccr gene complexes, bracketed by J1, J2 and J3 regions.Click here to view larger figure.
Figure 1. The essential structure of SCCmec elements demonstrating the mec and ccr gene complexes, bracketed by J1, J2 and J3 regions.Click here to view larger figure.
 Figure 2. Sample tube for DNA extraction. The microcentrifuge tube on the left contains sterile water while the tube on the right shows the desired cell turbidity for optimal PCR efficiency.Click here to view larger figure.
Figure 2. Sample tube for DNA extraction. The microcentrifuge tube on the left contains sterile water while the tube on the right shows the desired cell turbidity for optimal PCR efficiency.Click here to view larger figure.
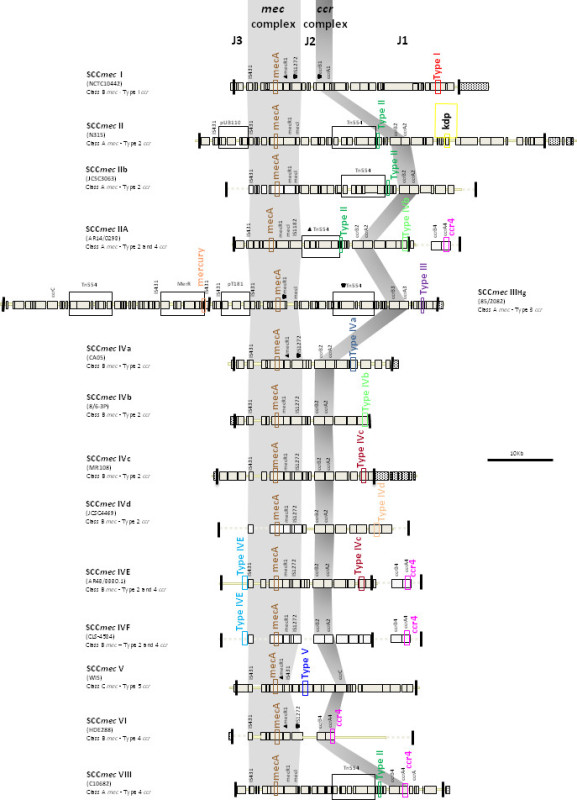 Figure 3. Representative SCCmec types I-VI and VIII and the locations of the specific targets used in the M-PCR.Click here to view larger figure.
Figure 3. Representative SCCmec types I-VI and VIII and the locations of the specific targets used in the M-PCR.Click here to view larger figure.
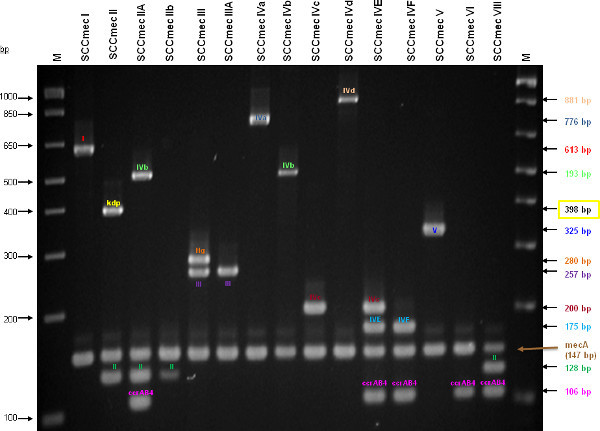 Figure 4. A representative gel demonstrating the expected PCR products for all representative strains of SCCmec types I-VI and VIII, and SCCmec IV subtypes a-F. Enhanced multiplex PCR assay identifies SCCmec types and subtype I-V. Lanes M: 1 Kb plus DNA ladder (Invitrogen). (Adapted from Zhang, et al.
9). Click here to view larger figure.
Figure 4. A representative gel demonstrating the expected PCR products for all representative strains of SCCmec types I-VI and VIII, and SCCmec IV subtypes a-F. Enhanced multiplex PCR assay identifies SCCmec types and subtype I-V. Lanes M: 1 Kb plus DNA ladder (Invitrogen). (Adapted from Zhang, et al.
9). Click here to view larger figure.
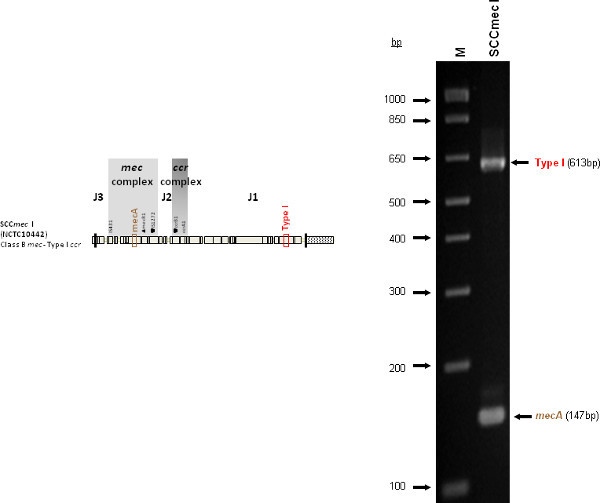 Figure 5. Representative SCCmec I (NCTC10442) sample showing the mecA and type I specific target.Click here to view larger figure.
Figure 5. Representative SCCmec I (NCTC10442) sample showing the mecA and type I specific target.Click here to view larger figure.
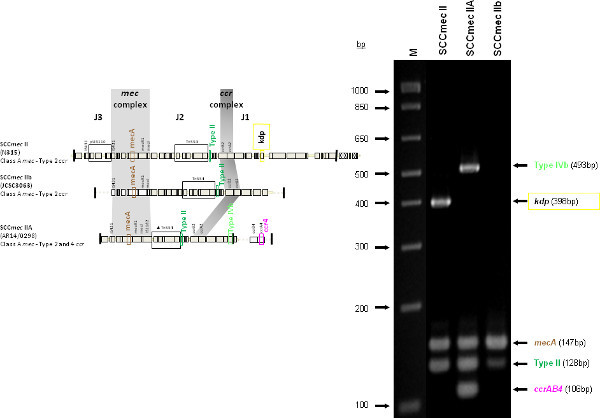 Figure 6. Representative SCCmec II (N315), SCCmec IIA (AR14/0298) and SCCmec IIb (PL150) samples. All samples are positive for mecA and type II specific target. The prototypic SCCmec II caries additional kdp gene, while type IIA carries the same J1 region as SCCmec IVb, as well as the ccrAB4 genes. Click here to view larger figure.
Figure 6. Representative SCCmec II (N315), SCCmec IIA (AR14/0298) and SCCmec IIb (PL150) samples. All samples are positive for mecA and type II specific target. The prototypic SCCmec II caries additional kdp gene, while type IIA carries the same J1 region as SCCmec IVb, as well as the ccrAB4 genes. Click here to view larger figure.
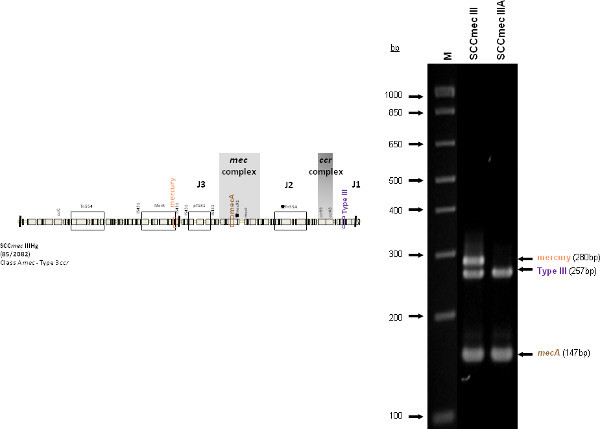 Figure 7. Representative SCCmec III (85/2082) and SCCmec IIIA (JCSC290) strains. Both are positive for mecA and the type III specific target. 85/2082 also bears the SCCHg element adjacent to the SCCmec III element. Click here to view larger figure.
Figure 7. Representative SCCmec III (85/2082) and SCCmec IIIA (JCSC290) strains. Both are positive for mecA and the type III specific target. 85/2082 also bears the SCCHg element adjacent to the SCCmec III element. Click here to view larger figure.
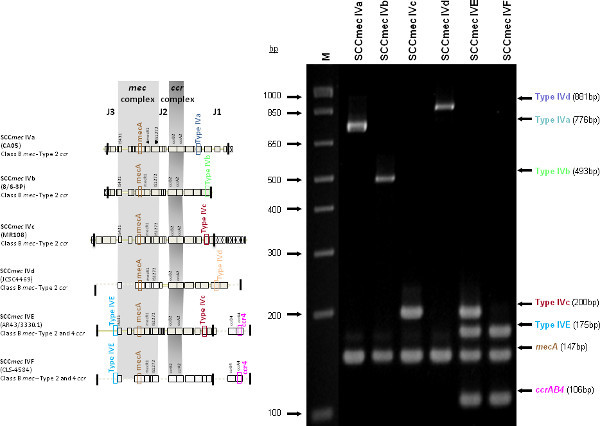 Figure 8. Representative SCCmec IV strains; SCCmec IVa (CA05), SCCmec IVb (8/6-3P), SCCmec IVc (MR108) and SCCmec IVd (JCSC4469), SCCmec IVE (AR43/3330.1) and SCCmec IVF (CLS-4584). All are positive for mecA and respective type specific targets. SCCmec IVE (AR43/3330.1) and SCCmec IVF (CLS-4584) both share the same J3 region and contain the ccrAB4 genes and are, therefore, positive for the IVE and ccrAB4 specific target. SCCmec IVE shares the same J1 region as type IVc and is, therefore, positive for type IVc target as well. Click here to view larger figure.
Figure 8. Representative SCCmec IV strains; SCCmec IVa (CA05), SCCmec IVb (8/6-3P), SCCmec IVc (MR108) and SCCmec IVd (JCSC4469), SCCmec IVE (AR43/3330.1) and SCCmec IVF (CLS-4584). All are positive for mecA and respective type specific targets. SCCmec IVE (AR43/3330.1) and SCCmec IVF (CLS-4584) both share the same J3 region and contain the ccrAB4 genes and are, therefore, positive for the IVE and ccrAB4 specific target. SCCmec IVE shares the same J1 region as type IVc and is, therefore, positive for type IVc target as well. Click here to view larger figure.
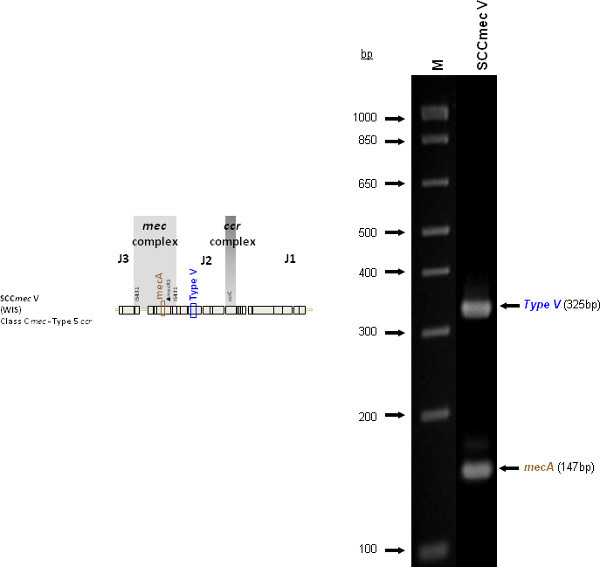 Figure 9. Representative SCCmec V (WIS [WBG8318]) showing the mecA and type V specific target.Click here to view larger figure.
Figure 9. Representative SCCmec V (WIS [WBG8318]) showing the mecA and type V specific target.Click here to view larger figure.
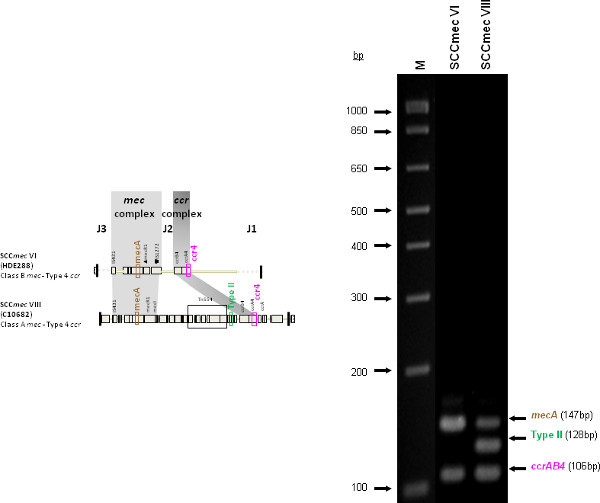 Figure 10. Representative SCCmec VI (HDE288) and SCCmec VIII (C10682). Both strains are positive for the mecA and ccrAB4 targets, while type VIII (which shares the same J2 region as type II) is positive for the type II target as well. Click here to view larger figure.
Figure 10. Representative SCCmec VI (HDE288) and SCCmec VIII (C10682). Both strains are positive for the mecA and ccrAB4 targets, while type VIII (which shares the same J2 region as type II) is positive for the type II target as well. Click here to view larger figure.
| Reagents: | Vol. (μl) | Oligonucleotide sequence | Amplicon size (bp) | Specificity |
| 10x PCR buffer | 2.5 | |||
| 50 mM MgCl2 | 1.25 | |||
| 2 mM dNTP | 2.5 | |||
| Platinum Taq | 0.2 | |||
| H2O | 2.28 | |||
| Primers (10 μM): | ||||
| Type I-F | 0.25 | GCTTTAAAGAGTGTCGTTACAGG | 613 | SCCmec I |
| Type I-R | 0.25 | GTTCTCTCATAGTATGACGTCC | ||
| Type II-F | 0.25 | CGTTGAAGATGATGAAGCG | 398 | SCCmec II |
| Type II-R | 0.25 | CGAAATCAATGGTTAATGGACC | ||
| Type II-F2 | 0.2 | TAGCTTATGGTGCTTATGCG | 128 | SCCmec II, VIII |
| Type II-R2 | 0.2 | GTGCATGATTTCATTTGTGGC | ||
| Type III-F | 0.33 | CCATATTGTGTACGATGCG | 280 | Mercury element of SCCmec III |
| Type III-R | 0.33 | CCTTAGTTGTCGTAACAGATCG | ||
| Type III-F5 | 0.4 | TTCTCATTGATGCTGAAGCC | 257 | SCCmec III and IIIA |
| Type III-R6 | 0.4 | GTGTAATTTCTTTTGAAAGATATGG | ||
| Type IVa-F | 0.25 | GCCTTATTCGAAGAAACCG | 776 | SCCmec IVa |
| Type IVa-R | 0.25 | CTACTCTTCTGAAAAGCGTCG | ||
| Type IVb-F | 0.7 | TCTGGAATTACTTCAGCTGC | 493 | SCCmec IVb, IIA, IIB, IIC, IIE |
| Type IVb-R | 0.7 | AAACAATATTGCTCTCCCTC | ||
| Type IVc-F2 | 0.25 | CCTGAATCTAAAGAGATACACCG | 200 | SCCmec IVc, IVE |
| Type IVc-R2 | 0.25 | GGTTATTTTCATAGTGAATCGC | ||
| Type IVd-F5 | 1.8 | CTCAAAATACGGACCCCAATACA | 881 | SCCmec IVd |
| Type IVd-R6 | 1.8 | TGCTCCAGTAATTGCTAAAG | ||
| Type IVE-F3 | 0.75 | CAGATTCATCATTTCAAAGGC | 175 | SCCmec IVE, IVF |
| Type IVE-R4 | 0.75 | AACAACTATTAGATAATTTCCG | ||
| Type V-F | 0.33 | GAACATTGTTACTTAAATGAGCG | 325 | SCCmec V |
| Type V-R | 0.33 | TGAAAGTTGTACCCTTGACACC | ||
| ccr4-Fd | 0.35 | ATCGCTCATTATGGATACYGC | 106 | SCCmec IIA, IIB, IIC, IIE, IVE, IVF, VI, VIII |
| ccr4-R5 | 0.35 | CCATTTTTTGATAACCTGAACG | ||
| ccr4-R6 | 0.35 | CTATTTTTTTATAGCCTGAACG | ||
| MecA147-F | 0.6 | GTGAAGATATACCAAGTGATT | 147 | mecA |
| MecA147-R | 0.6 | ATGCGCTATAGATTGAAAGGAT |
Table 1. PCR reagents and volumes of each per reaction.Note: Primers previously described 9.
Discussion
The multiplex PCR assay described here provides a simple, reliable method for classifying types and major subtypes of SCCmec I-V in Staphylococci, with simultaneous discrimination of MRSA from MSSA. The assay is robust, conveniently done in a single PCR reaction and results are easy to interpret. The assay does have limitations in that it targets individual loci within each SCCmec type, but does not offer comprehensive ccr and mec complex typing, and therefore cannot be considered a gold standard. Regardless, extensive validation of the protocol has proven that it remains a valuable molecular tool for scientific and epidemiological studies on MRSA.
Due to the sophisticated nature of the assay and the large number of primers present in the reaction, there is the potential for difficulties while adapting this assay to individual laboratories. Each laboratory has their own unique conditions and factors such as primer concentration or purity, as well as thermal cycler and PCR component variations which can affect the outcome of the assay. For that reason, it is important to establish and optimize this assay in their laboratory. There are, however, several steps that will assist in successful implementation.
Sample and template preparation is a major step in any PCR reaction and, as such, the first factor affecting the success of this PCR assay is the quality of the DNA template. While any purification method that produces pure DNA would be acceptable for this assay we opted for a simple, rapid boiling method to expedite sample handling. Experience led us to realize that there are critical steps in the heating method that affect the quality of the DNA. For example, the optimal sample volume was determined to be 75-100 μl, and volumes over that were less successful. As well, the heat block for bacterial cell lysis should be set to exactly 95 °C and the sample heated for exactly 10 min, as overheating was found to degrade the DNA. Allowing the heated tubes to cool on the bench prior to centrifugation was another step that was found to increase the efficiency of the M-PCR. While poorer quality DNA may still be sufficient as a template in this assay, taken together these steps dramatically increase the quality of the DNA and make the PCR reaction more efficient.
A second factor which was found to play a significant role in the success of the PCR was the nature of the Taq DNA polymerase. This may not be universal and only 2 types of Taq polymerase were tried in our lab, however, our experience was that only the Platinum Taq from Invitrogen (Carlsbad, CA, USA) worked well, while the Recombinant Taq (Invitrogen, Carlsbad, CA, USA) resulted in unreliable results with missing products. Laboratories should also pay close attention to the quantity and purity of their primers. Primer concentration is determined based on the nature of the equipment in the individual labs and the volumes (hence concentrations) listed in this method are based on our laboratory conditions. These concentrations may need to be varied slightly in order to optimally detect each PCR product, although the relative ratios between primers should remain relatively unchanged. Another caution is related to the amplification step; it is important to ensure that the PCR tubes being used are the correct ones for the thermal cycler. If the tubes do not fit tightly into the machine and optimal heating/cooling does not occur then the PCR will be unsuccessful and several targets may or may not be amplified.
Running the PCR products on the agarose gel is a final step that, while not significantly influencing the success of the reaction, does contain steps which can improve the interpretation of results. In particular, a 2.5% agarose gel was determined to result in superior separation of the PCR products over a 2% gel. Since the assay contains a large number of smaller products with small size differences, a 2.5% gel was particularly helpful in this regard. With respect to staining, gels can be run with ethidium bromide incorporated into them, or stained post running with whichever stain is used in a particular lab. One important note here is that gel red should not be incorporated into the gel prior to running the samples because it affects the mobility of the DNA and renders band interpretation difficult to meaningless.
Despite the apparent complexity of this multiplex PCR assay, it is actually quite simple to set up and offers reliable SCCmec typing results for most samples. Of course, as more SCCmec elements are discovered and described this assay will be limited in its ability to detect them and will require updating. Since initial publication of the assay, several new SCCmec types (VII, IX, X and XI) have been described. Type XI is also unique in that it contains a new mec resistance gene (mecC) 3. Due to the nature of this assay and the specific targets that it employs, these new SCCmec types remain untypeable with the assay. Future modifications to the assay would require the addition of targets specific to these SCCmec types in order to facilitate their detection. However the assay is limited in the number of targets that are realistically possible due to number of primers required and the need to discriminate PCR product sizes. Ultimately a comprehensive assay which targets the ccr and mec complex types would offer a more complete SCCmec typing scheme. However the goal of this assay was to provide a rapid method of detecting SCCmec types and subtypes I-V, which are the most common and major SCCmec types, with presumptive identification of types VI and VIII.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank T. Ito, K. Hiramatsu, R. Daum, H. de Lencastre, and D. Coleman for the kind gift of some reference strains used in this study. This work was in part supported financially by the Centre for Antimicrobial Resistance (CAR), Alberta Health Services-Calgary/Calgary Laboratory Services/University of Calgary.
References
- IWG-SCC. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrobial Agents and Chemotherapy. 2009;53(12):4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden M, et al. Guidelines for Reporting Novel mecA Gene Homologues. Antimicrobial Agents and Chemotherapy. 2012;56(10):4997–4999. doi: 10.1128/AAC.01199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. Journal of Clinical Microbiology. 2002;40(11):4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrobial Agents and Chemotherapy. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2002;46(7):2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2007;51(9):3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheirico C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: 'SCCmec IV multiplex'. Journal of Antimicrobial Chemotherapy. 2007;60(1):42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Journal of Clinical Microbiology. 2005;43(10):5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, McClure JA, Conly JM. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Molecular and Cellular Probes. 2012;26(5):218–221. doi: 10.1016/j.mcp.2012.04.002. [DOI] [PubMed] [Google Scholar]
- de Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one-step microbial DNA extraction method using "Chelex 100" suitable for gene amplification. Research in Microbiology. 1992;143(8):785–790. doi: 10.1016/0923-2508(92)90107-y. [DOI] [PubMed] [Google Scholar]
- Lee PY, Costumbrado J, Hsu CY, Kim YH. Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. 2012. p. e3923. [DOI] [PMC free article] [PubMed]
- McClure JA, Conly JM, Elsayed S, Zhang K. Multiplex PCR assay to facilitate identification of the recently described Staphylococcal cassette chromosome mec type VIII. Molecular and Cellular Probes. 2010;24(4):229–232. doi: 10.1016/j.mcp.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang K, McClure JA, Elsayed S, Conly JM. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2009;53(2):531–540. doi: 10.1128/AAC.01118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


