Abstract
The X-linked RHOX cluster encodes a set of homeobox genes that are selectively expressed in the reproductive tract. Members of the RHOX cluster regulate target genes important for spermatogenesis promote male fertility in mice. Studies show that demethylating agents strongly upregulate the expression of mouse Rhox genes, suggesting that they are regulated by DNA methylation. However, whether this extends to human RHOX genes, whether DNA methylation directly regulates RHOX gene transcription and how this relates to human male infertility are unknown. To address these issues, we first defined the promoter regions of human RHOX genes and performed gain- and loss-of-function experiments to determine whether human RHOX gene transcription is regulated by DNA methylation. Our results indicated that DNA methylation is necessary and sufficient to silence human RHOX gene expression. To determine whether RHOX cluster methylation associates with male infertility, we evaluated the methylation status of RHOX genes in sperm from a large cohort of infertility patients. Linear regression analysis revealed a strong association between RHOX gene cluster hypermethylation and three independent types of semen abnormalities. Hypermethylation was restricted specifically to the RHOX cluster; we did not observe it in genes immediately adjacent to it on the X chromosome. Our results strongly suggest that human RHOX homeobox genes are under an epigenetic control mechanism that is aberrantly regulated in infertility patients. We propose that hypermethylation of the RHOX gene cluster serves as a marker for idiopathic infertility and that it is a candidate to exert a causal role in male infertility.
INTRODUCTION
Homeobox genes were first identified in flies as encoding transcription factors important for body segment identity. Subsequent studies demonstrated that homeobox genes are ubiquitously present in eukaryotes ranging from S. cerevisiae to mammals, where they direct several conserved steps that are indispensable for a wide variety of embryonic developmental stages (1). Homeobox genes have undergone intense scrutiny for the past 30 years and many of their specific roles in embryonic development are relatively well understood. In contrast, much less is known about the roles of homeobox transcription factors in post-embryonic developmental events. To date, only a few homeobox transcription factors have been identified that control postnatal and adult developmental events (e.g. hematopoiesis, hair growth and gut homeostasis) and the molecular mechanisms by which they regulate these events are poorly understood (2,3). Even less is known about the role of homeobox genes in the subject of this report—gametogenesis—developmental process that is initiated in the embryo and occurs constitutively in adults.
The only homeobox genes that have known roles in gametogenesis in mice are the X-linked reproductive homeobox (Rhox) genes, all of which are selectively expressed in the male and female reproductive tract in mice, rats and humans (4–7). Knockout studies have demonstrated that some members of the Rhox cluster are crucial for normal spermatogenesis in mice (8,9). A number of the genes regulated by Rhox genes have known roles in spermatogenesis and germ cell survival and some of these target genes exist in conserved regulatory circuits with Rhox genes (10,11). While the functions of RHOX genes in humans are not known, they are expressed in a stage-specific manner in human male and female germ cells in the testis and ovary, respectively, thereby making them good candidates to direct transcriptional programs important for the development of human germ cells (12).
To understand the role of Rhox genes in both fertility and infertility, it is important to delineate how they are regulated. Increasing evidence suggests that a key regulator of mouse Rhox genes is DNA methylation. For example, mouse fibroblasts deficient in DNA methyltransferase 1 (DNMT1) exhibit dramatically upregulated Rhox gene expression (13). Likewise, mice deficient in DNA methyltransferases (DNMTs) exhibit demethylation and aberrant expression of some Rhox genes during embryogenesis (14). Treatment of various cell lines with DNA demethylating agents such as 5-Azacytidine (5-AzaC) also causes strong up-regulation of most mouse Rhox genes (13,15–17). Finally, depletion of the linker histone H1 genes triggers the demethylation and up-regulation of a specific subset of Rhox genes in mouse ES cells, which may contribute to their silencing by paternal imprinting in the placenta (17). Together, these studies clearly show that conditions that promote DNA demethylation increase the expression of mouse Rhox genes. However, it has not been clear whether this is the result of direct demethylation of these genes or indirect effects resulting, for example, from the demethylation and subsequent increased expression of regulators of Rhox genes.
In this report, we examine whether the human RHOX genes are directly regulated by DNA methylation. Given that mouse Rhox genes have known roles in male fertility, we also asked whether the methylation of human RHOX genes is aberrant in human male infertility patients. A remarkably high percentage (∼7%) of males who intend to father children are classified as infertile, yet there are surprisingly few markers that aid in diagnosing this condition (18–22). A semen analysis, which includes measuring sperm count, motility and morphology, is the first step in identifying the etiology of male infertility. However, the large natural variation in these parameters renders them relatively poor indicators for fertility. Furthermore, a normal measurement does not necessarily mean that a man is fertile (23–25). For most men, the only treatment for their infertility is in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI), which are both very costly, invasive for the female; and yield low pregnancy rates and even lower live birth rates (26). There is also evidence that the offspring born of IVF and ICSI procedures are at greater risk for developing a variety of conditions, including diabetes, obesity and imprinting disorders (27–35). These drawbacks highlight the need for better ways to identify and treat male infertility. In this article, we demonstrate that the human RHOX gene cluster serves as an excellent marker for male infertility. In particular, linear regression analysis of sperm from a large cohort of male infertility patients revealed a strong association between human RHOX cluster hypermethylation and the severity of the three semen defects typically analyzed in infertility clinics. We also demonstrate that DNA methylation directly represses the transcription of human RHOX gene family members in cultured cells, thereby providing evidence that the aberrant RHOX methylation that occurs in male infertility patients has the potential to have a causal role in their reproductive dysfunction.
RESULTS
Hypermethylation of RHOX gene promoters silences their transcription
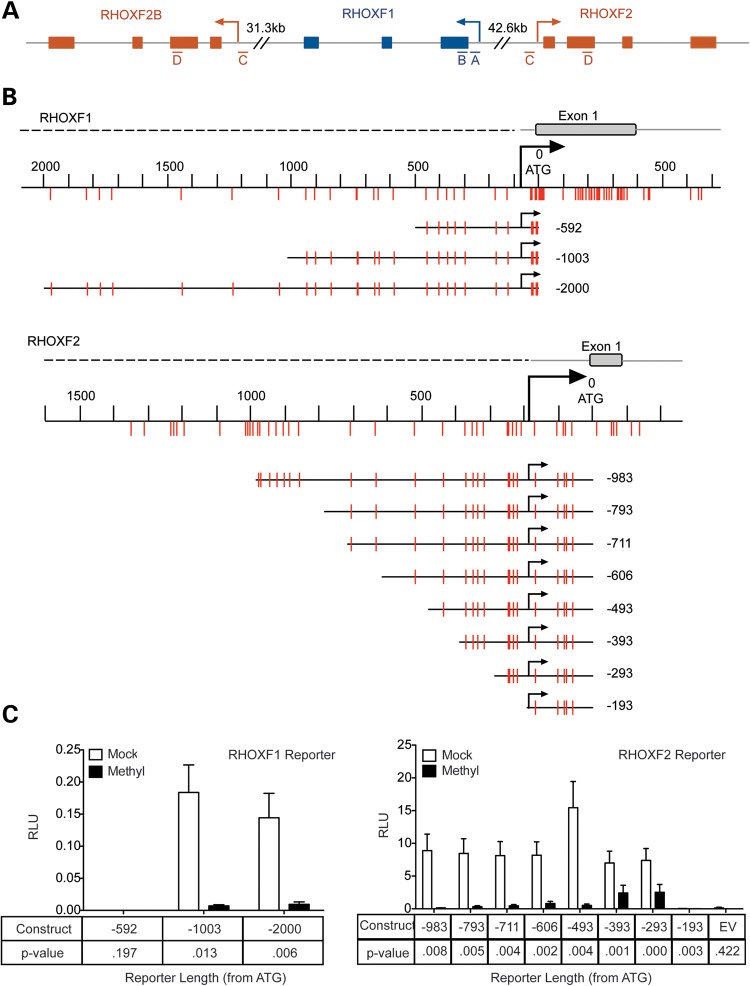
The human RHOX cluster contains three family members: RHOXF1 (also known as hPEPP1), RHOXF2 (hPEPP2) and RHOXF2B (36) (Fig. 1A). RHOXF2 and RHOXF2B are both 5479 in length and 99.9% identical in sequence (as annotated by the Ensembl databse), with only 8 nt differences: 2 in the exon regions, 6 in the intron regions and none in the 5′ or 3′ untranslated regions (UTRs). They are also nearly identical over a long stretch of their 5′ and 3′ flanking regions (19.7 and 34.1 kb, respectively), suggesting that one of them was copied from the other via a large duplication of 59.3 kb. These rare nucleotide differences make it impossible to distinguish RHOXF2 from RHOXF2B in our assays and thus we will refer to these two genes as simply ‘RHOXF2’ in this article.
Figure 1.
Methylation of RHOX promoters silences their transcription. (A) Schematic of the human RHOX cluster on Xq24. Exons (boxes), introns and UTRs (gray lines) are shown. The four regions analyzed in RHOXF1 and RHOXF2/2B by pyrosequencing (Fig. 3) are indicated as ‘A’ through ‘D’. (B) The portions of RHOXF1 and RHOXF2 upstream regions that were cloned into a CpG-free luciferase construct are shown, including the location of CpG residues relative to the translation start site. (C) Firefly luciferase expression from RHOXF1- and RHOXF2-reporter constructs that were mock treated (white bars) or in vitro methylated (black bars) and co-transfected in HEK-293 cells with a Renilla luciferase control construct. Data are plotted as the ratio of Firefly luciferase activity over Renilla luciferase activity. Error bars represent SEM.
To determine whether the human RHOX genes are regulated by DNA methylation, we first identified the approximate location of their transcription start sites (TSSs) by primer walking using quantitative polymerase chain reaction (qPCR) analyses with normal human testis RNA (Supplementary Material, Fig. S1). Once we identified the RHOXF1 and RHOXF2 TSSs, we cloned different lengths of the regions upstream into a CpG-free luciferase reporter construct. Transfection of these constructs identified minimal sequences sufficient for strong RHOXF1 and RHOXF2 transcription that extended 1003 and 293 nt upstream of their initiator ATGs, respectively (Fig. 1). Our finding that RHOXF2 transcription is driven by a minimal promoter containing only 293 nt upstream of the start ATG is consistent with the finding that numerous germ cell-specific genes have short core promoters (37). This was not the case for RHOXF1, perhaps due to the presence of a ∼400 nt C-rich repeat nestled between the region we found was required for transcription (−1003 to −593) and the TSS (data not shown).
To determine the effect of methylation on the RHOXF1 and RHOXF2 promoters, we methylated them in vitro using SssI methyltransferase. Because we inserted these promoters in a CpG-free vector, treatment with SssI results in methylation of CpG residues only in the RHOX promoter regions, thereby avoiding artifacts caused by methylation of nearby plasmid sequences. Transfection of these constructs into HEK-293 cells revealed that methylation of the RHOXF1 and RHOXF2 promoters reduced their transcription by ∼85 and ≥95%, respectively (Fig. 1). Methylation inhibited RHOXF2 promoter-driven transcription when as few as five CpGs in the RHOXF2 promoter were methylated (the −293 construct).
Methylation of RHOX promoters is sufficient to repress their transcription
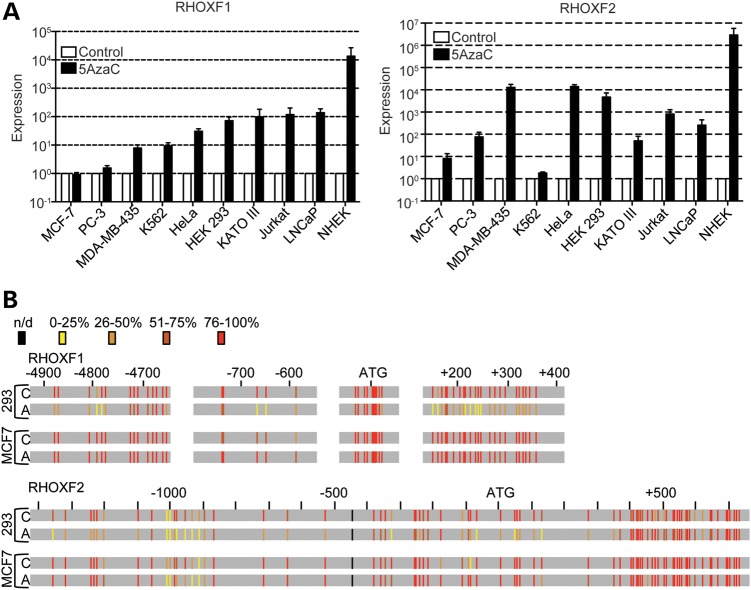
Human RHOX genes are expressed in a tissue-specific manner, with highest expression in the testis (5,12,36). Both the RHOXF1 and RHOXF2 proteins are primarily expressed in developing germ cells within the testis, where they exhibit distinct developmental patterns of expression (12). If DNA methylation is sufficient to dictate this tissue- and cell type-specific expression pattern, it leads to the prediction that DNA methylation inhibitors will be sufficient to activate RHOX gene expression in cell types that do not normally express RHOX genes. To test this, we treated a variety of non-testicular cell lines with the DNA methylation inhibitor 5-AzaC. We found that treatment with 10 μM 5-AzaC dramatically induced the expression of RHOXF1 and RHOXF2 in most cell lines that we tested (Fig. 2 and Supplementary Material, Fig. S2; note that mRNA levels are plotted on a log scale). An exception was K562 cells, which express extremely high constitutive levels of RHOXF2 (36), that was not significantly further increased by 5-AzaC treatment. Another exception was MCF-7 breast cancer cells, which express low-to-moderate basal levels of RHOXF1 and RHOXF2 mRNA, but did not exhibit a significant increase in these levels with our standard dose of 5-AzaC (10 μm). However, MCF-7 cells did up-regulate RHOXF2 expression in response to higher doses of 5-AzaC (Supplementary Material, Fig. S3), indicating these cells are not completely impervious to the effects of 5-AzaC.
Figure 2.
5-AzaC treatment is sufficient to demethylate RHOX genes in non-testicular cell lines. (A) qPCR analysis of RHOXF1 and RHOXF2 mRNA expression in cell lines seeded sub-confluently and treated with 5-AzaC. Data were normalized to β-actin mRNA expression and plotted relative to each cell line's control sample, which was set to 1. β-Actin mRNA level was not significantly affected by 5-AzaC treatment (data not shown). Error bars represent SEM. Data are plotted on a log-scale. (B) Bisulfite sequencing analysis of RHOXF1 and RHOXF2 from 293 and MCF-7 cell lines before (C) and after 48 h of treatment with 10 μm 5-AzaC (A). The degree of methylation at each CpG residue is indicated by vertical bars and was calculated by counting the number of sequences with a cytosine (representing 5-methylcytosine) divided by the total number of sequenced clones (see Supplementary Material, Figs S3 and S4). The degree of methylation is color-coded in a heat map style with yellow being the least methylated and red being the most methylated. The locations are based on the distance from translation start site. n/d, not determined.
To assess whether 5-AzaC acts directly on the RHOX genes, we analyzed the methylation status of the RHOXF1 and RHOXF2 promoters in a cell line that did (HEK-293) and did not (MCF-7) respond to 10 μm 5-AzaC. This revealed that 5-AzaC treatment demethylated regions in both RHOXF1 and RHOXF2 in HEK-293 cells (Fig. 2, Supplementary Material, Figs S3 and S4). RHOXF1 was most prominently demethylated in exon 1 (shown in green in Supplementary Material, Fig. S4). It was also significantly demethylated in the promoter region (shown in orange in Supplementary Material, Fig. S4) that we demonstrated above was necessary for RHOXF1 transcription (between −592 and −1003 relative to the ATG translation start site; Fig. 1C). RHOXF2 was demethylated in several regions, including regions upstream of the TSS (Supplementary Material, Fig. S5). Some regions of RHOXF2 were not significantly demethylated, including the body of the RHOXF2 gene (shown in lavender in Supplementary Material, Fig. S5). In MCF-7 cells, RHOXF1 was not significantly demethylated in any of the regions we analyzed (Supplementary Material, Fig. S4) and RHOXF2 was only significantly demethylated in two of the eight regions that we analyzed (Supplementary Material, Fig. S5); both of these regions were in the gene body and both of them were demethylated only modestly. The selective demethylation occurring at the RHOXF1 and RHOXF2 promoters in a cell line that expresses RHOX mRNAs in response to 5-AzaC and not in one that does not, coupled with our finding that in vitro methylation silenced both the RHOXF1 and RHOXF2 promoters, strongly suggests that DNA methylation is necessary and sufficient for directly repressing transcription of the RHOXF1 and RHOXF2 genes.
Hypermethylation of the RHOX gene cluster in sperm is highly associated with poor semen quality
Given that RHOX cluster gene transcription is tightly controlled by DNA methylation, we elected to examine whether RHOX methylation is altered in male infertility patients. We first used pyrosequencing to analyze the methylation status of RHOX genes. This analysis covered two relatively CpG-rich regions in both the RHOXF1 and RHOXF2 genes. Regions A and C are near the translation start sites of RHOXF1 and RHOXF2, respectively, while regions B and D are within downstream CpG islands (Fig. 1A). For this analysis, we compared the degree of methylation of the human RHOX genes in sperm from 140 men from couples seeking fertility treatment. These patients were selected on the basis of two criteria: (i) they had been unsuccessful in impregnating their partner for at least 12 months, and (ii) their infertility was not the result of obvious physical conditions, such as obstructive azoospermia or varicocele, or procedures and treatments that are known to affect male fertility, such as vasectomy or chemotherapy. All subjects underwent a complete clinical and biochemical exam that included a semen analysis and hormone measurements (Table 1). We classified 45 of the 140 patients as having ‘normal sperm’ because their semen parameters fell within the normal range as defined by the World Health Organization (38) and used this as our control population. The remaining 95 men were categorized as having ‘abnormal sperm’ since they had a sperm count, sperm motility and/or sperm morphology score below the normal range (Table 1). Because the presence of poor-quality semen does not always result in infertility, and conversely, a ‘normal’ semen analysis does not always result in fertility, our ‘normal’ and ‘patient’ pool may include infertile and fertile men, respectively, and thus, the results may, in fact, be an underestimate of the true differences between fertile and infertile men.
Table 1.
Patient parameters
| Clinical parameters | Normal (n = 45) | Abnormal (n = 95) | P-value |
|---|---|---|---|
| Age (years) | 36 (34–40) | 35 (32–39) | 0.2158 |
| Bitesticular volume (ml) | 54 (30–67) | 45 (34–56) | 0.0032 |
| Ejaculate volume (ml) | 4.2 (3.0–5.1) | 3.8 (3.0–5.0) | 0.9468 |
| Sperm concentration (mill./ml) | 53.0 (35.8–96.5) | 18.3 (9.3–33.7) | <0.0001 |
| Total sperm count (mill.) | 196.2 (158.2–278.3) | 65.6 (33.2–128.8) | 0.0026 |
| Progressive motility (%) | 55 (52–58) | 45 (39–51) | <0.0001 |
| Normal morphology (%) | 9 (7–11) | 6.0 (3.0–9.0) | <0.0001 |
| Testosterone (nmol/l) | 16.4 (12.6–20.1) | 16.0 (12.8–19.5) | 0.625 |
| LH (U/l) | 3.1 (2.2–4.0) | 3.7 (2.6–4.7) | 0.1633 |
| FSH (U/l) | 2.6 (2.0–3.98) | 4.2 (2.8–5.9) | 0.032 |
Median values and the 25th and 75th percentile are given for different parameters. P-values from unpaired t-test.
Normal and Abnormal refers to semen parameters for count, motility and morphology as defined by the WHO.
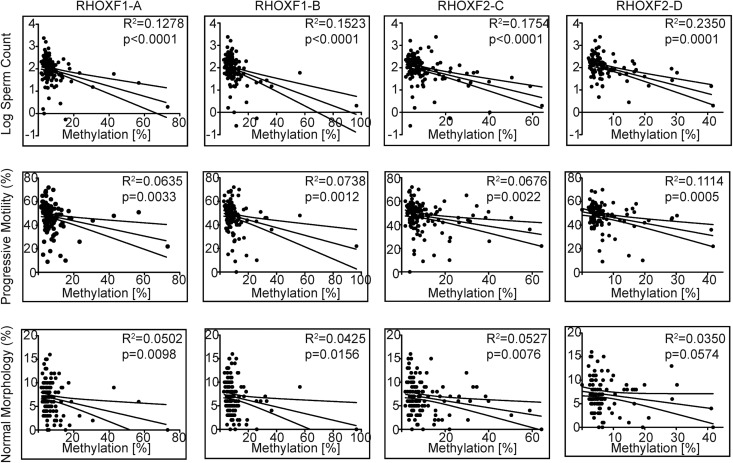
Our pyrosequencing data showed that the abnormal sperm group had significantly higher average methylation in all four RHOX regions relative to the normal sperm group (Table 2, Supplementary Material, Fig. S6). Remarkably, regression analysis revealed that the degree of RHOX gene methylation was significantly associated with all three sperm parameters: sperm count, sperm motility and sperm morphology (Fig. 3). To establish whether or not RHOX methylation has the potential to predict a particular semen defect, we subdivided the abnormal sperm group on the basis of the World Health Organization's definition of oligozoospermia (<39 million total sperm); asthenozoospermia (<32% progressive motility); and teratozoospermia (<4% normal morphology) (38) and compared them to each other as well as to the normal sperm group (Supplementary Material, Fig. S7). We found statistically significant differences among almost all of these groups with respect to total sperm count, progressively motile sperm and sperm with normal morphology (Fig. 3), indicating that RHOX methylation is a good predictor of all three of these semen defects.
Table 2.
Methylation levels in normal versus abnormal patient population
| Gene/location | Normal, % ± SEM | N-Normal | Abnormal, % ± SEM | N-Abnormal | P-value |
|---|---|---|---|---|---|
| RHOXF1 | |||||
| A | 5.1 ± 0.46 | 43 | 8.8 ± 1.1 | 91 | <0.0001 |
| B | 7.1 ± 0.45 | 45 | 12.0 ± 1.2 | 95 | 0.0006 |
| RHOXF2 | |||||
| C | 6.3 ± 0.74 | 45 | 12.0 ± 1.3 | 92 | <0.0001 |
| D | 4.9 ± 0.57 | 35 | 9.0 ± 1.10 | 69 | 0.0008 |
| MEST | 10.0 ± 0.93 | 45 | 16.0 ± 1.2 | 92 | 0.0045 |
P-values are for a comparison between normal samples and abnormal samples using a two-tailed t-test.
Normal and Abnormal refers to semen parameters for count, motility and morphology as defined by the WHO.
Figure 3.
RHOX hypermethylation is associated with male infertility. The severity of defective semen parameters from idiopathic infertile men was plotted against the percent RHOX methylation as determined by pyrosequencing at the four regions indicated in Figure 1A. The 95% confidence intervals are shown above and below the best-fit curve. See Table 2 for N values.
Hypermethylation at the RHOX locus does not extend to neighboring genes
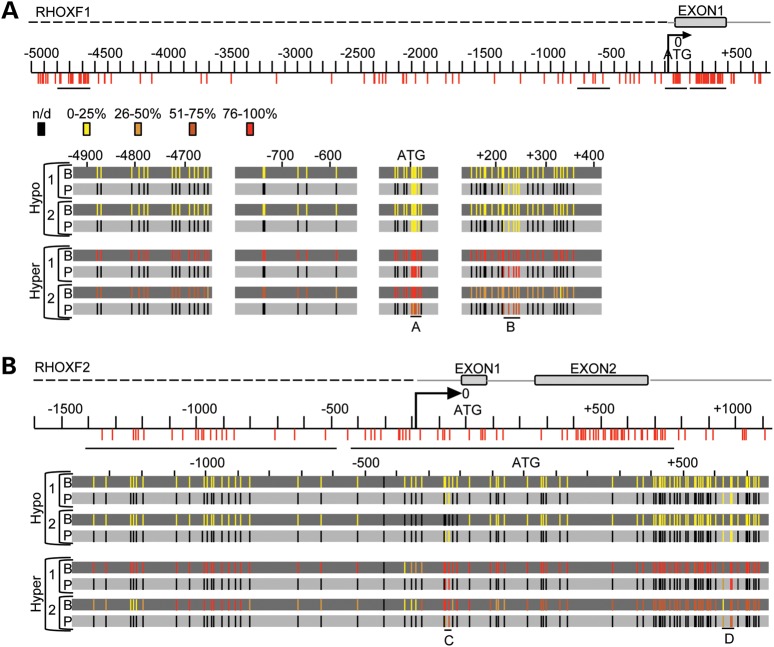
Because assaying short DNA stretches does not necessarily serve as a proxy for the methylation status of an entire gene locus (39), we elected to complement our pyrosequencing analysis with plasmid-based bisulfite sequencing for a few patients. Bisulfite sequencing permits analysis of larger stretches of nucleotide sequences by selecting multiple regions for amplification and sequencing after bisulfite conversion. We found that quantification of the percent methylation in the RHOX gene cluster by bisulfite sequencing was highly similar to the percent methylation as determined by pyrosequencing, supporting the latter method as a proxy for assessing regional methylation status. Bisulfite sequencing revealed that the normal group has remarkably little methylation in these regions. This is a common feature in CpG islands but is more rare in CpG-sparse regions, which tend to be uniformly methylated (40). These data also show that regions throughout the entire RHOX cluster, including sequences not detected by pyrosequencing, are strongly hypermethylated in the abnormal sperm parameter group (Fig. 4, Supplementary Material, Figs S8 and S9). This aberrant hypermethylation was nearly uniform in all gene elements, including both upstream and downstream CpG islands, as well as the CpG-sparse regions in the 5′-upstream region, which could also be considered intergenic. One exception was the −300 to −400 region in RHOXF2, which was less methylated than its neighboring regions in the group with abnormal semen parameters (Fig. 4B and Supplementary Material, Fig. S9, yellow region). Of note, approximately half of the clones analyzed in patient 2 from the Abnormal group exhibited methylation and half did not (for three of the four regions analyzed in RHOXF1; Supplementary Material, Fig. S8), suggesting that patient 2 has a mosaic phenotype in which roughly half of his sperm harbor a normal methylation pattern.
Figure 4.
The RHOX cluster is hypermethylated in men from infertile couples. (A) Bisulfite sequencing results are shown for four patients: two deemed hypermethylated and two deemed hypomethylated based on pyrosequencing results. Bisulfite sequencing results (B) are in the dark gray background and the corresponding pyrosequencing results (P) are in the light gray background. The RHOXF1 gene is displayed with CpG residues depicted (to scale) by vertical red lines according to their location from the translation start site. An arrow depicts the TSS. n/d, not-determined. (B) The RHOXF2 profile of the same four patients analyzed as described in (A).
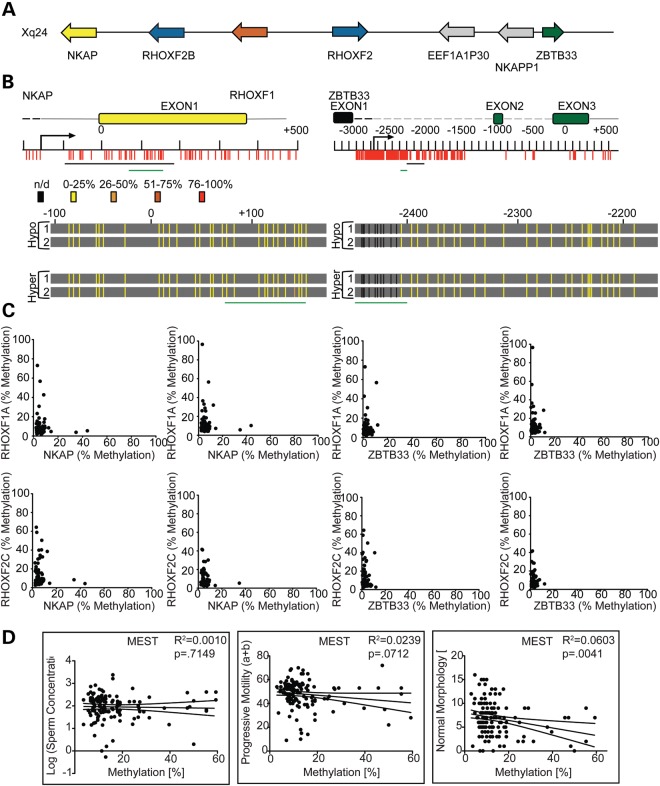
To determine whether hypermethylation is a promiscuous feature within the region of the X chromosome harboring the RHOX gene cluster, we analyzed the neighboring genes on either side of the RHOX cluster—NKAP and ZBTB33 (Fig. 5A)—both of which are hypomethylated in normal sperm (41) and whose expression we found did not change in most non-testicular cell lines when treated with 5-AzaC (Supplementary Material, Fig. S10). Pyrosequencing and bisulfite sequencing analyses revealed that the NKAP and ZBTB33 loci are hypomethylated in the vast majority of the patients, including those with abnormal sperm (Fig. 5B and C, and Supplementary Material, Fig. S11). Even patients with the highest degree of sperm RHOX methylation did not exhibit elevated methylation in the NKAP and ZBTB33 genes. These data indicate that the hypermethylation in this region of the X chromosome is restricted to the RHOX locus in male infertility patients.
Figure 5.
Hypermethylation in sperm from infertility patients is restricted to the RHOX cluster. (A) An expanded region of the X chromosome is shown depicting the location and direction of genes neighboring the RHOX cluster. Gray arrows indicate pseudogenes. (B) NKAP and ZBTB33 gene CpG residues and gene features are drawn to scale. The regions analyzed by bisulfite sequencing are shown as a black line and the regions analyzed by pyrosequencing are shown as a green line under the gene and/or under the bisulfite sequencing results, which are depicted in heat map style as in Figure 2. Black vertical lines indicate residues that were not analyzed by bisulfite sequencing. (C) DNA from the sperm of the entire infertile patient population was analyzed for percent methylation (number of Cs compared with number of Ts incorporated into the pyrosequencing run) in the NKAP and ZBTB33 genes at the regions indicated by green bars in (B). The results were plotted on the x-axis and compared with the percent methylation of each RHOX region, which is plotted on the y-axis. (D) MEST methylation was plotted against the severity of patient sperm defects as in Figure 1B. See Table 2 for N values.
RHOX hypermethylation versus MEST hypermethylation
We also analyzed the methylation status of MEST, an autosomal maternally imprinted gene previously reported to be aberrantly hypermethylated in males from infertile couples (42–50). Regression analysis revealed that unlike RHOX methylation, which associated with all three semen defect parameters, MEST methylation only associated with abnormal sperm morphology (Fig. 5D). It has been hypothesized that infertile males have global methylation defects in their sperm as a result of a general failure of epigenetic reprogramming in primordial germ cells (PGCs) (44). If such an event were responsible for MEST and RHOX hypermethylation in infertile patients, this would lead to the prediction that both of these genes would tend to be hypermethylated in the same patients. However, when we examined coincident methylation of MEST and RHOX using regression analyses, we found that few patients exhibited hypermethylation at both loci, as evidenced by low R2 values (Supplementary Material, Fig. S12). In contrast, different regions of the RHOX cluster exhibited higher R2 values when compared with each other, indicating that hypermethylation at one region of the RHOX cluster is very likely to be accompanied by hypermethylation at other regions of the gene cluster. This corroborates our finding that entire RHOX gene cluster is hypermethylated in a subset of patients.
DISCUSSION
In this report, we provide evidence that the RHOX homeobox gene cluster is more likely to be hypermethylated in male infertility patients with defective sperm than in patients with normal semen parameters (Fig. 3, Supplementary Material, Figs S5, S6 and S12 and Table 1). The magnitude of this methylation significantly associates with the severity of three independent sperm abnormalities (Fig. 4B). This association between RHOX methylation and sperm abnormalities is potentially functionally relevant, as we show that methylation has a causal role in regulating human RHOX gene transcription (Figs 1 and 2). While analysis of patients recruited from other fertility centers will be required to substantiate our results, our data support a model in which aberrant DNA methylation of the RHOX genes reduces their expression, which, in turn, disrupts the normal regulation of their downstream target genes and thereby negatively affects fertility.
There have been a number of recent studies reporting an association between aberrant DNA methylation and male infertility. Many of these studies have focused on genomically imprinted genes (42–55) since the mono-allelic expression of imprinted genes is dictated by DNA methylation. However, most of the imprinted genes that have been shown to display aberrant methylation associated with infertility are not known to be expressed in the male reproductive tract and do not have known roles in spermatogenesis. Thus, the physiological relevance of the aberrant methylation of these imprinted genes in sperm is unclear. In an effort to broaden the identification of genes that are mis-methylated in sperm from infertile men, two recent studies explored larger subsets of genes. Pacheco et al. (55) identified both imprinted and non-imprinted genes that were aberrantly methylated in men with low-motility sperm. Interestingly, the vast majority of these genes (∼80%) were hypomethylated, opposite to what we found for the RHOX cluster. Houshdaran et al. (44) identified nine genes that exhibited a statistically significant increase in methylation in poor quality sperm. It will be interesting to determine whether the mechanism that leads to one or more of these nine genes being hypermethylated also acts on the RHOX cluster. However, unlike the RHOX genes, none of these genes is known to be expressed in human male germ cells and most do not have known roles in spermatogenesis in model systems.
We suggest that the RHOX cluster locus is a good candidate to not only be aberrantly methylated in human infertility patients but also to play a direct role in their infertility. Rhox genes encode homeobox transcription factors that are highly expressed in the testis and regulate several genes that are involved in spermatogenesis (4,10,56). Their roles in spermatogenesis have begun to be dissected from mouse knockout studies that have established that their ablation leads to sperm defects and subfertility (4). Human RHOX genes are compelling candidates to have similar roles in human male infertility because they are also highly expressed in the testis (5,36). RHOXF2 is expressed in spermatogonia and early spermatocytes (12) and thus it may regulate genes important for the decision to self-renew or differentiate and/or early steps in meiosis. RHOXF1 is expressed mainly in mid-to-late meiotic spermatocytes and round spermatids (12), raising the possibility that it regulates genes important for later steps in meiosis, as well as functions in haploid spermatids. Intriguingly, human RHOX transcription factors have the ability to regulate some of the same target genes as mouse RHOX transcription factors, suggesting that this gene cluster has remained devoted to regulating some of the events during spermatogenesis (56,57).
To our knowledge, the RHOX genes are the only X-linked genes that have been thoroughly evaluated with regard to methylation status in infertile males. Interestingly, the X chromosome is enriched for large gene clusters expressed in the testes, as well as spermatogonia-expressed single-copy genes in mammals (58,59). A likely mechanism responsible for the enrichment of spermatogenesis-associated genes on the X chromosome derives from the fact that there is only one copy of this chromosome in males. This means that mutations that confer a competitive advantage for male functions can be rapidly fixed in the population. Thus, the X-linked RHOX gene cluster has the potential to have undergone very rapid selection during evolution. In support of this, RHOX genes have rapidly evolved in both sequence and copy number and there is considerable evidence that many of the sequence changes are the result of strong positive selection (60–62). However, this single-copy nature of X-linked genes in males also has potentially negative consequences. Since there is only one copy of X-linked genes, deleterious mutations and epimutations associated with them will have direct physiological penetrance and hence more likely to cause disease. We suggest that this makes the RHOX gene cluster and other X-linked genes expressed during spermatogenesis particularly strong candidates to have roles in male infertility.
While the RHOX genes are clearly hypermethylated in a subset of male infertility patients, we do not know the molecular mechanisms responsible for this defect. One possibility is that this group of patients aberrantly methylates RHOX genes at some point after PGC reprogramming. This could occur as a result of hyper-expression or -activity of enzymes involved in DNA methylation, such as DNMTs. In support of this theory, Pacheco et al. (55) found that the gene encoding the de novo methyl transferase DNMT3A shows statistically significant hypomethylation and increased expression in patients with low-motility sperm. If indeed the RHOX gene cluster is specifically targeted for hypermethylation in a subset of infertility patients, it will be fascinating to determine the underlying mechanism, as little is known about how DNMTs target specific genes for methylation. It is possible that some unique feature of the RHOX locus, such as a specific sequence or a certain aspect of its chromatin structure leads to their erroneous targeting by DNMTs in germ cells in a subset of infertile males. Because we observed that the RHOX locus is hypermethylated broadly across its entire length, but not in the neighboring genes on the X chromosome (Fig. 5, Supplementary Material, Fig. S11), the mechanism probably entails either regional targeting or a methylation boundary on either side of the RHOX locus. Because alterations in chromatin structure are often coupled with changes in DNA methylation, it is also possible that over-active DNA methylation at the RHOX locus is coupled to changes at the chromatin level. Male germ cells undergo a complex exchange of histone and histone-like proteins during spermatogenesis that could have a role in such a mechanism.
An alternative, non-mutually exclusive, hypothesis to explain aberrant hypermethylation at the RHOX locus is that some male infertility patients fail to completely demethylate the RHOX gene cluster during PGC reprogramming. A requirement of this hypothesis is that RHOX genes must be subject to PGC reprogramming. Many mammalian genes—particularly those with CpG islands—are never significantly methylated and thus are not subject to PGC reprogramming. In contrast, RHOX genes are methylated in adult somatic tissues that do not express the RHOX genes, such as the adrenal gland, lung and spleen (14,63). This raises the possibility that DNA methylation directs the tissue-specific regulation of RHOX gene expression, a notion that is also supported by our in vitro methylation analyses presented here (Figs 1 and 2), as well as the recent evidence that germline-specific genes are exclusively regulated by DNA methylation (64). It also suggests that RHOX genes are likely to be methylated in PGC progenitor cells, which harbor somatic levels of DNA methylation (65,66). Because we found that RHOX genes are unmethylated in male germ cells (Fig. 4, Supplementary Material, Figs S7 and S8), they must undergo demethylation at some point during or after germ cell specification. In support of the idea that this occurs during PGC reprogramming, it was recently shown that several Rhox genes are demethylated in PGCs (67,68) and the timing of this demethylation associates with their expression pattern in PGCs (69). In the future, it will be important to distinguish between whether aberrant PGC reprogramming or subsequent events are responsible for the hyper-methylation of the RHOX gene cluster in sperm from some male infertility patients and to determine whether this aberrant methylation has a causal role in the infertility of any of these patients.
MATERIALS AND METHODS
Primer walking and qPCR
RNA from normal human testis (Agilent Cat. No 540049) or cell lines treated with or without 5-AzaC was converted to cDNA using Bio-Rad's iScript kit (Cat. No. 170-8890). cDNA was subjected to real-time quantitative PCR using Bio-Rad's Sso Advantage system (Cat. No. 172-5264). Data were normalized to beta-actin expression. The primers can be found in Supplementary Material, Table S3.
Cloning
Primers were designed to amplify RHOXF1 and RHOXF2 5′-upsteram regions according to the primer-walking assay (provided upon request). Artificial restriction enzyme sites were added to the primers (BglII, NcoI or BamHI). Amplicons were cloned into pGEMT-Easy TA shuttling vector (Promega Cat. No. A1360) and digested and ligated into pCpGL-Luciferase (70) using a T4 DNA Ligase Kit (NEB Cat. No. M0202L). GT115 Competent Cells (Invivogen Cat. No. gt115-11) were transformed and plated on LB containing 100 μg/ml Zeocin (Invivogen Cat. No. ant-zn-1). Colonies were selected and grown in LB broth containing 100 μg/ml Zeocin. Positive clones were detected by sequencing using the primers provided upon request.
Luciferase assay
Cells were seeded at 60 000 cells per well in a 24-well format. After 24 h, cells were transfected using 1 μl per well of Lipofectamine 2000 (Invitrogen Cat. No. 11668-019) with 200 ng of reporter and 20 ng of TK-Renilla. 24 h after transfection, cells were lysed in 100 μl of 1X Passive Lysis Buffer and 10 μl of lysate was analyzed using 50 μl of Lucifearse Assay Reagent (2 s pause; 10 s read) and 50 μl of Stop ‘N Glo reagent (2 s pause; 10 s read) all provided with the dual luciferase kit (Promega Cat. No. E1960). Reagents were dispensed and detected with a Veritas Microplate Luminometer (Turner BioSystems).
In vitro methylation
Ten micrograms of DNA was either treated or mock-treated (omitting only enzyme) with SssI Methyltransferase (NEB Cat. No. M0226L) for 4 h at 37°C with the addition of more S-adenosylmethionine after 2 h. Plasmids were purified using the Qiagen PCR Purification Kit (Cat. No. 28106). Purified plasmids were diluted to an equal concentration and digested with MspI and HpaII to gauge full-methylation of each plasmid when compared with the mock-methylated plasmid.
Azacytidine treatment
MCF-7 (breast adenocarcinoma), PC-3 (prostate adenocarcinoma), MDA-MB-435 (breast carcinoma), K562 (myelogenous leukemia), HeLa (cervical carcinoma), HEK 293 (embryonic kidney), KATOIII (gastric carcinoma), LNCaP (prostate adenocarcinoma) and NHEK (normal human epidermal keratinocytes) cells were seeded at a density of 250 000 cells per well in six-well plates. Jurkat cells (T-lymphocyte) were maintained and treated in suspension in flasks. Cells were treated with 10 μm 5-AzaC for a total of 48 h with replacement of media containing 10 μm 5-AzaC after the first 24 h. Cells were harvested in 1 ml Trizol reagent (Invitrogen Cat. No. 15596-026) and RNA was harvested by chloroform extraction and nucleic acid precipitation. RNA was converted to cDNA using iScript kit (Cat. No. 170-8890). cDNA was subjected to real-time quantitative PCR using Bio-Rad's Sso Advantage system (Cat. No. 172-5264) and the primers provided upon request.
Bisulfite conversion and amplification of DNA
A maximum of 2 μg of DNA was converted using Epitect (Qiagen Cat. No. 59104). Converted DNA was amplified with Qiagen's PyroMark Kit (Cat. No. 978703) using the primers listed in Supplementary Material, Tables S1 and S2. Biotinylated primers were ordered from Eurofins MWG Operon.
Patient selection
The 140 men included in this study were recruited from couples presenting at the Centre of Reproductive Medicine and Andrology, Münster, Germany seeking advice for infertility. Exclusion criteria were history of or current condition such as maldescended testes, varicocele or infections of the genitourinary tract as well as chromosomal aberrations or Y-chromosomal AZF deletions, medication that is known to impair spermatogenesis and/or fertility, severe impairment of the endocrine system or any other known reasons for male infertility. Couples were diagnosed with infertility if they had been trying to conceive for 12 months or more. The fertility status of the female partner is unknown. All male participants underwent a complete physical examination including ultrasonographic analysis of the scrotal contents. Testicular volume was calculated using the ellipsoid method and summed as bi-testicular volume. A venous blood sample was drawn from the cubital vein in the morning. Mean hormone values are shown in Table 1. Serum concentrations of FSH and LH were determined by immunofluorometric assays (Autodelfia, Perkin Elmer, Freiburg, Germany) and serum testosterone by a commercial ELISA (DRG AURICA ELISA Testosterone Kit, DRG Instruments). Intra- and inter-assay coefficients of variation were <5% and <10%. Of these patients, the normal group (n = 45) consisted of those men possessing the following semen parameters: total sperm count of ≥100 million spermatozoa, ≥50% progressive sperm motility and ≥5% normal sperm morphology. All men provided written informed consent and agreed to the analysis of genetic material as approved by the Ethics Committee of the University of Muenster and the state medical board.
Semen analysis
Semen analysis and swim-up purification of spermatozoa were carried out according to the guidelines of the 2010 World Health Organization (WHO) laboratory manual for the examination and processing of human semen, which gives a comprehensive procedure on pate 164 (38). This is the preferred method for obtaining sperm from semen that have one or more abnormalities and it separates the sperm from any somatic cell contaminants in the semen.
Extraction of DNA from spermatozoa and cells
DNA from spermatozoa was extracted using the MasterPure DNA Purification Kit (EPICENTRE Biotechnologies, Madison, WI, USA) according to the manufacturer's instructions. DNA from cell lines was extracted using the DNEasy Kit from Qiagen (Cat. No. 69506) according to the manufacturer's protocol.
Pyrosequencing
Pyrosequencing was performed using Pyro Mark Q24, Thr PyroMark Q24 Software 2.06 and PyroMark Q24 reagents (Qiagen). For each run, the machine was calibrated by using serial dilutions of 100 and 0% methylated human genomic DNA. Percent methylation for individual CpG residues, which is a read-out of the ratio of Cs versus Ts that were incorporated in the pyrosequencing reaction, were normalized with a normalization coefficient from the calibration. Pyrosequencing primers can be found in Supplementary Material, Table S3.
Bisulfite sequencing
Amplicons were TA-cloned into Promega's pGEMT-Easy vector (Cat. No. A1360). Colonies were grown in LB-Ampicillin (50 μg/ml) and prepped with BioLine ISOLATE Plasmid Mini Kit (Cat. No. BIO-52027). Sequences were analyzed and dot-plots were generated using online software (QUMA- http://quma.cdb.riken.jp/) (71).
Data analysis and plotting
P-values were calculated using either the Mann–Whitney test for non-parametric data sets (all patient data) or Student's t-test when the population is assumed to be normally distributed (all cell line data). Box-plots of clinical data were plotted according to the Tukey method. All clinical calculations were performed and regression and box-plot images were generated with GraphPad Prism version 5.0d for Macintosh (GraphPad Software).
Venn diagrams
Venn diagrams were generated with an online tool called Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) (72).
AUTHOR CONTRIBUTIONS
M.E.R., M.F.W. and J.G. conceived and designed the study. J.G. and A.P. performed pyrosequencing, M.E.R. performed bisulfite sequencing and all in vitro studies. M.E.R., A.P., F.T. and J.G. performed pyrosequencing analysis and data interpretation. M.E.R. wrote the manuscript with input from all the other authors.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the German Research Foundation (grant number FOR1041 to J.G.); and the National Institutes of Health (R01-HD053808 and R01-HD45595 to M.F.W.; 5F32GM096722 to M.E.R.); German Research Foundation–International Collaboration (grant number GR 1547/15-1 to J.G.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank E. Lahrmann (UK Münster, Germany) for administrative help; S. Agarwal (UCSD) and H. Cook-Andersen (UCSD) for advice in manuscript preparation; and M. Klug (University Hospital, Regensburg, Germany) for the kind gift of the CpG-free luciferase reporter.
REFERENCES
- 1.Weatherbee S.D., Halder G., Kim J., Hudson A., Carroll S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awgulewitsch A. Hox in hair growth and development. Naturwissenschaften. 2003;90:193–211. doi: 10.1007/s00114-003-0417-4. [DOI] [PubMed] [Google Scholar]
- 3.Chiba S. Homeobox genes in normal hematopoiesis and leukemogenesis. Int. J. Hematol. 1998;68:343–353. doi: 10.1016/s0925-5710(98)00093-0. [DOI] [PubMed] [Google Scholar]
- 4.Maclean J.A., Chen M.A., Wayne C.M., Bruce S.R., Rao M., Meistrich M.L., Macleod C., Wilkinson M.F. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 5.Geserick C., Weiss B., Schleuning W., Haendler B. OTEX, an androgen-regulated human member of the paired-like class of homeobox genes. Biochem. J. 2002;375:367–375. doi: 10.1042/BJ20020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiti S., Doskow J., Li S., Nhim R.P., Lindsey J.S., Wilkinson M.F. The Pem homeobox gene. Androgen-dependent and -independent promoters and tissue-specific alternative RNA splicing. J. Biol. Chem. 1996;271:17536–17546. doi: 10.1074/jbc.271.29.17536. [DOI] [PubMed] [Google Scholar]
- 7.Lindsey J.S., Wilkinson M.F. An androgen-regulated homeobox gene expressed in rat testis and epididymis. Biol. Reprod. 1996;55:975–983. doi: 10.1095/biolreprod55.5.975. [DOI] [PubMed] [Google Scholar]
- 8.MacLean J.A., Wilkinson M.F. The Rhox genes. Reproduction. 2010;140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Tsai P., García A.-M., Logeman B., Tanaka T.S. A possible role of Reproductive Homeobox 6 in primordial germ cell differentiation. Int. J. Dev. Biol. 2011;55:909–916. doi: 10.1387/ijdb.113342cl. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z., Shanker S., MacLean J.A., Ackerman S.L., Wilkinson M.F. The RHOX5 homeodomain protein mediates transcriptional repression of the netrin-1 receptor gene Unc5c. J. Biol. Chem. 2008;283:3866–3876. doi: 10.1074/jbc.M706717200. [DOI] [PubMed] [Google Scholar]
- 11.Maclean J.A., Hayashi K., Turner T.T., Wilkinson M.F. The Rhox5 homeobox gene regulates the region-specific expression of its paralogs in the rodent epididymis. Biol. Reprod. 2012;86:189. doi: 10.1095/biolreprod.112.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song H.W., Anderson R.A., Bayne R.A., Gromoll J., Shimasaki S., Chang R.J., Parast M.M., Laurent L.C., De Rooij D.G., Hsieh T.C., et al. The RHOX homeobox gene cluster is selectively expressed in human oocytes and male germ cells. Hum. Reprod. 2013;28:1635–1646. doi: 10.1093/humrep/det043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 14.Oda M., Yamagiwa A., Yamamoto S., Nakayama T., Tsumura A., Sasaki H., Nakao K., Li E., Okano M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006;20:3382–3394. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki A.W., Doskow J., MacLeod C.L., Rogers M.B., Gudas L.J., Wilkinson M.F. The oncofetal gene Pem encodes a homeodomain and is regulated in primordial and pre-muscle stem cells. Mech. Dev. 1991;34:155–164. doi: 10.1016/0925-4773(91)90052-8. [DOI] [PubMed] [Google Scholar]
- 16.Li Q., Bartlett D.L., Gorry M.C., O'Malley M.E., Guo Z.S. Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Mol. Pharmacol. 2009;76:1072–1081. doi: 10.1124/mol.109.056291. [DOI] [PubMed] [Google Scholar]
- 17.Maclean J.A., Bettegowda A., Kim B.J., Lou C.-H., Yang S.-M., Bhardwaj A., Shanker S., Hu Z., Fan Y., Eckardt S., et al. The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol. Cell. Biol. 2011;31:1275–1287. doi: 10.1128/MCB.00734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tüttelmann F., Gromoll J., Kliesch S. Genetics of male infertility. Der Urologe. Ausg. A. 2008;47:1561–1567. doi: 10.1007/s00120-008-1804-4. [DOI] [PubMed] [Google Scholar]
- 19.De Jonge C. Semen analysis: looking for an upgrade in class. Fertil. Steril. 2012;97:260–266. doi: 10.1016/j.fertnstert.2011.12.045. [DOI] [PubMed] [Google Scholar]
- 20.Hamatani T. Human spermatozoal RNAs. Fertil. Steril. 2012;97:275–281. doi: 10.1016/j.fertnstert.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z., Xia Y., Guo X., Dai J., Li H., Hu H., Jiang Y., Lu F., Wu Y., Yang X., et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat. Genet. 2012;44:183–186. doi: 10.1038/ng.1040. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S.G., Marszalek J.D., Irenze K., Skaletsky H., Brown L.G., Oates R.D., Silber S.J., Ardlie K., Page D.C. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am. J. Hum. Genet. 2012;91:890–896. doi: 10.1016/j.ajhg.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker H.W., Kovacs G.T. Spontaneous improvement in semen quality: regression towards the mean. Int. J. Androl. 1985;8:421–426. doi: 10.1111/j.1365-2605.1985.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez C., Castilla J.A., Martínez L., Ramírez J.P., Vergara F., Gaforio J.J. Biological variation of seminal parameters in healthy subjects. Hum. Reprod. 2003;18:2082–2088. doi: 10.1093/humrep/deg430. [DOI] [PubMed] [Google Scholar]
- 25.Leushuis E., Van der Steeg J.W., Steures P., Repping S., Bossuyt P.M.M., Blankenstein M.A., Mol B.W.J., Van der Veen F., Hompes P.G.A. Reproducibility and reliability of repeated semen analyses in male partners of subfertile couples. Fertil. Steril. 2010;94:2631–2635. doi: 10.1016/j.fertnstert.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 26.Collins J. An international survey of the health economics of IVF and ICSI. Hum. Reprod. Update. 2002;8:265–277. doi: 10.1093/humupd/8.3.265. [DOI] [PubMed] [Google Scholar]
- 27.Katari S., Turan N., Bibikova M., Erinle O., Chalian R., Foster M., Gaughan J.P., Coutifaris C., Sapienza C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum. Mol. Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halliday J., Oke K., Breheny S., Algar E., Amor D.J. Beckwith-Wiedemann syndrome and IVF: a case-control study. Am. J. Hum. Genet. 2004;75:526–528. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher E.R. Imprinting and assisted reproductive technology. Hum. Mol. Genet. 2005;21:1009–1011. doi: 10.1093/hmg/ddi107. [DOI] [PubMed] [Google Scholar]
- 30.Amor D.J., Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum. Reprod. 2008;23:2826–2834. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 31.Ceelen M., Van Weissenbruch M.M., Vermeiden J.P.W., Van Leeuwen F.E., Delemarre-van de Waal H.A. Growth and development of children born after in vitro fertilization. Fertil. Steril. 2008;90:1662–1673. doi: 10.1016/j.fertnstert.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Manipalviratn S., DeCherney A., Segars J. Imprinting disorders and assisted reproductive technology. Fertil. Steril. 2009;91:305–315. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ørstavik K.H., Eiklid K., Van der Hagen C.B., Spetalen S., Kierulf K., Skjeldal O., Buiting K. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am. J. Hum. Genet. 2003;72:218–219. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox G.F., Bürger J., Lip V., Mau U.A., Sperling K., Wu B.-L., Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am. J. Hum. Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowdin S., Allen C., Kirby G., Brueton L., Afnan M., Barratt C., Kirkman-Brown J., Harrison R., Maher E.R., Reardon W. A survey of assisted reproductive technology births and imprinting disorders. Hum. Reprod. 2007;22:3237–3240. doi: 10.1093/humrep/dem268. [DOI] [PubMed] [Google Scholar]
- 36.Wayne C.M., MacLean J.A., Cornwall G., Wilkinson M.F. Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene. 2002;301:1–11. doi: 10.1016/s0378-1119(02)01087-9. [DOI] [PubMed] [Google Scholar]
- 37.Han S., Xie W., Kim S.H., Yue L., DeJong J. A short core promoter drives expression of the ALF transcription factor in reproductive tissues of male and female mice. Biol. Reprod. 2004;71:933–941. doi: 10.1095/biolreprod.104.030247. [DOI] [PubMed] [Google Scholar]
- 38.Cooper T.G., Noonan E., Von Eckardstein S., Auger J., Baker H.W.G., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., et al. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2009;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 39.Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 41.Molaro A., Hodges E., Fang F., Song Q., McCombie W.R., Hannon G.J., Smith A.D. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammoud S.S., Nix D.A., Hammoud A.O., Gibson M., Cairns B.R., Carrell D.T. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011;26:2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammoud S.S., Purwar J., Pflueger C., Cairns B.R., Carrell D.T. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil. Steril. 2010;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Houshdaran S., Cortessis V.K., Siegmund K., Yang A., Laird P.W., Sokol R.Z. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marques C.J., Costa P., Vaz B., Carvalho F., Fernandes S., Barros A., Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 46.Marques C.J., Carvalho F., Sousa M., Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700–1702. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 47.Marques C.J., Francisco T., Sousa S., Carvalho F., Barros A., Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil. Steril. 2010;94:585–594. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 48.Poplinski A., Tüttelmann F., Kanber D., Horsthemke B., Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int. J. Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi H., Sato A., Otsu E., Hiura H., Tomatsu C., Utsunomiya T., Sasaki H., Yaegashi N., Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum. Mol. Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 50.Boissonnas C.C., Abdalaoui H.E., Haelewyn V., Fauque P., Dupont J.M., Gut I., Vaiman D., Jouannet P., Tost J., Jammes H. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur. J. Hum. Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu W., Shen O., Qin Y., Niu X., Lu C., Xia Y., Song L., Wang S., Wang X. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PLoS ONE. 2010;5:e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Hajj N., Zechner U., Schneider E., Tresch A., Gromoll J., Hahn T., Schorsch M., Haaf T. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–69. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 53.Camprubí C., Pladevall M., Grossmann M., Garrido N., Pons M., Blanco J. Semen samples showing an increased rate of spermatozoa with imprinting errors have a negligible effect in the outcome of assisted reproduction techniques. Epigenetics. 2012;7:1115–1124. doi: 10.4161/epi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro-Costa P., Nogueira P., Carvalho M., Leal F., Cordeiro I., Calhaz-Jorge C., Gonçalves J., Plancha C.E. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum. Reprod. 2010;25:2647–2654. doi: 10.1093/humrep/deq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pacheco S.E., Houseman E.A., Christensen B.C., Marsit C.J., Kelsey K.T., Sigman M., Boekelheide K. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS ONE. 2011;6:e20280. doi: 10.1371/journal.pone.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Z., Dandekar D., O'Shaughnessy P.J., De Gendt K., Verhoeven G., Wilkinson M.F. Androgen-induced Rhox homeobox genes modulate the expression of AR-regulated genes. Mol. Endocrinol. 2010;24:60–75. doi: 10.1210/me.2009-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Z., MacLean J.A., Bhardwaj A., Wilkinson M.F. Regulation and function of the Rhox5 homeobox gene. Ann. N Y Acad. Sci. 2007;1120:72–83. doi: 10.1196/annals.1411.011. [DOI] [PubMed] [Google Scholar]
- 58.Mueller J.L., Mahadevaiah S.K., Park P.J., Warburton P.E., Page D.C., Turner J.M.A. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P.J., McCarrey J.R., Yang F., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 60.Niu A., Wang Y., Zhang H., Liao C., Wang J., Zhang R., Che J., Su B. Rapid evolution and copy number variation of primate RHOXF2, an X-linked homeobox gene involved in male reproduction and possibly brain function. BMC Evol. Biol. 2011;11:298. doi: 10.1186/1471-2148-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maiti S., Doskow J., Sutton K., Nhim R.P., Lawlor D.A., Levan K., Lindsey J.S., Wilkinson M.F. The Pem homeobox gene: rapid evolution of the homeodomain, X chromosomal localization, and expression in reproductive tissue. Genomics. 1996;34:304–316. doi: 10.1006/geno.1996.0291. [DOI] [PubMed] [Google Scholar]
- 62.Sutton K.A., Wilkinson M.F. Rapid evolution of a homeodomain: evidence for positive selection. J. Mol. Evol. 1997;45:579–588. doi: 10.1007/pl00006262. [DOI] [PubMed] [Google Scholar]
- 63.A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hackett J.A., Reddington J.P., Nestor C.E., Dunican D.S., Branco M.R., Reichmann J., Reik W., Surani M.A., Adams I.R., Meehan R.R. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development. 2012;139:3623–3632. doi: 10.1242/dev.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saitou M., Kagiwada S., Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 66.Popp C., Dean W., Feng S., Cokus S.J., Andrews S., Pellegrini M., Jacobsen S.E., Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guibert S., Forné T., Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22:633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daggag H., Svingen T., Western P.S., Van den Bergen J.A., McClive P.J., Harley V.R., Koopman P., Sinclair A.H. The rhox homeobox gene family shows sexually dimorphic and dynamic expression during mouse embryonic gonad development. Biol. Reprod. 2008;79:468–474. doi: 10.1095/biolreprod.107.067348. [DOI] [PubMed] [Google Scholar]
- 70.Klug M., Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 71.Kumaki Y., Oda M., Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oliveras J.C. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html. (accessed 16 August 2013) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.