Abstract
Aberrant imprinting of the insulin-like growth factor II (IGF2) gene is a molecular hallmark of many tumors. Reactivation of the normally suppressed maternal allele leads to upregulation of the growth factor that promotes tumor growth. However, the mechanisms underlying the loss of imprinting (LOI) remain poorly defined. We examined the epigenotypes at the gene promoters that control IGF2 allelic expression. Using chromatin immunoprecipitation, we found that in cells characterized by maintenance of IGF2 imprinting, three IGF2 promoters were differentially modified, with the suppressed allele heavily methylated at histone H3K27 while the active allele was unmethylated. In the LOI tumors, however, both alleles were unmethylated, and correspondingly there was no binding of SUZ12, the docking factor of the polycomb repressive complex 2 (PRC2), and of the zinc finger-containing transcription factor (CTCF) that recruits the PRC2. Using chromatin conformation capture, we found that the CTCF-orchestrated intrachromosomal loop between the IGF2 promoters and the imprinting control region was abrogated in cells with LOI. SUZ12, which docks the PRC2 to IGF2 promoters for H3K27 methylation, was downregulated in LOI cells. These data reveal a new epigenetic control pathway related to the loss of IGF2 imprinting in tumors.
INTRODUCTION
Reactivation of the normally suppressed (imprinted) maternal insulin-like growth factor II (IGF2) allele, known as loss of imprinting (LOI), is a hallmark of many human tumors, especially childhood tumors (1–8) and cancer stem cells (9). This aberrant biallelic expression may increase the production of IGF-II, promoting the malignant behavior of tumor cells through enhanced cell growth and self-renewal. IGF2-overexpressing tumors frequently display loss of PTEN, and they are often highly proliferative, exhibiting strong staining for phospho-Akt. These LOI tumors belong to a subclass of neoplasms characterized by poor survival (10). Detection of IGF2 LOI in circulating white blood cells represents a valuable biomolecular marker for predicting individuals with high risk for colorectal cancer (11).
However, the mechanism underlying loss of IGF2 imprinting in tumors remains elusive. The genes encoding IGF2 and H19 on human chromosome 11p15.5 are reciprocally imprinted, controlled by epigenetic modifications in the differentially methylated region (DMR) of the imprinting control region (ICR) located between these two adjacent genes. In the mouse, the exclusive binding of the insulating factor CTCF to the unmethylated maternal ICR creates a physical boundary, blocking the interaction of downstream enhancers with the remote IGF2 promoters and silencing the maternal allele (12–21). When this ICR is deleted (22) or mutated (23,24), the suppressed maternal IGF2 allele is reactivated, leading to LOI. In human tumors, however, a number of epigenetic modifications in the ICR have been described (25–27), and it is unclear whether this enhancer insulation is also causally involved in the abnormal regulation of IGF2 in tumors.
Using chromatin configuration capture (3C) methodology, it has been shown that CTCF participates in the formation of a long-range chromosomal loop to the upstream IGF2 DMRs when it is bound to the maternal ICR (19,28,29). A series of studies from our lab suggest that CTCF may not only function as a physical insulator but also actively participate in the regulation of the imprinted IGF2 allele (30–33). In this communication, we explored in detail the mechanisms by which the normally suppressed maternal promoters of IGF2 become reactivated during the process of loss of genomic imprinting.
RESULTS
Loss of promoter H3K27 methylation suppressive mark in tumors
To explore the mechanism underlying aberrant IGF2 imprinting in tumors, we examined the IGF2 epigenotypes in the promoter regions that control allelic expression. Unlike the mouse Igf2 (13,14), loss of human IGF2 imprinting in tumors may not necessarily be accompanied by changes in DNA methylation in known ICRs (11,25,34,35). We previously examined the DNA methylation status of IGF2 promoters in tumors and found that IGF2 promoters were unmethylated on both alleles, thus excluding a role of promoter DNA methylation in controlling allelic suppression (30).
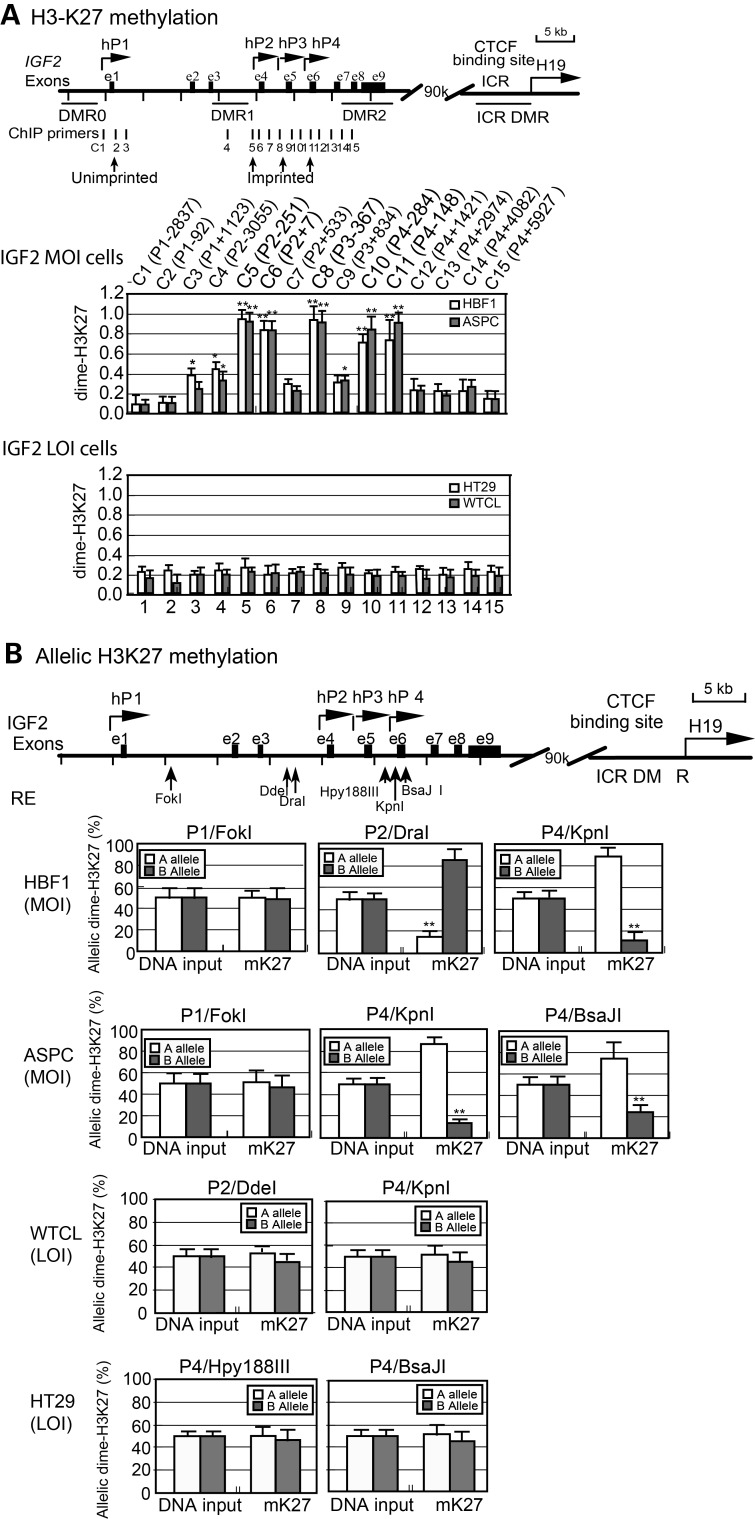
We thus focused on IGF2 promoter suppression by histone H3K27 methylation to determine whether this epigenetic suppressive mark was altered in tumors in association with LOI. Using chromatin immunoprecipitation (ChIP), we examined H3K27 methylation in the IGF2 promoters. Promoter 1 (hP1), located immediately downstream of the insulin gene, is not under the control of imprinting mechanisms and is normally biallelically expressed (36). In most tumor cells, hP1 is expressed at very low or undetectable levels, and it does not contribute substantially to IGF2 expression. We found that H3K27 in the hP1 region was unmethylated in both IGF2 LOI and maintenance of IGF2 imprinting (MOI) cells (Fig. 1A, primer set C1). Three downstream promoters (hP2–hP4) are imprinted in normal tissues but often exhibit LOI in tumors. In MOI cells, we found that there was increased H3K27 methylation in these three imprinted promoters (Fig. 1A, middle panel, primer sets C5, C6, C8, C10 and C11), correlating with the suppression of the imprinted allele. In LOI tumor cells, in contrast, H3K27 methylation suppressive mark was not observed in any of the IGF2 promoters (Fig. 1A, bottom panel), in parallel with the reactivation of the maternal IGF2 promoters in tumor cells.
Figure 1.
Histone 3 lysine 27 (H3K27) methylation at the IGF2 promoters. (A) H3K27 methylation in IGF2 promoters as measured by ChIP PCR. Vertical lines: PCR primers (C1–C15) used for ChIP assay. The specific location of each ChIP PCR primer pair was also specified in the parenthesis as the sequence number downstream (+) or upstream (−) of the transcription initiation site. DMR: differentially methylated regions; e1–e9: IGF2 exons; ICR: imprinted control region; MOI: maintenance of imprinting; LOI: loss of imprinting; hP1: non-imprinted promoter; hP2–hP4: imprinted promoters. Chromatin DNAs were immunoprecipitated with antisera against dimethyl H3K27 (dime-H3K27) and quantitated by ChIP primers. Primer sequences are listed in Supplementary Material, Table S2. The ChIP data are presented as the ratio of the dime-H3K27 ChIP signal over the input signal. Error bars represent the standard error of the average of three independent ChIP assays (each with three PCR reactions). *P < 0.05, **P < 0.01 as compared with the unimprinted C1. (B) Allele-specific H3K27 methylation. Allelic ChIP products were separated by polymorphic restriction enzymes (RE). The allelic interaction of CTCF with the IGF2 promoters (P1 and P4) was identified by using six polymorphic restriction enzymes (FokI, DraI, KpnI, DdeI, Hpy188III and BsaJ1), depending on cells used. The source of the specific parental alleles of ‘A’ and ‘B’ is not known. Allelic ChIP enrichment of dime-H3K27 is calculated as the percentage of the A or B allele over the total (A + B) after normalization with the input DNA.
We then examined whether the histones at the IGF2 promoters were differentially methylated at each parental allele. By mapping all available SNPs near the IGF2 promoters, we found two informative SNPs (Fok1 and Dde1) downstream of hP1 and three informative SNPs (Hyp188III, Kpn1 and BsaJ1) near hP3–hP4. With the aid of these SNPs, we found that there was no allelic difference in H3K27 methylation at the non-imprinted hP1 (Fig. 1B). At the imprinted hP2 and hP4 promoters, however, we found that H3K27 was monoallelically methylated in normal fibroblasts and in ASPC cells, a cancer cell line characterized by normal IGF2 imprinting. H3K27 hypermethylation at the promoter was associated with suppression of the maternal allele (30). In LOI tumor cell lines (WTCL and HT29), however, there was no allelic difference in H3K27 methylation, correlating with biallelic expression of IGF2.
Lack of SUZ12 binding to the IGF2 promoters
We then looked for factors that are associated with loss of H3K27 methylation at the IGF2 promoters. H3K27 methylation is catalyzed by methyltransferase EZH2, a critical component of polycomb repression complex 2 (PRC2). Using a mouse imprinting model, we previously demonstrated that PRC2 was recruited to the maternal promoter through the CTCF-mediated docking of a second PRC2 component SUZ12 (30). We therefore examined whether the SUZ12 interaction was also absent in parallel with LOI of IGF2 in tumors.
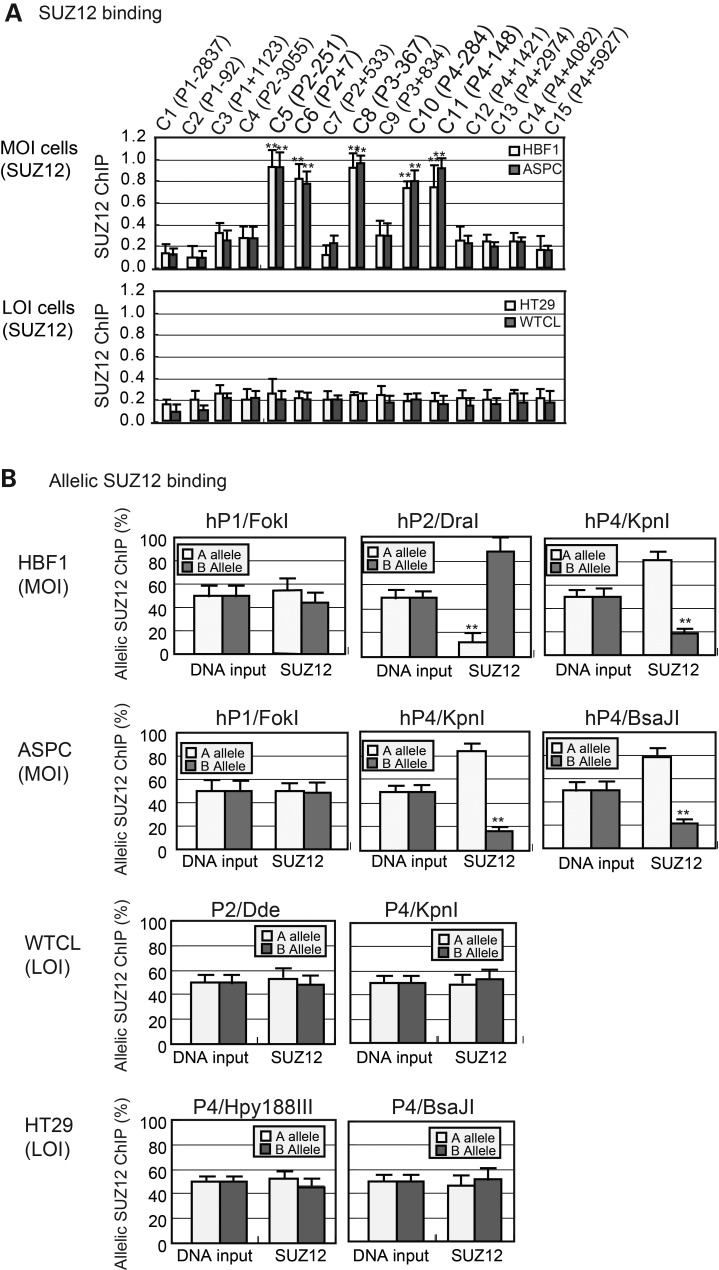
Using anti-SUZ12 ChIP, we found enrichment of SUZ12 at the three imprinted IGF2 promoters in IGF2 MOI cell lines (HBF1 and ASPC) (Fig. 2A, top panel), indicating an interaction of PRC2 with the imprinted IGF2 promoters. In the LOI tumor cell lines (HT29 and WTCL), however, this SUZ12 interaction was absent (bottom panel), indicating that PRC2 was not recruited to the three imprinted IGF2 promoters. As expected, there was no enrichment of SUZ12 at the unimprinted hP1 region (primer sets C1 and C2).
Figure 2.
(A) SUZ12–IGF2 promoter interaction. SUZ12 ChIP q-PCR quantitation in IGF2 MOI and LOI cells. Cells were immunoprecipitated with antisera against SUZ12, followed by PCR amplification with ChIP primers. Input DNA was DNA control before immunoprecipitation. See Figure 1A legend for details. (B) Allele-specific SUZ12 binding at the IGF2 promoters. The polymorphic restriction enzyme sites and PCR primers are the same as in Figure 1B. Allelic binding of SUZ12 is calculated as the percentage of the A or B allele over the total (A + B) after normalization with the input DNA.
Using available SNPs, we mapped the allelic binding of SUZ12 (Fig. 2B). In fibroblasts (HBF1) and ASPC cells that maintain normal IGF2 imprinting, SUZ12 interacted monoallelically with IGF2 promoters and hP4. In the LOI tumors, this differential hP4-SUZ12 allelic enrichment was not observed. Allelic binding of SUZ12 was also absent in the non-imprinted hP1 in both the IGF2 LOI or MOI cells. Thus, allelic recruitment of the PRC2 complex to the IGF2 promoters paralleled promoter H3K27 methylation, closely correlating with the imprinting status.
Altered allelic CTCF binding in IGF2 promoters
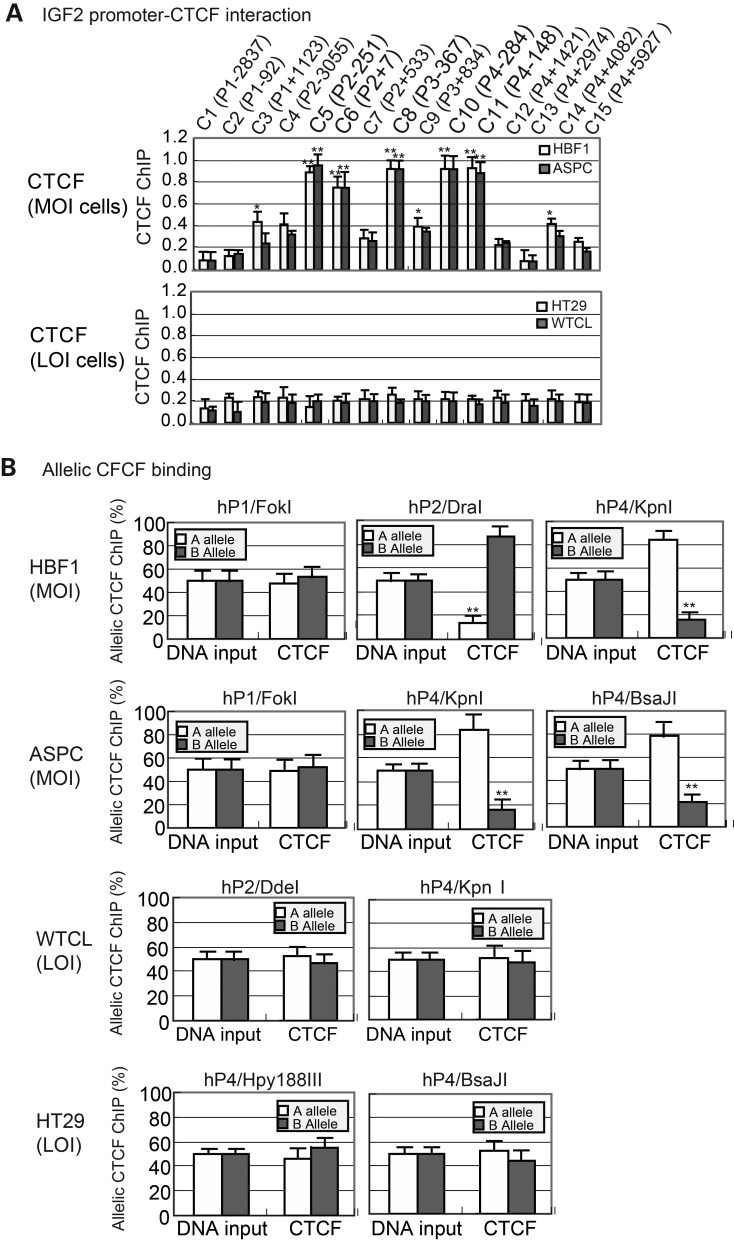
We previously demonstrated that the PRC2 complex was recruited to the maternal IGF2 promoters by CTCF, a zinc finger-containing transcription factor (31). CTCF binds to the IGF2 promoter and the ICR in front of the H19 gene, forming a long-range chromatin loop structure (28–30,33,37,38) that is essential for the recruitment of PRC2 and the subsequent establishment of the H3K27 methylation code in the maternal promoters. Using ChIP, we found that in sharp contrast to the MOI cell lines (HBF1 and ASPC) (Fig. 3A, top panel), CTCF binding to the imprinted promoters was lost in LOI tumor cell lines (HT29 and WTCL) (bottom panel).
Figure 3.
Binding of CTCF to the IGF2 promoters. (A) ChIP q-PCR of CTCF binding in the IGF2 promoters. Chromatin DNAs in MOI (HBF1 and ASPC) and LOI (HT29 and WTCL) cell lines were immunoprecipitated with antisera against CTCF and quantitated by q-PCR. Input DNA was DNA control before immunoprecipitation. Relative enrichments (fold) of CTCF binding across the promoter regions of human IGF2, which were calculated as described previously using input DNA, were plotted along P1–P4 regions of IGF2 DNA. See Figure 1A legend for details. (B) Quantitative allelic CTCF binding. The polymorphic restriction enzyme sites and PCR primers are the same as in Figure 1B. Allelic binding of CTCF is calculated as the percentage of the A or B allele over the total (A + B) after normalization with the input DNA.
We mapped the allelic binding of CTCF using several polymorphic restriction enzymes (Fig. 3B). In two IGF2 MOI cell lines (HBF1, ASPC), the unimprinted hP1 did not show significant allelic CTCF binding by using the polymorphic restriction enzyme (Fok1). However, using two restriction enzymes (Dra1 and Kpn1) near the imprinted promoters, we detected monoallelic binding of CTCF, in accord with the monoallelic expression of IGF2.
In IGF2 LOI tumor cells (HT29 and WTCL), however, no differential allelic enrichment of CTCF binding was observed. Using the polymorphic restriction enzymes (Kpn1 and BsaJ1), we could not detect allelic binding of CTCF to the imprinted promoters. These data suggest the requirement of CTCF promoter binding in the maintenance of genomic imprinting.
Loss of long-range intrachromosomal interaction in tumors
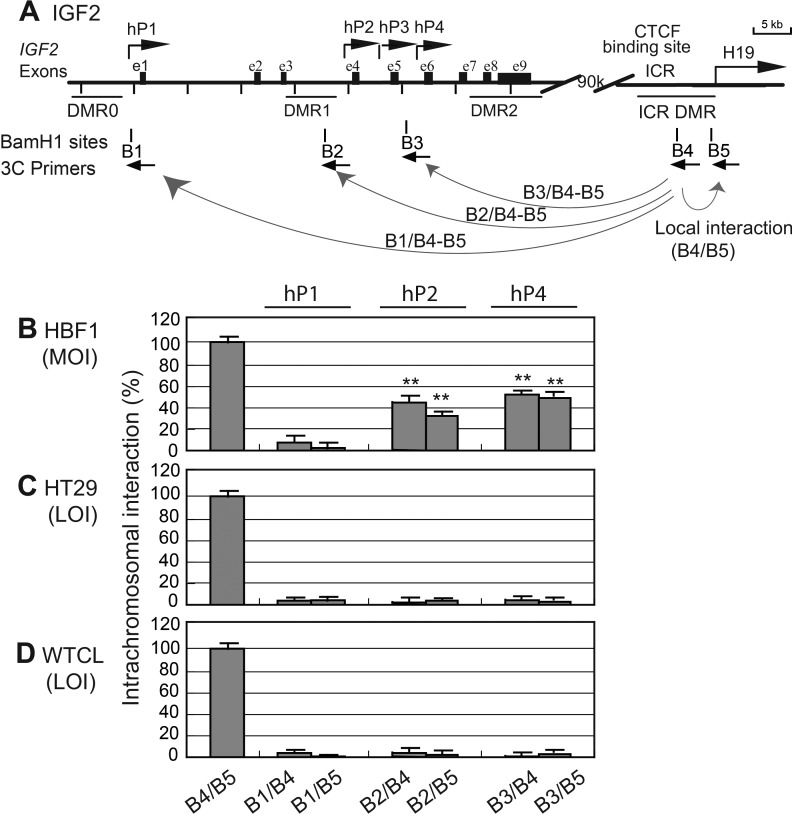
The CTCF–PRC2 promoter complex is formed through a long-range intrachromosomal loop structure between the promoter and the ICR. To further explore the mechanism underlying loss of IGF2 imprinting in some tumor cell lines, we first used a chromatin conformation capture (3C) method (30,39) to examine the intrachromosomal interaction in cells that have differential IGF2 imprinting. Cells were fixed with 2% formaldehyde and digested with restriction enzyme BamH1. Chromatin DNAs with spatial proximity were ligated with T4 DNA ligase and amplified with 3C primers that covering the IGF2 promoters (B1–B3) and the ICR (B4, B5) (Fig. 4A).
Figure 4.
Intrachromosomal interaction between the ICR and IGF2 promoters. (A) Schematic diagram of the IGF2/H19 gene locus. DMRs, differentially methylated regions; hP1–hP4, IGF2 promoters; ICR, imprinting control region; B1–B5, BamH1 sites used for 3C assay. The orientation and location of the 3C primers are shown by arrows under each BamH1 restriction site. (B–D) Quantitative intrachromosomal interactions. MOI, maintenance of IGF2 imprinting; LOI, loss of IGF2 imprinting. PCR primers B4 and B5 (ICR) were used in combination with each target primer in IGF2 promoters in (A). Each column represents the relative value of the intrachromosomal interaction. B4/B5 is local interaction used as the 3C control.
The hP1 promoter is not imprinted and contributes very little to the total IGF2 transcripts in tumors. Using 3C, we did not detect the presence of the long-range intrachromosomal interaction for the unimprinted hP1 (Fig. 4B, B1/B4 and B1/B5). In contrast, the IGF2 mRNA transcripts from the three most proximal promoters (hP2–hP4) are imprinted (36), and they form intrachromosomal interactions with the CTCF-binding sites in the ICR (30,31). We detected intrachromosomal loops between the ICR and the imprinted promoters hP2 (Fig. 4B, B2/B4 and B2/B5) and hP4 (B3/B4 and B3/B5) in normal skin fibroblasts (HBF1). In tumor cell lines with loss of IGF2 imprinting, however, these ICR promoter intrachromosomal complexes were not found (Fig. 4C and D). In a previous paper (32), we also showed that the CTCF promoter intrachromosomal loop was essentially preserved in normal tissues (adult liver, fetal liver and kidney) and in all IGF2 MOI cell lines (GM00498, HCT116, H146, ASPC), but was absent in all IGF2 LOI tumor cell lines (HT29, H522, WTCL). Taken together, it is clear that the long-range intrachromosomal loop is critical for the MOI (30,31,40).
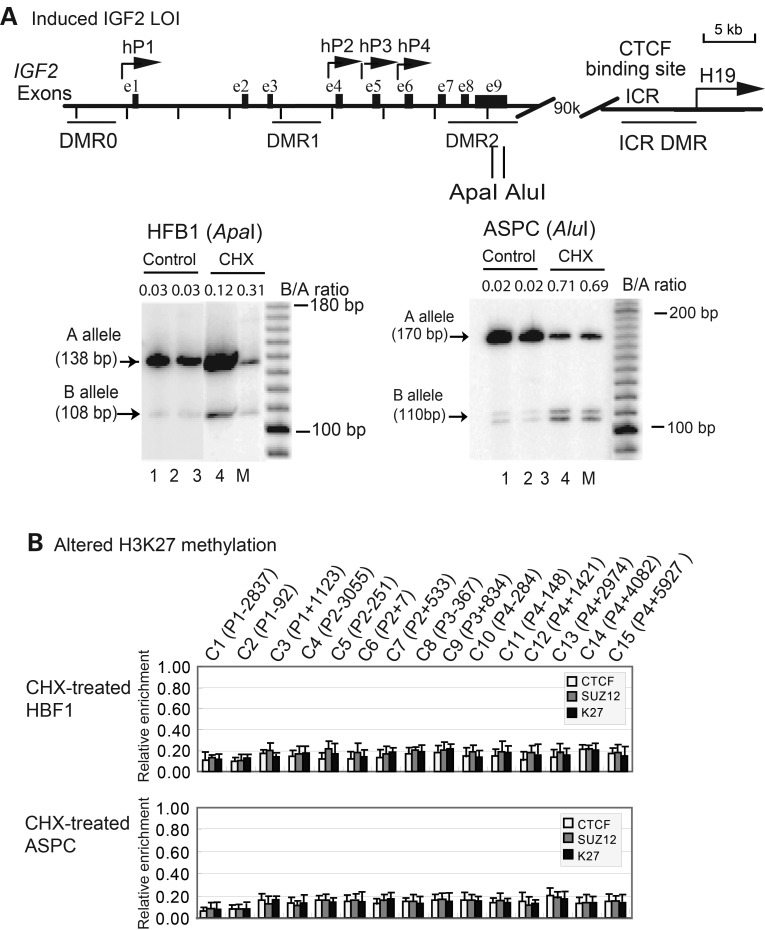
Induced IGF2 LOI by the protein synthesis inhibitor cycloheximide
To further delineate the molecular mechanisms underlying the LOI of IGF2, we inhibited protein synthesis in the MOI cell lines using cycloheximide, predicting that the reduced concentration of putative imprinting factors would alter intrachromosomal interactions, thus recapitulating the LOI as seen in the LOI tumor cell lines (25). We were particularly interested in whether the cycloheximide-treated human fibroblasts and ACPC cells also showed the loss of intrachromosomal interaction and H3K27 methylation.
Both HFB1 and ASPC expressed primarily the A allele, demonstrating a very typical pattern of the MOI (Fig. 5A, lanes 1–2). Treatment with cycloheximide induced various degrees of relaxation of the ‘B’ allele, depending on cells tested (lanes 3–4).
Figure 5.
Induced aberrant IGF2 imprinting by cycloheximide (CHX). (A) CHX-induced IGF2 LOI in human HBF1 fibroblasts and ASPC cells. Cell treated with 2.0 mg/ml CHX for 4 days. The parental alleles of IGF2 were separated by ApaI in HBF1 and AluI in ASPC cells. Lanes 1–2: HBF1 cDNA; lanes 3–4: cell treated with CHX. M, 100 bp marker. B/A ratio, quantitation of the normally imprinted ‘B’ allele as calculated by the ratio of the ‘B’ allele over the ‘A’ allele. Note the monoallelic expression of IGF2 (MOI) in untreated control cells and various levels of relaxation of the ‘B’ allele in CHX-treated cells. (B) ChIP q-PCR quantitation for altered CTCF and SUZ12 binding and H3K27 methylation. Cells were treated with CHX and immunoprecipitated with antisera against CTCF, Suz12 and dimethyl-H3-K27 (mK27), followed by q-PCR quantitation. Input DNA was DNA control before immunoprecipitation. Note the lack of CTCF and SUZ12 binding and H3K27 methylation of the IGF2 promoters in CHX-treated cells. See Figure 1A legend for details.
Using ChIP, we showed that in parallel with IGF2 LOI, the IGF2 promoters no longer were associated with SUZ12 and CTCF (Fig. 5B). As a result, H3K27 in the promoter region became unmethylated in these treated cells, indicating the importance of the imprinting factors in maintaining the intrachromosomal loop and the histone imprinting code.
Downregulation of SUZ12 in IGF2 LOI tumors
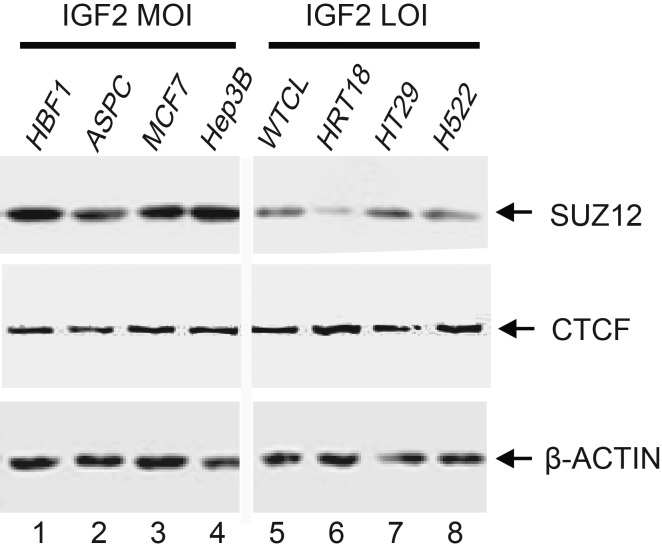
To further validate the role of chromatin factors in the MOI, we used Western blotting to compare the abundance of CTCF and SUZ12 proteins between the MOI and LOI cells. We did not detect any differences in CTCF abundance between the MOI and LOI cell lines. PRC2 docking factor SUZ12, however, was dramatically downregulated in all LOI tumor cell lines (Fig. 6B). Similarly, the abundance of SUZ12 protein was also low in IGF2 LOI cells induced by cycloheximide treatment, including HBF1, ASPC, MCF7 and Hep3B (Supplementary Material, Fig. S3). These data suggest that the downregulation of SUZ12 expression may play an important role in the loss of IGF2 imprinting in these tumors.
Figure 6.
Downregulation of SUZ12 in IGF2 LOI tumor cells. CTCF and SUZ12 proteins were quantitated by western blotting. Lanes 1–4: IGF2 MOI cells; lanes 5–8: IGF2 LOI cells. Note the reduced expression of SUZ12 in IGF2 LOI tumor cells.
DISCUSSION
In this study, we examined molecular mechanisms underlying the loss of IGF2 imprinting in several tumor cell lines. The three human IGF2 imprinted promoters contain CpG-rich sequences or CpG islands. However, unlike many other imprinted genes, where the CpG islands are often methylated in inactive promoters, the IGF2 promoter CpG islands are not differentially modified (25,30). Instead, imprinting of IGF2 is controlled by a DMR in the ICR (22,24,41).
The ICR region harbors-binding sites for CTCF, an insulator protein that demarcates active and inactive chromatin domains. CTCF binding is methylation sensitive and thus only the unmethylated maternal ICR is available for CTCF binding (13,14). The binding of CTCF to the ICR forms an insulator that prevents the IGF2 promoters from accessing endoderm-specific enhancers located downstream of the H19 gene. However, in human tumors, there is often a lack of correlation between DNA methylation and IGF2 imprinting status (11,25,34,35).
Studies using 3C demonstrate that the binding of CTCF at the ICR is required for intrachromosomal loop interactions on the maternal IGF2 allele (30,31). CTCF acts as a tethering protein, serving as a molecular glue to secure long-range intrachromosomal (28,30) and interchromosomal (42) interactions. Chromatin looping that brings the ICR and promoters into close contact and recruits PCR2, which induces the H3K27 methylation silencing (30,31). These data suggest a model whereby an intrachromosomal scaffold built with CTCF guides the imprinting signal in the remote ICR to establish the suppressive histone code in the distant IGF2 promoters.
Thus, the loss of IGF2 imprinting in human tumors could be caused by a defect in any of the steps during the formation of the CTCF–PRC2–ICR promoter intrachromosomal complex. One defect could be altered CpG DNA methylation in the ICR, where the imprinting signal resides in mouse (13,14). For example, hypermethylation at both parental ICRs could prevent the binding of CTCF and consequently the failure to form the intrachromosomal complex. A detailed analysis of DNA methylation at the IGF2 locus, however, shows that IGF2 LOI was not necessarily linked to, and may be independent of, epigenetic marks in the various DMRs, including the ICR (25). In some tumors, IGF2 LOI persists even when the ICR maintains its normally differentially methylated state. In some tumors, persistent IGF2 imprinting is accompanied by abnormal epigenetic modifications, for example, hypomethylation or hypermethylation, at CTCF-binding sites.
Data from this study, however, support the concept that aberrant biallelic expression of IGF2 in human tumors is associated with the loss of the CTCF-orchestrated intrachromosomal complex, which is required for the recruitment of the PRC2 via the co-interaction with SUZ12 (30,31). Without the formation of the intrachromosomal complex, the H3K27 methyltransferase EZH2 cannot be guided to the maternal IGF2 promoters, where it establishes the suppressive epigenotype. The H3K27 methylation-free promoters then become activated in a similar fashion as in the paternal promoters.
Through the quantitation of chromatin factors in IGF2 LOI cells either induced by cycloheximide treatment or as found in some malignancies, it appears that the downregulation of SUZ12 is key factor related to the loss of monoallelic expression of IGF2. Without SUZ12, the PRC2 cannot be recruited to the maternal IGF2 promoters, where EZH2 methylates H3K27 and induces the imprinting of the maternal allele (30).
It is also interesting to note that CTCF does not appear to be a key factor involved in loss of imprinting in tumors, as we did not detect significant difference in CTCF expression between the LOI and MOI cell lines (Fig. 6B). However, the binding of CTCF to the IGF2 promoter cannot be detected in IGF2 LOI cell lines (Fig. 3). Similarly, the intrachromosomal interaction between the ICR and the IGF2 promoter is also abolished in the LOI tumor cell lines. These data thus suggest that CTCF alone may not be able to orchestrate the intrachromosomal complex in this locus. Instead, the coexistence of SUZ12 is needed to coordinate the formation of the intrachromosomal complex in theMOI.
In support of this hypothesis, we have shown in a separate study that virally induced expression of SUZ12 is able to restore normal IGF2 imprinting in IGF2 LOI colon cancer cell lines (HT29, HRT18). Transfecting cells with a virally expressed SUZ12 cDNA, we found that SUZ12 bound to the IGF2 promoters and coordinated with CTCF to orchestrate a long-range intrachromosomal loop, leading to histone H3-K27 methylation in the IGF2 promoters and restoring monoallelic expression of IGF2 in tumor cells (Wang et al., unpublished data). It would also be interesting to explore if the downregulation of SUZ12 in tumors has global effects on other target genes and if those genes are also involved in the regulation of IGF2 allelic regulation. As SUZ12 is an essential docking factor in polycomb repressive complex 2 (PRC2), it would be of interest to learn if the downregulated SUZ12 in LOI tumors will affect its chromatin DNA binding and PRC2-mediated gene regulation at other target sites as well.
Recently, the potential role of IGF2 LOI itself has been explored as a therapeutic approach for tumor-specific gene therapy. Oncolytic adenoviruses were constructed by linking the IGF2/H19 enhancer-ICR promoter complex to diphtheria A toxin (DT-A) (43). The viruses induced tumor cell apoptosis specifically in IGF2 LOI (but not MOI) cell lines and suppressed tumor xenografts in nude mice. It will be interesting to examine if the restoration of IGF2 imprinting by viral expression of SUZ12 is able to alter tumorigenicity.
It should be noted that cycloheximide treatment is not a perfect model to induce IGF2 LOI in cells. In addition to inhibiting the synthesis of putative imprinting regulatory factors, like the chromatin factor SUZ12, cycloheximide may globally inhibit the synthesis of many factors involved in maintaining cell growth. Moreover, the degree of IGF2 LOI depends on the drug concentration and the duration of the treatment. Depending on the cell line tested, the drug may induce a partial loss of IGF2 imprinting (Fig. 5A, lane 3). More importantly, cycloheximide treatment cannot induce a permanent IGF2 LOI. Soon after the withdrawal of the drug, the cell assumes a normal IGF2 imprinting status (25). Thus, other models that specifically inhibit intrachromosomal looping are needed to further test the role of long-range interactions in IGF2 imprinting.
In summary, our data demonstrate for the first time that aberrant IGF2 imprinting in tumor cell lines is related to the loss of histone H3K27 methylation suppression mark in the gene promoter. SUZ12 is downregulated in LOI tumor cell lines. In the absence of SUZ12 binding, CTCF is unable to orchestrate a long-range intrachromosomal loop that juxtaposes the ICR close to the gene promoters, enabling the establishment of H3K27 methylation suppression through EZH2, a methyltransferase component of PRC2. In the absence of H3K27 methylation, the maternal allele becomes activated, leading to biallelic expression of IGF2.
MATERIALS AND METHODS
Cell lines and cell culture
Based on the status of IGF2 imprinting, human cell lines were divided into LOI and ‘maintenance of imprinting’ (MOI) groups (25,32). Four cell lines with differential IGF2 imprinting were selected for this study: HT29 (colorectal adenocarcinoma, IGF2 LOI), WTCL (Wilms' tumor, IGF2 LOI), HBF1 (human fibroblast, IGF2 MOI) and ASPC (pancreas adenocarcinoma, IGF2 MOI) (Supplementary Material, Table S1). Tumor cell lines HT29 and ASPC were purchased from ATCC (Rockville, MD, USA). WTCL was a kind gift from Dr Benjamin Tycko (44), and human fibroblast cells HBF1 were cultured from the skin of a human fetus (45,46).
HT29, ASPC and WTCL cells were maintained in RPMI 1640 and HBF1 in DMEM media (Invitrogen, Carlsbad, CA, USA). Both media were supplemented with 10% fetal bovine serum, and cells were cultured at 37°C, 5% CO2. Exponentially growing cells were collected by trypsin-EDTA for ChIP and PCR analyses.
Reverse transcription
Reverse transcription (RT) was performed with murine leukemia reverse transcriptase (Invitrogen) using both random hexamers and d(T)17 primers as described previously (45). To eliminate any residual genomic DNA, total RNAs (or total nucleic acids) were treated with DNase I (Takara) for 60 min (2 units/1 µg of RNA) and then extracted with phenol–chloroform before RT. Human multiple tissue panel cDNAs were purchased from CLONTECH (Palo Alto, CA, USA).
Allelic expression of IGF2
Tumor cell lines were first genotyped for heterozygosity of SNPs in IGF2 mRNA. Tumor cells with informative SNP sites were used for IGF2 imprinting studies (Supplementary Material, Table S1). IGF2 transcripts were amplified by RT–PCR (30 cycles of 95°C for 15 s and 60°C for 2 min, followed by a 5-min extension at 72°C) using primers specific for two polymorphic restriction enzymes (ApaI, AluI) in the last exon of human IGF2. PCR primers used to measure allelic expression of IGF2 included Apa1: #2505 (CTT GGA CTT TGA GTC AAA TTG GCC T) and #2506 (GAG GAG CCA GTC TGG GTT GTT GCT A); Alu1: #2949 (GTC CCC TCC TCT GCC ATC ACC TGA) and #2950 (GGA TTT TGC CGG AAA TAT TAG CGT). The amplified products were further labeled by primer extension using 32P end-labeled primer. The primer-extended products were digested with AluI and ApaI to distinguish two parental alleles and were separated on a 5% polyacrylamide–urea gel. The digested allele was quantitated as the relative value based on PhosphorImager scanning density (47).
ChIP
ChIP assays were performed with a ChIP assay kit ( Upstate Biotechnology, NY, USA) by following the protocol provided by the manufacturer with slight modifications as previously described (31). Briefly, 5 million cells were fixed with 1% formaldehyde and then sonicated for 180 s (10 s on and 10 s off) on ice with a Branson sonicator with a 2-mm microtip at 40% output control and 90% duty cycle settings. The sonicated chromatin (0.9 ml) was collected by centrifugation, aliquoted and snap-frozen in liquid nitrogen. To perform ChIP, sonicated chromatin (150 µl) was diluted 10-fold and purified with specific antiserum (2– 5 µl) and protein G-agarose (60 µl). Antibodies to CTCF, SUZ12 and dimethyl-H3-K27 (lysine 27 of histone H3) were obtained from Upstate Biotechnology. To reduce the ChIP background, we modified the manufacturer's protocol by additional two washing steps following immunoprecipitation. As previously reported (31), anti-IgG was used as the ChIP control in parallel with testing samples.
DNAs were released from the bound chromatins after cross-linking reversal and proteinase K treatment, and were precipitated and diluted in 100 µl of low-TE buffer (1 mm Tris, 0.1 mm EDTA). PCRs (3 µl under liquid wax) contained 1 µl ChIP (or input) DNA, 0.5 mm appropriate primer pairs, and 0.2 U Klen-Taq I (Ab Peptides, MO, USA). Standard PCR conditions were 95°C for 60 s, followed by 35 cycles of 95°C for 15 s, 65°C for 30 s of annealing, and 72°C for 1 min of extension. All primer sets were tested for the absence of primer–dimer products. To avoid heteroduplex formation that may interfere with restriction enzyme digestion, one of each set of primers was end-labeled with [γ-32P]ATP. The γ-32P-labeled primer was added to the PCR mixture (1 µl) at the last cycle of amplification. PCR products were separated on a 5% polyacrylamide–urea gel and quantified by a PhosphorImager scanner (Molecular Dynamics, Sunnyvale, CA, USA). For comparison, the ChIP data were presented as relative values by normalizing to PCR signals of input DNA (i.e. ratio of the ChIP over the input).
ChIP signals were relatively low in IGF2 LOI cells. Thus, a relatively high PCR cycle was used to amplify the ChIP DNA. After PCR, the DNA products were digested with 1 U of the appropriate polymorphic restriction enzymes in a total volume of 6 µl for 3 h. The digested products were separated on a 5% polyacrylamide–urea gel and quantified by a PhosphorImager scanner (Molecular Dynamics). The data were presented as the relative allele enrichment: the A or B allele/(A + B alleles) × 100%. ChIP was repeated in triplicate independently for each sample.
Allelic ChIP assay
Duplicate PCR reactions (3 µl under liquid wax) contained 1 µl ChIP (or input) DNA, 0.1 mm appropriate primer pairs (Supplementary Material, Tables S1 and S2), 50 mm deoxynucleotide triphosphate, and 0.2 U KlenTaq I (Ab Peptides, St. Louis, MO, USA). Standard PCR conditions were 95°C for 60 s, followed by 30 cycles of 95°C for 10 s and 65°C annealing (and extension) temperature for 90 s and finally 72°C for 10 min. All primer sets were tested for the absence of primer–dimer products. End-labeled PCR primers are listed in Supplementary Material, Table S3. The [γ-32P]-labeled primer was added in 1 µl PCR mixture at the last cycle of amplification. PCR products were digested with appropriate enzymes (New England Biolabs, MA, USA; 1 unit) listed in Supplementary Material, Table S3 in a total volume of 6 ml for 6–12 h under liquid wax. The digested products were separated on a 5% polyacrylamide–urea gel and quantified by a PhosphoImager (Molecular Dynamics). The relative enrichment of CTCF and SUZ12 at each specific site was determined as described previously (34). The allelic levels of modified histones in one parental allele (percentage of both alleles) at each specific site were calibrated with those from input DNA (DNA before ChIP).
Chromatin conformation capture assay
The 3C assay was performed as previously described (30,39). Briefly, MOI (HBF1) and LOI cells (WTCL and HT29) were cross-linked with 2% formaldehyde and lysed with cell lysis buffer (10 mm Tris, pH 8.0, 10 mm NaCl, 0.2% NP-40, protease inhibitors). Nuclei were collected, suspended in 1× restriction enzyme buffer in the presence of 0.3% sodium dodecyl sulfate (SDS). An aliquot of nuclei (2 × 106) was digested with 800 U BamH1 at 37°C overnight and ligated with 4000 U T4 DNA ligase at 16°C for 4 h. After the treatment with 10 mg/ml proteinase K at 65°C overnight to reverse cross-links and with 0.4 µg/ml RNase A for 30 min at 37°C, DNA was extracted with phenol–chloroform, ethanol precipitated, and used for PCR amplification for the ligated DNA products. PCR primers used in this study were previously described (32).
PCR polymorphism analysis
After genomic mapping, a total of eight available polymorphisms were used to cover the DNA sequence four IGF2 promoter and exon regions in a fibroblast cell (HFB1) and three tumor cell lines: ApaI (exon 4), AluI (exon 4), FokI (promoter hP1), DdeI (promoter hP2), DraI (promoter hP4), KpnI (promoter hP4), Hpy188III (promoter hP4) and BsaJI (promoter hP4). PCR reaction, restriction enzyme digestion, electrophoresis and quantitation analysis were performed using established methodology as described previously (30,48). Briefly, PCR amplification for polymorphism determination was performed in 96-well microtiter plates, each 3 ml reaction containing 10–20 ng of DNA for genotyping or cDNA for examining allelic expression, 50 nm dNTP, 0.2 mm corresponding primers, 0.1 µCi of [α-32P]dCTP, 0.125 U of Taq DNA polymerase with a hot start PCR. The PCR reaction solution was heated to 98°C for 2 min, amplified for 30–35 cycles at 95°C for 30 s and 65°C for 90 s, and followed by a 5-min extension at 72°C. PCR products were then digested with the polymorphic restriction enzymes (1 unit) in a 10 µl volume at 37°C for 4 h. Each digested product was detected or quantitated by PhosphorImager 445SI scanner (Molecular Dynamics) after electrophoresis in a 5% polyacrylamide urea gel.
Cell treatment with cycloheximide
Human fibroblasts cells HBF1 and pancreatic cell line ASPC were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 100 U/ml of penicillin and 100 mg/ml of streptomycin, and grown at 371°C with 5% CO2. ASPC, a pancreatic cancer cell line keeping normal IGF2 imprinting in cell culture, was maintained in RPMI 1640 medium as recommended by ATCC. Cells were seeded in six-well plates at the density of 2–105 cells/well. Twenty-four hours following plating, cells were replaced with fresh medium and were treated with the concentrations 2.0 mg/ml of cycloheximide as previously described (25). Tumor cells were collected after Day 4 and analyzed for IGF2 imprinting and ChIP assay.
Western blotting
Expression of CTCF and SUZ12 proteins were determined by western blotting as previously described (30). Cells were lysed with boiling 1% SDS, 10 mm Tris–HCI, pH 7.4, sonicated for 30 s and centrifuged at 15 000 g for 5 min. Supernatant lysates with equal amounts of protein were used for immunoblotting of CTCF protein. The proteins were examined by SDS–PAGE and western immunoblotting with the anti-CTCF and anti-Suz12 antibodies (1:1000; Upstate Biotechnology) and the ECL detection system (Amersham) by following the instructions of the manufacturer.
Statistical analysis
All experiments were performed in triplicate, and the data are expressed as mean ± SD. The comparative CT method was applied in the quantitative real-time RT–PCR assay according to the ΔΔCT method (25,30). The data were analyzed with one-way analysis of variance, and results were considered statistically significant at P ≤ 0.05.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by NIH Grant (1R43 CA103553-01), Department of Defense Grant (W81XWH-04-1-0597), California Institute of Regenerative Medicine (CIRM) Grant (RT2-01942), Jilin International Collaboration Grant (#20120720), NSFC Grant (#81272294) to J.F.H.; and NIH Grant (GM09031) and a Merit Review from the Medical Research Service of the Department of Veterans Affairs to A.R.H.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr Benjamin Tycko for providing the WTCL cell line.
REFERENCES
- 1.Ogawa O., Eccles M.R., Szeto J., McNoe L.A., Yun K., Maw M.A., Smith P.J., Reeve A.E. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature. 1993;362:749–751. doi: 10.1038/362749a0. doi:10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 2.Rainier S., Johnson L.A., Dobry C.J., Ping A.J., Grundy P.E., Feinberg A.P. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. doi:10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg A.P. Genomic imprinting and gene activation in cancer. Nat. Genet. 1993;4:110–113. doi: 10.1038/ng0693-110. doi:10.1038/ng0693-110. [DOI] [PubMed] [Google Scholar]
- 4.Randhawa G.S., Cui H., Barletta J.A., Strichman-Almashanu L.Z., Talpaz M., Kantarjian H., Deisseroth A.B., Champlin R.C., Feinberg A.P. Loss of imprinting in disease progression in chronic myelogenous leukemia. Blood. 1998;91:3144–3147. [PubMed] [Google Scholar]
- 5.Zhang L., Zhou W., Velculescu V.E., Kern S.E., Hruban R.H., Hamilton S.R., Vogelstein B., Kinzler K.W. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. doi:10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 6.Sohda T., Iwata K., Soejima H., Kamimura S., Shijo H., Yun K. In situ detection of insulin-like growth factor II (IGF2) and H19 gene expression in hepatocellular carcinoma. J. Hum. Genet. 1998;43:49–53. doi: 10.1007/s100380050036. doi:10.1007/s100380050036. [DOI] [PubMed] [Google Scholar]
- 7.Takeda S., Kondo M., Kumada T., Koshikawa T., Ueda R., Nishio M., Osada H., Suzuki H., Nagatake M., Washimi O., et al. Allelic-expression imbalance of the insulin-like growth factor 2 gene in hepatocellular carcinoma and underlying disease. Oncogene. 1996;12:1589–1592. [PubMed] [Google Scholar]
- 8.Ulaner G.A., Vu T.H., Li T., Hu J.F., Yao X.M., Yang Y., Gorlick R., Meyers P., Healey J., Ladanyi M., et al. Loss of imprinting of Igf2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 2003;12:535–549. doi: 10.1093/hmg/ddg034. doi:10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann W.K., Takeuchi S., Frantzen M.A., Hoelzer D., Koeffler H.P. Loss of genomic imprinting of insulin-like growth factor 2 is strongly associated with cellular proliferation in normal hematopoietic cells. Exp. Hematol. 2002;30:318–323. doi: 10.1016/s0301-472x(01)00797-4. doi:10.1016/S0301-472X(01)00797-4. [DOI] [PubMed] [Google Scholar]
- 10.Soroceanu L., Kharbanda S., Chen R., Soriano R.H., Aldape K., Misra A., Zha J., Forrest W.F., Nigro J.M., Modrusan Z., et al. Identification of IGF2 signaling through phosphoinositide-3-kinase regulatory subunit 3 as a growth-promoting axis in glioblastoma. Proc. Natl Acad. Sci. USA. 2007;104:3466–3471. doi: 10.1073/pnas.0611271104. doi:10.1073/pnas.0611271104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui H., Onyango P., Brandenburg S., Wu Y., Hsieh C.L., Feinberg A.P. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 12.Bartolomei M.S., Webber A.L., Brunkow M.E., Tilghman S.M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. doi:10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 13.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. doi:10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 14.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF Mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. doi:10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 15.Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. doi:10.1016/S0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 16.Arney K.L. H19 and Igf2 – enhancing the confusion? Trends Genet. 2003;19:17–23. doi: 10.1016/s0168-9525(02)00004-5. doi:10.1016/S0168-9525(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 17.Reik W., Constancia M., Dean W., Davies K., Bowden L., Murrell A., Feil R., Walter J., Kelsey G. Igf2 imprinting in development and disease. Int. J. Dev. Biol. 2000;44:145–150. [PubMed] [Google Scholar]
- 18.Engel N., Bartolomei M.S. Mechanisms of insulator function in gene regulation and genomic imprinting. Int. Rev. Cytol. 2003;232:89–127. doi: 10.1016/s0074-7696(03)32003-0. [DOI] [PubMed] [Google Scholar]
- 19.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. doi:10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 20.Mann J.R., Szabo P.E., Reed M.R., Singer-Sam J. Methylated DNA sequences in genomic imprinting. Crit. Rev. Eukaryot. Gene Expr. 2000;10:241–257. doi: 10.1615/critreveukargeneexpr.v10.i3-4.30. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki H., Ishihara K., Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000;127:711–715. doi: 10.1093/oxfordjournals.jbchem.a022661. doi:10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 22.Thorvaldsen J.L., Duran K.L., Bartolomei M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. doi:10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenherr C.J., Levorse J.M., Tilghman S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. doi:10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 24.Szabo P.E., Tang S.H., Silva F.J., Tsark W.M., Mann J.R. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell. Biol. 2004;24:4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. doi:10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H.L., Li T., Qiu X.W., Wu J., Ling J.Q., Sun Z.H., Wang W., Chen W., Hou A., Vu T.H., et al. Correction of aberrant imprinting of IGF2 in human tumors by nuclear transfer-induced epigenetic reprogramming. EMBO J. 2006;25:5329–5338. doi: 10.1038/sj.emboj.7601399. doi:10.1038/sj.emboj.7601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui H., Horon I.L., Ohlsson R., Hamilton S.R., Feinberg A.P. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat. Med. 1998;4:1276–1280. doi: 10.1038/3260. doi:10.1038/3260. [DOI] [PubMed] [Google Scholar]
- 27.Cui H., Niemitz E.L., Ravenel J.D., Onyango P., Brandenburg S.A., Lobanenkov V.V., Feinberg A.P. Loss of imprinting of insulin-like growth factor-II in Wilms’ tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 2001;61:4947–4950. [PubMed] [Google Scholar]
- 28.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. CTCF Binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. doi:10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon Y.S., Jeong S., Rong Q., Park K.Y., Chung J.H., Pfeifer K. Analysis of the H19ICR insulator. Mol. Cell. Biol. 2007;27:3499–34510. doi: 10.1128/MCB.02170-06. doi:10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Hu J.F., Qiu X., Ling J., Chen H., Wang S., Hou A., Vu T.H., Hoffman A.R. CTCF Regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex-2 intrachromosomal loop. Mol. Cell. Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. doi:10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Niu B., Hu J.F., Wang H., Ling J., Qian G., Ge S., Hoffman A.R. Interruption of intrachromosomal looping by CTCF decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J. Cell Biol. 2011;193:475–487. doi: 10.1083/jcb.201101021. doi:10.1083/jcb.201101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vu T.H., Nguyen A.H., Hoffman A.R. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum. Mol. Genet. 2010;19:901–919. doi: 10.1093/hmg/ddp558. doi:10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X., Vu T.H., Lu Q., Ling J.Q., Li T., Hou A., Wang S.K., Chen H.L., Hu J.F., Hoffman A.R. A complex deoxyribonucleic acid looping configuration associated with the silencing of the maternal igf2 allele. Mol. Endocrinol. 2008;22:1476–1488. doi: 10.1210/me.2007-0474. doi:10.1210/me.2007-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore T., Constancia M., Zubair M., Bailleul B., Feil R., Sasaki H., Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl Acad. Sci. USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. doi:10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan M.J., Taniguchi T., Jhee A., Kerr N., Reeve A.E. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene. 1999;18:7527–7534. doi: 10.1038/sj.onc.1203096. doi:10.1038/sj.onc.1203096. [DOI] [PubMed] [Google Scholar]
- 36.Vu T.H., Hoffman A.R. Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature. 1994;371:714–717. doi: 10.1038/371714a0. doi:10.1038/371714a0. [DOI] [PubMed] [Google Scholar]
- 37.Engel N., Raval A.K., Thorvaldsen J.L., Bartolomei S.M. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum. Mol. Genet. 2008;17:3021–3029. doi: 10.1093/hmg/ddn200. doi:10.1093/hmg/ddn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. doi:10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. doi:10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 40.Vu T.H., Nguyen A.H., Hoffman A.R. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum. Mol. Genet. 2009;19:901–919. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava M., Hsieh S., Grinberg A., Williams-Simons L., Huang S.P., Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 42.Ling J.Q., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. doi:10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 43.Pan Y., He B., Li T., Zhu C., Zhang L., Wang B., Xu Y., Qu L., Hoffman A.R., Wang S., et al. Targeted tumor gene therapy based on loss of IGF2 imprinting. Cancer Biol. Ther. 2010;10:290–298. doi: 10.4161/cbt.10.3.12442. doi:10.4161/cbt.10.3.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Keefe D., Dao D., Zhao L., Sanderson R., Warburton D., Weiss L., Anyane-Yeboa K., Tycko B. Coding mutations in p57KIP2 are present in some cases of Beckwith–Wiedemann syndrome but are rare or absent in Wilms tumors. Am. J. Hum. Genet. 1997;61:295–303. doi: 10.1086/514854. doi:10.1086/514854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J.F., Vu T.H., Hoffman A.R. Promoter-specific modulation of insulin-like growth factor II genomic imprinting by inhibitors of DNA methylation. J. Biol. Chem. 1996;271:18253–18262. doi: 10.1074/jbc.271.30.18253. doi:10.1074/jbc.271.30.18253. [DOI] [PubMed] [Google Scholar]
- 46.Hu J.F., Oruganti H., Vu T.H., Hoffman A.R. The role of histone acetylation in the allelic expression of the imprinted human insulin-like growth factor II gene. Biochem. Biophys. Res. Commun. 1998;251:403–408. doi: 10.1006/bbrc.1998.9401. doi:10.1006/bbrc.1998.9401. [DOI] [PubMed] [Google Scholar]
- 47.Hu J.F., Vu T.H., Hoffman A.R. Genomic deletion of an imprint maintenance element abolishes imprinting of both insulin-like growth factor II and H19. J. Biol. Chem. 1997;272:20715–20720. doi: 10.1074/jbc.272.33.20715. doi:10.1074/jbc.272.33.20715. [DOI] [PubMed] [Google Scholar]
- 48.Hu J.F., Oruganti H., Vu T.H., Hoffman A.R. Tissue-specific imprinting of the mouse insulin-like growth factor II receptor gene correlates with differential allele-specific DNA methylation. Mol. Endocrinol. 1998;12:220–232. doi: 10.1210/mend.12.2.0062. doi:10.1210/me.12.2.220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.