Abstract
Ciliopathies are a group of heterogeneous disorders associated with ciliary dysfunction. Diseases in this group display considerable phenotypic variation within individual syndromes and overlapping phenotypes among clinically distinct disorders. Particularly, mutations in CEP290 cause phenotypically diverse ciliopathies ranging from isolated retinal degeneration, nephronophthisis and Joubert syndrome, to the neonatal lethal Meckel–Gruber syndrome. However, the underlying mechanisms of the variable expressivity in ciliopathies are not well understood. Here, we show that components of the BBSome, a protein complex composed of seven Bardet–Biedl syndrome (BBS) proteins, physically and genetically interact with CEP290 and modulate the expression of disease phenotypes caused by CEP290 mutations. The BBSome binds to the N-terminal region of CEP290 through BBS4 and co-localizes with CEP290 to the transition zone (TZ) of primary cilia and centriolar satellites in ciliated cells, as well as to the connecting cilium in photoreceptor cells. Although CEP290 still localizes to the TZ and connecting cilium in BBSome-depleted cells, its localization to centriolar satellites is disrupted and CEP290 appears to disperse throughout the cytoplasm in BBSome-depleted cells. Genetic interactions were tested using Cep290rd16- and Bbs4-null mutant mouse lines. Additional loss of Bbs4 alleles in Cep290rd16/rd16 mice results in increased body weight and accelerated photoreceptor degeneration compared with mice without Bbs4 mutations. Furthermore, double-heterozygous mice (Cep290+/rd16;Bbs4+/−) have increased body weight compared with single-heterozygous animals. Our data indicate that genetic interactions between BBSome components and CEP290 could underlie the variable expression and overlapping phenotypes of ciliopathies caused by CEP290 mutations.
INTRODUCTION
The primary cilium is a subcellular organelle projecting from the plasma membrane of almost every vertebrate cell. Studies during the last decade show that primary cilia are involved in various fundamental signaling pathways, such as sonic hedgehog, receptor tyrosine kinases and Wnt (1–4). Consistent with their ubiquitous occurrence and diversity of function, ciliary defects result in a range of human genetic disorders, including Joubert syndrome (JBTS), Meckel–Gruber syndrome (MKS), Senior–Loken syndrome (SLSN), Bardet–Biedl syndrome (BBS) and nephronophthisis (NPHP), which are collectively called ‘ciliopathies’ (1,5,6).
Ciliopathies exhibit considerable variations in phenotype even between siblings with the same mutations. In addition, there is phenotypic overlap among clinically and genetically distinct ciliopathies. CEP290 (also known as BBS14, JBTS5, LCA10, MKS4, NPHP6 and SLSN6) is associated with multiple ciliopathies and displays extensive variable expressivity (7). For example, mutations in CEP290 are associated with Leber congenital amaurosis (LCA), NPHP, SLSN, JBTS, BBS and MKS. LCA is an inherited retinal dystrophy causing severe vision loss within the first year of life. SLSN involves cystic kidney as well as retinal degeneration. Patients with JBTS or JBTS-related disease such as cerebello–oculo–renal syndrome display cerebellar vermis aplasia/hypoplasia in addition to renal and ocular anomalies. Finally, MKS is a neonatal lethal disease characterized by central nervous system malformations, cystic kidney and polydactyly. Patients harboring the same CEP290 mutations often develop different degrees of neurological, ocular and renal involvement, and are clinically diagnosed with different diseases. Although more than 100 unique CEP290 mutations have been discovered thus far, no clear genotype–phenotype correlation has been established (7).
BBS is a pleiotropic disorder characterized by retinal degeneration, obesity, learning difficulties, polydactyly and renal abnormalities. Genetically, BBS is heterogeneous and 17 BBS genes (BBS1-BBS12, MKS1, CEP290, WDPCP, SDCCAG8 and LZTFL1) have been identified to date (6,8–12). Recent molecular and cellular studies have begun to determine molecular functions of BBS proteins. Seven core BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8 and BBS9) form a stable complex, called the BBSome, and this complex functions to sort membrane proteins to primary cilia (13,14). BBS6, BBS10 and BBS12 form another complex, the BBS/CCT chaperonin complex, that facilitates BBSome assembly (15). LZTFL1 (also known as BBS17) is shown to regulate the BBSome ciliary trafficking and hedgehog signaling (10,16). Four BBS genes (MKS1, CEP290, WDPCP and SDCCAG8) are primarily associated with other ciliopathies, including NPHP, JBTS and MKS, whose phenotypic components partly overlap with those of BBS (7,11,12,17).
At the molecular level, CEP290 is believed to be involved in controlling protein trafficking at the ciliary base. In Chlamydomonas reinhardtii, CEP290 localizes to the transition zone (TZ) of flagella and is proposed to function as a gatekeeper regulating flagellar protein content (18). Recent protein–protein interaction studies established the CEP290 interaction network at the ciliary TZ (19,20) and most proteins in this network are associated with ciliopathies.
In addition to the ciliary TZ, centriolar satellite localization of CEP290 has been described particularly in mammalian cells (21,22). Similar localization of the BBSome components to the centriolar satellite has been noticed (13,16,22), suggesting potential interactions between CEP290 and the BBSome. Furthermore, the wide clinical spectrum of CEP290-related ciliopathies cannot be explained by the CEP290 genotype alone, implying the presence of second-site modifier alleles. Here, we report physical and genetic interactions between the BBSome components and CEP290 and describe the contribution of BBS mutations to the phenotypic variation of CEP290-related ciliopathies.
RESULTS
Physical interaction between the BBSome and CEP290
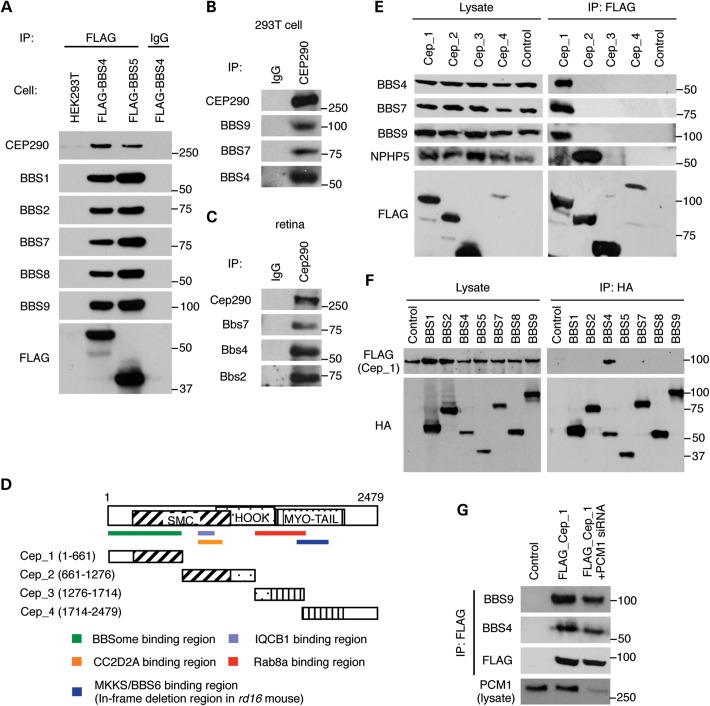
Previous studies have shown the localization of CEP290 to the ciliary TZ and centriolar satellites (18,19,21,22). BBS proteins are also found in these structures (16), suggesting possible interactions between CEP290 and the BBSome. To test this, we utilized stable cell lines expressing FLAG-BBS4 and FLAG-BBS5 and examined their association with endogenous CEP290. As shown in Figure 1A, endogenous CEP290 and other BBSome subunits were co-immunoprecipitated with both BBS4 and BBS5, indicating that CEP290 associates with the BBSome. Immunoprecipitation (IP) of endogenous CEP290 using HEK293T cell lysates and mouse retinal extracts confirmed the interaction of the BBSome and CEP290 in more physiological conditions (Fig. 1B and C). Interaction domain-mapping experiments showed that the BBSome (as represented by BBS4, BBS7 and BBS9) binds to the N-terminal part of CEP290 (Fig. 1D and E). Among the BBSome components, BBS4 is the CEP290-interacting subunit of the BBSome (Fig. 1F).
Figure 1.
Physical interaction between CEP290 and the BBSome. (A) Co-immunoprecipitation of CEP290 in HEK293T cells stably expressing FLAG-BBS4 and FLAG-BBS5. Lysates from stable cell lines and control (parental) cells were subjected to immunoprecipitation (IP) with the anti-FLAG antibody and precipitated proteins were analyzed by immunoblotting with indicated antibodies. Normal mouse IgG pull-down was used as a negative control. (B and C) Interaction of endogenous CEP290 and the BBSome in HEK293T cells (B) and mouse retina (C). Lysates from HEK293T cells and mouse retina were subjected to IP using antibodies against CEP290, and precipitated proteins were analyzed by immunoblotting with indicated antibodies. (D) Schematic representation of the CEP290 deletion mutants. Numbers indicate expressed portions of CEP290 in amino acid positions. Known IQCB1-, CC2D2A- and RAB8A-binding domains and the BBSome-interacting region are also summarized. SMC, structural maintenance of chromosomes; MYO-Tail, myosin-tail homology domain. (E) The BBSome binds to the N-terminal part of CEP290. CEP290 deletion mutants (FLAG-tagged) were transfected into HEK293T cells and lysates were analyzed by IP using anti-FLAG antibodies. Untransfected cells were used as a negative control. (F) BBS4 interacts with CEP290. HA-tagged, individual BBSome components were transiently transfected with FLAG-Cep_1 constructs. Lysates were subjected to IP with anti-HA antibodies. (G) PCM1-independent interaction between the BBSome and CEP290. HEK293T cells were co-transfected with Cep_1 fragment and siRNA against PCM1.
It has been shown that pericentriolar material 1 (PCM1) also localizes to the centriolar satellite and physically interacts with BBS4 and CEP290 (21,22). To test whether PCM1 mediates the interaction between CEP290 and the BBSome, we blocked the expression of PCM1 by transfecting small interfering RNAs (siRNAs) and evaluated the BBSome–CEP290 interaction. In this experiment, depletion of PCM1 resulted in only a modest decrease in the association between CEP290 and the BBSome (Fig. 1G), indicating that the BBSome–CEP290 interaction is at least partly independent of PCM1.
Proper localization of CEP290 to the centriolar satellite and photoreceptor connecting cilium is BBSome-dependent
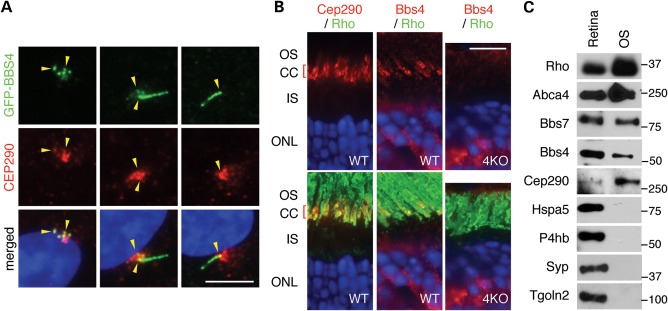
We next examined localization of the BBSome and CEP290. As previously reported (21,22), CEP290 localizes to the ciliary base (the TZ) and centriolar satellites in hTERT-RPE1 cells (Fig. 2A). BBS4 shows extensive overlap with CEP290 in the centriolar satellite and the TZ but also localizes to the cilium, where CEP290 is not detected. BBS9, another component of the BBSome, also shows localization patterns similar to BBS4 (Supplementary Material, Fig. S1A). We next investigated the localization of Cep290 and Bbs4 in mouse retinas by immunofluorescence microscopy. Consistent with previous reports (23), Cep290 was primarily detected in the connecting cilium of photoreceptor cells (Fig. 2B). Bbs4 was also found in the connecting cilium of photoreceptor cells. Bbs4 staining in the outer nuclear layer (ONL) is considered non-specific because staining is also observed in Bbs4-null retinas (Fig. 2B, right panel). Co-localization of the BBSome and Cep290 in the photoreceptor outer segment (OS) was further supported by subcellular fractionation experiments (Fig. 2C). For example, Cep290 and the BBSome components, Bbs4 and Bbs7, were highly enriched in the OS fraction together with OS markers such as rhodopsin (Rho) and Abca4 (24). In contrast, markers of the endoplasmic reticulum (Hspa5/Grp78 and P4hb/PDI) (25,26), Golgi (Tgoln2/Tgn38) (27) and synaptic vesicles (Syp) (28) were not detected in the OS fractions, demonstrating that contamination from the remaining part of the retina is minimal in our OS preparations.
Figure 2.
Co-localization of CEP290 and the BBSome. (A) Co-localization of GFP-tagged BBS4 and CEP290 in RPE1 cells (yellow arrowheads). Localization of CEP290 (red) was probed with the anti-CEP290 antibody in hTERT-RPE1 after 24 h of serum withdrawal, whereas BBS4-GFP was probed with the anti-GFP antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Note the variable localization of BBS4, although these cells were cultured in the same condition (from a single well). Scale bar, 5 μm. (B) Co-localization of Bbs4 and Cep290 in the mouse retina. Antibodies against Cep290 (red) and rhodopsin (green; a marker of the OS) were used in WT photoreceptors in the left panel, whereas in the middle and right panels Bbs4 (red) and rhodopsin (green) localizations were probed in WT and Bbs4−/− (4KO) photoreceptors. Both Cep290 and Bbs4 localize to the connecting cilium. CC, connecting cilium; ONL, outer nuclear layer. Scale bar, 10 μm. (C) Co-fractionation of the BBSome and Cep290 in the photoreceptor OS fraction. The photoreceptor OS was isolated from the mouse retina and probed with antibodies against multiple subcellular marker proteins, Bbs4, Bbs7 and Cep290.
We next evaluated functional interactions between the BBSome and CEP290. We first tested whether the BBSome is required for proper localization of CEP290. To this end, we examined CEP290 localization in BBS1-, BBS4- and BBS9-depleted hTERT-RPE1 cells. Efficient depletion of BBS1, BBS4 and BBS9 by siRNA transfection was confirmed at the protein or mRNA level (Supplementary Material, Fig. S2). In control siRNA-transfected cells, CEP290 localized to the TZ and the centriolar satellite, particularly near the centriole (Fig. 3A and B). BBS1, BBS4 or BBS9 knockdown resulted in a significant decrease in the centriolar satellite pool of CEP290. A similar but more substantial reduction was observed in PCM1 knockdown cells. These results indicate that association of CEP290 with the centriolar satellite is both BBSome- and PCM1-dependent. Of note, the CEP290 pool in the TZ and centrosomes was not affected by depletion of BBS or PCM1 proteins. Interestingly, CEP290 localization to the centrosome and TZ was also not affected by microtubule depolymerization, whereas centriolar satellite localization of CEP290, PCM1 and BBS4 was disturbed upon depolymerization of microtubules (21). These findings suggest that localization of CEP290 to the TZ and centrosomes may be independent of centriolar satellites. Alternatively, centrosome-associated CEP290 may be stable with a slow rate of turn over. A previous study by Craige et al. (18) reported that BBS proteins were moderately increased in Chlamydomonas cep290 mutant flagella. In contrast, a more recent study using RPE1 cells showed that CEP290 is required for the BBSome ciliary localization (29). In our experiments, ciliary localization of the BBSome was not affected by CEP290 depletion. Furthermore, BBSome localization to the centriolar satellite was also not affected by CEP290 depletion (Supplementary Material, Fig. S1).
Figure 3.
The CEP290 localization in the centriolar satellite and the connecting cilium is BBSome-dependent. (A) The BBSome is required for centriolar satellite localization of CEP290 (red). RPE1 cells were transfected with siRNAs against CEP290, BBS1, BBS4, BBS9 and PCM1. Antibodies against γ-tubulin and acetylated tubulin (green) were used to mark the basal body and cilia, respectively. (B) Quantification of CEP290 mis-localization in BBSome-depleted RPE1 cells. Cells with concentrated CEP290 staining around the centrosome (within 2 μm from the centrosome) were counted as positive, whereas cells with CEP290 only in the TZ and centrosome were considered negative. At least 120 cells per experiment were counted and graphs represent averages of three independent experiments. Error bars represent SEM. One-way ANOVA followed by Tukey's post-test was used for statistical analysis. **P < 0.01 compared with Ctrl KD cells. (C) Localization of Cep290 (green; left) in WT (top), Bbs1M390R/M390R (middle) and Bbs4−/− (bottom) mouse retinas. OS localization of Prph2 (red) with respect to acetylated tubulin (green) is not affected in BBS mutant retinas (right panels). Scale bars, 10 μm.
We also examined whether the BBSome is required for Cep290 localization in mouse retina. To minimize potential secondary effects of retinal degeneration in BBS mice, we examined Cep290 localization in young animals (P21–P30) before retinal degeneration is apparent. At this age, the ONL thickness in Bbs1M390R/M390R and Bbs4−/− retina is comparable with that of wild-type animals (Fig. 3C). In addition, OS proteins peripherin 2 (Prph2; also known as Rds) and the vast majority of rhodopsin localize properly to the OS, demonstrating that protein trafficking to the OS is not completely disrupted. However, in Bbs1M390R/M390R retina, Cep290 was found dispersed throughout the photoreceptor cell, including the entire OS, inner segment (IS) and nuclear layer (Fig. 3C). A similar phenotype was also observed in Bbs4−/− mice. Consistent with the results obtained from RPE1 cells, a subset of Cep290 persisted at the connecting cilium in BBS mutant retinas. These results suggest that the BBSome limits Cep290 to the connecting cilium and prevents improper localization of Cep290 in other locations in photoreceptor cells.
Increased obesity in mice with combined loss of Cep290 and Bbs4 genes
Given the physical interaction between CEP290 and the BBSome and the requirement of the BBSome for proper localization of CEP290, we investigated the genetic interaction between Cep290 and BBS genes. To this end, we used Cep290rd16 (23) and Bbs4 knock-out (30) mouse lines. Cep290rd16 is a hypomorphic allele with an internal in-frame deletion of 299 residues (amino acids 1599–1897; NP_666121) from the Cep290 protein (23). In humans, the phenotypes associated with CEP290 mutations range from LCA, which manifests only photoreceptor degeneration, to the neonatal lethal MKS, which involves severe cystic kidney and neural tube patterning defects as well as photoreceptor degeneration. Cep290rd16 homozygous mutant mice display only the photoreceptor degeneration phenotype but not other components of the CEP290-associated ciliopathy phenotypes such as obesity, cystic kidney and neural tube patterning defects. We examined whether the phenotype of Cep290rd16 mice is affected by the presence of Bbs4 mutations.
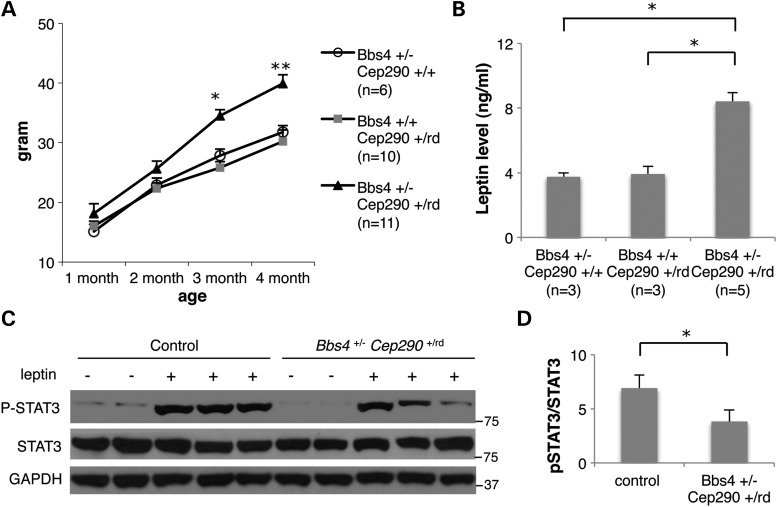
First, we measured the body weight of progeny from Cep290rd16- and Bbs4-null mouse crosses (Fig. 4 and Supplementary Material, Fig. S3). In general, body weight correlated with the number of mutant alleles in Cep290 and Bbs4 genes. For example, Cep290rd16/rd16 single-homozygous mutant mice did not develop obesity on this genetic background (Supplementary Material, Fig. S3). However, loss of one copy of Bbs4 (i.e. Cep290rd16/rd16; Bbs4+/−) resulted in a significant increase in body weight. Cep290rd16/rd16; Bbs4−/− double-homozygous mutant mice were the most obese among all genotypes examined (Supplementary Material, Fig. S3). Interestingly, double-heterozygous mice (Cep290+/rd16; Bbs4+/−) showed significantly increased body weight compared with their sex-matched wild-type or single-heterozygous littermates (Fig. 4A and Supplementary Material, Fig. S3). Previously, higher blood leptin levels and leptin resistance were observed in obese BBS mice (31–33). Consistent with this, Cep290+/rd16; Bbs4+/− double-heterozygous animals had higher blood leptin levels than wild-type or single-heterozygous littermates (Fig. 4B). We further compared the responsiveness of the hypothalamic anorexic circuitry to exogenous leptin by measuring phosphorylated STAT3 upon leptin injection (33). In control single-heterozygous mice, intra-peritoneal (i.p.) injection of leptin results in a robust increase in STAT3 phosphorylation (Fig. 4C and D). However, the phosphorylation level of STAT3 was significantly reduced in double-heterozygous mice injected with leptin. Combined, these data suggest that the obesity component of the CEP290-associated ciliopathy can be modified by the presence of mutations in BBS genes.
Figure 4.
Increased body weight and higher leptin levels in Bbs4+/−; Cep290+/rd16 mice. (A) Weight gains in male animals versus age (minimum of six animals per group). Values are expressed as mean + SEM. By month 3, double-heterozygous mice are significantly heavier than single-heterozygous littermates. *P < 0.05 versus single-heterozygous animals; **P < 0.01 versus single-heterozygous animals. (B) Serum leptin levels of single- and double-heterozygous mice. One-way ANOVA and t-test were used for statistical analysis. *P < 0.05 versus single-heterozygous animals. (C) STAT3 phosphorylation upon leptin administration was reduced in double-heterozygous mice. Hypothalamic protein extracts were analyzed by western blotting. (D) Quantification of STAT3 phosphorylation. Band intensities of phospho-STAT3 (P-STAT3) were normalized with those of total STAT3 and induction ratios were calculated by comparison with vehicle-injected samples. Data represent mean + SEM. n = 5 for vehicle and n = 10 for leptin. *P < 0.05.
Accelerated retinal degeneration in mice with combined loss of Cep290 and Bbs4 alleles
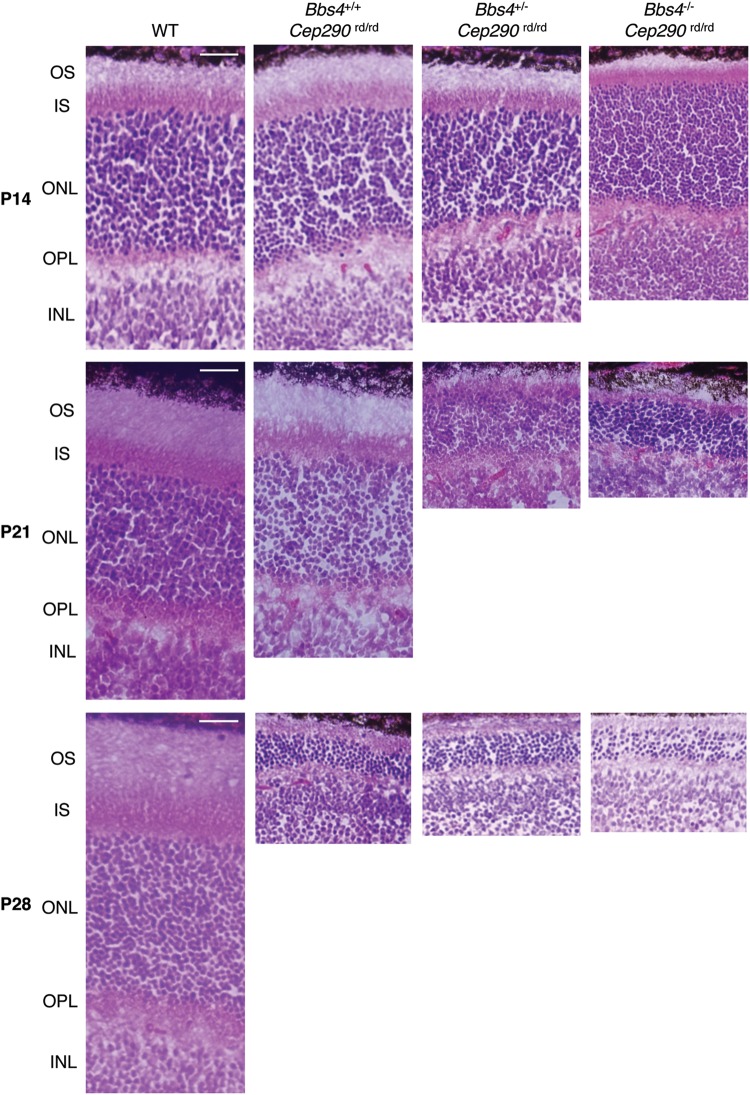
The Cep290rd16/rd16 and Bbs4−/− mice display retinal abnormalities (23,30,34). Like the obesity phenotype, mice carrying a combination of three or four mutations in Cep290 and Bbs4 genes displayed more severe phenotypes in retinal photoreceptor cells (Fig. 5). For example, although all layers of the retina were present and appeared grossly normal with IS lengths comparable with the WT and Cep290rd16/rd16 single homozygotes at P14, OS elongation was clearly stalled and beginning to regress in Cep290rd16/rd16; Bbs4+/− and Cep290rd16/rd16; Bbs4−/− mouse retinas (Fig. 5). Almost complete loss of the OS was observed in these retinas at P21, with thinner ONL in Cep290rd16/rd16;Bbs4−/− mouse retinas. Cep290rd16/rd16 single homozygotes also showed severe photoreceptor degeneration at P28.
Figure 5.
Accelerated photoreceptor degeneration with additional loss of Bbs4 alleles in Cep290rd16/rd16 mice. H&E-stained sections of the superior central retina at the level of the optic nerve head from P14, P21 and P28 mice are shown. Scale bars, 10 μm.
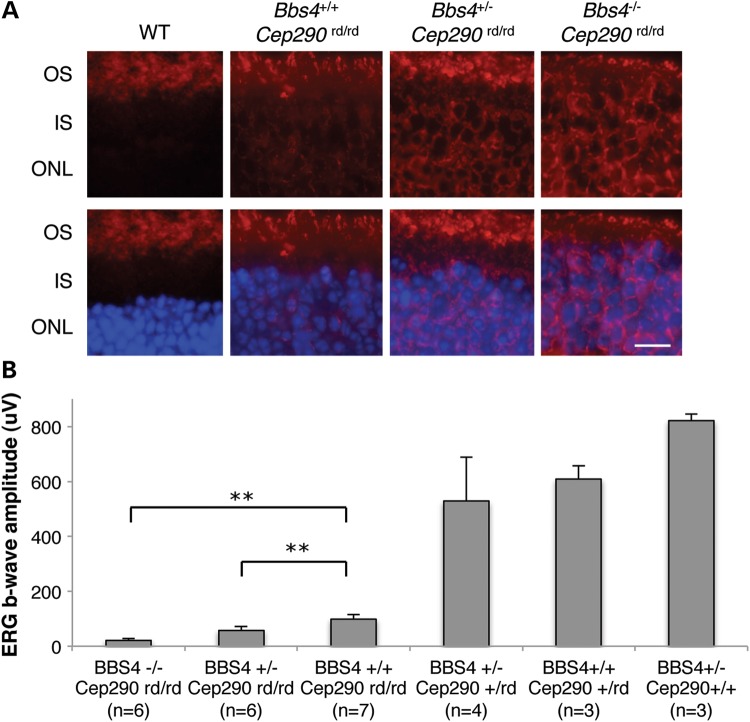
Next, we sought to determine the effect of combined loss of Cep290 and Bbs4 alleles on the trafficking of phototransduction proteins in the retina. Whereas no mis-localization of rhodopsin was detected in wild-type photoreceptor cells, rhodopsin mis-localization to the ONL was apparent in Cep290rd16/rd16 single-homozygous retinas at P21 (Fig. 6A). Losing one or two copies of Bbs4 in this background further impaired rhodopsin trafficking and more rhodopsin was found in the ONL and IS. Consistent with these findings, dark-adapted standard combined response (DA-SCR) electroretinography (ERG) b-wave of Cep290rd16/rd16; Bbs4+/− and Cep290rd16/rd16; Bbs4−/− mice at 1 month of age showed considerable decline compared with that of Cep290rd16/rd16 single homozygotes (Fig. 6B). In addition to the reduction of the ERG b-wave in Cep290rd16/rd16 mice, the oscillatory potentials were also markedly reduced (Supplementary Material, Fig. S4). This reduction was more pronounced when one or two Bbs4-null alleles were present, suggesting more severe photoreceptor dysfunction by loss of Bbs4 alleles in Cep290rd16/rd16 retinas. Alternatively, this reduction in ERG b-wave may be due to more inner retina dysfunction in these animals. Together, these data suggest that although homozygous Cep290rd16 alleles cause severe photoreceptor degeneration in mice, loss of one or two copies of Bbs4 on this background further disrupts retinal morphology and protein trafficking in the retina, and thus results in accelerated photoreceptor degeneration.
Figure 6.
Impaired rhodopsin trafficking and diminished ERG responses in mice with combined loss of Cep290 and Bbs4 alleles. (A) Immunohistochemical analysis of WT, Bbs4+/+;Cep290rd/rd, Bbs4+/−;Cep290rd/rd and Bbs4−/−; Cep290rd/rd retinas at P21 with antibodies against rhodopsin. Scale bar, 5 μm. (B) DA-SCR ERG b-wave amplitudes in the indicated mouse genotypes (at the age of 1 month). Removing one or two Bbs4 alleles on a Cep290rd/rd background results in a lower ERG response. **P < 0.01 versus Bbs4+/+;Cep290rd/rd mice. Error bars are SD.
DISCUSSION
Ciliopathies are characterized by significant inter- and intra-familial phenotypic variability, as well as phenotypic overlap among clinically and genetically distinct syndromes. This variability suggests the presence of modifiers and genetic interactions among ciliopathy genes. Indeed, modifiers and genetic interactions among ciliopathy genes have been reported in several studies. For example, a JBTS gene Ahi1 genetically interacts with Nphp1 to modulate the retinal degeneration phenotype and functions as a modifier for the ocular component of NPHP (35). Synergetic interactions between NPHP and MKS genes were also observed in Caenorhabditis elegans (36). Finally, variable expression of neurological phenotypes that span MKS and JBTS has been reported in Tmem67 mutant mice (37), which suggests a contribution of modifier alleles in different genetic backgrounds to the variable cilia-related phenotypes. In this study, we demonstrate physical and genetic interactions between BBS genes and CEP290. Our findings indicate that the BBSome is required for proper localization of CEP290 in cultured ciliated cells and photoreceptor cells. We also demonstrate that the presence of BBS mutations in Cep290 hypomorphs exacerbates the phenotype of Cep290 mutants. Interestingly, our mouse model exhibits phenotypic features with tissue-specific penetrance. For example, compared with single-heterozygous mice, increased body weight but not retinal degeneration was found in double-heterozygous mice (Cep290+/rd16; Bbs4+/−). However, Cep290rd16/rd16; Bbs4+/− mice showed both increased obesity and more severe retinal degeneration compared with the Cep290rd16/rd16; Bbs4+/+ mice. Cep290rd16/rd16; Bbs4−/− double-homozygous mice showed the most severe phenotypes in both body weight and retinal degeneration among all genotypes. In addition, neither kidney cysts nor polydactyly was observed in any of the mice we examined, including Cep290rd16/rd16; Bbs4−/− double-homozygous mutant mice (n = 6; data not shown). These findings suggest a modifying effect of BBS alleles on the CEP290 phenotype and that this modifying effect is tissue-specific. Importantly, our data also indicate that specific combinations of heterozygous alleles could contribute to individual common human phenotypes such as obesity.
Many ciliopathy proteins are components of the BBSome, IFT complexes and the NPHP and MKS modules (13,19,20,36,38). These proteins participate in specific steps or aspects of pathways that control protein trafficking to and from cilia. For example, whereas the BBSome and the IFT complexes are involved in transporting ciliary proteins, NPHP and MKS proteins form distinct modular complexes at the ciliary TZ and function as a gatekeeper. Coordinated functions of these proteins are essential for primary cilia formation, maintenance and function. In a study using C. elegans as a model, synergetic genetic interactions were observed between members of the NPHP and the MKS modules but not within each module (36). The BBSome and CEP290 also belong to different modules/complexes and we observe a genetic interaction. It will be interesting to test whether BBS genes genetically interact with other members of TZ-localizing ciliopathy genes and whether these genetic interactions contribute to the phenotypic variability and overlap among ciliopathies.
In mouse models, combined loss of Bbs4 and hypomorphic impairment of Cep290 functions result in exacerbated disease phenotypes, including obesity and photoreceptor degeneration. However, a recent study using Mkks (also known as Bbs6)-null mice and Cep290rd16 mice showed that compared with single homozygotes, Cep290rd16 and Mkks−/− double-homozygous and triple-allelic combinations had a less severe phenotype or even rescued the defects observed in sensory cells (39). Considering the requirement of Mkks for BBSome assembly (15), these contrasting results in these two studies are interesting. However, it is noteworthy that, in Mkks−/− mice, several BBSome components (e.g. Bbs2, Bbs7 and Bbs9) are unstable and degraded, leading to failure of BBSome assembly (15), whereas in Bbs4−/− mice the BBSome is formed normally except for the absence of Bbs4 (40). The presence of the BBSome (with or without BBS4) in Cep290rd16/rd16 or Cep290rd16/rd16; Bbs4−/− mice may cause disease in these animals, whereas the lack of the BBSome in Cep290rd16/rd16; Mkks−/− mice may alleviate the mutant phenotype.
Another possibility to explain the differences between the Cep290-Mkks study and ours is the functional independence of individual domains in Cep290 and specific loss of a protein–protein interaction in rd16 mutant mice. In the Rachel et al. study (39), Cep290 was shown to interact with Mkks via its Myosin-tail homology domain, which is deleted in the rd16 mouse, whereas our data show that the N-terminal region of CEP290 interacts with the BBSome. Therefore, the rd16 mutant form of Cep290 is not able to interact with Mkks but still maintains its interaction with the BBSome. In Cep290rd16/rd16 mice, the inability of Mkks to associate with Cep290 may be deleterious to photoreceptor cells. Reduction of Mkks activity in this background may result in alleviation of the photoreceptor degeneration phenotype. In contrast, loss of BBSome components in rd16 mutant mice will compromise residual Cep290 function, resulting in the more severe phenotypes observed in our mouse model. Notably, these findings show that genetic interactions between BBS genes and CEP290 can influence the phenotypic variations in CEP290-related ciliopathies.
MATERIALS AND METHODS
Animal studies
Mice were bred and maintained in standardized conditions at the University of Iowa. The use of mice was approved by the University Animal Care and Use Committee. Cep290rd16/rd16 mutant mice (23) on a BXD24 background and Bbs4−/− mice (30) on a 129/Sv background were used to generate double mutants on a mixed BXD24 and 129 background. Weights of mice was measured monthly beginning at weaning, and those of double-heterozygous mice were compared with single heterozygote from 1 to 5 months of age, using an average of 10 animals per group. Circulating leptin levels were analyzed in blood collected from adult double-heterozygous mutant mice (n = 5) and control mice (n = 3). Plasma was obtained by centrifuging blood at 2040g for 8 min. Concentration of murine leptin was measured by ELISA using the mouse leptin ELISA kit (Crystal Chem).
Antibodies, plasmids and reagents
Expression vectors for BBS genes were published (15). The four CEP290 fragments encoding about 600 amino acids (1–661 amino acids, 661–1276 amino acids, 1276–1714 amino acids and 1714–2479 amino acids) of human wild-type CEP290 (NCBI Reference Sequence: NG_008417.1) were amplified from human retina cDNA (Clontech), cloned into the pSS-FS2 vector (41) and sequences verified. The PCR primers used for these clonings are
CEP290—primer (1–661 amino acids) F: 5′-ATGCCACCTAATATAAACTG-3′;
CEP290—primer (1–661 amino acids) R: 5′-CATTTCCTTAATTGCTT-3′;
CEP290—primer (661–1276 amino acids) F: 5′-ATGCAGAAAGATCCTG-3′;
CEP290—primer (661–1276 amino acids) R: 5′-TCCACTAAACTGTC-3′;
CEP290—primer (1276–1714 amino acids) F: 5′-GGAGCTTTACCCTTGGCAC-3′;
CEP290—primer (1276–1714 amino acids) R: 5′-TGCTTCTTTTTGAGCCTGAAG-3′;
CEP290—primer (1714–2479 amino acids) F: 5′-GCAAATTCAAGAGCTCC-3′;
CEP290—primer (1714–2479 amino acids) R: 5′-TTAGTAAATGGGGAAATTAACAGG-3′.
siRNAs were purchased from Dharmacon (ON-TARGETplus SMARTpool) and transfected at 50 nm concentration for all experiments with RNAiMAX (Invitrogen) following the manufacturer's protocol.
Antibodies against BBS1, BBS2, BBS4 and BBS7 were described previously (13). Other antibodies used were purchased from the following sources: mouse monoclonal antibodies against ABCA4 (3F4; Abcam), acetylated tubulin (6–11B-1; Sigma), γ-tubulin (GTU-88; Sigma), FLAG (M2; Sigma), HA (F-7; Santa Cruz Biotechnology), GFP (3E6; Invitrogen), beta-actin (AC-15; Sigma), rhodopsin (RET-P1; Santa Cruz Biotechnology), and synaptophysin (Santa Cruz Biotechnology; sc-55507); mouse polyclonal antibodies against IQCB1 (NPHP5; Abcam; ab69927); rabbit polyclonal antibodies against BBS8 (Sigma; HPA003310), BBS9 (Sigma; HPA021289), CEP290 (for immunoprecipitation, immunoblotting and RPE1 cell immunofluorescence microscopy; Bethyl Lab; IHC-00365), HSPA5/GRP78/BiP (Cell Signaling; 3177), PCM1 (Sigma; HPA023374), PDI (Sigma; P7372), PRPH2 (Proteintech Group; 18109), STAT3 (Santa Cruz Biotechnology; SC-483), phospho-STAT3 (Y705; Cell Signaling; 9131) and Tgoln2/Tgn46 (Abcam; ab16059). Rabbit monoclonal antibody GAPDH (14C10) conjugated with HRP was purchased from Cell Signaling. The anti-Cep290 antibody used for mouse photoreceptor cell immunofluorescence microscopy was a gift from Dr Robert Mullins (University of Iowa).
Cell culture, transfection and co-immunoprecipitation assays
hTERT-RPE1 cells were maintained in DMEM/F12 media (Invitrogen) supplemented with 10% FBS. Cells were transfected with siRNAs using RNAiMAX for 48 h and further incubated in serum-free medium for 24 h for ciliation. HEK293 T cells were maintained in DMEM (Invitrogen), supplemented with 10% fetal bovine serum and penicillin/streptomycin (Invitrogen) and grown at 37°C in 5% CO2. Stable cell lines expressing FLAG-tagged BBS4 and FLAG-tagged BBS5 were generated by co-transfecting pSS-FS-BBS4 and pSS-FS-BBS5 with pCS2-puro into HEK293T cells followed by puromycin selection (1.5 μg/ml; Invitrogen). Cells were grown in six-well dishes, transfected with FLAG-tagged CEP290 or BBSome subunits using Lipofectamine 2000 (Invitrogen) and harvested after 24 h by scraping into 500 μl ice-cold lysis buffer (50 mm HEPES, 200 mm KCl, 2 mm EGTA, 1 mm MgCl2, 10% glycerol, 1% Triton X-100) supplemented with protease inhibitors (Roche). Co-IP was performed as previously described (15).
Immunoprecipitation
Human HEK293T cells were lysed in ice-cold lysis buffer supplemented with protease inhibitors (Roche) and spun at 20 000g for 15 min at 4°C. The supernatants were incubated with antibodies against CEP290 overnight. Protein A beads (Thermo Scientific) were then added and incubated for 2 h. The beads were washed four times with lysis buffer and the interactions were detected by western blotting. TrueBlot anti-rabbit IgG secondary antibodies (Rockland) were used to avoid detecting IgG heavy and light chains. For the immunoprecipitation in the mouse retina, the mouse eyes were enucleated and homogenized in ice-cold lysis buffer (50 mm HEPES, 200 mm KCl, 2 mm EDTA, 1% NP-40, 0.5 mm DTT) supplemented with protease inhibitors (Roche). The lysates were spun at 20 000g for 15 min at 4°C, and the supernatants were pre-cleared by incubation with protein A and proteins G beads (Thermo Scientific) for 2 h. IP was performed on the cleared lysates through the procedure described above.
Histology and ERG
After euthanasia by carbon dioxide asphyxiation, eyes were collected and immersed in a solution of 4% paraformaldehyde in PBS. After 2 to 4 h of fixation, eyes were washed three times in PBS followed by infiltration and embedding in acrylamide (42), and then were frozen in the optimal cutting temperature (OCT) compound (Tissue-Tek). Cryostat sections were collected at a thickness of 7 μm, and stored at −20°C until use. Sections were stained following the standard hematoxylin and eosin (H&E) staining protocol. Mouse ERGs were measured using the Diagnosis ColorDome system after dark adaptation (>6 h) and anesthesia (ketamine, 65 mg/kg, xylazine, 5 mg/kg) as previously described (43,44).
Immunofluorescence microscopy
For immunofluorescence microscopy, cultured cells were seeded on glass coverslips in 24-well plates and transfected with siRNAs or with plasmid DNAs as described above. Cells were cultured for 72 h before fixation with the last 24 h in serum-free medium to induce ciliogenesis, and then fixed with 4% paraformaldehyde in PBS followed by cold methanol. Samples were blocked with 5% BSA and 2% normal goat serum in PBST (0.1% Triton X-100) and incubated with primary antibodies in the blocking buffer. For immunofluorescence microscopy of retinal sections, eyes were fixed as above and embedded and frozen in OCT. Sections (7 μm) were collected and blocked with 5% BSA in PBST for 20 min at 25°C before incubation with primary antibody at 4°C overnight. Primary antibodies were visualized by Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) and Alexa Fluor 568 goat anti-mouse IgG (Invitrogen). Coverslips were mounted on VectaShield mounting medium with DAPI (Vector Laboratories), and images were taken with Olympus IX71 microscope.
Photoreceptor OS isolation
The mouse eyes were enucleated and placed in ice-cold homogenization buffer (50% w/v sucrose in PBS, complete protease inhibitor). The retina was dissected from enucleated eyes and then disrupted by repeated pipetting through a P1000 tip with ∼1.5–2.0 mm orifice followed by 30 s vortexing. The homogenate was spun at 200g for 3 min and the supernatant was further spun at 13 000g for 20 min at 4°C. After centrifugation, the supernatant was diluted 1 : 1 in PBS without sucrose. The diluted OS fraction was applied on top of a 50% (w/w) sucrose/PBS cushion and centrifuged at 13 000g for 30 min at 4°C. The OS fraction was collected at the interface and diluted with five volumes of PBS. ROS was collected as pellet after the diluted fraction was spun at 6000g for 10 min at 4°C. For SDS–PAGE and western, the pellet was re-suspended in 50 μl of 2× LDS buffer (Invitrogen).
Leptin resistance study
Sex-, age- and body weight-matched control (single-heterozygous) and double-heterozygous mice were housed in individual cages at 6–10 weeks of age. Food was removed 18 h before i.p. leptin injection. Leptin (1 μg/g body weight; R&D Systems) or vehicle (PBS) was injected i.p. and animals were sacrificed 2 h later by CO2 asphyxiation. Hypothalami were quickly dissected and homogenized in the lysis buffer containing protease and phosphatase inhibitors. Protein extracts were loaded onto 4–12% NuPAGE Bis–Tris gels and analyzed by immunoblotting.
SUPPLEMENTARY MATERIAL
AUTHORS' CONTRIBUTIONS
Y.Z. performed experiments, interpreted data and wrote the manuscript. S.S. designed the study, performed experiments, interpreted data and wrote the manuscript. S.B. and A.V.D. performed ERG measurements and interpreted data. K.B. maintained the mouse colony and contributed to the histological study. C.C.S. assisted in subcloning, and Q.Z. initially conceived the project and assisted in gene cloning and geno-typing. E.M.S. analyzed data and wrote the manuscript. V.C.S. initiated the project, designed experiments, analyzed data and wrote the manuscript.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by US National Institutes of Health grants (R01EY022616 to S.S.; R01EY110298 and R01EY017168 to V.C.S.) and the Roy J. Carver Charitable Trust. E.M.S. and V.C.S. are investigators of the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Robert Mullins (University of Iowa) for the anti-Cep290 antibody.
REFERENCES
- 1.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 2.Singla V., Reiter J.F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 3.Goetz S.C., Anderson K.V. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen S.T., Clement C.A., Satir P., Pedersen L.B. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J. Pathol. 2012;226:172–184. doi: 10.1002/path.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 6.Sheffield V.C. The blind leading the obese: the molecular pathophysiology of a human obesity syndrome. Trans. Am. Clin. Climatol. Assoc. 2010;121:172–181. Discussion 181–172. [PMC free article] [PubMed] [Google Scholar]
- 7.Coppieters F., Lefever S., Leroy B.P., De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 2010;31:1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 8.Zaghloul N.A., Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R.A., Eyaid W., Banin E., et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 10.Marion V., Stutzmann F., Gerard M., De Melo C., Schaefer E., Claussmann A., Helle S., Delague V., Souied E., Barrey C., et al. Exome sequencing identifies mutations in LZTFL1, a BBSome and smoothened trafficking regulator, in a family with Bardet–Biedl syndrome with situs inversus and insertional polydactyly. J. Med. Genet. 2012;49:317–321. doi: 10.1136/jmedgenet-2012-100737. [DOI] [PubMed] [Google Scholar]
- 11.Otto E.A., Hurd T.W., Airik R., Chaki M., Zhou W., Stoetzel C., Patil S.B., Levy S., Ghosh A.K., Murga-Zamalloa C.A., et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.K., Shindo A., Park T.J., Oh E.C., Ghosh S., Gray R.S., Lewis R.A., Johnson C.A., Attie-Bittach T., Katsanis N., et al. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 14.Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., Nachury M.V. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo S., Baye L.M., Schulz N.P., Beck J.S., Zhang Q., Slusarski D.C., Sheffield V.C. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl Acad. Sci. USA. 2010;107:1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., Sheffield V.C. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 2011;7:e1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyttala M., Tallila J., Salonen R., Kopra O., Kohlschmidt N., Paavola-Sakki P., Peltonen L., Kestila M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat. Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- 18.Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., Witman G.B. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M., et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J., et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Krishnaswami S.R., Gleeson J.G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes C.A., Prosser S.L., Romio L., Hirst R.A., O'Callaghan C., Woolf A.S., Fry A.M. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J. Cell Sci. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Illing M., Molday L.L., Molday R.S. The 220-kDa rim protein of retinal rod outer segments is a member of the ABC transporter superfamily. J. Biol. Chem. 1997;272:10303–10310. doi: 10.1074/jbc.272.15.10303. [DOI] [PubMed] [Google Scholar]
- 25.Yang L.P., Wu L.M., Guo X.J., Tso M.O. Activation of endoplasmic reticulum stress in degenerating photoreceptors of the rd1 mouse. Invest. Ophthalmol. Vis. Sci. 2007;48:5191–5198. doi: 10.1167/iovs.07-0512. [DOI] [PubMed] [Google Scholar]
- 26.Vasireddy V., Jablonski M.M., Mandal M.N., Raz-Prag D., Wang X.F., Nizol L., Iannaccone A., Musch D.C., Bush R.A., Salem N., Jr., et al. Elovl4 5-bp-deletion knock-in mice develop progressive photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 2006;47:4558–4568. doi: 10.1167/iovs.06-0353. [DOI] [PubMed] [Google Scholar]
- 27.Evans R.J., Schwarz N., Nagel-Wolfrum K., Wolfrum U., Hardcastle A.J., Cheetham M.E. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum. Mol. Genet. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- 28.Brandstatter J.H., Lohrke S., Morgans C.W., Wassle H. Distributions of two homologous synaptic vesicle proteins, synaptoporin and synaptophysin, in the mammalian retina. J. Comp. Neurol. 1996;370:1–10. doi: 10.1002/(SICI)1096-9861(19960617)370:1<1::AID-CNE1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Stowe T.R., Wilkinson C.J., Iqbal A., Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell. 2012;23:3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mykytyn K., Mullins R.F., Andrews M., Chiang A.P., Swiderski R.E., Yang B., Braun T., Casavant T., Stone E.M., Sheffield V.C. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl Acad. Sci. USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis R.E., Swiderski R.E., Rahmouni K., Nishimura D.Y., Mullins R.F., Agassandian K., Philp A.R., Searby C.C., Andrews M.P., Thompson S., et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc. Natl Acad. Sci. USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahmouni K., Fath M.A., Seo S., Thedens D.R., Berry C.J., Weiss R., Nishimura D.Y., Sheffield V.C. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J. Clin. Invest. 2008;118:1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo S., Guo D.F., Bugge K., Morgan D.A., Rahmouni K., Sheffield V.C. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum. Mol. Genet. 2009;18:1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichers E.R., Abd-El-Barr M.M., Paylor R., Lewis R.A., Bi W., Lin X., Meehan T.P., Stockton D.W., Wu S.M., Lindsay E., et al. Phenotypic characterization of Bbs4 null mice reveals age-dependent penetrance and variable expressivity. Hum. Genet. 2006;120:211–226. doi: 10.1007/s00439-006-0197-y. [DOI] [PubMed] [Google Scholar]
- 35.Louie C.M., Caridi G., Lopes V.S., Brancati F., Kispert A., Lancaster M.A., Schlossman A.M., Otto E.A., Leitges M., Grone H.J., et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet. 2010;42:175–180. doi: 10.1038/ng.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell. Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelhamed Z.A., Wheway G., Szymanska K., Natarajan S., Toomes C., Inglehearn C., Johnson C.A. Variable expressivity of ciliopathy neurological phenotypes that encompass Meckel-Gruber syndrome and Joubert syndrome is caused by complex de-regulated ciliogenesis, Shh and Wnt signalling defects. Hum. Mol. Genet. 2013;22:1358–1372. doi: 10.1093/hmg/dds546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenbaum J.L., Witman G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 39.Rachel R.A., May-Simera H.L., Veleri S., Gotoh N., Choi B.Y., Murga-Zamalloa C., McIntyre J.C., Marek J., Lopez I., Hackett A.N., et al. Combining Cep290 and Mkks ciliopathy alleles in mice rescues sensory defects and restores ciliogenesis. J. Clin. Invest. 2012;122:1233–1245. doi: 10.1172/JCI60981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q., Yu D., Seo S., Stone E.M., Sheffield V.C. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet-Biedl syndrome protein complex, the BBSome. J. Biol. Chem. 2012;287:20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humbert M.C., Weihbrecht K., Searby C.C., Li Y., Pope R.M., Sheffield V.C., Seo S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl Acad. Sci. USA. 2012;109:19691–19696. doi: 10.1073/pnas.1210916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson L.V., Blanks J.C. Application of acrylamide as an embedding medium in studies of lectin and antibody binding in the vertebrate retina. Curr. Eye Res. 1984;3:969–974. doi: 10.3109/02713688409167215. [DOI] [PubMed] [Google Scholar]
- 43.Aleman T.S., LaVail M.M., Montemayor R., Ying G., Maguire M.M., Laties A.M., Jacobson S.G., Cideciyan A.V. Augmented rod bipolar cell function in partial receptor loss: an ERG study in P23H rhodopsin transgenic and aging normal rats. Vision Res. 2001;41:2779–2797. doi: 10.1016/s0042-6989(01)00157-2. [DOI] [PubMed] [Google Scholar]
- 44.Cheng H., Aleman T.S., Cideciyan A.V., Khanna R., Jacobson S.G., Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet. 2006;15:2588–2602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.