Abstract
Mutations in the lamin A/C gene (LMNA) encoding A-type nuclear lamins cause dilated cardiomyopathy with variable muscular dystrophy. These mutations enhance mitogen-activated protein kinase signaling in the heart and pharmacological inhibition of extracellular signal-regulated kinase (ERK) 1 and 2 improves cardiac function in LmnaH222P/H222P mice. In the current study, we crossed mice lacking ERK1 to LmnaH222P/H222P mice and examined cardiac performance and survival. Male LmnaH222P/H222P/Erk1−/− mice lacking ERK1 had smaller left ventricular end systolic diameters and increased fractional shortening (FS) at 16 weeks of age than LmnaH222P/H222P/Erk1+/+ mice. Their mean survival was also significantly longer. However, the improved cardiac function was abrogated at 20 weeks of age concurrent with an increased activity of ERK2. LmnaH222P/H222P/Erk1−/− mice treated with an inhibitor of ERK1/2 activation had smaller left ventricular diameters and increased FS at 20 weeks of age. These results provide genetic evidence that ERK1 and ERK2 contribute to the development of cardiomyopathy caused by LMNA mutations and reveal interplay between these isoenzymes in maintaining a combined pathological activity in heart.
INTRODUCTION
Mutations in LMNA, which encodes A-type nuclear lamins, intermediate filament proteins of the nuclear envelope expressed in most differentiated somatic cells, cause a diverse range of diseases often called laminopathies (1,2). Laminopathies selectively affect different tissues and organ systems, leading to striated muscle disease, partial lipodystrophy, peripheral neuropathy and multisystem disorders such as Hutchinson–Gilford progeria syndrome that have features of accelerated aging. Of the laminopathies, the most prevalent affect striated muscle and manifest as cardiomyopathy with or without different types of skeletal muscular dystrophy (3).
The striated muscle condition caused by LMNA mutations was originally defined as distinct, clinical disorders based on the collections of patients originally studied and included autosomal Emery–Dreifuss muscular dystrophy (4), isolated cardiomyopathy and conduction defects with minimal skeletal muscle involvement (5) and limb-girdle muscular dystrophy 1B (6). It is now apparent that any of these classical clinical phenotypes, as well as overlapping phenotypes, can be caused by LMNA mutations, even by the same mutation within the same family (7,8). The common feature is cardiomyopathy characterized by left ventricular dilatation and systolic dysfunction of one or both ventricles. Affected subjects also typically have early heart block and a course complicated by other arrhythmias. While sudden death from arrhythmias may be prevented by implantation of a pacemaker and/or defibrillator, heart failure occurs in 64% of patients by the age 50 years, eventually becoming resistant to symptomatic treatment and necessitating cardiac transplantation (9).
We have discovered an abnormal activation of the extracellular signal-regulated kinase (ERK) 1/2 branch of the mitogen-activated protein kinase signaling cascade in hearts of knock-in mice with a Lmna H222P mutation (10). The orthologous mutation causes Emery–Dreifuss muscular dystrophy in humans and homozygous Lmna H222P knock-in mice develop dilated cardiomyopathy and skeletal muscle disease mimicking affected human subjects (11). We have further demonstrated that small molecule inhibitors of mitogen-activated protein/extracellular signal-regulated kinase (MEK) 1/2, the enzymes that phosphorylate and activate ERK1/2, have beneficial effects on heart function as well as survival in LmnaH222P/H222P mice (12–14).
The discovery that treatment with MEK1/2 inhibitors ameliorates cardiomyopathy in LmnaH222P/H222P mice suggests that ERK1/2 hyperactivation plays an underlying role in the pathogenesis of cardiomyopathy caused by LMNA mutations. However, unknown off-target effects could potentially confuscate the interpretation of results obtained with small molecule enzymes inhibitors. Results obtained using pharmacological inhibitors could further be influenced by factors such as the time of treatment initiation, duration of treatment and pharmacokinetic variables such as alterations in drug distribution in animals with heart failure. MEK1/2 inhibitors are also not selective in terms of blocking activation of the ERK1 and ERK2 isoenzymes. Here, we examine the role of the ERK1 in the development of cardiomyopathy caused by LMNA mutation by depleting this isoenzyme from LmnaH222P/H222 mice. The results obtained using a genetic approach, independent of pharmacological inhibitors, confirm the role of ERK1/2 in pathogenesis. They also reveal interplay between ERK1 and ERK2 in maintaining a combined pathologic activity in heart.
RESULTS
Generation of LmnaH222P/H222P mice lacking ERK1
To definitively prove that ERK1/2 plays a central role in the pathogenesis of cardiomyopathy caused by LMNA mutations, we crossed mice lacking ERK1 to LmnaH222P/H222P mice. As Erk2−/− mice die during embryonic development (15,16), we focused on ERK1, which could be completely depleted genetically. Erk1−/− mice are viable and fertile, and only thymocyte proliferation and maturation has been reported to be abnormal (17). Erk1−/− null mice have normal heart structure and function (18).
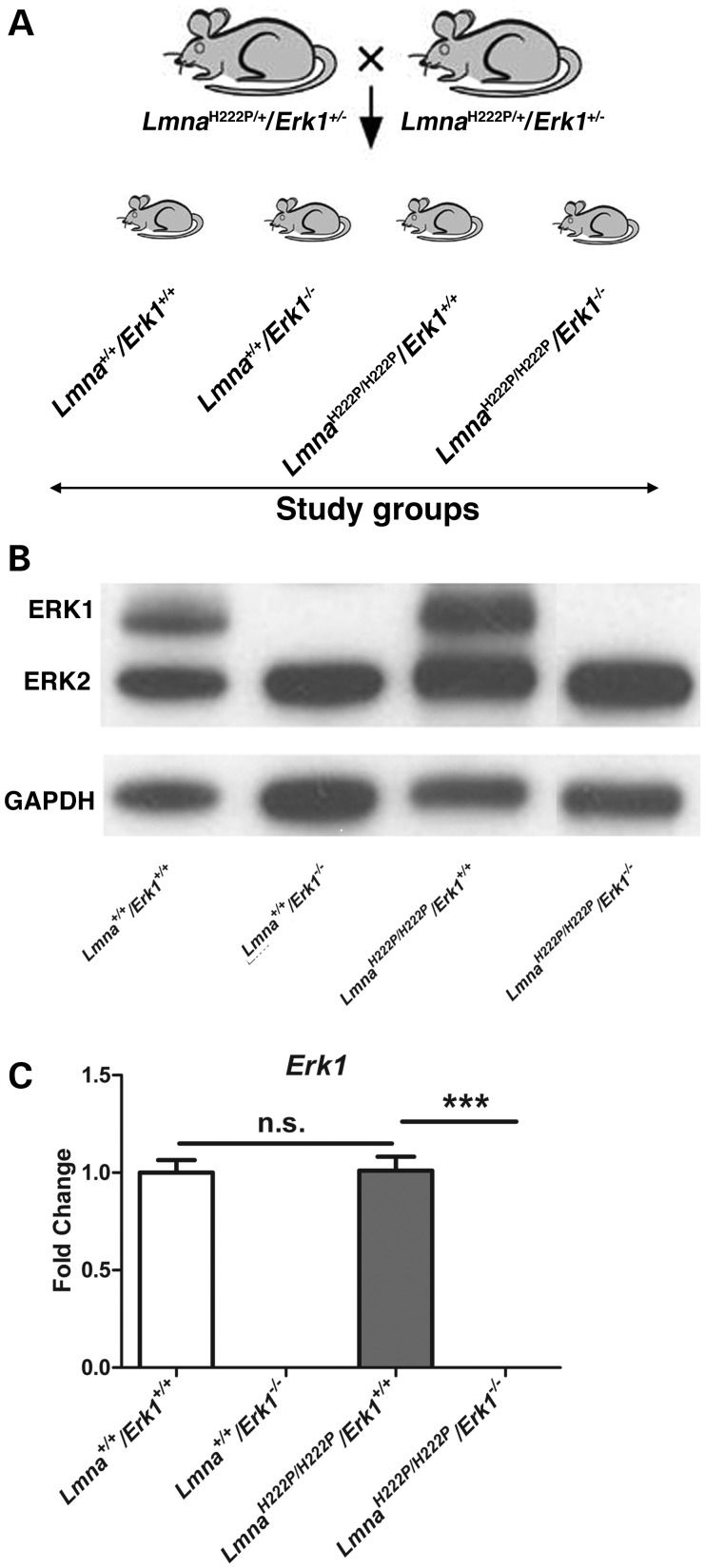
We crossed male and female LmnaH222P/+/Erk1+/− mice on a 129 genetic background to generate four study groups: Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice (Fig. 1A). To validate the loss of ERK1, we used antibodies that recognized ERK1 and ERK2 for immunoblotting of proteins extracted hearts. There was total absence of ERK1 in hearts from Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice, which was present in Lmna+/+/Erk1+/+ and LmnaH222P/H222P/Erk1+/+ mice (Fig. 1B). Erk1 mRNA was also absent from heart tissue of Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice analyzed by reverse transcription followed by real-time quantitative polymerase chain reaction (Fig. 1C).
Figure 1.
Generation of mice lacking Erk1. (A) Schematic diagrams showing sibling mating of LmnaH222P/+/Erk1+/− mice to generate the study groups. (B) Representative immunoblots using antibodies against ERK1 and ERK2 and against GAPDH to probe proteins extracted from hearts of 16-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. (C) Expression of Erk1 mRNA in hearts from Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. Values represent fold change compared with Erk1 mRNA in hearts from Lmna+/+/Erk1+/+ mice and are given as means ± standard errors in Lmna+/+/Erk1+/+ (n = 6), Lmna+/+/Erk1−/− (n = 6), LmnaH222P/H222P/Erk1+/+ (n = 5) and LmnaH222P/H222P/Erk1−/− (n = 6) mice. ***P < 0.0005; n.s., not significant.
Genetic deletion of Erk1 improves left ventricular function in LmnaH222P/H222P mice at 16 weeks of age
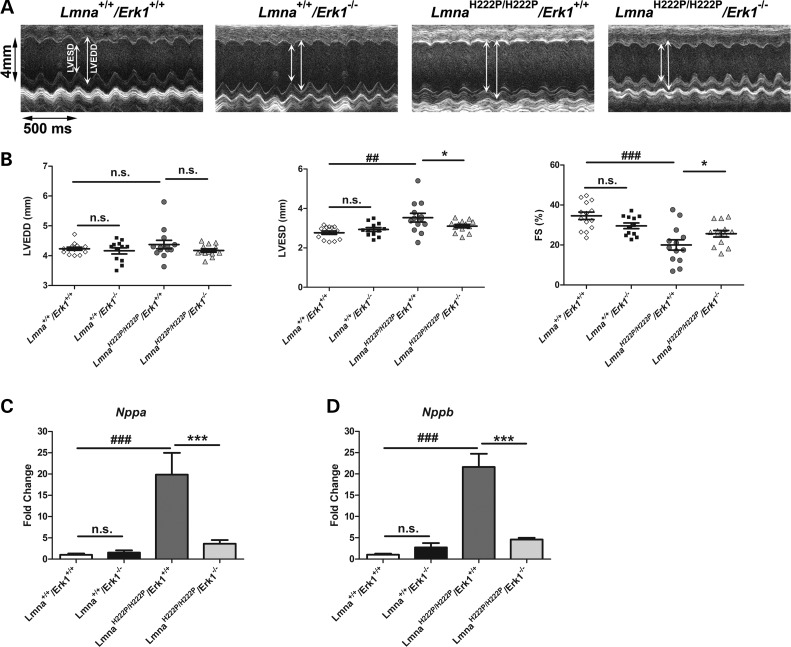
At 16 weeks of age, male LmnaH222P/H222P mice have increased left ventricular diameters, decreased fractional shortenings (FSs) and decreased ejection fractions (11). We used transthoracic echocardiography to determine if genetic deletion of Erk1 prevents these abnormalities. Echocardiograms were performed on 16-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice (Fig. 2A). From these, mean heart rate, left ventricular end diastolic diameter (LVEDD), left ventricle end systolic diameter (LVESD) and FS were determined for each geno type (Table 1). Compared with Lmna+/+/Erk1+/+ mice, LmnaH222P/H222P/Erk1+/+ mice had a significantly increased mean LVESD and decreased FS at 16 weeks (Fig. 2B). Left ventricular diameters and FS were not significantly different between Lmna+/+/Erk1+/+ and Lmna+/+/Erk1−/− mice (Fig. 2B), confirming that the lack of ERK1 was not apparently detrimental to the heart (18). However, at 16 weeks of age, LmnaH222P/H222P/Erk1−/− mice had a significant 12% smaller mean LVESD and a significant increase of ∼20% in FS compared with LmnaH222P/H222P/Erk1+/+ mice (Fig. 2B). We also examined the expression of genes encoding natriuretic peptide precursors, which are upregulated by left ventricular dilatation (19). Compared with LmnaH222P/H222P/Erk1+/+ mice, hearts from LmnaH222P/H222P/Erk1−/− mice lacking ERK1 had significantly reduced expression of Nppa, which encodes atrial natriuretic peptide A (Fig. 2C). The expression of Nppb, which encodes brain natriuretic peptide B, was also significantly reduced in LmnaH222P/H222P/Erk1−/− mice compared with LmnaH222P/H222P/Erk1+/+ (Fig. 2D). Hence, depletion of ERK1 prevented or delayed the development of significant left ventricular dilatation and dysfunction in LmnaH222P/H222P mice.
Figure 2.
Genetic deletion of Erk1 improves left ventricular function in LmnaH222P/H222P mice at 16 weeks of age. (A) Representative M-mode transthoracic echocardiographic tracings from 16-week-old male Lmna+/+/Erk1+/+ (diamonds), Lmna+/+/Erk1−/− (squares), LmnaH222P/H222P/Erk1+/+ (circles) and LmnaH222P/H222P/Erk1−/− (triangles) mice. (B) Graphs showing mean LVEDD, LVESD and FS in 16-week-old male Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. Values for each individual mouse as well as means (long horizontal bars) and standard errors are shown. ##P < 0.005; ###P < 0.0005; *P < 0.05; n.s., not significant. (C) Expression of Nppa mRNA in hearts from 16-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. Values represent fold change compared with Nppa mRNA in hearts of Lmna+/+/Erk1+/+ mice and are given as means ± standard errors in Lmna+/+/Erk1+/+ (n = 6), Lmna+/+/Erk1−/− (n = 6), LmnaH222P/H222P/Erk1+/+ (n = 5) and LmnaH222P/H222P/Erk1−/− (n = 6) mice. ###P < 0.0005; ***P < 0.0005; n.s., not significant. (D) Expression of Nppb mRNA in hearts from 16-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. Values represent fold change of Nppb mRNA in hearts of Lmna+/+/Erk1+/+ mice and are given as means ± standard errors in Lmna+/+/Erk1+/+ (n = 6), Lmna+/+/Erk1−/− (n = 6), LmnaH222P/H222P/Erk1+/+ (n = 5) and LmnaH222P/H222P/Erk1−/− (n = 6) mice. ###P < 0.0005; ***P < 0.0005; n.s., not significant.
Table 1.
Echocardiographic data for mice at age of 16 weeks of age
| Genotype | n | Heart rate (beats/min) | LVEDD (mm) | LVESD (mm) | FS (%) |
|---|---|---|---|---|---|
| Lmna+/+/Erk1+/+ | 14 | 506 ± 9 | 4.2 ± 0.1 | 2.8 ± 0.1 | 34.6 ± 1.8 |
| Lmna+/+/Erk1−/− | 12 | 534 ± 7 | 4.2 ± 0.1 | 2.9 ± 0.1* | 29.6 ± 1.4** |
| LmnaH222P/H222P/Erk1+/+ | 13 | 490 ± 9 | 4.4 ± 0.1 | 3.5 ± 0.2## | 20.0 ± 2.6### |
| LmnaH222P/H222P/Erk1−/− | 13 | 513 ± 8 | 4.2 ± 0.1 | 3.1 ± 0.1*# | 25.6 ± 1.6*# |
Values are means ± standard errors. Compared with LmnaH222P/H222P/Erk1+/+ *P < 0.05 and **P < 0.005; compared with Lmna+/+/Erk1+/+ male mice #P < 0.05 and ###P < 0.0005.
Effect of genetic deletion of Erk1 on survival of LmnaH222P/H222P mice
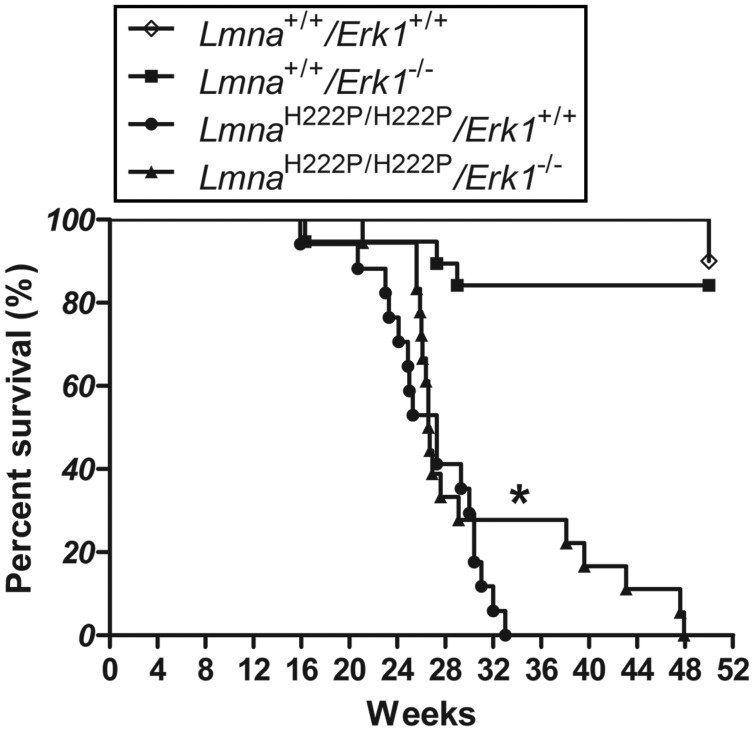
LmnaH222P/H222P mice have a significantly shorter lifespan than wild-type mice with all of the male mice dying between 16 and 40 weeks of age (11,20). We have previously shown that treatment with the MEK1/2 inhibitor selumetinib, which blocks activation of both ERK1 and ERK2, prolonged the median survival of LmnaH222P/H222P mice by >10% (14). We therefore examined if the lack of ERK1 altered survival (the time to death or significant distress requiring euthanasia) in LmnaH222P/H222P mice. Compared with LmnaH222P/H222P/Erk1+/+ mice, LmnaH222P/H222P/Erk1−/− mice had a significantly prolonged mean but not median survival (Fig. 3). The maximum survival time of LmnaH222P/H222P/Erk1−/− was 48 weeks, whereas no LmnaH222P/H222P/Erk1+/+ mouse lived longer than 33 weeks. Deficiency of ERK1 therefore appeared provides a modest albeit not robust survival benefit to LmnaH222P/H222P mice.
Figure 3.
Mouse survival analysis. Kaplan–Meier survival curves for Lmna+/+/Erk1+/+ (n = 10), Lmna+/+/Erk1−/− (n = 19), LmnaH222P/H222P/Erk1+/+ (n = 18) and LmnaH222P/H222P/Erk1−/− (n = 17). *P < 0.05 for difference in mean survival between LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice.
Abrogation of improved left ventricular function at 20 weeks of age in LmnaH222P/H222P mice with deletion of Erk1
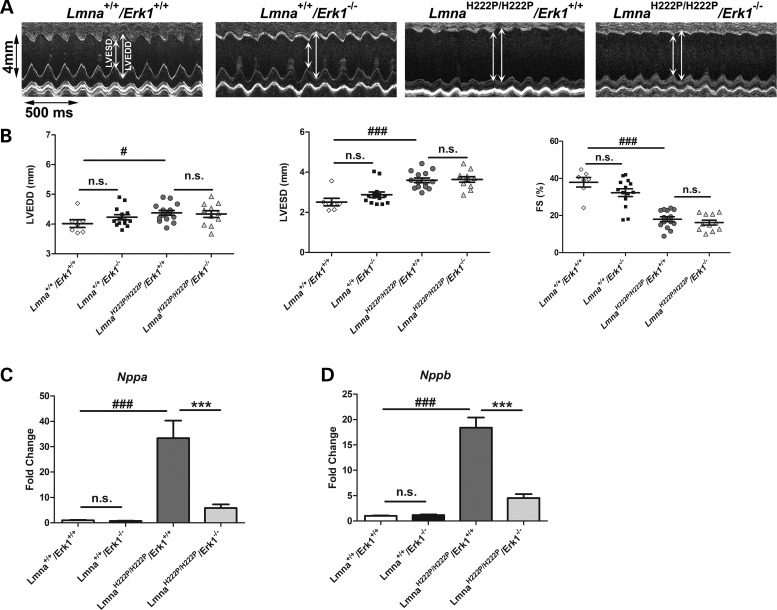
Because deficiency of ERK1 led to only a modest survival benefit in LmnaH222P/H222P mice, we asked if the improved left ventricular function observed at 16 weeks of age was maintained as the animals aged. To answer this question, we performed echocardiograms on 20-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice (Fig. 4A). From these, mean heart rate, LVEDD, LVESD and FS were determined for each genotype (Table 2). Compared with Lmna+/+/Erk1+/+ mice, LmnaH222P/H222P/Erk1+/+ mice had a significantly increased mean LVEDD, LVESD and decreased FS at 20 weeks (Fig. 4B). At this age, however, there were no significant differences between these parameters in LmnaH222P/H222P/Erk1−/− and LmnaH222P/H222P/Erk1+/+ mice (Fig. 4B). LmnaH222P/H222P/Erk1−/− mice lacking ERK1 still had reduced expression of Nppa (Fig. 4C) and Nppb (Fig. 4D) at 20 weeks of age, possibly suggesting a lag time in their enhanced synthesis. Nonetheless, at 20 weeks of age, most of the beneficial effects of Erk1 deletion on cardiac function in LmnaH222P/H222P are lost.
Figure 4.
Abrogation of improved left ventricular function at 20 weeks of age in LmnaH222P/H222P mice with deletion of Erk1. (A) Representative M-mode transthoracic echocardiographic tracings from 20-week-old male Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. (B) Graphs showing mean LVEDD, LVESD and FS in 20-week-old male Lmna+/+/Erk1+/+ (diamonds), Lmna+/+/Erk1−/− (squares), LmnaH222P/H222P/Erk1+/+ (circles) and LmnaH222P/H222P/Erk1−/− (triangles) mice. Values for each individual mouse as well as means (long horizontal bars) and standard errors are shown. #P < 0.05; ###P < 0.0005; n.s., not significant. (C) Expression of Nppa mRNA in hearts from 20-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. Values represent fold change compared with Nppa mRNA in hearts of LmnaH222P/H222P/Erk1+/+ mice and are given as means ± standard errors in Lmna+/+/Erk1+/+ (n = 4), Lmna+/+/Erk1−/− (n = 4), LmnaH222P/H222P/Erk1+/+ (n = 3) and LmnaH222P/H222P/Erk1−/− (n = 5) mice. ###P < 0.0005; ***P < 0.0005; n.s., not significant. (D) Expression of Nppb mRNA in hearts from 20-week-old Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. Values represent fold change compared with Nppb mRNA in hearts of LmnaH222P/H222P/Erk1+/+ mice and are given as means ± standard errors in Lmna+/+/Erk1+/+ (n = 4), Lmna+/+/Erk1−/− (n = 4), LmnaH222P/H222P/Erk1+/+ (n = 3) and LmnaH222P/H222P/Erk1−/− (n = 5) mice. ###P < 0.0005; ***P < 0.0005; n.s., not significant.
Table 2.
Echocardiographic data for mice at age of 20 weeks of age
| Genotype | n | Heart rate (beats/min) | LVEDD (mm) | LVESD (mm) | FS (%) |
|---|---|---|---|---|---|
| Lmna+/+/Erk1+/+ | 7 | 491 ± 5 | 4.0 ± 0.1 | 2.5 ± 0.2 | 37.9 ± 2.6 |
| Lmna+/+/Erk1−/− | 14 | 500 ± 2 | 4.2 ± 0.1 | 2.9 ± 0.1** | 32.3 ± 2.1*** |
| LmnaH222P/H222P/Erk1+/+ | 14 | 504 ± 1 | 4.4 ± 0.1 | 3.6 ± 0.1### | 18.0 ± 1.3### |
| LmnaH222P/H222P/Erk1−/− | 11 | 509 ± 6 | 4.3 ± 0.1 | 3.6 ± 0.1### | 16.2 ± 1.3### |
Values are means ± standard errors. Compared with LmnaH222P/H222P/Erk1+/+ mice **P < 0.005 and ***P < 0.0005; compared with Lmna+/+/Erk1+/+ mice ###P < 0.0005.
Enhanced ERK2 activity in hearts of 20-week-old LmnaH222P/H222P mice lacking ERK1
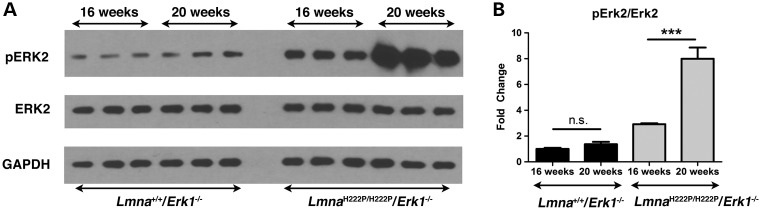
To explain the loss of benefit Erk1 deletion on left ventricular function in LmnaH222P/H222P mice, we hypothesized that there was an increase in ERK2 activity. We tested this by examining the expression and activation of ERK2 in hearts from LmnaH222P/H222P/Erk1−/− mice at 16 and 20 weeks of age. We performed immunoblotting to detect phosphorylated (activated) and total ERK2 in hearts of Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice (Fig. 5A). In hearts from Lmna+/+/Erk1−/− mice lacking ERK1 without the Lmna H222P mutation, there was no significant change in ERK2 activity between 16 and 20 weeks of age (Fig. 5B). In hearts from LmnaH222P/H222P/Erk1−/− mice, however, there was a significant increase in ERK2 activity at 20 weeks compared with 16 weeks of age (Fig. 5A and B).
Figure 5.
Increased activation of ERK2 in hearts of 20-week-old LmnaH222P/H222P mice lacking Erk1. (A) Representative immunoblots using antibodies against phosphorylated ERK2, total ERK2 and GAPDH to probe proteins extracted from hearts of 16-week-old and 20-week-old Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice. (B) Bar graphs showing quantification of pERK2/ERK2 in hearts of 16-week-old and 20-week-old Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice. Values are means ± standard errors (n = 3). ***P < 0.0005; n.s., not significant.
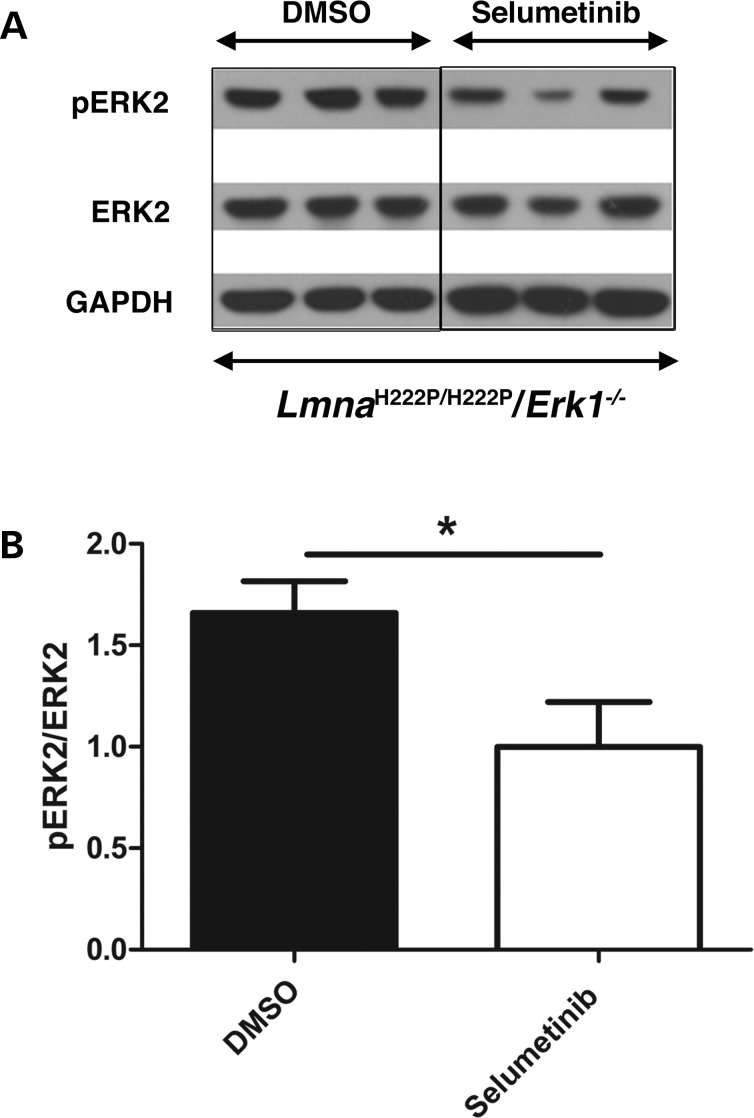
The finding of increased ERK2 activity after 16 weeks of age in hearts of LmnaH222P/H222P/Erk1−/− mice suggested that it ‘pathologically compensates’ for the loss of ERK1 in these animals. To test this hypothesis, we treated 16-week-old LmnaH222P/H222P/Erk1−/− mice with selumetinib, which inhibits MEK1/2, the kinase that phosphorylates ERK2. As shown by immunoblotting of proteins in tissue homogenates (Fig. 6A), hearts of 20-week-old mice treated with selumetinib had significantly reduced levels of phosphorylated ERK2 compared with those in hearts from mice treated with dimethyl sulfoxide (DMSO) as a placebo (Fig. 6B).
Figure 6.
Treatment with selumetinib blocks ERK2 phosphorylation in hearts of LmnaH222P/H222P mice lacking Erk1. (A) Representative immunoblots using antibodies against phosphorylated ERK2, ERK2 and GAPDH to probe proteins extracted from hearts of 20-week-old LmnaH222P/H222P/Erk1−/− mice treated with DMSO placebo or selumetinib. (B) Bar graphs showing quantification of pERK2/ERK2 in hearts 20-week-old LmnaH222P/H222P/Erk1−/− mice treated with DMSO placebo or selumetinib. Values are means ± standard errors (n = 3). *P < 0.05.
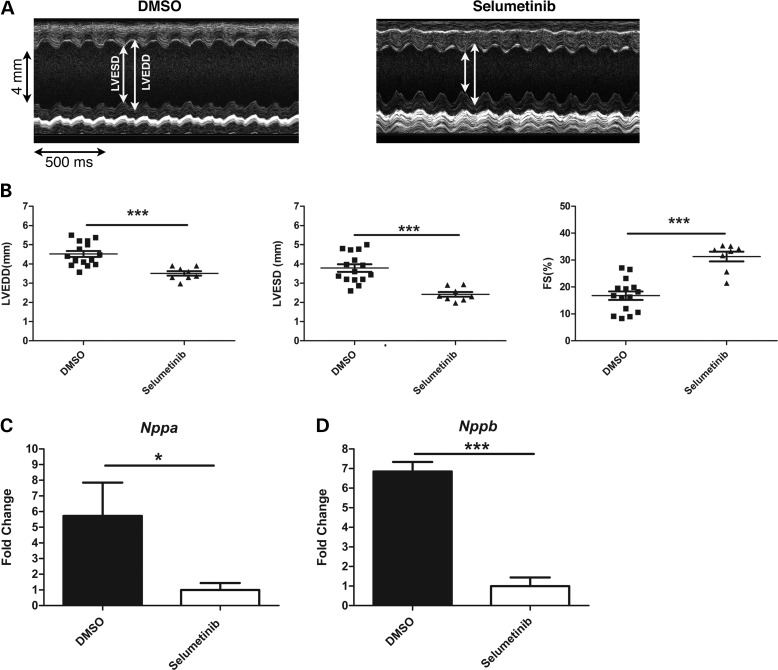
We performed transthoracic echocardiograms on LmnaH222P/H222P/Erk1−/− treated with selumetinib for placebo from 16 to 20 weeks of age (Fig. 7A). From the echocardiograms, we calculated mean heart rate, LVEDD, LVESD and FS (Table 3). LVEDD and LVESD were both significantly smaller and FS significantly greater in LmnaH222P/H222P/Erk1−/− mice treated with selumetinib compared with placebo-treated mice (Fig. 7B). Hearts from the selumetinib-treated LmnaH222P/H222P/Erk1−/− mice also had significantly reduced expression of Nppa (Fig. 7C) and Nppb (Fig. 7D) compared with placebo-treated controls. These results showed that inhibition of ERK2 in hearts of LmnaH222P/H222P lacking ERK1 improves left ventricular performance, at least in part reversing the effects of the observed increase in ERK2 activity that occurs after 16 weeks of age in these animals.
Figure 7.
Treatment with selumetinib improves cardiac function in LmnaH222P/H222P mice lacking Erk1. (A) Representative M-mode transthoracic echocardiographic tracings from 20-week-old male LmnaH222P/H222P/Erk1−/− mice treated with DMSO placebo or selumetinib. (B) Graphs showing mean LVEDD, LVESD and FS in 20-week-old male LmnaH222P/H222P/Erk1−/− mice treated with DMSO placebo or selumetinib. Values for each individual mouse (squares and triangles) as well as means (long horizontal bars) and standard errors are shown. ***P < 0.0005. (C) Expression of Nppa mRNA in hearts from 20-week-old LmnaH222P/H222P/Erk1−/− mice treated with DMSO placebo or selumetinib. Values represent fold change compared with Nppa mRNA in hearts of selumetinib-treated LmnaH222P/H222P/Erk−/− mice and are given as means ± standard errors (n = 4). ***P < 0.0005. (D) Expression of Nppb mRNA in hearts from 20-week-old LmnaH222P/H222P/Erk1−/− mice treated with DMSO placebo or selumetinib. Values represent fold change compared with Nppb mRNA in hearts of selumetinib-treated LmnaH222P/H222P/Erk−/− mice and are given as means ± standard errors (n = 4). ***P < 0.0005.
Table 3.
Echocardiographic data for 20-week-old male LmnaH222P/H222P/Erk1−/− mice treated with DMSO or selumetinib
| Treatment | n | Heart rate (beats/ min) | LVEDD (mm) | LVESD (mm) | FS (%) |
|---|---|---|---|---|---|
| DMSO | 15 | 500 ± 1 | 4.5 ± 0.2 | 3.8 ± 0.2 | 16.8 ± 1.6 |
| Selumetinib | 8 | 500 ± 1 | 3.5 ± 0.1*** | 2.4 ± 0.1*** | 31.3 ± 1.8*** |
Values are means ± standard errors; ***P < 0.0005 for comparison between DMSO and selumetinib-treated LmnaH222P/H222P/Erk1−/− mice.
Enhanced p38α and c-Jun NH2-terminal kinase activity in hearts of 20-week-old LmnaH222P/H222P mice lacking ERK1
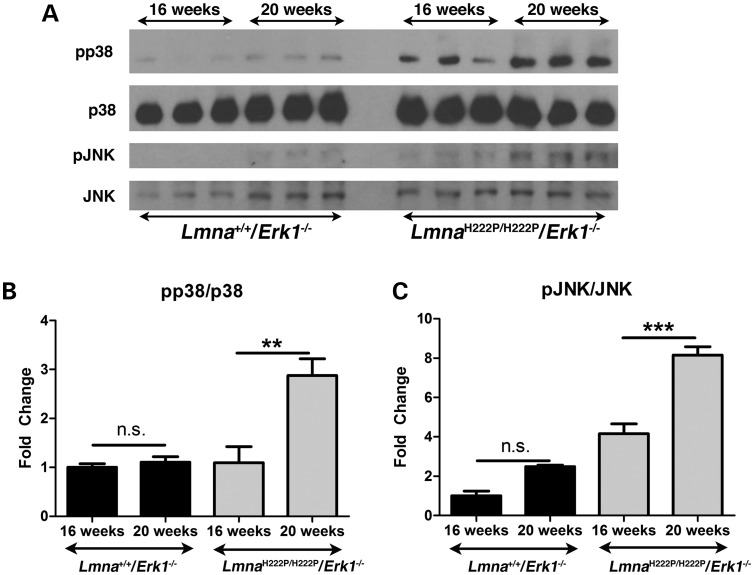
We have previously shown that in addition to ERK1/2 the mitogen-activated protein kinases p38α and c-Jun NH2-terminal kinase (JNK) have increased activities in hearts of LmnaH222P/H222P mice (10,13,21). We therefore examined their activities in hearts from LmnaH222P/H222P/Erk1−/− mice at 16 and 20 weeks of age. We performed immunoblotting to detect phosphorylated (activated) and total p38α and phosphorylated (activated) and total JNK in hearts of Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice (Fig. 8A). In hearts of Lmna+/+/Erk1−/− mice lacking ERK1 without the Lmna H222P mutation, there was no significant change p38α (Fig. 8B) or JNK (Fig. 8C) activities between 16 and 20 weeks of age. In hearts from LmnaH222P/H222P/Erk1−/− mice, however, there were significant increases in both p38α (Fig. 8B) and JNK (Fig. 8C) activities at 20 weeks compared with 16 weeks of age.
Figure 8.
Increased activation p38α and JNK in hearts of 20-week-old LmnaH222P/H222P mice lacking Erk1. (A) Representative immunoblots using antibodies against phosphorylated p38α (pp38), total p38α (p38), phosphorylated JNK (pJNK) and total JNK (JNK) to probe proteins extracted from hearts of 16-week-old and 20-week-old Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice. (B) Bar graphs showing quantification of the ratio of phosphorylated p38α to total p38α (pp38/p38) in hearts of 16-week-old and 20-week-old Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice. Values are means ± standard errors (n = 3). **P < 0.0005; n.s., not significant. (C) Bar graphs showing quantification of the ratio of phosphorylated JNK to total JNK (pJNK/JNK) in hearts of 16-week-old and 20-week-old Lmna+/+/Erk1−/− and LmnaH222P/H222P/Erk1−/− mice. Values are means ± standard errors (n = 3). ***P < 0.0005; n.s., not significant.
DISCUSSION
Abnormally increased activation of mitogen-activated protein kinases, including ERK1/2, has been implicated in the pathogenesis of cardiomyopathy caused by LMNA mutations (10). We have now shown that germline deletion of Erk1 from mice with the cardiomyopathy-causing LmnaH222P/H222P mutation leads to improved heart function at 16 weeks of age. At this age, male LmnaH222P/H222P mice have ∼30% reduced left ventricular FS (11). Genetic deletion of Erk1 leads to a significant improvement in FS of ∼30%.
The improvement in heart function in LmnaH222P/H222P mice lacking Erk1 disappeared at 20 weeks of age concurrent with a greater than 2-fold increase in ERK2 activation in the LmnaH222P/H222P mice lacking ERK1. Blocking cardiac ERK2 activity by treatment with the MEK1/2 inhibitor selumetinib leads to a significant improvement in FS at 20 weeks, strongly suggesting that the increased ERK2 activity after 16 weeks of age leads to the deterioration in heart function in the LmnaH222P/H222P mice lacking Erk1. In embryonic fibroblasts and thymocytes from Erk1−/− mice, the amount of ERK2 is unchanged but there is a more sustained activation than in wild-type cells after stimulation with extracellular factors (17). Brains from Erk1−/− mice similarly have no change in the amount of ERK2 but neurons in primary culture have enhanced phosphorylation of ERK2 after stimulating glutamate receptors or inducing membrane depolarization with potassium chloride (22). Removal of ERK1 from cultured fibroblasts by RNA interference also increases activation of ERK2 (23,24). In hearts of LmnaH222P/H222P mice lacking Erk1, however, the increased ‘pathological’ activation of ERK2 did not occur until after 16 weeks of age. Hence, germline depletion of Erk1 had a transient beneficial effect on cardiomyopathy in these animals. This enhanced ERK2 activity that occurs at a later age in hearts of mice lacking Erk1 has not been previously reported. In a study of a mouse model of Noonan syndrome in which the animals develop hypertrophic cardiomyopathy, depletion of ERK1 by crossing to Erk1−/− mice resulted in improved cardiac function at 15 weeks of age; however, heart function at later ages was not reported (25). Studies examining the role of ERK1/2 in protecting myocardium from ischemic injury and in cardiac hypertrophy also did not look at ERK2 activity in Erk1−/− mice older than 10 weeks (18,26).
The precise roles of ERK1/2 in normal physiology and pathology are not entirely clear despite considerable research. Enhanced activation of ERK1/2 occurs in dilated hearts of human subjects with late-stage cardiac failure resulting from several different causes (27,28). In contrast, hearts of male LmnaH222P/H222P mice have detectable increased activity of these kinases as early as 4 weeks of age, when cardiac structure and function are normal and mice have no evidence of cardiomyopathy (10,29). Primary activation of ERK1/2 signaling has most often been associated with a hypertrophic response. Transgenic expression in mice of the ERK1/2 kinase MEK1 has been reported to lead to compensated hypertrophy without signs of cardiomyopathy or lethality up to 12 months of age (30). Expression in transgenic mice of Ras, which activates MEK1/2, has been reported to cause cardiac hypertrophy and selective diastolic dysfunction (31). However, genetic deletion of both ERK1 and ERK2 from mouse hearts does not block the cardiac hypertrophic response but does affect the balance between eccentric and concentric growth (32). In humans, activation mutations affecting components of the Ras-Raf-MEK-ERK pathway that activate the ERK1/2 signaling cause several syndromatic disorders that often have cardiac hypertrophy as a feature (33–39). However, LmnaH222P/H222P mice and most human subjects with cardiomyopathy caused by LMNA mutations have left ventricular dilation without a preceding hypertrophic phase (9,11). Therefore, the pathogenic response of the heart to early, enhanced ERK1/2 activity is complex, likely resulting in hypertrophic or primary dilated cardiomyopathy depending upon factors such as the degree of activation, time of onset of abnormal activity and concurrent activities of other pathways. In addition to enhanced ERK1/2 signaling, hearts from LmnaH222P/H222P mice also have enhanced activity of the mitogen-activated protein kinases JNK and p38α early in the course of disease (10,13,21), which we observed in the hearts of these mice lacking Erk1. However, LmnaH222P/H222P/Erk1−/− appeared to have increased p38α activity compared with ‘control’ Lmna+/+/Erk1−/− mice at 20 weeks but not 16 weeks of age (Fig. 8), whereas on an Erk1+/+ background LmnaH222P/H222P mice have increased activity of this kinase compared with Lmna+/+ mice at earlier ages (21). This result suggests some type of ‘cross talk’ between ERK1/2 and p38α signaling. There is also enhanced AKT-mTOR signaling prior to the onset of cardiomyopathy in hearts of LmnaH222P/H222P mice, which is reduced by blocking ERK1/2 activity (40), suggesting ‘cross talk’ between ERK1/2 and this pathway as well in hearts of LmnaH222P/H222P mice. Future experiments using inhibitors of p38α, JNK and mTOR in LmnaH222P/H222P/Erk1−/− mice and examining their effects on heart function could help elucidate the separate or overlapping contributions of these different signaling pathways to heart pathology caused by Lmna mutation.
While we have previously shown benefits of MEK1/2 inhibitors in LmnaH222P/H222P mice (12–14), the present experimental results using a purely genetic approach definitively confirm ERK1 as a molecular target for therapeutic interventions. While treatment with MEK1/2 inhibitors provide some survival benefit to LmnaH222P/H222P mice (14), they still have a shortened lifespan compared with wild-type animals. Early death of LmnaH222P/H222P mice even after blocking ERK1/2 activity may be secondary to involvement of the diaphragm, which does not occur in human subjects with LMNA mutations. The early death may also be due to cardiac arrhythmias, which in human could be prevented by implantation of a pacemaker and defibrillator. In addition, attenuating other contributing pathogenic mechanisms in addition to ERK1/2 signaling may be required to obtain a robust therapeutic response. Nonetheless, the fact that inhibition of ERK1/2 signaling has clear beneficial effects on cardiac function and confers survival benefits in LmnaH222P/H222P mice provide strong support for a clinical trial of a MEK1/2 in human subjects. Only this will determine if such treatment can slow the progression of heart damage, improve cardiac function and delay the development of severe heart failure in patients with cardiomyopathy caused by LMNA mutations.
MATERIALS AND METHODS
Mice
LmnaH222P/H222P mice (11) were obtained from Dr Gisèle Bonne (Institut de Myologie, Paris) and Erk1+/− mice (17) from Drs Gilles Pagès and Jacques Pouysségur (Université de Nice). A female Erk1+/− mouse (C57BL/6 genetic background) was crossed to a male LmnaH222P/H222P mouse (129 genetic background) to obtain LmnaH222P/+/Erk1+/− mice. LmnaH222P/+/Erk1+/− mice were then backcrossed to LmnaH222P/H222P for five generations to generate the sixth generation of LmnaH222P/+/Erk1+/− on a 129 genetic background. These mice were then crossed to generate Lmna+/+/Erk1+/+, Lmna+/+/Erk1−/−, LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice. These mice and offspring from interbreeding of siblings of each of these genotypes were used for the experiments reported in this paper. Genotyping was performed by PCR using genomic DNA isolated from tail clippings. Mice were fed a chow diet and housed in a disease-free barrier facility with 12/12 h light/dark cycles. The Institutional Animal Care and Use Committee at Columbia University Medical Center approved the use of animals and all experimental protocols.
Protein extraction and immunoblotting
Hearts were excised from sacrificed mice and tissue homogenized in extraction buffer as described previously (10,12). Proteins were separated by to SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes and blotted with primary antibodies against ERK1/2 (No. Sc-94, Santa-Cruz), phosphorylated ERK1/2 (No. 9101, Cell Signaling) and GAPDH (No. AM4300, Ambion), p38 (No. 9212, Cell Signaling), phosphorylated p38 (No. 4511, Cell Signaling), JNK1/3 (No. Sc474, Santa-Cruz) and phosphorylated JNK (No. 9251, Cell Signaling). Secondary antibodies were horseradish peroxidase-conjugated (Amersham). Recognized proteins were visualized with SuperSignal West Pico Cheminescent Substrate (Thermo Scientific). Quantification of immunoblots was performed using ImageJ64 software.
Reverse transcription and quantitative polymerase chain reaction
Total RNA was extracted from heart tissue using the RNeasy isolation kit (Qiagen) as described previously (12,13). The cDNA was synthesized using Superscript First Strand Synthesis System according to the manufacturer's instructions (Invitrogen) on total RNA. For each replicate in each experiment, RNA from tissue samples of different animals was used. Primers were designed correspond to mouse RNA sequences using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) for Nppa (forward: 5′-gcttccaggccatattggag-3′; reverse: 5′-ccctgcttcctcagtctgct-3′), Nppb (forward: 5′-ggaccaaggcctcacaaaag-3′; reverse: 5′-tacagcccaaacgactgacg-3′), Erk1 (forward: 5′-cctgaagccttccaatctgc-3′; reverse: 5′-atgatctctggggctcggta-3′), Gapdh (forward: 5′-tgcaccaccaactgcttag -3′; reverse: 5′-ggatgcagggatgatgttc-3′). Quantitative polymerase chain reaction reactions were carried out on an ABI 7300 Real-Time PCR System (Applied Biosystems) using HotStart-IT SYBR green qPCR Master Mix (Affymetrix). Relative levels of mRNA expression were calculated using the ΔΔCT method (41). Individual expression values were normalized by comparison with Gapdh mRNA.
Thansthoracic echocardiography
Mice were anesthetized with isoflurane/oxygen (1.5%) and body temperature was maintained at ∼37°C. Transthoracic 2D and M-mode echocardiography was performed using a Vevo 770 imaging system (Visualsonics) equipped with a 30-MHz linear transducer applied to the chest wall. Measurements of LVESD, LVEDD and FS were averaged from at least three separate cardiac cycles for the number of animals indicated in each experiment. The echocardiography was blinded to the genotype or treatment of the mice.
Mouse survival analysis
End points for survival analysis were death or significant distress requiring euthanasia. Significant distress included (i) difficulty with normal ambulation, (ii) failure to eat or drink, (iii) loss of >20% body mass, (iv) depression, (v) rough or unkempt hair coat and (vi) significant respiratory distress. These were confirmed by consulting with veterinarians at the Institute of Comparative Medicine at Columbia University Medical Center. Euthanasia was performed in a CO2 chamber followed by cervical dislocation, according to the protocol of the Institute of Comparative Medicine. Euthanasia was confirmed by checking for lack of response to limb and tail pinch.
Selumetinib treatment
Selumetinib (Selleck Chemicals) was dissolved in DMSO (Sigma) at a concentration of 0.5 mg/ml. The placebo control consisted of the same volume of DMSO. Selumetinib was administered at a dose of 1 mg/kg/day by intraperitoneal injection using a 27 G 5/8 syringe starting when mice were 16 weeks of age and continuing until 20 weeks of age.
Statistical analysis
Statistical significance comparison between two groups was determined using the Student t-test. For comparisons between more than two groups, significance was determined using Welch ANOVA and Tukey adjustment for post hoc multiple comparisons. Survival curves were generated using the Kaplan–Meier estimator method (42). Survival data for LmnaH222P/H222P/Erk1+/+ and LmnaH222P/H222P/Erk1−/− mice passed D'Agostino and Pearson omnibus normality test (43) and Student t-test was then used to determine statistical significance between mean survivals of these two groups. Statistical analyses were performed using GraphPad (Prism Software).
Conflict of Interest statement. H.J.W. and A.M. are inventors on a pending United States Patent Application ‘Methods for Treating and/or Preventing Cardiomyopathies by ERK or JNK Inhibition’ (US 20110110916 A1) filed by The Trustees of Columbia University in the City of New York.
FUNDING
This work was supported by the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (grant number R01AR048997) to H.J.W. A.M. was supported by L'Association Française contre les Myopathies.
ACKNOWLEDGEMENTS
We are grateful to Dr Gisèle Bonne (Institut de Myologie, Paris) for providing Lmna H222P knock-in mice and Dr Gilles Pagès and Dr Jacques Pouysségur (Université de Nice) for providing Erk1 knockout mice.
REFERENCES
- 1.Dauer W.T., Worman H.J. The nuclear envelope as a signaling node in development and disease. Dev. Cell. 2009;17:626–638. doi: 10.1016/j.devcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Worman H.J., Fong L.G., Muchir A., Young S.G. Laminopathies and the long strange trip from basic cell biology to therapy. J. Clin. Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J., Muchir A., Nagy P.L., Worman H.J. LMNA cardiomyopathy: cell biology and genetics meet clinical medicine. Dis. Model. Mech. 2011;4:562–568. doi: 10.1242/dmm.006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 5.Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Spudich S., De Girolami U., et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 6.Muchir A., Bonne G., van der Kooi A.J., van Meegen M., Baas F., Bolhuis P.A., de Visser M., Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 7.Bonne G., Mercuri E., Muchir A., Urtizberea A., Bécane H.M., Recan D., Merlini L., Wehnert M., Boor R., Reuner U., et al. Clinical and molecular genetic spectrum of autosomal dominant emery–dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol. 2000;48:170–180. [PubMed] [Google Scholar]
- 8.Brodsky G.L., Muntoni F., Miocic S., Sinagra G., Sewry C., Mestroni L. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation. 2000;101:473–476. doi: 10.1161/01.cir.101.5.473. [DOI] [PubMed] [Google Scholar]
- 9.van Berlo J.H., de Voogt W.G., van der Kooi A.J., van Tintelen J.P., Bonne G., Yaou R.B., Duboc D., Rossenbacker T., Heidbüchel H., de Visser M., et al. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J. Mol. Med. 2005;83:79–83. doi: 10.1007/s00109-004-0589-1. [DOI] [PubMed] [Google Scholar]
- 10.Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in emery–dreifuss muscular dystrophy. J. Clin. Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacène E., Fromes Y., Toussaint M., Mura A.M., Keller D.I., et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 12.Muchir A., Shan J., Bonne G., Lehnart S.E., Worman H.J. Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet. 2009;18:241–247. doi: 10.1093/hmg/ddn343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W., Muchir A., Shan J., Bonne G., Worman H.J. Mitogen activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by lamin A/C gene mutation. Circulation. 2011;123:53–61. doi: 10.1161/CIRCULATIONAHA.110.970673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muchir A., Reilly S.A., Wu W., Iwata S., Homma S., Bonne G., Worman H.J. Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc. Res. 2012;93:311–319. doi: 10.1093/cvr/cvr301. [DOI] [PubMed] [Google Scholar]
- 15.Yao Y., Li W., Wu J., Germann U.A., Su M.S., Kuida K., Boucher D.M. Extracellular signal-regulated kinase 2 is necessary for mesoderm differentiation. Proc. Natl Acad. Sci. USA. 2003;100:12759–12764. doi: 10.1073/pnas.2134254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saba-El-Leil M.K., Vella F.D., Vernay B., Voisin L., Chen L., Labrecque N., Ang S.L., Meloche S. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 2003;4:964–968. doi: 10.1038/sj.embor.embor939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagès G., Guerin S., Grall D., Bonino F., Smith A., Anjuere F., Auberger P., Pouysségur J. Defective thymocyte maturation in p44 MAP kinase (Erk1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 18.Purcell N.H., Wilkins B.J., York A., Saba-El-Leil M.K., Meloche S., Robbins J., Molkentin J.D. Genetic inhibition of cardiac ERK1/2 promotes stress-induced apoptosis and heart failure but has no effect on hypertrophy in vivo. Proc. Natl Acad. Sci. USA. 2007;104:14074–14079. doi: 10.1073/pnas.0610906104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi T., Allen P.D., Izumo S. Expression of A-, B-, and C-type natriuretic peptide genes in failing and developing human ventricles: correlation with expression of the Ca(2+)-ATPase gene. Circ. Res. 1992;71:9–17. doi: 10.1161/01.res.71.1.9. [DOI] [PubMed] [Google Scholar]
- 20.Arimura T., Sato R., Machida N., Bando H., Zhan D.Y., Morimoto S., Tanaka R., Yamane Y., Bonne G., Kimura A. Improvement of left ventricular dysfunction and of survival prognosis of dilated cardiomyopathy by administration of calcium sensitizer SCH00013 in a mouse model. J. Am. Coll. Cardiol. 2010;55:1503–1505. doi: 10.1016/j.jacc.2009.10.065. [DOI] [PubMed] [Google Scholar]
- 21.Muchir A., Wu W., Choi J.C., Iwata S., Morrow J., Homma S., Worman H.J. Abnormal p38α mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 2012;21:4325–4333. doi: 10.1093/hmg/dds265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzucchelli C., Vantaggiato C., Ciamei A., Fasano S., Pakhotin P., Krezel W., Welzl H., Wolfer D.P., Pagès G., Valverde O., et al. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron. 2002;34:807–820. doi: 10.1016/s0896-6273(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 23.Lefloch R., Pouysségur J., Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol. Cell. Biol. 2008;28:511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vantaggiato C., Formentini I., Bondanza A., Bonini C., Naldini L., Brambilla R. ERK1 and ERK2 mitogen-activated protein kinases affect Ras-dependent cell signaling differentially. J. Biol. 2006;5:14. doi: 10.1186/jbiol38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura T., Colbert M., Krenz M., Molkentin J.D., Hahn H.S., Dorn G.W., Robbins J. Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J. Clin. Invest. 2007;117:2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lips D.J., Bueno O.F., Wilkins B.J., Purcell N.H., Kaiser R.A., Lorenz J.N., Voisin L., Saba-El-Leil M.K., Meloche S., Pouysségur J., et al. MEK1-ERK2 Signaling pathway protects myocardium from ischemic injury in vivo. Circulation. 2004;109:1938–1941. doi: 10.1161/01.CIR.0000127126.73759.23. [DOI] [PubMed] [Google Scholar]
- 27.Haq S., Choukroun G., Lim H., Tymitz K.M., del Monte F., Gwathmey J., Grazette L., Michael A., Hajjar R., Force T., et al. Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation. 2001;103:670–677. doi: 10.1161/01.cir.103.5.670. [DOI] [PubMed] [Google Scholar]
- 28.Takeishi Y., Huang Q., Abe J., Che W., Lee J.D., Kawakatsu H., Hoit B.D., Berk B.C., Walsh R.A. Activation of mitogen-activated protein kinases and p90 ribosomal S6 kinase in failing human hearts with dilated cardiomyopathy. Cardiovasc. Res. 2002;53:131–137. doi: 10.1016/s0008-6363(01)00438-2. [DOI] [PubMed] [Google Scholar]
- 29.Choi J.C., Wu W., Muchir A., Iwata S., Homma S., Worman H.J. Dual specificity phosphatase 4 mediates cardiomyopathy caused by lamin A/C (LMNA) gene mutation. J. Biol. Chem. 2012;287:40513–40524. doi: 10.1074/jbc.M112.404541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bueno O.F., De Windt L.J., Tymitz K.M., Witt S.A., Kimball T.R., Klevitsky R., Hewett T.E., Jones S.P., Lefer D.J., Peng C.F., et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 2000;19:6341–6350. doi: 10.1093/emboj/19.23.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter J.J., Tanaka N., Rockman H.A., Ross J., Jr, Chien K.R. Ventricular expression of a MLC-2v-ras fusion gene induces cardiac hypertrophy and selective diastolic dysfunction in transgenic mice. J. Biol. Chem. 1995;270:23173–23178. doi: 10.1074/jbc.270.39.23173. [DOI] [PubMed] [Google Scholar]
- 32.Kehat I., Davis J., Tiburcy M., Accornero F., Saba-El-Leil M.K., Maillet M., York A.J., Lorenz J.N., Zimmermann W.H., Meloche S., et al. Extracellular signal-regulated kinases 1 and 2 regulate the balance between eccentric and concentric cardiac growth. Circ. Res. 2011;108:176–183. doi: 10.1161/CIRCRESAHA.110.231514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S., et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 34.Aoki Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., Filocamo M., Kato K., Suzuki Y., Kure S., et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 35.Estep A.L., Tidyman W.E., Teitell M.A., Cotter P.D., Rauen K.A. HRAS Mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am. J. Med. Genet. A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- 36.Niihori T., Aoki Y., Narumi Y., Neri G., Cavé H., Verloes A., Okamoto N., Hennekam R.C., Gillessen-Kaesbach G., Wieczorek D., et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 37.Schubbert S., Zenker M., Rowe S.L., Böll S., Klein C., Bollag G., van der Burgt I., Musante L., Kalscheuer V., Wehner L.E., et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 38.Pandit B., Sarkozy A., Pennacchio L.A., Carta C., Oishi K., Martinelli S., Pogna E.A., Schackwitz W., Ustaszewska A., Landstrom A., et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 39.Razzaque M.A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., Kamisago M., Momma K., Katayama H., Nakagawa M., et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 40.Choi J.C., Muchir A., Wu W., Iwata S., Homma S., Morrow J.P., Worman H.J. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 2012;4:144ra102. doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponchel F., Toomes C., Bransfield K., Leong F.T., Douglas S.H., Field S.L., Bell S.M., Combaret V., Puisieux A., Mighell A.J., et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- 43.D'Agostino R., Pearson E.S. Tests for departure from normality. Empirical results for the distributions of b2 and √ b1. Biometrika. 1973;60:613–622. [Google Scholar]