Abstract
Human perception of bitterness displays pronounced interindividual variation. This phenotypic variation is mirrored by equally pronounced genetic variation in the family of bitter taste receptor genes. To better understand the effects of common genetic variations on human bitter taste perception, we conducted a genome-wide association study on a discovery panel of 504 subjects and a validation panel of 104 subjects from the general population of São Paulo in Brazil. Correction for general taste-sensitivity allowed us to identify a SNP in the cluster of bitter taste receptors on chr12 (10.88– 11.24 Mb, build 36.1) significantly associated (best SNP: rs2708377, P = 5.31 × 10−13, r2 = 8.9%, β = −0.12, s.e. = 0.016) with the perceived bitterness of caffeine. This association overlaps with—but is statistically distinct from—the previously identified SNP rs10772420 influencing the perception of quinine bitterness that falls in the same bitter taste cluster. We replicated this association to quinine perception (P = 4.97 × 10−37, r2 = 23.2%, β = 0.25, s.e. = 0.020) and additionally found the effect of this genetic locus to be concentration specific with a strong impact on the perception of low, but no impact on the perception of high concentrations of quinine. Our study, thus, furthers our understanding of the complex genetic architecture of bitter taste perception.
INTRODUCTION
Taste is one of the key adaptive physiological factors driving personal food preferences, dietary habits and ultimately health balance. Specifically, interindividual variation in the perceived bitterness of certain foods has been linked to individual disliking and avoidance of these foods (1,2). Understanding the link between genetics, bitter taste sensitivity and food choice has been complicated by the relative complexity of mammalian bitter taste physiology. Humans perceive several hundred chemically diverse compounds as bitter (3) and perception of these compounds is mediated via about 25 G-protein-coupled receptors from the T2R family (4–6). Some of these T2Rs are highly selective for specific chemical compounds or molecular groups, while others are broadly tuned (7). The mammalian T2R family of receptors shows pronounced signs of recent population-specific selection (8–10) that might reflect adaptation to the food-specific risks and opportunities found in ancestral food environments. These evolutionary processes included a succession of gene-duplication and pseudogenization events (11) that led to the clustering of the majority of the human T2Rs in specific regions of chromosomes 7 (141–142.6 Mb, build 36.1) and 12 (10.88–11.24 Mb, build 36.1) (6). This diversity at the receptor level is contrasted by a relatively strict conservation of the down-stream signaling chain. T2R-mediated taste perception shares the same cellular signaling and neuronal pathways (12) and as a consequence all known bitter compounds generate essentially the same basic taste sensation.
RESULTS
To better understand the extent and the mechanism through which genetic variation impacts human bitter taste perception, we conducted a genome-wide association study (GWAS). In designing this study, we decided to perform broad and precise phenotyping of our panelists at the expense of a modest panel size of ∼500 individuals. While such a small sample size has limited statistical power to detect mild effects we reasoned that this would be compensated, at least in part, by the accuracy of our measurements and the ability to disentangle compound-, concentration- and modality-specific taste variation.
Panelists were (Table 1) healthy adult subjects between 18 and 45 years old recruited from the general population of São Paulo in Brazil. The principal component analysis (Supplementary Material, Fig. S1) of the panelists' genotypic data shows that the study panel is characterized by a continuous admixture of European with African and Asian genotypes, respectively, that is typical of the general São Paulo population. We recruited two panels, a discovery panel of 504 subjects and a validation panel of 104 subjects. All procedures were approved by the Institutional Review Board of the Sírio Libanês Hospital, where tests were administered, and by the National Committee of Research Ethics at the Brazilian Ministry of Health (HSL 2007/25 Process no. 25000.114841/2007-17). Each participant of both panels underwent an extensive 2-week program of taste tests on 13 pure compounds representing the five basic taste modalities (sweet, sour, bitter, salty and umami). Here, we report the results on the perception of the three bitter compounds included in these tests (PROP (N-propylthiouracil), quinine and caffeine).
Table 1.
Demographic composition of subject panels
| Discovery panel | Replication panel | |
|---|---|---|
| N | 503 | 104 |
| Male | 253 | 53 |
| Female | 250 | 51 |
| Age (avg., stdev, min, max) | 32.5, 7.5, 18, 46 | 32.8, 7.3, 19, 46 |
| BMI (avg., stdev, min, max) | 25.7, 4.9, 16.7, 50.7 | 25.1, 4.2, 17.9, 36.6 |
We determined individual taste detection thresholds for the 13 tested compounds with the well-established staircase method (13,14). This method determines the compound concentration at which a subject is able to distinguish consistently a compound-containing solution from pure water. In parallel, we measured the perceived intensity of suprathreshold compound concentrations. For this, subjects were presented, in a pseudorandomized order, with five samples of aqueous solutions of the tested compound spanning a two orders of magnitude concentration range plus a 6th sample containing only water. The subjects rated the taste intensity of each solution on a continuous quasi-logarithmic scale (gLMS, general labeled magnitude scale) (13).
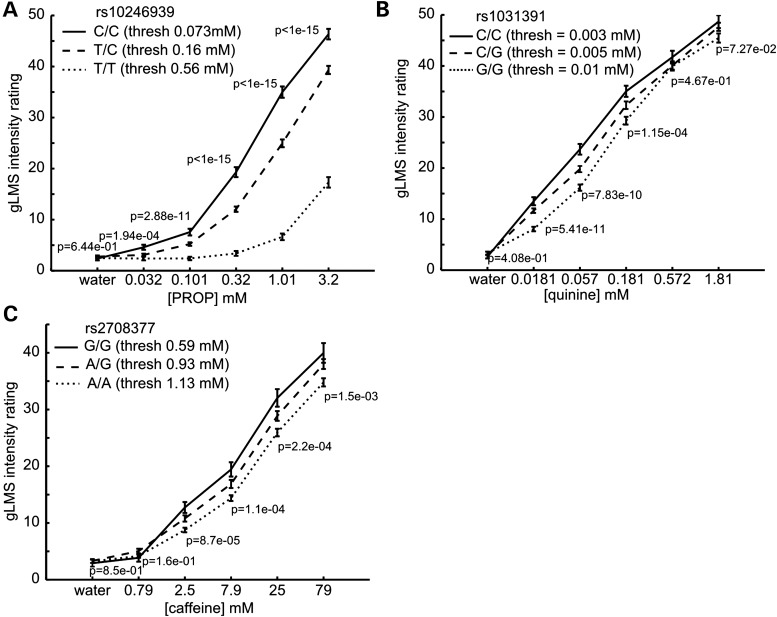
To identify polymorphisms influencing taste sensitivity, our discovery cohort (n = 504) was genotyped by SNP chip. As a validation of data integrity, we performed a GWAS on the panelists' detection thresholds for the bitter compound PROP. The detection thresholds for this bitter compound and the related bitter compound PTC are strongly driven by genetic polymorphisms in the gene for the bitter taste receptor Tas2R38 (15,16) and should therefore be easily detectable in our data set. Our results perfectly replicate these previous reports identifying the same set of SNPs in Tas2R38 (top SNP rs10246939, P = 1.7 × 10−62, r2 = 34.3%, β = −0.39, s.e. = 0.023) (Fig. 1a) using the same detection-threshold-based phenotype. Consistent with previous reports (15,16), we further find that the same SNPs associated with variation in detection threshold are also associated with variation in the perceived intensity of suprathreshold concentrations of PROP. Notably, this genotype effect extends across the entire 100-fold concentration range tested in our study (Fig. 2a).
Figure 1.
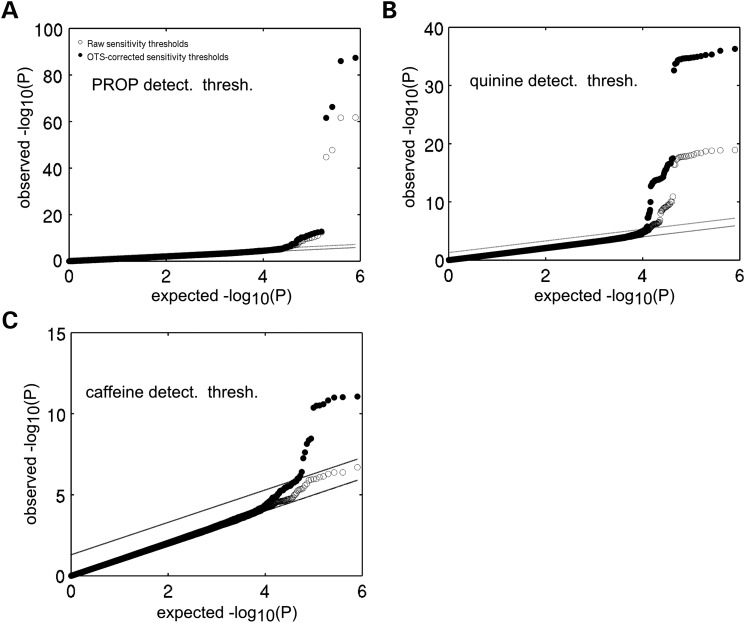
Across-phenotype correction boosts association signal for taste detection thresholds. QQ-plots of the GWAS for detection thresholds of PROP (A), quinine (B) and caffeine (C) showing the boost in association strength afforded by cross-phenotype correction. Open circles indicate P-values obtained from GWASs conducted for taste detection thresholds that were corrected only for demographics (gender, age, BMI) and the first 10 principal components of a genetic ancestry analysis. Solid circles indicate P-values obtained when detection thresholds were additionally corrected for overall taste sensitivity.
Figure 2.
Effect of detection-threshold-associated SNPs on the perceived intensity of suprathreshold compound concentrations. Perceived taste intensities for suprathreshold concentrations of PROP (A), quinine (B) and caffeine (C) as a function of compound concentration and subjects' genotypes at the top-associated SNP for the respective compound-detection-threshold GWASs. Error bars show the standard error of intensity ratings and a P-value is given for ANOVA tests of across-genotype differences at each compound concentration.
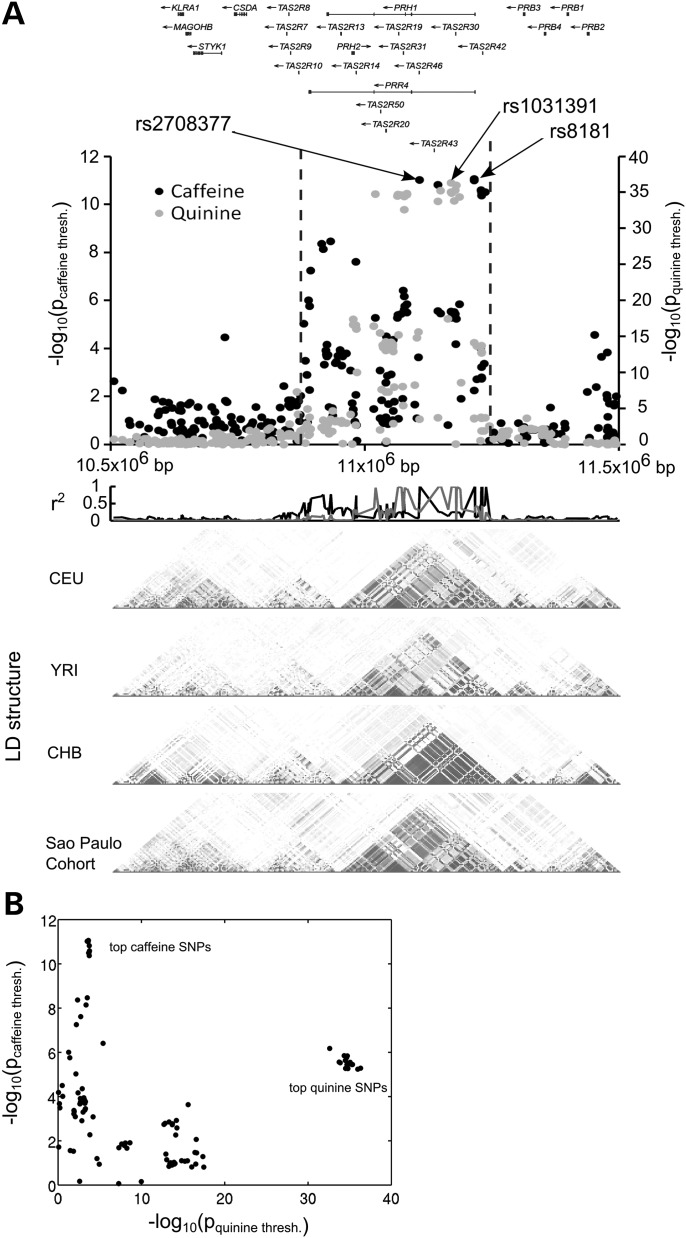
We performed the same analysis for quinine-detection thresholds and replicated the previously reported association (15) between quinine bitterness and the cluster of T2R receptor genes on chr12 (10.88–11.24 Mb, build 36.1) with the top hit being rs1031391 (P = 1.93 × 10−19, r2 = 13.2%, β = 0.23, s.e. = 0.026). The local Manhattan plot (Fig. 3) of the association is characterized by a broad association peak, characteristic of a pronounced linkage-disequilibrium (LD) block that was also observed by Reed et al. (15). Yet, the genotyping chip used in our study provides enhanced coverage across the distal side of this plateau. The pairwise LD (measured as r2) between SNPs in different populations is shown in the bottom panel of Figure 3a. Judging by this plot, the addition of subjects having African and Asian ancestry, which characterizes our study population, appears to have led to a partial breakdown of the LD block. While the panel above illustrates the LD of each SNP with the top associated SNP pointing out that the two phenotype association signals are only partially overlapping. Notably, the association peak is now split into two parts separated by a cleft centered on Tas2R46. Together, these factors allow a more precise mapping of the quinine-detection-threshold locus and thereby reduces the number of candidate bitter taste receptor genes that fall within the association peak (Tas2Rs 19[48], 20[49], 30[47], 31[44], 43, 50) (numbers in squared brackets indicate alternative naming conventions). Interestingly, two of those candidate receptors (Tas2R31 and Tas2R43) are known to be activated by quinine in in vitro assays (7).
Figure 3.
Caffeine and quinine detection thresholds are associated with distinct loci. (A) Local Manhattan plot of caffeine (black) and quinine (gray) detection-threshold loci showing local genes and genotype r2 of each SNP to the top-associated SNP for caffeine- (black) and quinine-detection thresholds (gray), respectively. Below are shown the LD block structures of the region in the central European (CEU) West African Yoruba (YRI) and Han Chinese (CHB) populations from the Hapmap data set together with the LD block structure found in the São Paulo cohort from our study. (B) Scatter plot of association P-values for quinine and caffeine for each SNP in the chromosome region between the dotted vertical lines on the local Manhattan plot (A).
Unlike the situation observed for PROP, the effect of the top quinine-detection-threshold SNP (rs1031391) on the perceived intensity of suprathreshold quinine solutions is clearly concentration dependent (Fig. 2b). In the low-concentration range, the rs1031391 genotype shows a very pronounced effect on the perceived intensity of quinine bitterness (ANOVA, P-value at 0.0181 mm quinine = 5.41 × 10−11). But no significant effect is observed on the perceived intensity of the two highest concentrations of quinine (P > 0.05). The suprathreshold data further indicate that this lack of an effect of rs1031391 genotype on high quinine concentrations is not the result of intensity-saturation effects: First, the taste intensity curves of quinine show no signs of leveling off and secondly, we do see clear genotype effects at iso-intense concentrations of PROP and caffeine (Fig. 2a and c). One possible explanation for these observations is that low and high concentrations of quinine may be perceived by at least two distinct molecular receptors. In this model, the receptor putatively tagged by rs1031391 (due to LD, the true causal receptor cannot be identified with certitude) is responsible for the perception of low concentrations of quinine and as a result perception of low quinine concentrations varies with rs1031391 genotype. By contrast, the perception of high quinine concentrations, which, in this model, is mediated by a different receptor, is not affected by rs1031391 genotype. Based on the results of in vitro studies of bitter taste receptors (7) a multiple-receptor model for quinine perception appears plausible. In those studies, nine of the 25 Tas2R bitter taste receptors were activated by quinine, while many other bitter compounds activate only one or a small number of receptors.
Comparison of threshold data across all tested compounds, including sweet, umami, sour and salty compounds (17), revealed pronounced positive across-compound correlations. In other words, subjects who were sensitive to any specific compound also tended to be sensitive to all other tested compounds. To capture this effect we defined an overall taste-sensitivity parameter OTS (see Supplementary Material, Fig. S2 for a graphical representation of the overall taste-sensitivity concept and the methods section for a mathematical definition of this parameter). In brief, this parameter is calculated as the average of the normalized taste-sensitivity scores across all compounds. The z-score normalization ensures that the concentration levels of different compounds are on a comparable scale. This overall sensitivity parameter proved to be remarkably robust relative to the inclusion or omission of data for specific compounds. For example, inclusion or omission of PROP, quinine and caffeine detection thresholds in these calculations did not significantly alter individuals' overall sensitivity parameters, nor did they significantly impact the results of subsequent analyses based on this parameter. The resulting overall sensitivity parameter explains 8.8, 33.9 and 27.4% of the respective overall variance in detection thresholds for PROP, quinine and caffeine. Notably, the overall sensitivity parameter is not primarily driven by overall demographic parameters such as age, gender, ethnicity or BMI and a GWAS on the overall sensitivity parameter did not generate genome-wide significant associations.
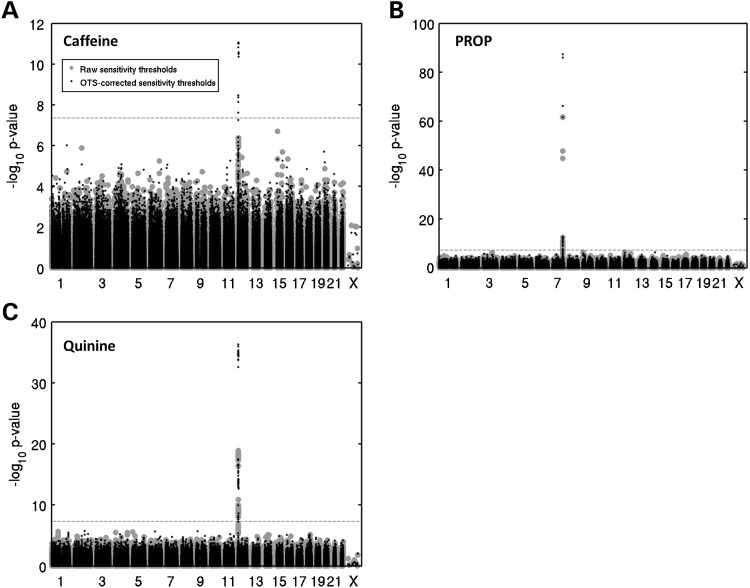
While our current data set did not allow us to pinpoint the biological mechanism that drives overall taste sensitivity it is nevertheless clear that this parameter will act as a confounding factor for compound-specific taste sensitivity. Correcting for overall taste sensitivity should therefore boost compound-specific genotype–phenotype association signals. To test the utility of this correction procedure, we included the overall taste-sensitivity parameter as a covariate in a GWAS on PROP detection threshold alongside routine covariates such as age and gender. Correction for overall taste sensitivity indeed provides a very substantial boost to the PROP-TAS2R38 association signal (Figs. 1a and 4b) from P = 1.70 × 10−62, r2 = 34.3% to pcorr = 1.06 × 10−86, r2corr = 42.2%. Furthermore, this boost occurs specifically for those SNPs that already showed genome-wide significant associations prior to correction (Fig. 4b; Supplementary Material, Fig. S3a) and is not associated with a general inflation of P-values (lambda before OTS correction = 1.0129, lambda after OTS correction = 0.9531). Correction for overall taste sensitivity thus shows exactly those changes in the P-value profile that would be expected following the elimination of a confounding variable. Encouraged by the results of this analysis, we corrected all detection thresholds for overall taste sensitivity and used them as input phenotypes for further GWASs. For quinine detection thresholds, OTS correction results in the same type of specific boost (Figs. 1b and 4c; Supplementary Material, Fig. S3b) in the strength of the association signal (P = 1.93 × 10−19, r2 = 13.2% to pcorr = 4.97 × 10−37, r2corr = 23.2%) again without an overall inflation of P-values (lambda before = 0.9524, lambda after = 1.0595).
Figure 4.
Caffeine detection thresholds are associated with a locus on chromosome 12 and OTS correction P-value boosts are loci specific. Manhattan plots from the GWAS analysis of log10-transformed (A) caffeine, (B) PROP and (C) quinine detection thresholds for the 504 subjects of the discovery panel shows a pronounced association at the beginning of chromosome 12 for both caffeine and quinine and a locus on chromosome 7 for PROP. Correction for OTS boosted SNPs only within associated loci for all three bitter compounds. Gray points and black triangles represent, respectively, P-values before and after correction for OTS while dashed lines represent thresholds for GWAS significance.
Correction for overall taste sensitivity also lead to a new genome-wide association signal between the detection threshold for caffeine and a locus on chromosome 12 (Fig. 4a and Table 2) (top SNP rs8181, P = 8.01 × 10−12, r2 = 9.4%, β = −0.12, s.e. = 0.018, second best rs2708377, pcorr = 8.75 × 10−12, r2 = 9.4%, β = −0.12, s.e. = 0.018). As previously observed for the PROP and quinine-detection-threshold phenotypes, OTS correction only boosted the most significant SNPs (Fig. 4a and Supplementary Material, Fig. S3c) and no overall P-value inflation was observed (lambda before = 1.0272 lambda after = 1.0493). To exclude the possibility that OTS correction might simply boost the most strongly associated SNPs – regardless of whether they are truly linked to the phenotype – we performed a bootstrap analysis using data sets in which caffeine thresholds were randomized across subjects (100 repeats). This analysis confirmed that for datasets lacking an underlying association between genotype and phenotype OTS correction does not result in boosted association P-values (Supplementary Material, Fig. S3d).
Table 2.
Association statistics for SNPs in the bitter receptor cluster on chromosome 12 with the caffeine detection threshold phenotype
| SNP namea | Chrb | Positionb | MAFc (%) | Call ratec | 504 subjects |
504 subjects OTS corrected |
104 subjects |
104 subjects OTS corrected |
Meta-analyzed | Meta-analyzed OTS corrected | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | r2 (%) | P-value | r2 (%) | P-value | r2 (%) | P-value | r2 (%) | P-value | r2 (%) | P-value | r2 (%) | |||||
| rs10772397 | 12 | 11029950 | 48.9 | 1.00 | 1.24E−02 | 1.37 | 7.91E−04 | 2.45 | 1.49E−01 | 2.33 | 1.81E−01 | 2.01 | 2.17E−03 | 1.72 | 1.71E−04 | 2.57 |
| rs10845271 | 12 | 11035068 | 28.2 | 1.00 | 6.08E−01 | 0.06 | 1.11E−01 | 0.56 | 5.76E−01 | 0.36 | 2.00E−01 | 1.86 | 2.38E−01 | 0.26 | 6.28E−02 | 0.64 |
| rs1450839 | 12 | 11040799 | 28.0 | 1.00 | 5.61E−01 | 0.08 | 1.01E−01 | 0.60 | 2.41E−01 | 1.56 | 5.05E−01 | 0.51 | 2.18E−01 | 0.28 | 5.77E−02 | 0.67 |
| rs10845279 | 12 | 11040978 | 28.1 | 1.00 | 5.90E−01 | 0.06 | 1.07E−01 | 0.57 | 3.66E−01 | 0.93 | 6.07E−01 | 0.30 | 2.30E−01 | 0.27 | 6.09E−02 | 0.65 |
| rs10845280 | 12 | 11040987 | 28.1 | 1.00 | 5.90E−01 | 0.06 | 1.07E−01 | 0.57 | 2.79E−01 | 1.33 | 5.55E−01 | 0.40 | 2.30E−01 | 0.27 | 6.09E−02 | 0.65 |
| rs12226919 | 12 | 11041300 | 28.9 | 1.00 | 5.64E−01 | 0.07 | 7.16E−02 | 0.72 | 2.22E−01 | 1.68 | 4.95E−01 | 0.53 | 2.18E−01 | 0.28 | 4.05E−02 | 0.78 |
| rs12226920 | 12 | 11041313 | 28.0 | 0.99 | 5.59E−01 | 0.08 | 9.38E−02 | 0.62 | 3.06E−01 | 1.19 | 6.32E−01 | 0.26 | 2.33E−01 | 0.26 | 5.93E−02 | 0.66 |
| SNP12–11041321 | 12 | 11041321 | 14.0 | 1.00 | 9.32E−01 | 0.00 | 8.71E−01 | 0.01 | 6.82E−01 | 0.19 | 3.29E−01 | 1.08 | 8.00E−01 | 0.01 | 8.06E−01 | 0.01 |
| rs7135018 | 12 | 11041507 | 18.9 | 1.00 | 2.13E−02 | 1.17 | 2.59E−03 | 1.98 | 5.31E−01 | 0.45 | 4.50E−01 | 0.65 | 2.73E−02 | 0.90 | 3.08E−03 | 1.61 |
| rs10772408 | 12 | 11042866 | 47.5 | 1.00 | 1.36E−02 | 1.34 | 3.16E−05 | 3.71 | 1.36E−01 | 2.49 | 2.78E−01 | 1.34 | 2.51E−03 | 1.68 | 1.16E−05 | 3.46 |
| rs12581501 | 12 | 11045061 | 19.2 | 0.96 | 1.42E−02 | 1.32 | 1.40E−03 | 2.22 | 6.80E−01 | 0.20 | 5.69E−01 | 0.37 | 2.39E−02 | 0.94 | 2.26E−03 | 1.71 |
| rs11054155 | 12 | 11051659 | 19.3 | 1.00 | 1.75E−02 | 1.24 | 1.19E−03 | 2.29 | 7.19E−01 | 0.15 | 5.71E−01 | 0.37 | 2.39E−02 | 0.94 | 1.58E−03 | 1.83 |
| rs11054160 | 12 | 11053800 | 28.0 | 1.00 | 5.61E−01 | 0.08 | 1.01E−01 | 0.60 | 9.69E−02 | 3.07 | 1.79E−01 | 2.03 | 2.18E−01 | 0.28 | 5.77E−02 | 0.67 |
| rs4763606 | 12 | 11054258 | 27.9 | 1.00 | 5.56E−01 | 0.08 | 1.06E−01 | 0.58 | 2.48E−01 | 1.51 | 4.27E−01 | 0.72 | 2.17E−01 | 0.28 | 6.07E−02 | 0.65 |
| rs7304447 | 12 | 11055369 | 28.0 | 0.99 | 6.34E−01 | 0.05 | 1.13E−01 | 0.56 | 3.09E−01 | 1.18 | 6.18E−01 | 0.29 | 2.53E−01 | 0.24 | 6.42E−02 | 0.64 |
| rs7304579 | 12 | 11055462 | 28.1 | 1.00 | 5.90E−01 | 0.06 | 1.07E−01 | 0.57 | 2.37E−01 | 1.58 | 4.66E−01 | 0.61 | 2.30E−01 | 0.27 | 6.09E−02 | 0.65 |
| rs7304936 | 12 | 11055749 | 28.2 | 0.97 | 3.72E−01 | 0.18 | 3.99E−02 | 0.93 | 2.47E−01 | 1.52 | 4.85E−01 | 0.56 | 1.25E−01 | 0.44 | 2.27E−02 | 0.96 |
| rs7315843 | 12 | 11056018 | 19.8 | 1.00 | 1.66E−03 | 2.15 | 4.49E−05 | 3.58 | 1.96E−01 | 1.88 | 5.32E−01 | 0.45 | 1.21E−04 | 2.68 | 3.69E−06 | 3.84 |
| rs10845289 | 12 | 11058493 | 20.9 | 0.99 | 9.80E−03 | 1.46 | 1.56E−02 | 1.29 | 5.23E−01 | 0.47 | 2.08E−01 | 1.79 | 8.04E−03 | 1.29 | 6.05E−03 | 1.39 |
| rs10743937 | 12 | 11064722 | 41.4 | 0.94 | 5.89E−05 | 3.47 | 4.81E−06 | 4.45 | 3.77E−01 | 0.89 | 9.00E−02 | 3.20 | 9.81E−05 | 2.75 | 1.14E−06 | 4.23 |
| rs10772420 | 12 | 11065543 | 41.9 | 1.00 | 2.70E−05 | 3.78 | 3.56E−06 | 4.57 | 4.12E−01 | 0.77 | 9.91E−02 | 3.03 | 4.89E−05 | 2.98 | 8.72E−07 | 4.32 |
| rs7297949 | 12 | 11079407 | 40.3 | 0.99 | 2.66E−05 | 3.78 | 1.62E−06 | 4.87 | 8.18E−01 | 0.06 | 7.75E−01 | 0.09 | 4.32E−05 | 3.03 | 7.15E−07 | 4.38 |
| rs34836514 | 12 | 11098452 | 21.5 | 0.99 | 1.64E−02 | 1.27 | 2.06E−02 | 1.18 | 7.79E−01 | 0.09 | 5.00E−01 | 0.52 | 1.51E−02 | 1.09 | 8.45E−03 | 1.28 |
| rs2708381 | 12 | 11105412 | 17.4 | 0.96 | 7.13E−03 | 1.59 | 2.30E−04 | 2.93 | 6.23E−01 | 0.28 | 5.01E−01 | 0.52 | 1.45E−02 | 1.10 | 5.02E−04 | 2.21 |
| rs2708377 | 12 | 11107582 | 31.2 | 1.00 | 4.18E−07 | 5.40 | 8.75E−12 | 9.40 | 6.70E−02 | 3.71 | 4.70E−02 | 4.34 | 2.68E−08 | 5.45 | 5.31E−13 | 8.86 |
| rs2597972 | 12 | 11144640 | 41.8 | 0.98 | 9.76E−06 | 4.17 | 2.52E−06 | 4.70 | 7.12E−01 | 0.16 | 3.13E−01 | 1.16 | 1.78E−05 | 3.32 | 5.81E−07 | 4.45 |
| rs2597963 | 12 | 11149278 | 42.1 | 0.99 | 1.95E−05 | 3.90 | 3.22E−06 | 4.61 | 4.86E−01 | 0.56 | 1.96E−01 | 1.88 | 3.80E−05 | 3.07 | 7.29E−07 | 4.38 |
MAF, minor allele frequency; pHW, Hardy–Weinberg P-value.
aSNPs are filtered for MAF >5%, pHW >10−5 and call rate >90%, only SNP observed in both discovery and replication cohort are shown.
bBased on human genome build 36.1.
cBased on meta-analyzed data (608 subjects).
To validate the new association, we recruited an additional panel of 104 subjects from the same general São Paulo population. Subjects from this subpanel underwent the same genotyping and phenotyping procedure as subjects from the main panel but their genotype surrounding the caffeine and quinine loci was also determined using next-generation sequencing. Since rs8181 was not available in the replication set, we used rs2708377 as proxy (r2 = 0.996). The tests on this second panel replicate the association found with the main panel (rs2708377, pcorr = 0.047, r2corr = 4.3%, βcorr = −0.07, s.e.corr = 0.036) with the combined panels giving a top association at SNP rs2708377 (pcorr = 5.31 × 10−13, r2corr = 8.9%, βcorr = −0.12, s.e.corr = 0.016). Notably, meta-analysis of the combined discovery and replication cohorts results in a genome-wide significant (pmeta = 2.68 × 10−8, r2meta = 5.5%, βmeta = −0.11, s.e.meta = 0.020) association between rs2708377 and caffeine detection threshold even without overall taste-sensitivity correction, thus providing further confirmation that OTS correction acts by boosting an underlying association signal.
The newly identified locus for caffeine detection threshold falls into a cluster of bitter taste receptors that partially overlaps with the receptor cluster (chr12 position 10.88–11.24 Mb, build 36.1) found to be associated with the quinine detection threshold (Figs. 3 and 4a and c). The top SNP, rs2708377, is located just adjacent to the coding region of the taste receptor gene Tas2R46. Incidentally, this receptor has been shown to be activated by caffeine in vitro (7). Further validation of the association between the sensitivity to the taste of caffeine and the new locus on chromosome 12 comes from the taste intensity ratings of suprathreshold solutions of caffeine. Participants who carry the low-threshold-associated allele rated suprathreshold concentrations of caffeine to be more intense (Fig. 2) than participants who do not carry this allele.
A local Manhattan plot of the quinine and caffeine associations indicates that the identified associations represent two independent, though partially overlapping loci (Fig. 3). Notably, the r2 between the genotype of the top quinine-associated SNP (rs1031391) and the top caffeine-associated SNP (rs8181) is only 25%. A scatter plot of quinine versus caffeine detection-threshold P-values (Fig. 3b) for the SNPs located in this region of chromosome 12 underlines the independence of the two loci further – SNPs that are strongly associated with quinine detection thresholds are weakly associated with caffeine detection thresholds and vice versa. To further illustrate independence, we performed conditional association analysis. For this, we tested caffeine association with rs8181 conditional on rs1031391 and then inverted the role of the two SNPs. The results revealed that rs8181 remained significantly associated with caffeine detection threshold (P = 3.37 × 10−7) irrespective of rs1031391, while rs1031391 ceased to be associated with caffeine detection (P = 0.06) once corrected for rs8181. The opposite was observed for quinine detection, rs1031391 being the major factor. We assume that the genetic diversity of our population, which is characterized by the continuous admixture of different genetic ancestries (Supplementary Material, Fig. S1), contributed positively to the ability to separate these two loci.
Association analysis conditional on the top hit did not reveal additional genome-wide significant signals for any of the bitter traits. Using sequencing technology for the replication panel could have enabled us to narrow down the causal variant; however, due to the small sample size, we do not have statistically significant evidence for a better-associated (untyped) marker. It therefore remains open whether the SNPs identified here are the causal SNPs or if they simply act as proxies for the functional variant. It therefore also remains open if the genetic changes underlying variation in caffeine and quinine perception exert their effect by changing the amino acid sequence of the receptor itself, as is the case for the mutations in the TAS2R38 gene that change PROP perception, or if the effect is more indirect (e.g. changes in receptor expression levels).
Consistent with caffeine's negligible contribution to the overall bitterness of coffee (18) the discovered caffeine detection-threshold-associated locus (using the best available SNP rs3741843) shows no association with coffee consumption neither in the largest-to-date meta-analysis of 47 338 individuals (P = 0.51) (19) nor in our panel.
DISCUSSION
In the current study, we have identified a genetic locus that contributes to the variation in human perception of caffeine bitterness. As Figure 3a shows this locus partially overlaps with the adjacent locus for quinine taste sensitivity. However, analysis of the relative strength of the association (Fig. 3b) between the SNPs in this region and the sensitivity to the two bitter compounds shows that the implicated loci are essentially independent from one another. Comparison of our results with the results obtained on a panel of predominantly central European ancestry (15) and inspection of the loci's LD structure (bottom panel of Fig. 3a) in the founder populations that dominate the diversity of our Brazilian study panel indicates that the extensive ethnic diversity of our study panel was instrumental for disentangling these two loci. From an evolutionary perspective, this data further suggest that caffeine and quinine taste sensitivity are genetic traits that could and have been selected upon independently in some of the world's populations. Further studies will be needed to pinpoint the causal genes and to investigate their mechanisms of effect.
Together with the findings in other studies, the results presented here provide an interesting insight into human variation in bitter taste sensitivity. On the one hand, there are genetic variations in specific bitter taste receptor genes that drive compound- and, as we have shown here, concentration-specific differences in taste sensitivity. On the other hand, we observe a pronounced correlation of taste sensitivities across all five taste modalities. Interestingly, this overall taste sensitivity is not driven primarily by either a single genetic factor or by a demographic factor (age, gender, BMI, ethnicity). Notably, absent are genetic factors that impact bitter taste perception as a whole (i.e. bitter versus sweet, sour or umami tastes).
The study also highlights that broad and high-quality phenotyping has the potential to offset the loss of statistical power incurred by a smallstudy panel. The design of GWAS studies usually involves a tradeoff between the extent and precision of phenotyping and the size of the study panel (for a detailed discussion of these tradeoffs in GWAS on sensory phenotypes (20)). In the current study, very extensive phenotyping—each participant underwent an 8-day full-time taste phenotyping program—was chosen at the expense of a much reduced panel size (500 subjects). A previous study on bitter taste genetics (15) reflected a different choice in this tradeoff and used a panel of nearly three times the size (1400 subjects) but employed a very streamlined sensory testing protocol. A comparison of the two studies indicates that improvements in phenotyping quality and breadth can indeed compensate for a very substantial reduction in panel size. For example, Reed et al. (15) found that the top SNP for quinine sensitivity explained 5.8% of phenotypic variation, while in the current study, the same locus explained 23% of variance. As can be seen from the P-values of association (current study P = 4.97 × 10−37, Reed et al. P = 1.8 × 10−15), this boost in explained variance more than compensated for the reduced panel size. It appears that the boost in explained variance observed in our study can be attributed in equal parts to two factors: by assessing taste sensitivity to quinine at multiple concentrations, it was evident that the genetic effect on quinine perception is concentration dependent. This allowed us to focus the analysis on the detection-threshold data where the genetic effect is maximal. In contrast, Reed et al. assessed quinine sensitivity at a concentration of 0.2 mm where the size of the genetic effect is already much diminished (see Fig. 2). An equally large boost can be attributed to across-phenotype correction which has allowed us to eliminate overall taste sensitivity as a confounding factor for quinine-specific taste sensitivity.
Overall, the results of the current study bode well for future attempts to map genetic variations that drive interindividual differences in human chemosensory perception. The responsible chemosensory receptors, notably bitter and olfactory receptors (21), display pronounced genetic diversity (8,9,22). But these receptors also tend to cluster together in genomic regions with high LD, which was thought to make it difficult to map the genetic drivers of interindividual variations in sensory perception with the help of association studies. Chemosensory phenotypes also have a reputation for being unstable over time and difficult to assess experimentally. The results from the current study indicate that these challenges can be overcome by the use of ethnically diverse study panels and careful phenotyping.
MATERIALS AND METHODS
Ethics statement
The research described here was approved by the Institutional Review Board of the Sírio Libanês Hospital, where tests were administered, and by the National Committee of Research Ethics at the Brazilian Ministry of Health (HSL 2007/25 Process no. 25000.114841/2007-17). Each participant gave informed consent for their participation to the study and indicated so by signing a formal informed consent statement.
Subjects and methods
Subjects between 18 and 45 years old were recruited from the general population of São Paulo, Brazil. Subjects had to be in good general health as assed by a health questionnaire and by an on-site medical exam. Pregnancy, food allergies a history of sensory or other neurological disorders or recent changes in food habits were used as exclusion criteria. Smokers were accepted in the study but were instructed not to smoke at least 3 h prior to the start of the taste tests. A breakdown of subjects according to age, gender and BMI is provided in Table 1.
Sensory testing
The testing procedures used in the current study were previously described (13,20). Briefly, detection thresholds and suprathreshold intensities were determined for 13 taste compounds (bitter: PROP, quinine and caffeine; sweet: sucrose, maltitol, aspartame and saccharin; salty: NaCl and KCl; sour: hydrochloric acid and calcium citrate; umami: monosodium glutamate and inosine monophosphate) using the staircase and gLMS techniques, respectively. For gLMS testing, subjects underwent thorough training on the use of the scale. Detection-thresholds were recorded in duplicate or triplicate and suprathreshold intensity data were recorded in triplicate or quadruplicate.
Overall taste-sensitivity parameter
To account for across-compound correlations in detection thresholds, we calculated for each participant k an overall taste sensitivity parameter zk. This parameter represents the across-compound averaged z-scores of log10-transformed detection thresholds.
with
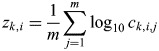 |
where n is the number of taste compounds, m is the number of repeat measurements available for a given individual and compound, ck,i,j is the detection threshold for individual k, for compound i during measurement j and 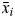 and e.s.d.i is the mean and estimated standard deviation of xk,i across all subjects. To obtain overall sensitivity for as many individuals as possible, sporadic missing detection-threshold values were ignored for the mean calculation.
and e.s.d.i is the mean and estimated standard deviation of xk,i across all subjects. To obtain overall sensitivity for as many individuals as possible, sporadic missing detection-threshold values were ignored for the mean calculation.
Genotyping
DNA for genotyping was extracted from blood donated by the participants. Genotyping was outsourced to Expression Analysis Inc. (Durham, NC, USA) and conducted using the Illumina (Illumina Inc., San Diego, CA, USA) omni-quad chip that genotypes ∼1 million SNPs per subject. Subjects from the replication panel were further genotyped by sequence enrichment of the 12p13.2 locus on Sure Select arrays (Agilent Inc., Santa Clara, CA, USA) followed by HiSEQ (Illumina Inc.) sequencing performed by Fasteris SA (Geneva, Switzerland)). Genotype calling was performed with Beadstudio software (Illumina). Calls with a genotyping score <0.2 were excluded from further analysis. Single nucleotide polymorphisms (SNPs) with a call rate <90% and individuals with a call rate <95% were also excluded. Genotyping data were of high quality with an average call rate of 99.8% for all SNPs. 99.4% of SNPs had a call rate of greater than the cutoff value (95%) set for the rejection of individual SNPs. The average Q-score for all SNPs was 0.71 and for 99.6% of called SNPs the Q-score passed the cutoff (0.2) for inclusion.
GWAS
Population stratification and relatedness was assessed using the ancestry principal components as previously described (23,24). Input phenotypes were corrected for essential covariates via a multiple-linear regression. We used age, sex, body mass index and the first 10 principal components of the ancestry analysis as covariates if they showed statistical significance in single linear regression versus the phenotype (cutoff P < 0.01). When indicated, overall taste-sensitivity scores were added as a covariate in this correction model. The residuals from this multiple-linear regression, i.e. the corrected phenotype parameters were then regressed against SNP allele dosage using a linear, additive model implemented as in-house Matlab code (The Mathworks Inc., Natick, MA, USA). The resulting P-values underwent genomic control (25). SNPs were further filtered for Hardy–Weinberg equilibrium P-value (>10−5), Q-score (>0.2) and minor allele frequency (>5%). Log-transformed detection thresholds (caffeine, quinine, PROP) were normally distributed according to the Kolmogorov–Smirnov test (26), with P = 0.73, 0.22, 0.11, respectively.
Conflict of Interest statement. None declared.
FUNDING
The study was supported by the Nestlé Research Center. Mirko Ledda, Nathalie Martin, Johannes le Coutre and Ulrich Genick are, or were, full-time employees of the Nestlé Research Center. Sven Bergmann received financial support from the Nestlé Research Center. Funding to pay the Open Access publication charges for this article was provided by the University of Lausanne.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Christoph Hartmann, Veronica Galindo-Cuspinera, Sidney Simon, Alan Rudolph and Jennifer Brower for their role in planning and organizing the project on which this manuscript is based. We further acknowledge Silvia Rozsa, Gabriela Silva, Denise Monzani, Marina Monzani, Agnes Curto and Ana Elisa Sestini for their contribution to data collection. We thank the authors of the GWAS of caffeine consumption (19), especially Marilyn Cornelis, for kindly providing results for the association between our top locus and caffeine consumption.
REFERENCES
- 1.Drewnowski A., Henderson S.A., Barratt-Fornell A. Genetic taste markers and food preferences. Drug Metab. Dispos. 2001;29:535–538. [PubMed] [Google Scholar]
- 2.Drewnowski A., Henderson S.A., Shore A.B. Taste responses to naringin, a flavonoid, and the acceptance of grapefruit juice are related to genetic sensitivity to 6-n-propylthiouracil. Am. J. Clin. Nutr. 1997;66:391–397. doi: 10.1093/ajcn/66.2.391. [DOI] [PubMed] [Google Scholar]
- 3.Wiener A., Shudler M., Levit A., Niv M.Y. BitterDB: a database of bitter compounds. Nucleic Acids Res. 2012;40:D413–D419. doi: 10.1093/nar/gkr755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockhoff A., Behrens M., Roudnitzky N., Appendino G., Avonto C., Meyerhof W. Receptor agonism and antagonism of dietary bitter compounds. J. Neurosci. 2011;31:14775–14782. doi: 10.1523/JNEUROSCI.2923-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrashekar J., Hoon M.A., Ryba N.J., Zuker C.S. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 6.Chandrashekar J., Mueller K.L., Hoon M.A., Adler E., Feng L., Guo W., Zuker C.S., Ryba N.J. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 7.Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 8.Kosiol C., Vinar T., da Fonseca R.R., Hubisz M.J., Bustamante C.D., Nielsen R., Siepel A. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wooding S. Signatures of natural selection in a primate bitter taste receptor. J. Mol. Evol. 2011;73:257–265. doi: 10.1007/s00239-011-9481-0. [DOI] [PubMed] [Google Scholar]
- 10.Parry C.M., Erkner A., le Coutre J. Divergence of T2R chemosensory receptor families in humans, bonobos, and chimpanzees. Proc. Natl Acad. Sci. USA. 2004;101:14830–14834. doi: 10.1073/pnas.0404894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara T., Go Y., Udono T., Morimura N., Tomonaga M., Hirai H., Imai H. Diversification of bitter taste receptor gene family in western chimpanzees. Mol. Biol. Evol. 2011;28:921–931. doi: 10.1093/molbev/msq279. [DOI] [PubMed] [Google Scholar]
- 12.Simon S.A., de Araujo I.E., Gutierrez R., Nicolelis M.A. The neural mechanisms of gustation: a distributed processing code. Nat. Rev. Neurosci. 2006;7:890–901. doi: 10.1038/nrn2006. [DOI] [PubMed] [Google Scholar]
- 13.Galindo-Cuspinera V., Waeber T., Antille N., Hartmann C., Stead N., Martin N. Reliability of threshold and suprathreshold methods for taste phenotyping: characterization with PROP and sodium chloride. Chemosens. Percept. 2009;2:214–228. doi: 10.1007/s12078-009-9059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartoshuk L.M. The psychophysics of taste. Am. J. Clin. Nutr. 1978;31:1068–1077. doi: 10.1093/ajcn/31.6.1068. [DOI] [PubMed] [Google Scholar]
- 15.Reed D.R., Zhu G., Breslin P.A., Duke F.F., Henders A.K., Campbell M.J., Montgomery G.W., Medland S.E., Martin N.G., Wright M.J. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum. Mol. Genet. 2010;19:4278–4285. doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim U.K., Jorgenson E., Coon H., Leppert M., Risch N., Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 17.Ledda M., Kutalik Z., Godinot N., Galindo-Cuspinera V., Antilles N., Destito M.C.S., Souza M.M., A., Cirillo C., Zamboni A., Morya E., et al. Inter-individual variation in human taste perception. 2013 In press. [Google Scholar]
- 18.Frank O., Blumberg S., Kunert C., Zehentbauer G., Hofmann T. Structure determination and sensory analysis of bitter-tasting 4-vinylcatechol oligomers and their identification in roasted coffee by means of LC-MS/MS. J. Agric. Food Chem. 2007;55:1945–1954. doi: 10.1021/jf0632280. [DOI] [PubMed] [Google Scholar]
- 19.Cornelis M.C., Monda K.L., Yu K., Paynter N., Azzato E.M., Bennett S.N., Berndt S.I., Boerwinkle E., Chanock S., Chatterjee N., et al. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet. 2011;7:e1002033. doi: 10.1371/journal.pgen.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genick U.K., Kutalik Z., Ledda M., Destito M.C., Souza M.M., Cirillo C.A., Godinot N., Martin N., Morya E., Sameshima K., et al. Sensitivity of genome-wide-association signals to phenotyping strategy: the PROP-TAS2R38 taste association as a benchmark. PLoS ONE. 2011;6:e27745. doi: 10.1371/journal.pone.0027745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buck L.B. Unraveling the sense of smell (Nobel lecture) Angew. Chem. Int. Ed. Engl. 2005;44:6128–6140. doi: 10.1002/anie.200501120. [DOI] [PubMed] [Google Scholar]
- 22.Hasin-Brumshtein Y., Lancet D., Olender T. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 2009;25:178–184. doi: 10.1016/j.tig.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R., et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 26.Dicks J., Savva G. Comparative Genetics. In: Balding D.J., Bishop M.J., Cannings C., editors. Handbook of Statistical Genetics. Chichester, UK: John Wiley and Sons; 2007. pp. 160–199. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.