Abstract
We present the analysis of a prospective multicentre study to investigate genetic effects on the prognosis of newly treated epilepsy. Patients with a new clinical diagnosis of epilepsy requiring medication were recruited and followed up prospectively. The clinical outcome was defined as freedom from seizures for a minimum of 12 months in accordance with the consensus statement from the International League Against Epilepsy (ILAE). Genetic effects on remission of seizures after starting treatment were analysed with and without adjustment for significant clinical prognostic factors, and the results from each cohort were combined using a fixed-effects meta-analysis. After quality control (QC), we analysed 889 newly treated epilepsy patients using 472 450 genotyped and 6.9 × 106 imputed single-nucleotide polymorphisms. Suggestive evidence for association (defined as Pmeta < 5.0 × 10−7) with remission of seizures after starting treatment was observed at three loci: 6p12.2 (rs492146, Pmeta = 2.1 × 10−7, OR[G] = 0.57), 9p23 (rs72700966, Pmeta = 3.1 × 10−7, OR[C] = 2.70) and 15q13.2 (rs143536437, Pmeta = 3.2 × 10−7, OR[C] = 1.92). Genes of biological interest at these loci include PTPRD and ARHGAP11B (encoding functions implicated in neuronal development) and GSTA4 (a phase II biotransformation enzyme). Pathway analysis using two independent methods implicated a number of pathways in the prognosis of epilepsy, including KEGG categories ‘calcium signaling pathway’ and ‘phosphatidylinositol signaling pathway’. Through a series of power curves, we conclude that it is unlikely any single common variant explains >4.4% of the variation in the outcome of newly treated epilepsy.
INTRODUCTION
In clinical epidemiology, prognosis refers to the future course and outcome of a disease. A notable aspect of the epilepsies is their highly variable prognosis, even among individuals with the same seizure types and epilepsy syndrome. Approximately 60% of people with epilepsy achieve long-term remission of seizures very shortly after starting antiepileptic drug (AED) treatment, while 20–30% have a chronic disorder without ever experiencing significant periods of remission (1). Seizure control is an important factor in minimizing the risk of death from epilepsy, and remission of seizures is associated with improvements in quality of life (2). Therefore, a key issue for clinical practice in epilepsy, and the development of new therapeutic approaches, is the extent to which genetic variation contributes to variation in treatment response. While a great deal is becoming known about genetic susceptibility to epilepsy (3), very little is known about genetic influences on the prognosis of epilepsy, and to date, genetic effects on epilepsy prognosis are unexplored at a genome-wide level.
For most pharmacogenetic research, attempts to identify genetic factors governing individual response to treatment are founded on a usually untested assumption—namely that genetically determined individual responses exist (4). For epilepsy however, the clinical observation that therapeutic response to the first AED predicts response to subsequent AEDs (5) supports the presence of individual effects on broad treatment response. Twin studies suggest that such individual effects on the outcome of treated epilepsy are mediated, at least in part, by epilepsy genetic susceptibility factors (6).
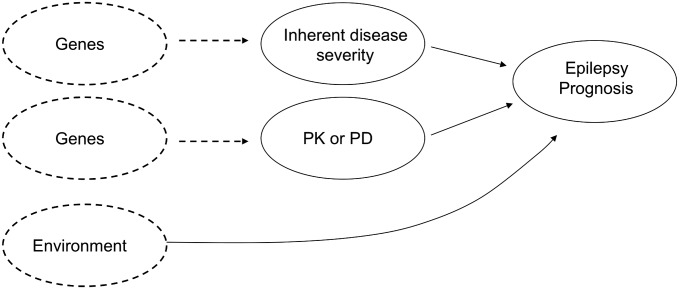
From a mechanistic point of view, genetic effects on the prognosis of newly treated epilepsy can be envisaged to operate on a number of levels including effects on inherent disease severity (7), or via pharmacodynamic (PD) or pharmacokinetic (PK) mechanisms of pharmacological effect (8) (Fig. 1). Determining the precise mechanism of effect for any given genetic association with epilepsy prognosis requires downstream experimental investigation, but from an epidemiological perspective, genetic associations with epilepsy prognosis via any mechanism may have important implications for the development of new therapeutic approaches and could contribute increased precision to prediction of AED response.
Figure 1.
Mechanistic pathways for prognosis of newly treated epilepsy. PK, pharmacokinetic; PD, pharmacodynamic.
Another important consideration in the investigation of genetic effects on the prognosis of epilepsy is whether the study should be conducted in the retrospective case–control setting or using a prospective cohort design. While the retrospective case–control design has been the standard approach for disease susceptibility genome-wide association studies (GWASs), in the study of disease prognosis, the prospective cohort design confers a number of important advantages. These include the ability to characterize clinical risk factors before treatment is initiated (mitigating concerns regarding the retrospective ascertainment of exposure), improved accuracy of measurement of clinical exposure and outcome, the ability to minimize bias in the selection of cases and controls, improved understanding of gene-exposure interactions and improved accuracy of predictive modelling (9,10). These strengths led us to adopt a prospective cohort design for our study despite its disadvantages in terms of time duration and cost compared with the retrospective case–control design.
Here, we report the first GWAS of prognosis of epilepsy using two independent, prospective cohorts of newly treated epilepsy. We report single SNP association P-values for each cohort and total evidence from a meta-analysis of the two cohorts. In addition, we sought evidence that particular classes of biological pathways are associated with epilepsy prognosis, which is an important next step in translating GWAS information to knowledge of disease processes underlying prognosis of epilepsy, as well as the development of future multi-genic predictors for use in clinical settings.
RESULTS
Patients with a new clinical diagnosis of epilepsy requiring medical treatment were recruited to independent prospective cohorts of newly treated epilepsy in the UK and Australia. The UK cohort consisted of 916 subjects who participated in the Standard and New AED (SANAD) trial (11,12). The Australian (AUS) cohort consisted of 380 subjects recruited from epilepsy clinics at two hospitals in Australia; the Royal Melbourne Hospital and the Austin Hospital in Victoria. The distribution of clinical characteristics for all subjects and for those included in the GWAS is detailed in Table 1.
Table 1.
Baseline patient demographics
| UK cohort |
AUS cohort |
|||
|---|---|---|---|---|
| ALL | GWAS | ALL | GWAS | |
| Total patients | 916 | 654 | 380 | 235 |
| One-year remission | ||||
| Yes | 562 (61.4) | 436 (66.7) | 244 (64.2) | 188 (80.0) |
| No | 342 (37.3) | 218 (33.3) | 96 (25.3) | 47 (20.0) |
| Not available (exclude) | 12 (1.3) | 0 | 40 (10.5) | 0 |
| Gender | ||||
| Male | 499 (54.5) | 346 (52.9) | 210 (55.3) | 120 (51.1) |
| Female | 417 (45.5) | 308 (47.1) | 170 (44.7) | 115 (48.9) |
| Age at treatment in years, mean (IQR) | 39 (23–53) | 39 (22–53) | 41 (25–55) | 43 (26–56) |
| Neurological impairment | ||||
| Yes | 69 (7.5) | 37 (5.7) | 9 (2.4) | 7 (3.0) |
| No | 847 (92.5) | 617 (94.3) | 264 (69.5) | 228 (97.0) |
| Not available (exclude) | 0 | 0 | 107 (28.2) | 0 |
| Number of seizures ever before treatment | ||||
| One or not available (exclude) | 2 (0.2) | 0 | 53 (13.9) | 0 |
| 2 | 113 (12.3) | 93 (14.2) | 88 (23.2) | 64 (27.2) |
| 3 | 88 (9.6) | 71 (10.9) | 40 (10.5) | 31 (13.2) |
| 4 | 67 (7.3) | 57 (8.7) | 25 (6.6) | 16 (6.8) |
| 5 | 35 (3.8) | 26 (4.0) | 12 (3.2) | 10 (4.3) |
| >5 | 611 (66.7) | 407 (62.2) | 162 (42.6) | 114 (48.5) |
| Epilepsy type | ||||
| Generalized | 140 (15.3) | 107 (16.4) | 73 (19.2) | 41 (17.4) |
| Focal | 647 (70.6) | 455 (69.6) | 273 (71.8) | 185 (78.7) |
| Unclassified | 125 (13.6) | 92 (14.1) | 18 (4.7) | 9 (3·8) |
| Not available | 4 (0·4) | 0 | 16 (4.2) | 0 |
| EEG results | ||||
| Normal/non-specific abnormality | 606 (66.2) | 431 (65.9) | 229 (60.3) | 152 (64.7) |
| Epileptiform abnormality | 238 (26.0) | 174 (26.6) | 142 (37.4) | 81 (34.5) |
| Not done | 72 (7.9) | 49 (7.5) | 9 (2.4) | 2 (0.9) |
| CT/MRI results | ||||
| Normal | 519 (56.3) | 381 (58.2) | 279 (73.4) | 185 (78.2) |
| Abnormal | 186 (20.3) | 119 (18.2) | 84 (22.1) | 46 (20.2) |
| Not done | 211 (23.0) | 154 (23.5) | 17 (4.5) | 4 |
| Initial AED treatment | ||||
| CBZ | 159 (17.4) | 107 (16.4) | 172 (45.3) | 111 (47.2) |
| Gabapentin (GBP) | 156 (17.0) | 105 (16.1) | 1 (0.3) | 0 |
| Levetiracetam (LEV) | 0 | 0 | 25 (6·6) | 18 (7·7) |
| Lamotrigine (LTG) | 208 (22.7) | 152 (23.2) | 21 (5.5) | 16 (6.8) |
| Phenytoin (PHT) | 0 | 0 | 14 (3.7) | 8 (3.4) |
| Zonisamide (ZNS) | 0 | 0 | 4 (1.1) | 3 (1.3) |
| Oxcarbazepine (OXC) | 99 (10.8) | 70 (10.7) | 0 | 0 |
| Topiramate (TPM) | 225 (24.6) | 165 (25.2) | 0 | 0 |
| Sodium valproate (VPS) | 69 (7.5) | 55 (8.4) | 141 (37.1) | 79 (33.6) |
| Not available | 0 | 0 | 2 (0.5) | 0 |
Values in the table are actual number with percentages in brackets. ALL refers to all patients recruited to the study, and GWAS to those patients included in the genome-wide association study.
The recent consensus statement from the International League Against Epilepsy (ILAE) proposes that treatment success in epilepsy should be defined as freedom from seizures for a minimum of 12 months, as this outcome is consistently associated with improved quality of life (13). Therefore, patients achieving 12-month (365 days or longer) remission of seizures were defined as “responders”, and patients failing to achieve 12-month remission were defined as “non-responders”. Patients followed for <1 year were excluded from the study. A potential difficulty with this outcome is that it is not possible to know whether non-responders might become responders if followed for long enough. However, based on the empirical distribution of time to 12-month remission for patients achieving 12-month remission (Supplementary Material, Fig. S1), we observed that the likelihood of remission falls sharply with time, such that for all patients in the study who achieved 12-month remission, 90% did so within 2.03 years of starting AED therapy. As the median follow-up of non-responders in our study was 2·4 years (IQR 1.7–3.6 years), the number of patients misclassified due to inadequate follow-up is expected to be small.
To date, no validated genetic association with epilepsy prognosis has been reported. However, a number of clinical factors such as the number of seizures pre-treatment and to a lesser extent the results of electroencephalographic (EEG) and brain-imaging investigations have been shown to be associated with chance of remission of seizures after starting treatment (14). We reasoned that if genetic factors influence epilepsy prognosis via clinical factors associated with prognosis, then adjustment for clinical prognostic factors may result in no evidence for genetic association, as in the case of the metabolic trait fasting glucose and the FTO gene, where there is no evidence for association when adjusting for the body mass index (15). Conversely, failure to adjust for clinical prognostic factors may reduce the power to detect genetic associations. A comprehensive analysis of genetic influences on remission of seizures after starting AED treatment therefore requires statistical analysis with and without adjustment for clinical factors informative for epilepsy prognosis.
We identified clinical factors informative for epilepsy prognosis using a univariate logistic regression model. Univariate odds ratios (ORs) for association of clinical factors with 12-month remission of seizures in the UK and AUS cohorts are shown in Table 2.
Table 2.
Univariate ORs and 95% CIs for 12-month remission of seizures in newly treated epilepsy
| UK cohort (ALL) |
AUS cohort (ALL) |
|||
|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P- value | |
| Age at treatment (per ten years) | 1.12 (1.03–1.22) | 0.01 | 1.08 (0.91–1.27) | 0.39 |
| Sex (female) | 1.35 (0.98–1.87) | 0.07 | 1.24 (0.65–2.35) | 0.52 |
| Epilepsy type (focal) | ||||
| Generalized | 1.73 (1.05–2.85) | 0.02 | 1.66 (0.65–4.22) | 0.29 |
| Unclassified | 1.73 (1.07–2.78) | 0.03 | (0-Inf) | 1 |
| Neurological impairment (no) | 0.45 (0.23–0.87) | 0.02 | 1.52 (0.18–12.9) | 0.70 |
| Number seizures before treatment (>5) | ||||
| 2 | 3.20 (1.83–5.60) | 0.00005 | 2.78 (1.14–6.77) | 0.02 |
| 3 | 3.12 (1.66–5.88) | 0.0004 | 1.42 (0.53–3.81) | 0.48 |
| 4 | 1.38 (0.77–2.47) | 0.28 | 0.71 (0.24–2.34) | 0.62 |
| 5 | 3.79 (1.28–11.19) | 0.02 | (0-Inf) | 0.98 |
| EEG results (normal) | ||||
| Epileptiform abnormality | 1.07 (0.73–1.56) | 0.72 | 1.12 (0.57–2.24) | 0.73 |
| Not done | 0.46 (0.25–0.83) | 0.01 | 0.26 (0.02–4.21) | 0.34 |
| CT/MR results (normal) | ||||
| Abnormal | 0.88 (0.60–1.30) | 0.51 | 0.42 (0.20–0.86) | 0.02 |
| Not done | 1.28 (0.82–2.01) | 0.28 | 0.60 (0.06–6.00) | 0.67 |
| AED treatment (LTG) | ||||
| OXC | 0.75 (0.41–1.34) | 0.32 | NA | NA |
| VPS/VPA | 1.64 (0.79–3.38) | 0.18 | 1.17 (0.29–4.70) | 0.82 |
| CBZ | 0.94 (0.56–1.59) | 0.82 | 0.68 (0.18–2.58) | 0.57 |
| TPM | 1.10 (0.68–1.77) | 0.69 | NA | NA |
| GBP | 0.54 (0.32–0.90) | 0.02 | NA | NA |
| LEV | NA | NA | 3.92 (0.36–42.2) | 0.26 |
| PHT | NA | NA | 0.69 (0.09–5.29) | 0.72 |
| ZNS | NA | NA | (0-Inf) | 0.99 |
For categorical covariates, ORs are relative to the reference state provided in brackets. In three cases, the number of occurrences of a state was too few to permit meaningful estimation of OR (shown as 0-Inf). Covariate states nominally significant in the UK cohort were included in the adjusted GWAS.
Clinical prognostic factors chosen for inclusion in the GWAS based on the significance at P < 0.05 in the UK cohort were as follows: age at starting treatment, number of seizures before treatment, EEG result, epilepsy type, presence or absence of neurological impairment and treatment with gabapentin (GBP).
After sample and genotyping quality control (QC, Materials and Methods), a total of 889 newly treated epilepsy patients followed for at least 1 year and for whom remission status and complete clinical covariate information was available were included in the analysis. The duration of follow-up is reported in Supplementary Material, Table S1. There was no difference in proportions of responders and non-responders among patients included or excluded in the GWAS (P = 0.91, 0.75 and 1.0 for UK, AUS and combined, respectively).
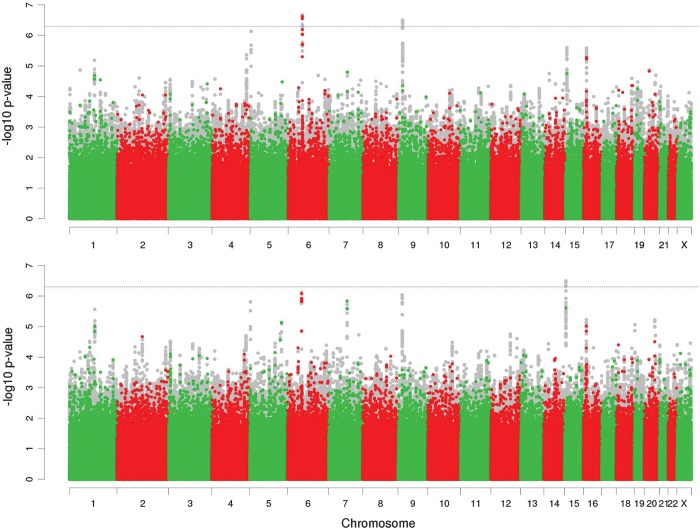
Association analysis of 12-month remission of seizures was performed for the UK and AUS cohorts separately, and the results from each cohort were combined using a fixed-effect meta-analysis (Materials and Methods). The genomic inflation factors for the two pairs of GWAS and their meta-analyses were UKAdjusted = 1.01, UKUnadjusted = 1.00, AUSAdjusted = 1.06, AUSUnadjusted = 1.01, MetaAdjusted = 0.98, MetaUnadjusted = 0.98. Quantile–quantile (QQ) plots of the expected versus observed P-value distributions for these are shown in Supplementary Material, Figure S3. Manhattan plots of –log10-transformed P-values from the meta-analyses with and without adjustment for significant clinical prognostic factors are shown in Figure 2.
Figure 2.
Plot of –log10 transformed P-values of SNP associations with 12-month remission of seizures from the meta-analysis of the UK and AUS cohorts. Top, unadjusted for significant clinical prognostic factors; bottom, adjusted for significant clinical prognostic factors. Coloured dots correspond to genotyped SNPs and grey dots imputed SNPs. The dashed horizontal line marks a P-value significance threshold of 5.0 × 10−7.
Supplementary Material, Table S2, reports all SNPs with associated P-values <1.0 × 10−4 from the meta-analyses. No variant achieved genome-wide significance (defined as Pmeta < 5.0 × 10−8). Two loci (indexed by rs492146 and rs72700966) showed suggestive evidence (defined as Pmeta < 5.0 × 10−7) for association in the unadjusted analysis, and a third locus (indexed by rs143536437) showed suggestive evidence for association in the adjusted analysis (Table 3).
Table 3.
Loci associated with remission of seizures in newly treated epilepsy at Pmeta < 5.0 × 10−7 (with and without adjusting for clinical prognostic factors)
| Index SNP | Imp | Gene | Chr | Position (BP) | Test allele | Ref allele | Test allele freq. (UK) | Test allele freq. (AUS) | Adjusted for clinical prognostic factors |
Unadjusted for clinical prognostic factors |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UK P-value | AUS P-value | Meta P-value | OR (meta) | UK P-value | AUS P-value | Meta P-value | OR (meta) | |||||||||
| rs492146 | Yesa,b | GSTA4 | 6 | 52835895 | G | A | 0.48 | 0.44 | 2.6 × 10−5 | 0.007 | 7.9 × 10−7 | 0.56 | 5.9 × 10−6 | 0.01 | 2.1 × 10−7 | 0.57 |
| rs72700966 | Yes | PTPRD | 9 | 10505224 | C | T | 0.92 | 0.92 | 3.7 × 10−7 | 0.41 | 9.1 × 10−7 | 2.8 | 8.2 × 10−8 | 0.47 | 3.1 × 10−7 | 2.7 |
| rs143536437 | Yesb | ARHGAP11B | 15 | 30898332 | C | T | 0.72 | 0.74 | 4.9 × 10−6 | 0.022 | 3.2 × 10−7 | 1.9 | 2.6 × 10−5 | 0.035 | 2.6 × 10−6 | 1.73 |
SNP, single nucleotide polymorphism; Chr, chromosome; freq, frequency; OR, odds ratio; bp, base-pair; Imp, imputed.
aAdditional genotyped SNPs also exceeded the 5·0 × 10−7 significant threshold.
bAssociation present in both the UK and AUS cohorts independently.
MAF for responders and non-responders for each cohort (UK and AUS) for these SNPs and all SNPs with P < 1.0 × 10−4 are given in Supplementary Material, Table S2.
A further eight previously unreported loci were tentatively (defined as P < 1.0 × 10−5) associated with epilepsy prognosis (Supplementary Material, Table S2). The full list of SNP IDs and associated P-values are available online at https://www.gwascentral.org/.
The two loci indexed by rs492146 and rs143536437 were independently associated in both the UK and AUS cohorts, fulfilling the Wellcome Trust Case Control Consortium 2 (WTCCC2) criteria for ‘strong’ evidence for association (16), while rs72700966 was only significant in the UK cohort (although the observed direction of effect was the same in AUS as that in the UK study).
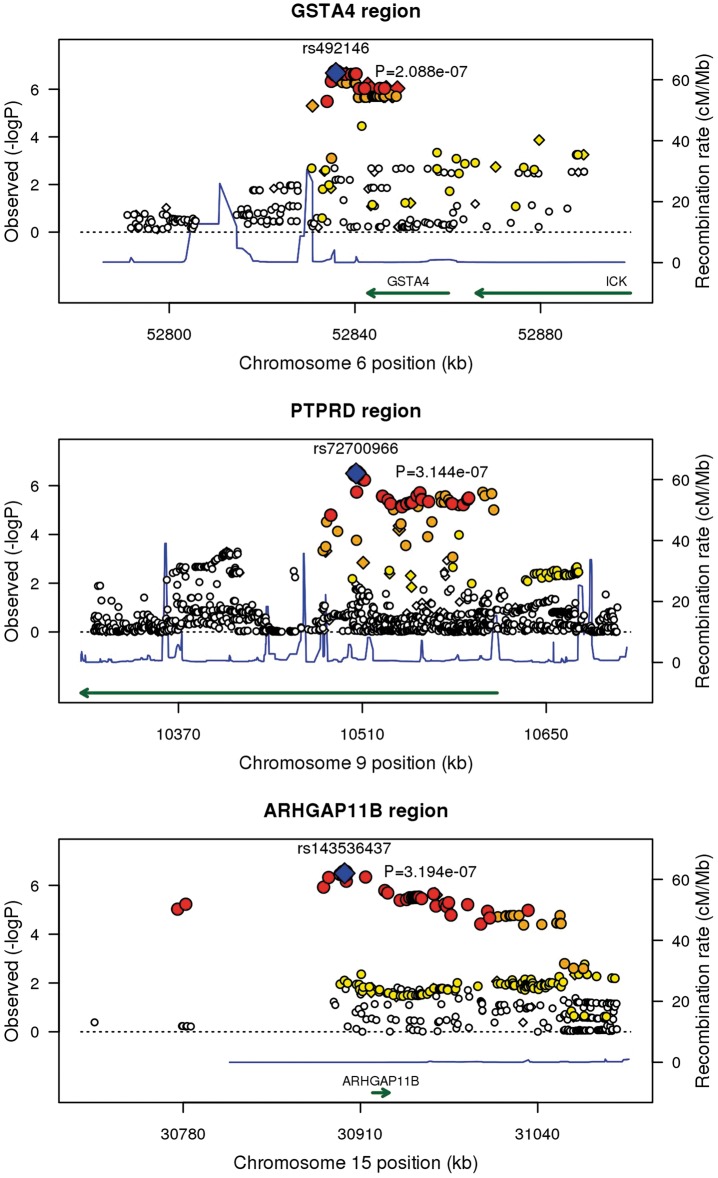
All the three variants indexed by SNPs with Pmeta < 5.0 × 10−7 exhibited similar effect sizes in the adjusted and unadjusted analyses, suggesting that the associations were not mediated by the identified clinical prognostic factors. Regional association plots for the three loci showing strongest association with remission of seizures are shown in Figure 3.
Figure 3.
Regional plots for loci associated with remission of seizures at P < 5.0 × 10−7. SNPs are represented by diamonds and plotted by –log10 transformed P-value and genomic position. Estimated recombination rates are shown by the blue peaks, and gene annotations are indicated by green arrows. Plots are for the meta-analysis results adjusted (ARHGAP11B) and unadjusted (GSTA4, PTPRD) for clinical prognostic factors.
In order to provide an approximate guide to the credibility of the three suggestive associations, we calculated a posterior probability of association (PPA) as described in Stephens and Balding (17). The PPAs were calculated using the combined genotypic data for UK and AUS, without clinical covariates, assuming a normal prior for effect sizes and a 4:1 weighting of additive and general models. We assigned a prior probability of association of 10−4, corresponding to an assumption that ∼300 kb is tightly linked with a variant associated with remission of seizures in newly treated epilepsy. The PPAs obtained were 0.72 for rs492146, 0.16 for rs72700966 and 0.18 for rs143536437, suggesting that the first of these is more likely than not a true association, while the other two are less secure but have a non-negligible probability and are worthy of further consideration.
Genes of biological interest at the three loci with index SNPs associated with epilepsy prognosis at Pmeta < 5.0 × 10−7 are considered briefly below:
6p12.2 (rs492146): GSTA4 encodes glutathione S-transferase (GST) alpha 4. GSTs are a superfamily of phase-II drug-metabolizing enzymes. The alpha class of GSTs encode enzymes with glutathione peroxidase activity that function in the detoxification of lipid peroxidation products and are implicated in the protection of neurons following injury (18).
9p23 (rs72700966): PTPRD encodes protein tyrosine phosphatase receptor type D. PTPRD is implicated in the regulation of synapse development and function (19). Rare structural variants in PTPRD have been associated with attention deficit disorder (20) and autism (21) and ptprd−/− mice exhibit impaired spatial learning and enhanced long-term potentiation (22). Variants in the 5′ UTR of PTPRD are associated with the neurological disorder restless legs syndrome (23).
15q13.2 (rs143536437): ARHGAP11B (alias FAM7B1) encodes rho GTPase activating protein 11B. Rho GTPases play an essential role in neuronal development (24). ARHGAP11B is one of the seven genes (the others being MTMR15, MTMR10, TRPM1, KLF13, OTUD7A and CHRNA7) deleted in 15q13.3 microdeletion syndrome associated with mental retardation and refractory epilepsy (25), and rare ARHGAP11B deletions are observed in autism spectrum disorder (26).
A focussed examination of 280 genes involved in drug absorption, distribution, metabolism and excretion (ADME) defined according to the PharmaADME database (27), as well as ADME genes for AEDs (Supplementary Material, Table S3), revealed no significant association between these genes and prognosis of newly treated epilepsy other than GSTA4. Similarly, we observed no significant association between the outcome of newly treated epilepsy and the human leukocyte antigen genes or genes associated with epilepsy susceptibility by GWASs (28,29) (Supplementary Material, Table S3).
For the three loci indexed by SNPs below the Pmeta < 5.0 × 10−7 threshold, we investigated whether there was an interaction between the genotype and the treatment type by conditioning on each AED in turn (see Materials and Methods and Supplementary Material, Fig. S4). We first performed the analysis according to the initial AED treatment, but since patients may change AED during their course of treatment we repeated the analysis for the 355 patients who made no change in AED during the study. After adjusting for epilepsy type, we observed a nominally significant interaction between rs72700966 (PTPRD) and oxcarbazepine (P = 7.0 × 10−4) and between rs492146 (GSTA4) and topiramate (P = 0.04). However, only rs72700966 survived correction for multiple tests (P = 0.035), and the interaction results should be further interpreted with caution, given the small numbers in each treatment category.
For all SNPs associated with prognosis of epilepsy at P < 1.0 × 10−4, we tested whether the observed allele frequencies in UK and AUS cohorts were significantly different from those in the 381 European samples of the 1000 Genomes project (Supplementary Material, Table S2). The results were not significant for rs72700966 (P = 0.53), and only nominally significant for rs492146 and rs143536437 (P = 0.01 and 0.02 respectively), providing little evidence that these variants may be epilepsy susceptibility SNPs. In keeping with this conclusion, none of the top three loci showed evidence for being epilepsy susceptibility SNPs in GWAS reporting loci associated with epilepsy at genome-wide significance (28,29).
We estimated the power of our study to detect genetic associations with prognosis of newly treated epilepsy following the methodology of Bacanu et al. (30), modified for an additive test. Our power calculations (Supplementary Material, Fig. S5) indicated that we had 80% power to detect causal variants at genome-wide significance (P < 5.0 × 10−8) which individually explained ≥4.4% of the variance of outcome of newly treated epilepsy, and 50% power for variants explaining ≥3.3% of the variance. The findings from our GWAS therefore suggest that there are unlikely to exist common variants that individually have a strong influence on a patient's likelihood of achieving remission after starting AED treatment.
Our results do not exclude a model in which epilepsy prognosis is a polygenic trait of multiple common variants of small effect. Under such a model, sets of genes representing causal biological pathways may be enriched among genes with moderate association P-values. To date, pathway analysis of prognosis of epilepsy has been unexplored, and pathway analysis is an under-utilized approach to analysing treatment response phenotypes in general (31). Yet such an approach might inform biological processes underling disease prognosis, and act as a starting point for the development of multi-SNP predictors of outcome. We therefore sought evidence from the GWAS that particular classes of biological pathways are associated with epilepsy prognosis using two independent pathway methods, ALIGATOR (32,33) and pathMaster (34) (Materials and Methods). ALIGATOR and pathMaster methods can be considered complementary; rather than testing for enrichment of significant genes within pathways as ALIGATOR does, pathMaster tests the absolute association between a pathway and an outcome by aggregating all information within a pathway into a single test statistic. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathways associated with epilepsy prognosis by both ALIGATOR and pathMaster are reported in Table 4, with the full results from either method given in Supplementary Material, Table S4. GWAS P-value thresholds for ALIGATOR were chosen in accordance with the summary statistics for ALIGATOR (Supplementary Material, Table S5).
Table 4.
Functional categories significantly enriched for genes associated with prognosis of newly treated epilepsy by both ALIGATOR and pathMaster
| GWAS P-value | Category ID | Pathway length | Expected overlap | Observed overlap | ALIGATOR P-value | pathMaster P-value | Biological function |
|---|---|---|---|---|---|---|---|
| 0.0001 | hsa04020 | 174 | 0.7 | 4 | 0.0052 | 0.03 | Calcium signalling pathway |
| 0.0001 | hsa04070 | 78 | 0.3 | 3 | 0.005 | 0.043 | Phosphatidylinositol signalling system |
| 0.001 | GO:0008270 | 209 | 5.3 | 16 | 0.0002 | 0.021 | Zinc ion binding |
| 0.001 | GO:0046870 | 8 | 0.2 | 8 | 0.0002 | 0.045 | Cadmium ion binding |
| 0.0001 | GO:0030262 | 27 | 0.0 | 2 | 0.001 | 0.001 | Apoptotic nuclear change |
ALIGATOR and pathMaster P-value = pathway enrichment P-value.
Proposed biological functions were provided by the Gene Ontology (prefixed by GO) and KEGG databases (prefixed by hsa).
Pathway length, expected overlap and observed overlap refer to ALIGATOR statistics.
Taken together, our results suggest that prognosis of newly treated epilepsy is potentially influenced by multiple genetic factors. In such situations, heritability analysis, when applied to distantly related individuals in a GWAS, can be used to estimate the total phenotypic variance of a trait by considering the variance explained by a linear mixed-effects model involving all SNPs (35,36). Using the software LDAK (36), we computed kinship matrices for each cohort, both with and without adjustment for linkage disequilibrium, then using GCTA (35) we estimated variance explained, both with and without adjustment for clinical prognostic factors. Unfortunately, the standard error of the heritability estimates were too high (a minimum of 50% for both UK and AUS cohorts) to provide reliable estimates of heritability.
Finally, for the three loci indexed by SNPs with Pmeta < 5.0 × 10−7, we examined whether the SNPs could be tagging copy number variation (CNV) using cnvHap (37). Several rare CNVs were identified in genes at or within 20 kb of the index SNPs (Supplementary Material, Table S3), none of which were associated with remission of seizures at significance ≤5%.
DISCUSSION
To date, the focus of genetic efforts in treatment response to epilepsy has been on candidate gene studies. However, the biological mechanisms by which AEDs act are poorly understood, constraining candidate gene investigations to the existing knowledge base. In contrast, the genome-wide association method offers a hypothesis-free approach to systematically investigate genetic effects. Yet despite the advantages of the genome-wide approach in pharmacogenetics, fewer than 5% of GWASs catalogued by the US National Human Genome Research Institute are studies of treatment response (http://www.genome.gov/gwastudies/), perhaps reflecting the difficulties of studying prognosis as opposed to susceptibility.
Here, we report the genome-wide association and biological pathway analysis of prognosis of newly treated epilepsy. Uniform baseline clinical characteristics were collected on all patients at study entry and seizure outcomes were measured prospectively. This prospective design overcomes many of the problematic aspects of analysing disease prognosis in the retrospective case–control setting, and permits an unbiased incorporation of clinical prognostic factors in the statistical analysis.
How to define responders and non-responders is a key issue in any study of treatment response. We chose 12-month remission of seizures as the clinical outcome in accordance with the recent consensus statement from the ILAE and because this epilepsy outcome is consistently associated with improved quality of life (and in many countries, including the UK and Australia, is the minimum seizure-free period that allows a patient with epilepsy to drive legally). While this definition is not without limitations, international acceptance of an outcome definition is essential to facilitate replication and future meta-analysis, and external clinical validity is important for developing clinically relevant predictive models of response.
We considered that the inclusion of clinical covariates informative for epilepsy prognosis could either help or hinder the detection of genetic effects on prognosis, depending on whether the genetic factors acted via one or more intermediate clinical prognostic factor. Therefore, we performed the analyses both with and without adjustment for clinical prognostic factors. This approach has the advantage of being agnostic about whether including clinical prognostic factors in the model can increase or decrease the power to detect genetic effects on outcome.
In our study, no single SNP achieved the WTCCC2 cut-off for genome-wide significance (Pmeta < 5.0 × 10−8), although suggestive evidence for association (Pmeta < 5.0 × 10−7) was observed for three loci, and calculation of posterior probabilities of association suggested that rs492146 is more likely than not a true association. Although our moderate sample size does limit our ability to detect variants of small effect, we have sufficient power to conclude it is unlikely that any single common variant explains >4.4% of the variance of outcome of newly treated epilepsy.
The finding that no single common variant has a major influence on the chance of remission of seizures in newly treated epilepsy is important, since it has been suggested that drug response phenotypes might be mediated by higher effect size common variants due to little negative evolutionary pressure on drug response variants. While large effect sized SNPs have been reported for hypersensitive drug reactions (e.g. 38), our study suggests that this may not be the case for drug efficacy phenotypes, where there is a more complex interplay between inherent disease severity and pharmacological effectiveness. Our results suggest that for epilepsy, the genetic architecture of treatment response more closely aligns with complex traits than expected, and so would be improved by the inclusion of additional samples. This conclusion is supported by the large standard error associated with our estimates of heritability, but to our knowledge, the cohorts reported here are the only genotyped prospective cohorts of newly treated epilepsy currently available worldwide.
As a result of these insights, we reasoned that the analysis of sets of genes representing biological pathways may have greater power to detect genetic effects on the outcome of newly treated epilepsy. We used two methods of pathway analysis (ALIGATOR and pathMaster) that could be considered complimentary; ALIGATOR tests for enrichment of significant genes within pathways whereas pathMaster aggregates all information within pathways. A number of candidate pathways informative for epilepsy outcome were identified in both ALIGATOR and pathMaster analyses, including KEGG categories ‘calcium signaling pathway’ and ‘phosphatidylinositol signaling pathway,’ which may warrant further investigation.
In conclusion, the findings from our GWAS represent a first step in the comprehensive analysis of genetic effects on the prognosis of newly treated epilepsy. Our results suggest a limited role for common variants of strong effect and prompt efforts directed at increasing the sample size through additional prospective cohorts of newly treated epilepsy, the development of geneset analyses and exploring the role of rare variant effects in epilepsy outcome.
MATERIALS AND METHODS
Patients
Epilepsy patients were recruited to UK and Australian prospective cohorts of newly treated epilepsy. The UK cohort consisted of 916 patients who participated in the Standard and New AED (SANAD) trial (11,12). SANAD was a randomized, controlled trial consisting of two treatment arms. Arm A included patients for whom carbamazepine (CBZ) was considered the first-line treatment, most of whom had focal epilepsy. Patients in Arm A were randomly assigned to receive CBZ, GBP, lamotrigine, oxcarbazepine or topiramate. Arm B included patients for whom sodium valproate was considered the first-line treatment, most of whom had generalized epilepsy. Patients in Arm B were randomly assigned to receive sodium valproate, topiramate or lamotrigine. Inclusion criteria for the study were (i) epilepsy patients aged ≥5 years, (ii) two or more spontaneous seizures requiring AED treatment, (iii) not previously treated with AED, (iv) monotherapy considered the most appropriate treatment option and (v) willing to provide consent. Exclusion criteria were (i) provoked seizures (e.g. alcohol), (ii) acute symptomatic seizures (e.g. acute brain injury) and (iii) progressive neurological disease (e.g. brain tumour). Patients were classified according to clinician's judgement and classification of epilepsy and seizure outcomes were re-assessed at final data entry. Database checks highlighting inconsistencies were queried with the investigator. The Australian (AUS) cohort consisted of 380 treatment naïve patients prospectively recruited from epilepsy clinics at two hospitals in Australia: the Royal Melbourne Hospital and the Austin Hospital in Victoria. Inclusion and exclusion criteria for the AUS cohort were identical to the UK cohort, except that the AUS study excluded patients <10 years and the choice of AED was determined by physician's preference.
Clinical covariates
Baseline clinical covariates were gender, age at starting treatment, cranial computed tomography (CT) or magnetic resonance imaging (MRI) result, EEG result, total number of seizures pre-treatment, type of epilepsy, neurological impairment (defined as localizing neurological signs resulting in functional impairment) and initial AED. CT and MRI scans were classified as abnormal, not done or normal/non-specific abnormality. EEGs were classified as epileptiform abnormality (defined as focal or generalized spike or spike and slow wave activity), not done or normal/non-specific abnormality. Seizure types and epilepsy syndromes were classified according to the ILAE Classification (39). Epilepsy type was classified as focal, generalized or unclassified (unclassified where there was uncertainty between focal or generalized onset epilepsy). The UK cohort, which was more than twice the size of AUS, was chosen as the discovery cohort for the purpose of selecting clinical prognostic factors for inclusion in the GWAS; clinical factors which showed association with 12-month remission of seizures in a univariate logistic regression model at P < 0.05 were included.
Outcome definition
Epilepsy patients achieving 12-month (365 days or longer) remission of seizures were defined as “responders”, and patients failing to achieve 12-month remission were defined as “non-responders”. Patients followed for <1 year were excluded from the study.
Sample and genotyping QC
The UK samples were genotyped at the Wellcome Trust Sanger Institute on Illumina 660. QC of samples was based on the following criteria, with inclusion/exclusion thresholds for each determined empirically: samples were removed if they displayed heterozygosity outside the interval [0.281,0.299] (28 samples failed), sample call rate <0.98 (11 additional samples failed), gender discordance (3 additional samples failed), pairwise relatedness >0.9 (i.e. accidental duplicates, in which case the lowest quality sample was excluded) (28 samples failed). The presence of highly related individuals can cause confounding in association studies, so a second filtering was then applied to ensure that no pair had estimated relatedness >0.1, a threshold set just below that expected for first cousins (12 samples removed). Principal component analysis (PCA) was performed using a subset of SNPs in approximate linkage disequilibrium in order to identify ancestry outliers: individuals with extreme values on principal component axes 1 or 2 were removed (two samples) (Supplementary Material, Fig. S2). Of the remaining 822 samples, the UK GWAS was performed using the 654 treatment naïve patients observed for at least 1 year after starting AED therapy and for whom the remission status and complete clinical covariate information was available (436 responders and 218 non-responders).
The AUS cohort of 380 samples was genotyped on the same platform and at the same institution (but at a different time) as the UK cohort, and genotype QC followed the same procedures: heterozygosity outside the interval [0.291, 0.309] (10 samples failed), call rate <0.98 (8 additional samples failed), accidental duplication (2 additional samples failed), pairwise relatedness >0.1 (7 samples failed) and PCA (4 samples removed). For the remaining 349 samples, remission status and complete clinical covariate information were available for 235 treatment naïve patients followed for at least 1 year (188 responders and 47 non-responders) who were included in the AUS GWAS.
SNP imputation
The Illumina 660 chip interrogates genotype values for 594 398 SNPs. This number was first reduced to 540 497 for the UK cohort and 533 985 for AUS, by applying (QC) thresholds based on minor allele frequency (MAF > 0.01), call rate (CR > 0.95) and a P-value from a test for Hardy-Weinberg Equilibrium (HWE > 10−6). We imputed against the 1000 Genome reference panel using IMPUTE2 (40) (first dividing the genome into approximately 5 Mb regions) resulting in (expected) genotypic values for ∼40 M SNPs. We then performed SNP QC for a second time, keeping only SNPs with (expected) MAF > 0.01, (expected) CR > 0.95, and INFO > 0.8 (the latter an imputation quality score calculated by the IMPUTE2 algorithm); 6 923 995 SNPs passed these criteria in both cohorts.
GWAS analysis
Supplementary Material, Figure S2, presents plots of the first two principal component axes for the UK and AUS cohorts separately, and when combined. When compared against samples from the HapMap project, it is evident that the UK and AUS cohorts are predominantly of Caucasian ancestry, but also that AUS appears to be more heterogeneous than the UK cohort. For this reason, we decided to analyse each cohort separately, then combine the two sets of results by meta-analysis.
Association analyses were performed using a logistic regression model; letting p denote the probability of becoming a responder, the model supposes logit(p) = log(p/1 − p) = μ + Xβ + Cλ + e, where μ reflects the baseline odds, X represents the genotype of the SNP under examination (with effect size β) and C denotes the covariates (with effect sizes λ). Each SNP's genotypes were coded under an additive model, which assumes that the SNP's effect on the log odds is determined by the count of the alternative allele (0, 1 or 2). For imputed SNPs, X could be non-integer, equal to the expected allele count provided by the imputation; however, it is advised to perform the analysis using the expected values rather than replace them with, say, the most likely genotype value (41). C includes covariates representative of population structure, obtained through PCA of the genotype matrix; the leading axes with eigenvalues significant at 5% using the Tracy–Widom test were included (four for UK, three for AUS). The regression analysis was performed using the logistic option of PLINK (42). For both cohorts, the analysis was performed once with significant clinical prognostic factors included in C (“adjusted”), and once without (“unadjusted”). The effect size estimates for each SNP from the UK and AUS GWASs were corrected for genomic inflation (43) and combined using a fixed-effect meta-analysis, weighting the effect size estimates from each study by their standard deviation using the PLINK option—meta-analysis. Regional association plots (“Broad Plots”) for the most strongly associated regions were prepared using R code provided by the Broad Institute (44).
To examine evidence for an interaction between the genotype and the treatment type, for the top SNPs from the meta-analysis we enlarged the logistic regression model to include a drug-specific effect size. For example, when considering the possible effect of CBZ, we included β_CBZ to allow for an SNP's effect to be different across patients administered CBZ relative to those on other treatments. We considered each of the AEDs in turn, computing a P-value based on whether β_DRUG was significantly non-zero. Because patients may change AED during the course of the study, we repeated this analyses restricted to the 355 patients in the study whose AED remained unchanged.
Pathway analysis
We used two independent methods, ALIGATOR (32,33) and pathMaster (34), to test the results of the meta-analysis for over-representation of biological pathways obtained from GO (downloaded from http://www.geneontology.org/, restricting to pathways containing between 5 and 600 genes) and KEGG. ALIGATOR corrects for varying numbers of SNPs per gene and multiple overlapping functional pathways. To apply ALIGATOR, it is necessary to specify a GWAS P-value threshold; each pathway is scored by counting the number of its genes that contain one or more SNPs with P-value below this threshold. This score is then tested for significance by permutation. The choice of GWAS P-value threshold is arbitrary, since it depends on the sample size and the distribution of genetic effect sizes which is usually unknown; the most informative threshold will therefore balance confidence that the identified pathways have a true causal relationship with the phenotype and not missing any genuine pathway associations. As a pragmatic solution to the problem of choosing a P-value threshold ALIGATOR recommends exploring a range of thresholds to determine which gives the most significant increase in overrepresented functional categories. We therefore considered P-value thresholds at 0.01, 0.001 and 0.0001. Analyses were undertaken before and after adjusting for clinical prognostic factors. All ALIGATOR analyses used 5000 simulated replicate gene lists and 2000 simulated replicate studies. PathMaster (34) evaluates the overall genetic contribution of a given pathway via a cumulative trend test statistic; this is the sum of the Armitage trend test statistic over all of the SNPs in the pathway. The null distribution of the pathway statistic is estimated by a skew normal or gamma distribution; the distribution chosen is determined by the Kolmogorov–Smirnof test statistic. The parameters of the chosen distribution are estimated from 100 random permutations of case–control labels. The null distribution is estimated separately for each pathway (see (42) for details). Analyses were undertaken before adjusting for clinical prognostic factors. For both ALIGATOR and pathMaster, SNPs were assigned to genes if they were within gene boundaries or 20 kb either side.
Copy number variation (CNV) analysis
For loci indexed by SNPs below the P < 5.0 × 10−7 threshold, we examined whether the SNPs were tagging CNV using cnvHap (37). cnvHap is an integrative multi-platform haplotype-based method which uses population distribution of allele frequency to train its haplotype hidden Markov model and has been shown to be more accurate than other methods in calling CNVs from SNP data. We extracted Log R Ratio (LRR) and B allele frequency (BAF) from the intensity files and corrected for the GC content and long-range autocorrelation. CNV calls were generated in cnvHap using LRR, BAF and Illumina platform-specific parameters and any potential CNV call was visually inspected. We searched each gene using a 50 Kb window, testing each CNV discovered for association with 12-month remission using a logistic regression framework with population genotype derived principal components and age and gender included as covariates.
Power plots
Power plots were estimated following the methodology of Bacanu et al. (30) modified for an additive test. Power was calculated based on the significance threshold of 5.0 × 10−8 (genome-wide significance), given the number of responders and non-responders in our meta-analysis, for MAFs between 1 and 50%.
Ethics
This study was conducted under MREC 02/8/45. Consents were obtained according to the Declaration of Helsinki (BMJ 1991; 302: 1194).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was funded by the Wellcome Trust (WT066056 to M.R.J.), The NIHR Biomedical Research Centres Scheme (P31753 to M.R.J.), Department of Health NHS Chair of Pharmacogenetics (to M.P.), The Medical Research Council (REF: 25105 to D.B. and M.R.J.), The Royal Melbourne Hospital Foundation Lottery Grants (REF: 604955 to S.P.) and The RMH Neuroscience Foundation (to T.J.O'B.). M.R.J. conceived the UK study and coordinated the UK DNA collection. T.J.O'B. conceived the Australian study and coordinated the Australian DNA collection. M.R.J., D.C., M.P. and D.B. obtained the funding to collect the UK DNA samples. T.J.O'B. obtained the funding to collect Australian samples. M.R.J., D.C., M.P., D.B., A.P., A.M., S.P. and T.J.O'B. obtained the funding to genotype the samples. Genomic data were analysed by D.S., C.H., I.T., S.P., A.J., A.P., M.D., T.D. and M.R.J. DNA samples and phenotypic information was collected and analysed by S.P., A.J., M.T., D.S., P.E.S., P.C., M.K., S.H., M.R., G.E.S., D.C., M.P., I.S., S.B., A.M., I.S., S.B., M.N., R.Y., M.T., C.F. and M.R.J. D.S., C.H., S.P. and M.R.J. drafted the manuscript. All authors reviewed and edited the manuscript. M.R.J. confirms that he has full access to all the data in the study and has final responsibility for the decision to submit for publication.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the clinical and non-clinical staff who supported the UK DNA collection: Alwaidh M (Whiston Hospital, St Helens and Knowsley Hospitals NHS Trust), Andrews J (New Cross Hospital), Appleton RE (Alder Hey Hospital, Royal Liverpool Children's NHS Trust), Bell S (Craigavon Area Hospital), Bucknall J (Homerton Hospital), Cleland P (Sunderland Royal Hospital), Cock H (St Georges Hospital), Cockerell OC (Royal London Hospital), Corston R (New Cross Hospital), Cramp C (Princess Royal Telford), Crawford P (York Hospitals NHS Trust), Dafalla BEA (Calderdale & Huddersfield NHS Trust), Thompson C (Calderdale & Huddersfield NHS Trust), Doherty C (Imperial College London), Doran M (The Walton Centre for Neurology and Neurosurgery), Duncan S (Hope Hospital), Esmonde TFG (Royal Victoria, Belfast), Goulding P (S James's University Hospital, The Leeds Teaching Hospitals NHS Trust), Gurtin D (Homerton Hospital), Harrington B (Wexham Maelor), Hinde F (Princess Royal Telford), Howell S (Doncaster and Bassetlaw Hospital), Hughes A (Arrowe Park Hospital, Wirral Hospitals NHS Trust), Hulme A (Hope Hospital), Kindley AD (Royal Aberdeen Children's Hospital), Lawden M (Leicester Royal Infirmery), Litherland G (Whiston Hospital, St Helens and Knowsley Hospitals NHS Trust), MacDonald S (Ninewells Hospital, Dundee), McLean B (Royal Cornwall Hospital), Middleton C (Imperial College London), Minchom P (Wexham Maelor), Newton R (Manchester Royal Infirmary), Nicholl D (Queen Elizabeth, Birmingham), Owens G (Wexham Maelor), Parret M (Royal Cornwall Hospital), Reuber M (Royal Hallamshire Hospital, Sheffield & Chesterfield & North Derbyshire Royal Hospital), Roberts R (Ninewells Hospital, Dundee), Sharrack B (Lincoln County Hospital), Silver N (North Cheshire Hospitals NHS Trust), Sood R (Homerton Hospital), Stewart J (Arrowe Park Hospital, Wirral Hospitals NHS Trust), Tidswell P (Blackburn Royal Infirmary), Von Oertzen T (St Georges Hospital), Waite P (Lincoln County Hospital), Weishmann U (The Walton Centre for Neurology and Neurosurgery) and White K (Ninewells Hospital, Dundee).
REFERENCES
- 1.Annegers J.F., Hauser W.A., Elverback L.R. Remission of seizures and relapse in patients with epilepsy. Epilepsia. 1979;20:729–737. doi: 10.1111/j.1528-1157.1979.tb04857.x. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby A., Lane S., Marson A., Baker G.A. MESS Study Group. Relationship of clinical and quality of life trajectories following the onset of seizures: findings from the UK MESS study. Epilepsia. 2011;52:965–974. doi: 10.1111/j.1528-1167.2010.02973.x. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrand M.S., Dahl H.H., Damiano J.A., Smith R.J., Scheffer I.E., Berkovic S.F. Recent advances in the molecular genetics of epilepsy. J. Med. Genet. 2013;50:271–279. doi: 10.1136/jmedgenet-2012-101448. [DOI] [PubMed] [Google Scholar]
- 4.Senn S. Individual response to treatment: is it a valid assumption. BMJ. 2004;329:966–968. doi: 10.1136/bmj.329.7472.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan P., Brodie M.J. Early identification of refractory epilepsy. N. Engl. J. Med. 2000;342:314–319. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- 6.Johnson M.R., Milne R.L., Torn-Broers Y., Hopper J.L., Scheffer I.E., Berkovic S.F. A twin study of genetic influences on epilepsy outcome. Twin Res. 2003;6:140–146. doi: 10.1375/136905203321536263. [DOI] [PubMed] [Google Scholar]
- 7.Rogawski M.A., Johnson M.R. Intrinsic severity as a determinant of antiepilepsy drug refractoriness. Epilepsy Curr. 2008;8:127–130. doi: 10.1111/j.1535-7511.2008.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwan P., Schachter S.C., Brodie M.J. Drug resistant epilepsy. N. Engl. J. Med. 2011;365:919–926. doi: 10.1056/NEJMra1004418. [DOI] [PubMed] [Google Scholar]
- 9.Manolio T.A., Bailey-Wilson J.E., Collins F.S. Genes, environment and the value of prospective cohort studies. Nat. Rev. Genet. 2006;7:812–820. doi: 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]
- 10.Leschziner G., Jorgensen A., Andrew T., Pirmohamed M., Williamson P., Marson A.G., Coffey A.J., Middleditch C., Rogers J., Bentley D.R., et al. Clinical factors and ABCB1 polymorphisms in prediction of antiepileptic drug response: a prospective cohort study. Lancet. Neurol. 2006;5:668–676. doi: 10.1016/S1474-4422(06)70500-2. [DOI] [PubMed] [Google Scholar]
- 11.Marson A.G., Al-Kharusi A.M., Alwaidh M., Appleton R., Baker G.A., Chadwick D.W., Cramp C., Cockerell O.C., Cooper P.N., Doughty J., et al. The SANAD study of effectiveness of carbamazepine, gabapentin, lamotrigine, oxcarbazepine, or topiramate for treatment of partial epilepsy: an unblinded randomised controlled trial. Lancet. 2007;369:1000–1015. doi: 10.1016/S0140-6736(07)60460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marson A.G., Al-Kharusi A.M., Alwaidh M., Appleton R., Baker G.A., Chadwick D.W., Cramp C., Cockerell O.C., Cooper P.N., Doughty J., et al. The SANAD study of effectiveness of valproate, lamotrigine, or topiramate for generalised and unclassifiable epilepsy: an unblinded randomised controlled trial. Lancet. 2007;369:1016–1026. doi: 10.1016/S0140-6736(07)60461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwan P., Arzimanoglou P., Berg A.T., Brodie M.J., Hauser A.W., Mathern G., Moshe S.L., Perucca E.L., Wiebe S., French J. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2010;51:1069–1077. doi: 10.1111/j.1528-1167.2009.02397.x. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald B.K., Johnson A.L., Goodridge D.M., Cockerell O.C., Sander J.W., Shorvon S.D. Factors predicting prognosis of epilepsy after presentation with seizures. Ann. Neurol. 2000;48:833–841. [PubMed] [Google Scholar]
- 15.Freathy R.M., Timpson N.J., Lawlor D.A., Pouta A., Ben-Shlomo Y., Ruokonen A., Ebrahim S., Shields B., Zeggini E., Weedon M.N., et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–1426. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International Multiple Sclerosis Genetics Consortium & The Wellcome Trust Case Control Consortium 2. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens M., Balding D. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 18.Al Nimer F., Strom M., Lindblom R., Aeinehband S., Bellander B.M., Nyengaard J.R., Lidman O., Piehl F. Naturally occurring variation in the glutathione-S-transferase 4 gene determines neurodegeneration after traumatic brain injury. Antioxid. Redox. Signal. 2013;18:784–794. doi: 10.1089/ars.2011.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi H., Katayama K., Sohya K., Miyamoto H., Prasad T., Matsumoto Y., Ota M., Yasuda H., Tsumoto T., Aruga J., Craig A.M. Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat. Neurosci. 2012;13:389–398. doi: 10.1038/nn.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elia J., Gai X., Xie H.M., Perin J.C., Geiger E., Glessner J.T., D'arcy M., de Berardinis R., Frackelton E., Kim C., et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes R.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uetani N., Kato K., Ogura H., Mizuno K., Kawano K., Mikoshiba K., Yakura H., Asano M., Iwakura Y. Impaired learning with enhanced hippocampal long-term potentiation in PTPδ-deficient mice. EMBO J. 2000;19:2775–2785. doi: 10.1093/emboj/19.12.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schormair B., Kemlink D., Roeske D., Eckstein G., Xiong L., Lichtner P., Ripke S., Trenkwalkder C., Zimprich A., Stiasny-Kolster K., et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat. Genet. 2008;40:946–948. doi: 10.1038/ng.190. [DOI] [PubMed] [Google Scholar]
- 24.Govek E.E., Newey S.E., Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 25.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leblond C.S., Heinrich J., Delorme R., Proepper C., Betancur C., Huguet G., Konyukh M., Chaste P., Ey E., Rastam M., Anckarsater H., et al. Genetic and functional analysis of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Zhang L., Zhou H., Stoneking M., Tang K. Global patterns of genetic diversity and signals of natural selection for human ADME genes. Hum. Mol. Genet. 2011;20:528–540. doi: 10.1093/hmg/ddq498. [DOI] [PubMed] [Google Scholar]
- 28.Gou Y., Baum L.W., Sham P.C., Wong V., Ng P.W., Lui C.H.T., Sin N.C., Tsoi T.H., Tang C.S.M., Kwan J.S.H., et al. Two-stage genome-wide association study identifies variants in CAMSAP1L1 as suceptibility loci for epilepsy in Chinese. Hum. Mol. Genet. 2012;21:1184–1189. doi: 10.1093/hmg/ddr550. [DOI] [PubMed] [Google Scholar]
- 29.Steffans M., Leu C., Ruppert A-K., Zara F., Striano P., Robbiano A., Capovilla G., Tinuper A., et al. EPICURE Consortium., EMINet Consortium. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum. Mol. Genet. 2012;21:5359–5372. doi: 10.1093/hmg/dds373. [DOI] [PubMed] [Google Scholar]
- 30.Bacanu S-A., Whittaker J.C., Nelson M.R. How informative is a negative finding in a small pharmacogenetic study? Pharmacogenomics J. 2012;12:93–95. doi: 10.1038/tpj.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou K., Pearson E.R. Insights from genome-wide association studies of drug response. Annu. Rev. Pharmacol. Toxicol. 2012;53:299–310. doi: 10.1146/annurev-pharmtox-011112-140237. [DOI] [PubMed] [Google Scholar]
- 32.Holmans P., Green E.K., Pahwa J.S., Ferreira M.A.R., Purcell S.M., Sklar P. Owen M.J., O'Donovan M.C., Craddock N. Wellcome Trust Case-Control Consortium. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmans P., Moskvina V., Jones L., Sharma M. Vedernikov A., Buchel F., Sadd M., Bras J.M., Bettella F., et al. The International Parkinson's Disease Genomics Consortium (IPDGC). A pathway-based analysis provides additional support for an immune-related genetic susceptibility to Parkinson's disease. Hum. Mol. Genet. 2012;22:1039–1049. doi: 10.1093/hmg/dds492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eleftherohorinou H., Hoggart C.J., Wright V.J., Levin M., Coin L.J. Pathway-driven gene stability selection of two rheumatoid arthritis GWAS identifies and validates new susceptibility genes in receptor mediated signaling pathways. Hum. Mol. Genet. 2011;20:3494–3506. doi: 10.1093/hmg/ddr248. [DOI] [PubMed] [Google Scholar]
- 35.Jang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speed D., Hemani G., Johnson M.R., Balding D.J. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coin L.J., Asher J.E., Walters R.G., Moustafa J.S., de Smith A.J., Sladek R., Balding D.J., Froguel P., Blakemore A.I. cnvHap: an integrative population and haplotype-based multiplatform model of SNPs and CNVs. Nat. Methods. 2010;7:541–546. doi: 10.1038/nmeth.1466. [DOI] [PubMed] [Google Scholar]
- 38.McCormack M., Alfirevic A., Bourgeois S., Farrell J.F., Kasperaviciute D., Carrington M.N., Sills G., Marson A., de Bakker P.I., Chinthapalli K., et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N. Engl. J. Med. 2011;364:1134–1143. doi: 10.1056/NEJMoa1013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berg A.T., Berkovic S.F., Brodie M.J., Buchhalter J., Cross J.H., van Emde Boas W., Engel J., French J., Glauser T.A., Mathern G.W. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2009;51:676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 40.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Skiar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 44.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT., Lund University., and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.