Abstract
Loss of cholesterol homeostasis and altered vesicle trafficking have been detected in Huntington's disease (HD) cellular and animal models, yet the role of these dysfunctions in pathophysiology of HD is unknown. We demonstrate here that defects in caveolar-related cholesterol trafficking directly contribute to the mechanism of HD in vivo. We generated new mouse models that express mutant Huntington's protein (mhtt), but have partial or total loss of caveolin-1 (Cav1) expression. Fluorescence resonance energy transfer dequenching confirms a direct interaction between mhtt and Cav1. Mhtt-expressing neurons exhibited cholesterol accumulation and suppressed caveolar-related post-Golgi trafficking from endoplasmic reticulum/Golgi to plasma membrane. Loss or reduction of Cav1 expression in a knock-in HD mouse model rescues the cholesterol phenotype in neurons and significantly delays the onset of motor decline and development of neuronal inclusions. We propose that aberrant interaction between Cav1 and mhtt leads to altered cholesterol homeostasis and plays a direct causative role in the onset of HD pathophysiology in vivo.
INTRODUCTION
Huntington's disease (HD) gene was identified more than a decade ago (1). The mutation, a CAG repeat expansion, lies at the 5′ end of the gene and codes for a long and toxic tract of polyglutamines in the N-terminus of the huntingtin (htt) protein (1). Knowledge of the genetic defect, however, has not led directly to the understanding of the underlying mechanisms of toxicity. The major barrier is the functional complexity. The normal htt is a large and multifunctional protein, and mutation results in aberrant protein–protein interactions that affect diverse functions including vesicle trafficking (2), lipid metabolism (3), mitochondrial function (4), autophagy (5,6), transcription (7) and lysosomal biogenesis (8–10). Whether all of these defects have equivalent impacts on toxicity is unknown.
Accumulating evidence, however, suggests that altered lipid homeostasis is a nexus for dysfunction and that the events at endoplasmic reticulum (ER)/Golgi subcellular compartment are relevant to HD pathophysiology. The ER/Golgi compartment is a hub for vesicle/lysosome biogenesis, the center for trafficking of membrane-encapsulated vesicles/organelles, and the site of cholesterol biosynthesis. Indeed, alterations in cholesterol homeostasis in HD models have been reported both at the trafficking (11) and biosynthetic levels (12–15). Since cholesterol levels are tightly regulated in the cell, ER sensing of high levels of cholesterol suppresses its biosynthesis (16). Thus, cholesterol biosynthesis in the ER and its traffic away from the ER are likely to be linked. In that regard, one of the key modulators of cholesterol trafficking is caveolin-1 (Cav1). Cav1 is a cholesterol-binding protein and the major structural component of caveolae (17,18). Lipid rafts in both the plasma and Golgi membranes contain Cav1, which is responsible for cholesterol transport in and out of the cell (17,18). Consistent with those functions, caveolins are located in cis- and trans-Golgi complex, the ER/Golgi network, and in cytosolic lipid bodies (18).
We have previously demonstrated that mhtt inhibits lipid internalization through a Cav1-related pathway, and causes significant accumulation of cholesterol in striatal neurons from HD72 YAC mouse model (11). In neurons from these animals, knockdown of Cav1 using siRNA restores lipid endocytosis and prevents cholesterol accumulation (11). Our findings support the hypothesis that Cav1 in mhtt-expressing neurons promotes the cholesterol dysfunction and loss of homeostasis. However, the mechanisms by which Cav1 might be involved, or whether Cav1-related lipid transport is relevant for HD toxicity in vivo is unknown.
To address these issues, we have created new mouse models in which mhtt is expressed at endogenous levels (19), but expression of Cav1 is reduced or eliminated. HdhQ150/Cav1(+/−) and HdhQ150/Cav1(−/−) mice were derived from the original HdhQ150 line that is characterized by early onset of motor decline (19). We find that a deleterious interaction between mhtt and Cav1 causes post-Golgi trafficking defects, accumulation of cholesterol, and motor decline. Remarkably, loss or reduction of Cav1 is sufficient to rescue these phenotypes. We find that a direct interaction of Cav1 and mhtt confers a novel ‘gain of function’, that promotes toxicity in vivo. The results provide direct evidence that trafficking defects in mhtt-expressing cells impart deleterious effects by perturbing lipid homeostasis.
RESULTS
Cav1 and mhtt directly interact in cells
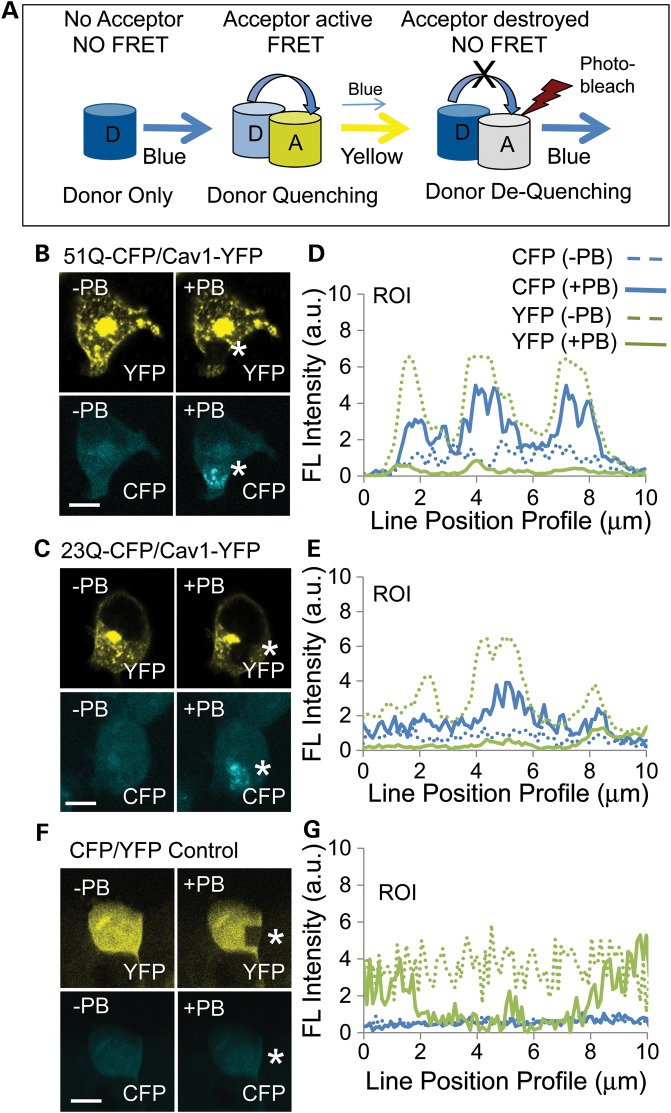
We have previously reported that Cav1 interacts with htt and mhtt in primary neurons isolated from HD72 YAC animals based on the co-immunoprecipitation (IP) and glutathione S-transferase (GST)-pull-down experiments in vitro (11). Here, we used fluorescence resonance energy transfer (FRET) to test whether there is a direct interaction between mhtt and Cav1 in cells. In FRET, energy transfer depends on the distance between two fluorophores (efficiency = 1/(1 + (r/R0)6) (20). Thus, a FRET signal occurs only when the donor and acceptor fluorophores are in close physical proximity (5–10 nm) and mutually oriented (Fig. 1A). We created Cav1 and htt/mhtt FRET pairs by fusing the C-terminus of each protein with either the yellow fluorescent protein (YFP) or with a cerulean fluorescent protein (CFP). Full-length Cav1 (amino acid 1–178)–YFP served as the acceptor, and full-length htt or mhtt (amino acid 1–3144)–CFP served as the donor. Htt–CFP comprised 26 CAG repeats, while mhtt–CFP comprised 51 CAG repeats. Vectors harboring the donor and acceptor pairs were cotransfected into human embryonic kidney cells (HEK293 T) and tested for FRET using a dequenching assay (Fig. 1A).
Figure 1.
Htt and mhtt directly interact with Cav1 in cells. (A) Schematic of a FRET dequenching experiment. The CFP donor alone emits blue in the absence of an acceptor. FRET occurs when emission of the donor excites the acceptor. Under these conditions, the CFP signal decreases (quenching) and the YFP acceptor signal increases. If the YFP acceptor is photobleached, then there is no FRET and the intensity of the CFP donor emission increases (dequenching). Blue cylinder is the CFP donor and the yellow cylinder is the YFP acceptor; the white cylinder indicates photodestruction of the acceptor and loss of its signal intensity after repeated laser scanning at 514 nm (red lightening bolt). (B and C) Images of the mhtt 51Q–CFP (B), or htt 23Q–CFP (C), or Cav1–YFP expression before (−PB) and after (+PB) photobleaching. HEK293 T cells were transfected with plasmids to coexpress fusion proteins Cav1–YFP (acceptor) and the 51Q–CFP (B) or the 23Q–YFP donors (C). FRET is measured by the increase in the 51Q–CFP (B) or 26Q–CFP (C) donor signal (blue) after photobleaching (+PB) of Cav1–YFP. The white star indicates the site of the photobleaching, which appears as a black hole in the yellow signal or an elevated blue signal. (E and F) Quantification of the results from (B and C). Donor dequenching was determined by line pixel analysis of images (pixels in the ROI highlighted by white stars in YFP panels). The solid blue line is CFP after photobleaching (+PB); the dashed blue line is 51Q–CFP (E) or 26Q–CTF (F) before photobleaching (−PB), as indicated; the solid green line is Cav1–YFP after photobleaching (+PB); the dashed green line is Cav1–YFP before photobleaching (−PB). (D) As a negative control, plasmids encoding only CFP and YFP were cotransfected into HEK293 T cells. The signal of CFP did not increase upon photobleaching of YFP, indicating no specific interaction between the two fluorophores. (G) Quantification of the results from (F). Parameters are the same as in (E) and (F). Scale bar is 10 µm in all image panels.
Typically, energy transfer from an excited donor to an acceptor results in a decrease of the donor signal (Fig. 1A, donor quenching) and an increase in the acceptor signal (Fig. 1A, FRET). In a dequenching assay, photobleaching (+PB) destroys the acceptor and blocks FRET, leading to a rise (dequenching) in the donor intensity (Fig. 1A, NO FRET). Indeed, repeated scanning of the transfected cells at 514 nm photobleached the Cav1–YFP acceptor, and led to a remarkable increase in blue signal intensity of the mhtt–CFP (Fig. 1B and E) or htt–CFP (Fig. 1C and F) donor within the irradiated region of interest (ROI) (Fig. 1B and C, white star). More than 90% of cells observed to coexpress both Cav1–YFP and htt–CPF (or mhtt–CFP) displayed FRET, implying that Cav1 directly interacted with both htt and mhtt in cells, consistent with our previously reported (IP) and GST-pull-down experiments in vitro (11). As a negative control, plasmids expressing only CFP and YFP were cotransfected into HEK293 T cells (Fig. 1D and G). After photobleaching (+PB), the YFP signal in the negative control sample decreased, but there was no change in CFP signal, indicating a random interaction. Collectively, these results provided evidence that Cav1 resided within 5–10 nm of mhtt, consistent with a physical interaction in cells.
Progeny of Cav1(+/−) mice crossed to HdhQ150(+/−) miceare viable and healthy
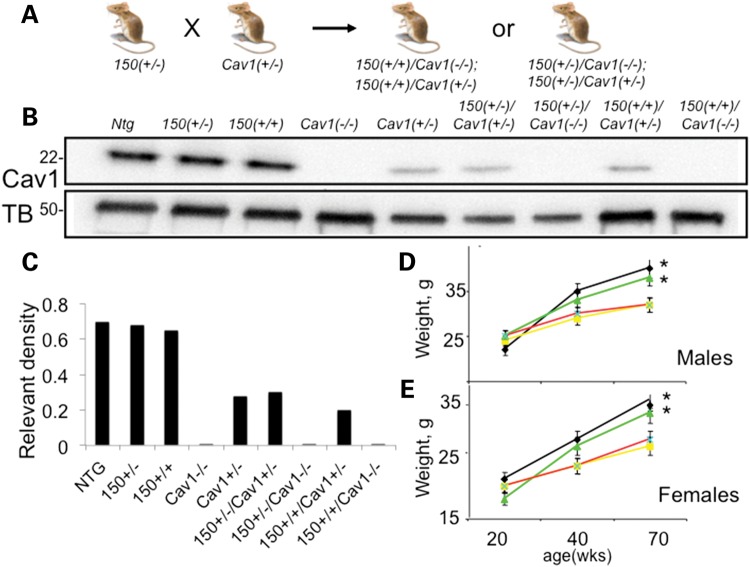
We tested whether the interaction of mhtt and Cav1 had consequences in vivo. We crossed Cav1 (+/−) and HdhQ150(+/−) animals to generate mice that expressed mhtt but had a reduction or entirely lacked expression of Cav1 (Fig. 2). We generated nine genotypes of mice with every combination of htt, mhtt and Cav1 alleles (Fig. 2B). The expression of htt, mhtt and Cav1 in these animals matched the expected genotypes (Fig. 2B and C). In each mouse line, Cav1 was expressed in the cytoplasm and at the plasma membrane of primary neurons (Supplementary Material, Fig. S1), and migrated on a SDS–PAGE gel as a 22-kDa protein that was indistinguishable from Cav1 characterized in other cell types (Fig. 2B). Loss or reduction of Cav1 in HD mice had no effect on the survival or fertility of these animals relative to the controls. We did not observe age-dependent differences in levels of expression of mhtt (Supplementary Material, Fig. S2A) or Cav1 (Supplementary Material, Fig. S2B) in HdhQ150(+/−) or HdhQ150(+/+) animals between E17 and 60 weeks (Supplementary Material, Fig. S2). Thus, these animals expressed a relatively constant ratio of htt/mhtt and Cav1 throughout their life, and were suitable models for testing the effect of an mhtt–Cav1 interaction in vivo. In the following results, the most useful lines will be discussed: C57BL6 control, also referred to Cnt (black), Cav1(−/−) (light gray), HdhQ150(+/+) (striped), HdhQ150(+/−) (dark gray), HdhQ150(+/+)/Cav1 (−/−) (dark purple), HdhQ150(+/+)/Cav1(+/−) (light purple), HdhQ150(+/−)/Cav1 (−/−) (dark cyan) and HDHQ150(+/−)/Cav1(+/−) (blue). The Cav1(+/−) animals had similar properties to the Cav1 (−/−) and other control animals.
Figure 2.
Characterization of novel mouse models generated by crossing HdhQ150(+/−) and Cav1(+/−) mice. 150 denotes HdhQ150. (A) Schematic diagram of breeding strategy. (B) Antibody detection of Cav1 expression (22 kDa) in the striatal tissue from 20-week mice in each animal line. Ntg – control non-transgenic littermates. The proteins were resolved on an SDS–PAGE gel with molecular weight markers shown. Tubulin (TB) (55 kDa) was used as a loading control. (C) Quantification of the Cav1 expression relative to the expression of tubulin from western blot presented in (B) using densitometry. (D and E) Weight as a function of age in male and female animals: 150(+/−) (yellow); 150(+/+) (red); 150(+/−)/Cav1 (−/−) (green); 150(+/+)/Cav1 (−/−) (black). Error bars are standard deviation. *, statistical significance was achieved in the separation between 150(+/−) and 150(+/−)/Cav1 (−/−) (P < 0.05); and with 150(+/+) and 150(+/+)/Cav1 (−/−) (P < 0.05) in both male and female groups at 70 weeks of age.
Loss or reduction of Cav1 suppresses age-dependent weight loss and accumulation of cholesterol observed in the HdhQ150(+/−) or HdhQ150(+/+) animals
Weight loss is a feature of human HD pathology. Although well groomed and indistinguishable by visual appearance, the HdhQ150(+/+) and HdhQ150(+/−) animals, on the average, gained less weight with age and were roughly 15% lighter relative to their control littermates by 70 weeks (Fig. 2D and E). However, loss of Cav1 in the HdhQ150(+/−)/Cav1 (−/−) or HdhQ150(+/+)/Cav1 (−/−) crosses led to an increase in body weight, which was equivalent to Cav1 (−/−) controls or non-transgenic littermates (Ntg) (Fig. 2D and E). Thus, loss or reduction of Cav1 had a beneficial effect on growth in mhtt-expressing animals.
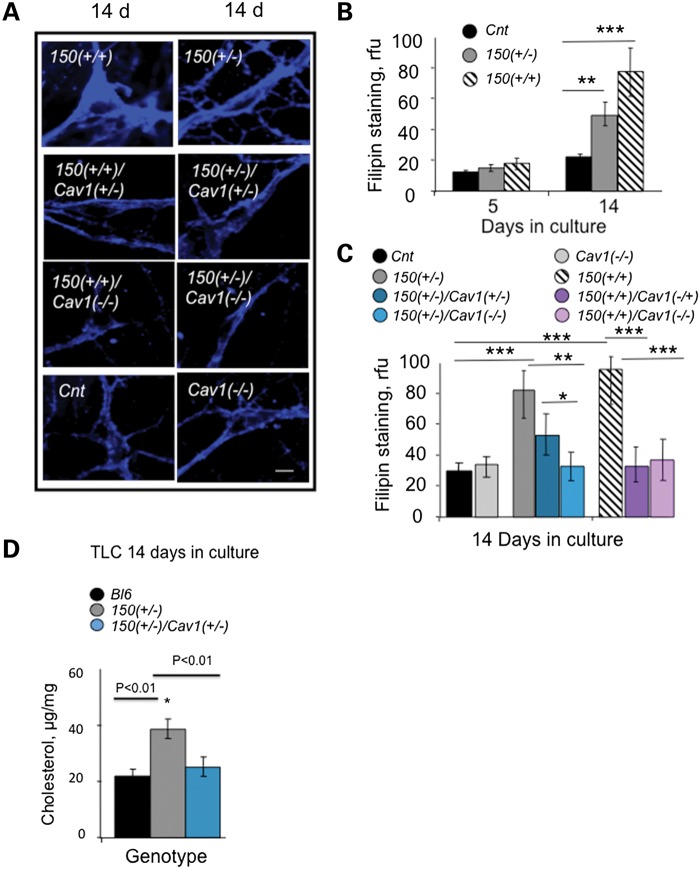
Cav1 is responsible for cholesterol transport in the cell. Since weight loss was Cav1-dependent, we tested whether it was accompanied by any changes in caveolar-related cholesterol trafficking. The cholesterol level was measured by thin-layer chromatography (TLC), mass spectrometry, or staining with filipin (a fluorescent polyene antibiotic that directly binds free cholesterol (21)). Since cholesterol binds to Cav1, a trafficking defect was measured directly by a block in caveolar lipid transport, and visualized by the accompanying alterations in cholesterol distribution. The trafficking defect was obvious. Consistent with our previous findings (11), trafficking of BODIPY-lactosylceramide (LacCer) (a fluorescent lipid marker for caveolar trafficking) was inhibited in primary neurons from HdhQ150(+/−) or HdhQ150(+/+) animals relative to controls (Supplementary Material, Fig. S3). Moreover, the caveolar trafficking block altered the cholesterol distribution (Fig. 3A and B). Five days after plating, cholesterol locally accumulated in neurons from HdhQ150(+/−) or HdhQ150(+/+) animals, primarily in the ER/Golgi region (Supplementary Material, Fig. S4A), which is enriched in Cav1 (18). By 14 days in culture, the caveolar trafficking block resulted in widespread cholesterol relative to Cnt (Fig. 3A and B). Early in development, neurons are still synthesizing cholesterol (22,23). Thus, blocking trafficking in E17 neurons from HdhQ150(+/−) or HdhQ150(+/+) animals resulted in elevation of cholesterol relative to controls, as quantified by both filipin staining (Fig. 3B; Supplementary Material, Fig. S4B) and TLC (Fig. 3D). In mammals, cholesterol synthesis is suppressed if the cellular cholesterol level is high (16,24). Thus, we expected that cholesterol levels would decline in adult animals if the trafficking defect and accumulation of cholesterol persisted. Indeed, as measured by mass spectrometry, the levels of cholesterol, zymosterol, stigmasterol and lathosterol progressively declined by 15–40% in the brains of HdhQ150(+/+) animals relative to controls by 80 weeks (Table 1; Supplementary Material, Fig. S5).
Figure 3.
Loss of Cav1 reduces cholesterol accumulation in striatal neurons from HdhQ150(+/+) and HdhQ150(+/−) mice. 150 denotes HdhQ150. Cnt denotes C57BL6. (A) Fluorescence images of filipin staining in primary striatal neurons from control (Cnt); 150(+/+); 150(+/−); 150 (+/+)/Cav1(+/−); 150 (+/+)/Cav1 (−/−); 150 (+/−)/Cav1(+/−) and 150(+/−)/Cav1 (−/−) animals in culture. Neurons were plated side-by-side, cultured for 3 days in serum-containing medium and switched to cholesterol- and serum-free medium. The cells were fixed at 5 or 14 days, and stained with filipin. Cholesterol accumulation at 14 days in culture is visualized by blue staining. Images were acquired using LSM 510 confocal microscope with ×100 (1.4 NA) oil DIC lens. Scale bar is 10 µm. (B) Quantification of filipin fluorescence staining intensity in neurons from Cnt (black), 150(+/−) (gray) or 150 (+/+) (striped) mice at 5 and 14 days in culture. (C) Quantification of filipin staining in striatal neurons from Cnt (black); 150(+/−) (gray); 150(+/+) (striped); Cav1 (−/−) (light gray); 150(+/−)/Cav1(+/−) (dark teal); 150 (+/)/Cav1 (−/−) (light teal); 150(+/+)/Cav1(+/−) (dark purple); 150(+/+)/Cav1 (−/−) (light purple) mice at 14 days in culture. Values represent relative fluorescence units (rfu), shown is the mean ± SD of at least 60 cells in each of three independent experiments. Statistical analysis was calculated using one-way ANOVA followed by the Newman–Keuls post hoc test for multiple comparisons; *P = 0.05; **P < 0.01; ***P < 0.001. (D) Quantification of cholesterol resolved by TLC in neuronal cultures from Bl6 (black bars), 150(+/−) (gray bars) and 150(+/−)/Cav1(+/−) (teal bars) animals. Cholesterol was extracted as described (58). The extracted lipids were separated by TLC using CHCl3/C2H5OC2H5/CH3COOH, 65:15:1 (v/v/v) as the developing solvent. Cholesterol content was normalized with protein level and expressed as µg/mg of protein. *P = 0.01 estimated using Student's t-test.
Table 1.
Ratio of steroids in the striatum of HdhQ150(+/+)(HD) animals relative to controls measured by mass spectrometry
| Name | KEGGa | PubChemb | 12-16wks HD/Cntc | t-testd | 80wks HD/Cntc | t-testd |
|---|---|---|---|---|---|---|
| Zymosterol | C05437 | 92746 | 1.07 | 0.46 | 0.85* | 0.01 |
| Stigmasterol | C05442 | 5280794 | 0.95 | 0.67 | 0.59* | 0.001 |
| Squalene | C01751 | 638072 | 0.75 | 0.66 | 0.87 | 0.51 |
| Cholesterol | C00187 | 5997 | 1.25 | 0.11 | 0.72* | 0.03 |
| β-Sitosterol | C01753 | 222284 | 1.09 | 0.21 | 1.09 | 0.79 |
| Lathosterol | C01189 | 25201212 | 0.92 | 0.64 | 0.82* | 0.01 |
The number of animals: N = 6. The normalized peak area for each ratio is listed in Supplementary Material, Figure S5.
a,bCompound identifiers from the KEGG and PubChem databases.
cHD/Cnt is the ratio of peak areas of the indicated steroids in HD to control animals; the dry weight of protein analyzed for 12–16 weeks animals is HD = 0.015 g; Cnt = 0.02 g and 80 weeks is HD = 0.02 g; Cnt = 0.016 g.
dt-test is the P-values from Student's t-test.
*Significant reduction.
Remarkably, loss or reduction of Cav1 in the HdhQ150(+/−)/Cav1 (−/−), HdhQ150(+/+)/Cav1 (−/−), HdhQ150(+/−)/Cav1 (+/−) and HdhQ150(+/+)/Cav1 (+/−) crosses prevented the trafficking block, and reinstated cholesterol homeostasis (Fig. 3A and C). Loss or reduction of Cav1 restored trafficking of LacCer (Supplementary Material, Fig. S3), prevented cholesterol accumulation (Fig. 3A), and the distribution of cholesterol in the HdhQ150(+/+)/Cav1 (−/−), HdhQ150(+/−)/Cav1 (+/−) crosses was similar to those of Cnt (Fig. 3A). Furthermore, in the crosses, the reduction in Cav1 restored the cholesterol level to that of controls, as quantified by either filipin staining (Fig. 3C) or TLC (Fig. 3D). Collectively, these analyses implied that mhtt inhibited Cav1-dependent trafficking, which led to both accumulation of cholesterol in the ER/Golgi and subsequent suppression of new cholesterol synthesis. Both of these phenotypes were reversed if Cav1 was reduced in cells.
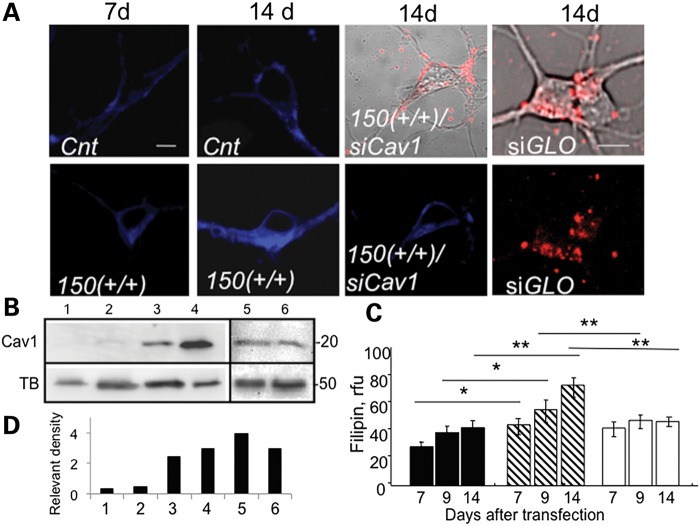
Cav1 siRNA (siCav1) also blocked cholesterol accumulation in neurons from HdhQ150(+/−) and HdhQ150(+/+) animals (Fig. 4). We transfected siCav1 into primary neurons from these animals and measured the intensity of filipin staining 7, 9 and 14 days posttransfection (Fig. 4A and C). All siCav1 transfections included a visual marker siGlo to monitor transfection efficiency and to identify the transfected cells (Fig. 4A). The siCav1 uptake was efficient, as judged by siGLO intensity (Fig. 4A). siCav1 effectively knocked down Cav1 expression in Cnt or HdhQ150(+/+) neurons, as detected by specific antibodies for Cav1 (Fig. 4B and D, lanes 1–4). In the absence of the siCav1, cholesterol accumulation by 14 days in culture doubled in neurons from HdhQ150(+/+) animals compared with Cnt animals (Fig. 4A and C). If an interaction of mhtt and Cav1 caused cholesterol accumulation, then loss of Cav1 should reduce filipin staining. Indeed, treatment with siCav1 reduced the degree of filipin staining in primary HdhQ150(+/+) neurons to levels observed in control cells by 9 days posttransfection (Fig. 4A and C). Knockdown was not observed, if the bases of the siRNA were scrambled (Fig. 4B and D, lanes 5 and 6).
Figure 4.
Cav1 siRNA (siCav1) suppresses cholesterol accumulation observed in striatal neurons from HdhQ150(+/+) mice. 150(+/+) denotes HdhQ150(+/+). Cnt denotes C57BL6. (A) Cholesterol levels visualized by filipin staining (blue) in neurons from control Cnt and 150(+/+) mice transfected with siCav1 at 7 and 14 days in culture. Red is siGLO (Dharmacon), a fluorescent nonspecific siRNA marker to identify transfected cells. Shown are transmission images of Cnt or 150(+/+) neurons 14 days posttransfection with siCav1 and siGLO (in upper panels) and fluorescence images of 150(+/+) neurons transfected with siGLO (red) (lower panel). Images were acquired using LSM510 confocal microscope with ×100 (1.4 NA) DIC lens. Scale bar is 5 µm. siGLO intensity was not due to surface binding, since visualization of siGLO intensity was performed on thin, internal optical sections (∼5 µm). (B) siCav1 significantly suppresses Cav1 expression in neurons. Western blot analysis of Cav1 expression in neurons from control Cnt mice 8 days posttransfection with siCav1 (lane 1); neurons from 150(+/+) mice 8 days posttransfection with siCav1 (lane 2); untransfected Cnt mice at 8 days in culture (lane 3); epithelial A431 cell extract stained with a Cav1 Ab as a positive control for Cav1 expression (lane 4); neurons from Cnt (lane 5) and HdhQ151 (lane 6) mice transfected with scrambled siRNA. Tubulin (TB) was used as loading control. (C) Quantification of free cholesterol using filipin staining in neurons from Cnt (black bars), 150(+/+) (striped bars) and 150(+/+) animals transfected with siCav1 (white bars) at the indicated days in culture. Results represent data from three independent experiments. At least 50 cells per bar were analyzed by the one-way ANOVA followed by the Newman–Keuls post hoc test for multiple comparisons; *P = 0.01; **P < 0.001. Data are expressed as mean ± SEM. (D) Quantification of the Cav1 expression relevant to the expression of tubulin from western blot presented in (B) using densitometry.
Thus, both in vitro and in vivo, reduction of Cav1 was sufficient to relieve the trafficking defect and to suppress cholesterol accumulation in mhtt-expressing animals. No trafficking abnormalities in neurons were observed if mhtt was expressed in the absence of Cav1 (in HdhQ150(+/−)/Cav1 (−/−) animals), if Cav1 was expressed in the absence of mhtt (Cnt), or if mhtt was expressed in the absence of Cav1 (in Cav1 (−/−) or Cav1(+/−) animals). Thus, Cav1 and mhtt together were required to confer the trafficking defect, which was an abnormal ‘gain of function’.
Cholesterol accumulation in HdhQ150(+/+) neurons does not result from enhanced clathrin-mediated endocytosis
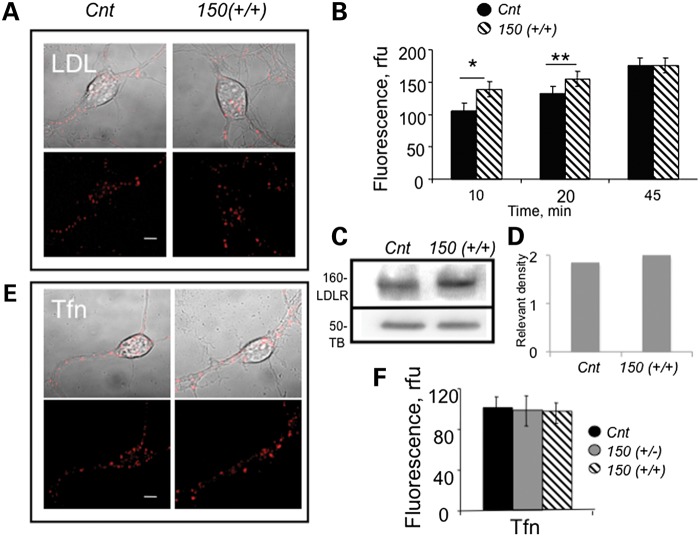
One pathway that could lead to cholesterol accumulation in the cells is via internalization of low-density lipoprotein receptor (LDLR) through a clathrin-dependent mechanism (22). To test whether mhtt enhanced LDLR endocytosis, we plated neurons from HdhQ150(+/+) animals side by side with those of Cnt, and monitored entry of a fluorescently labeled DiI-LDL (3,3′-dioctadecylindocarbocyanine-low-density lipoprotein) with time (Fig. 5A and B). Endocytosis of DiI-LDL was faster in HdhQ150(+/+) neurons within the first 20 min of observation (Fig. 5B). However, the initial effect was modest, and by 45 min, the level of intracellular DiI-LDL was identical in neurons from HdhQ150(+/+) and Cnt animals (Fig. 5B, 45 min). The overall level of expressed LDLRs was similar in Cnt and HdhQ150(+/+) neurons (Fig. 5C and D). Thus, the expression of mhtt did not significantly alter the expression of LDLRs or internalization of LDL.
Figure 5.
Expression of mhtt does not enhance cholesterol endocytosis through clathrin-dependent or clathrin-independent pathways in neurons from HdhQ150(+/+) mice. 150 denotes HdhQ150(+/+). Cnt denotes C57BL6. (A) Internalization of DiI-LDL (red) in neurons from Cnt and 150(+/+) mice 20 min after incubation. Confocal images (5 µm optical sections) were acquired using LSM510 confocal microscope with ×100 oil DIC lens (1.4 NA). Scale bar is 5 µm. (B) Quantification of DiI-LDL in control and 150(+/+) neurons from (A) at the indicated time after internalization. Fluorescence intensity in arbitrary units (rfu) is calculated from at least 30 cells for each point and analyzed using Student's t-test. *P < 0.002; **P < 0.01. Error bars are standard errors. (C) Western blot analysis of LDL receptors in striatal tissue extracts from Cnt and 150(+/+) animals at E17. Proteins were resolved on 12% polyacrylamide gels, and bands were detected using rabbit polyclonal LDLR Ab (1:1000, BioVision). Monoclonal tubulin Ab (DM1A 1:6000, Sigma) was used for loading control. (D) Quantification of the LDLR expression relevant to the expression of tubulin (TB) from western blot presented in (C) using densitometry. (E) Internalization of fluorescent Tfn is not enhanced in neurons from 150(+/+) mice. Confocal images of live neurons 15 min after incubation with Tfn. Scale bar is 5 μm. (F) Quantification of Tfn internalization described in (E). At least 15 cells were analyzed per point. Error bars are standard deviation. All images were acquired using LSM 510 confocal microscope with ×100 oil DIC lens (1.4 NA).
As a second measure, we monitored the uptake of Alexa Fluor 594-labeled transferrin (Tfn) to test whether expression of mhtt influenced clathrin-mediated endocytosis (Fig. 5E and F). Consistent with the LDL results, internalization of Tfn in neurons from HdhQ150(+/−) or HdhQ150(+/+) animals was indistinguishable from that of the Cnt (Fig. 5E and F) or Cav1 (−/−) neurons (Supplementary Material, Fig. S6). Thus, clathrin-mediated internalization of neither LDL nor Tfn was enhanced in neurons from HdhQ150(+/−) or HdhQ150(+/+) animals relative to Cnt cells. Cholesterol can be internalized by transcytosis through a clathrin independent, caveolar pathway, but uptake of LacCer was also not increased in neurons from HdhQ150(+/−) animals relative to controls (Supplementary Material, Figs. S3 and S6). Therefore, the interaction of mhtt and Cav1 did not alter the activity of major endocytic pathways known to internalize cholesterol.
Loss or reduction of Cav1 rescues the post-Golgi trafficking defect observed in HdhQ150(+/+) and HdhQ150(+/−) primary neurons
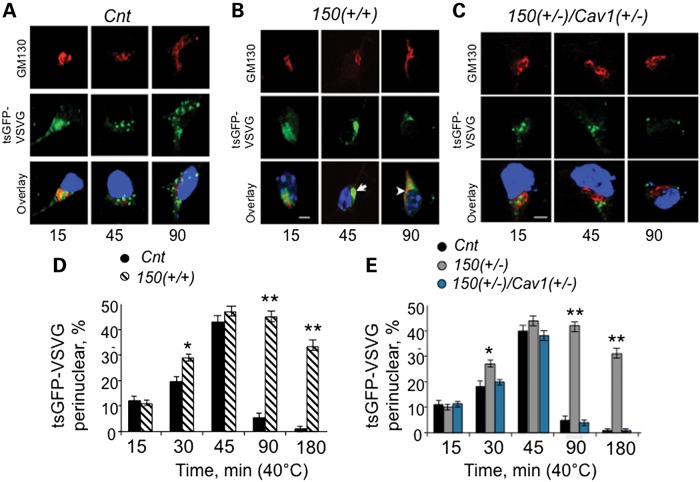
Since mhtt did not increase entry of lipids, we tested whether cholesterol accumulation arose from defective exit of cholesterol. Cav1 directly binds cholesterol (25) and mediates its transport from the ER to the plasma membrane via the trans-Golgi network (17,18). To test whether exit of cholesterol was impaired in primary neurons from HdhQ150(+/+) animals, we expressed a temperature-sensitive form of green fluorescent protein (tsGFP)-tagged vesicular stomatitis virus G protein (tsGFP-VSVG) in primary HdhQ150(+/+) neurons, and used the subcellular location of the virus as a visual probe for export (Fig. 6A–C, green). VSVG is a well-characterized secretory protein, which is expressed in rough ER where the Glc3-Man9-GlcNac2 oligosaccharide is added and sugars are removed as the protein travels to the Golgi apparatus (26). VSVG is transported from the trans-Golgi network to the plasma membrane through a Cav1-related pathway (27). If mhtt suppresses non-clathrin, caveolar-related exocytosis, then tsGFP-VSVG should remain in the Golgi.
Figure 6.
ER/Golgi trafficking is inhibited in neurons expressing mhtt. Fluorescence micrographs of cultured primary striatal neurons (E17) from C57BL6 mice (Cnt) (A), 150(+/+) mice (B) and 150(+/−)/Cav1(+/−) mice (C) transfected with a plasmid encoding tsGFP-VSVG. Transfected neurons were allowed to recover for 16 h at 40°C to ensure ER/Golgi retention. The temperature was lowered to 32°C (the permissive temperature), and trafficking of tsGFP-VSVG was observed for the period of 3 h. The tsGFP-VSVG (green) was transported out of the Cnt neurons in large distinctive vesicles at the indicated times (A); tsGFP-VSVG (green) was not transported out of the 150(+/+) neurons, and remained localized near the Golgi-like perinuclear compartment for up to 180 min (B, arrows). Reduction of mhtt–Cav1 interaction in 150(+/−)/Cav1(+/−) neurons restored post-Golgi trafficking to the extent observed in Cnt mice (C). The tsGFP-VSVG is green, cis-Golgi is identified using GM130 antibody (red) and the nuclear staining by Hoechst is blue. Images were taken on LSM510 confocal microscope with ×100 DIC lens (1.4 NA). Scale bar is 5 µm. (D and E) Quantification of cells with perinuclear localization of tsGFP-VSVG at different time at permissive temperature in neurons from Cnt, 150(+/+), 150(+/−) and 150(+/−)/Cav1(+/−) animals. At least 20 cells were taken for every time point. Results are expressed as mean ± SEM. Student's t-test was applied to analyze results represented in (D); one-way ANOVA followed by the Newman–Keuls post hoc test for multiple comparisons was applied to analyze results in (E). *P < 0.01; **P < 0.0001.
In the trafficking assay, cells were transfected with tsGFP-VSVG plasmid at 37°C for 4–6 h to allow expression and transport of the viral protein to the Golgi. The temperature was raised to the non-permissive one (40°C) to ensure retention of the expressed tsGFP-VSVG in the Golgi. The temperature was then lowered to the permissive temperature (32°C), and trafficking from the trans-Golgi network to the plasma membrane resumed. We visualized exit of tsGFP-VSVG from the Golgi with time. VSVG localized to the cis-Golgi as marked by colocalizaton with the cis-Golgi GM130 antibody (Fig. 6A–C). In Cnt cells, within the first 15 min at the permissive temperature, colocalization of GFP with the red GM130 marker was lost, as tsGFP-VSVG (Fig. 6A, green) moved away from the ER/Golgi compartments (Fig. 6A, red). By 45 min, the tsGFP-VSVG protein in Cnt neurons moved outside of the perinuclear Golgi-like compartment in distinctive vesicles (Fig. 6A, 45 min), which migrated towards the plasma membrane (Fig. 6A, 90 min).
Trafficking of tsGFP-VSVG in HdhQ150(+/+) neurons displayed a distinctly different profile (Fig. 6B). Similar to control cells, after 15 min at the permissive temperature, the tsGFP-VSVG in HdhQ150(+/+) cells was transported to the ER-Golgi-like perinuclear compartment (Fig. 6B, 15 min, green and red). However, the majority of the tsGFP-VSVG remained near the Golgi for over 180 min (Fig. 6B, white arrows at 45′ and 90′). Moreover, in HdhQ150(+/+) neurons, the number of vesicles formed after exiting the Golgi was significantly lower relative to control neurons (Fig. 6B, green). To quantify these results, we counted the number of cells in which the majority of tsGFP-VSVG was retained in the perinuclear Golgi-like subcellular compartment (Fig. 6D). Indeed, tsGFP-VSVG-containing vesicles were retained at the Golgi for significantly longer time in HdhQ150(+/+) neurons compared with Cnt cells (Fig. 6D). Similar results were obtained in human skin fibroblasts from control individuals and HD patients (Table 2; Supplementary Material, Fig. S7). Thus, expression of mhtt suppressed post-Golgi trafficking to the plasma membrane and exit of cholesterol without altering its import via the clathrin pathway.
Table 2.
Description of human primary fibroblasts
| Coriell no. | Definition | Gender/age | Allele 1 | Allele 2 | Onset |
|---|---|---|---|---|---|
| 04212 | HD | F/50 | 16 | 41 | Early symptoms |
| 01516 | Control | F/49 | 16 | 25 | None |
We repeated experiments comparing the post-Golgi trafficking of tsGFP-VSVG in neurons from HdhQ150(+/−)/Cav1(+/−) mice relative to HdhQ150(+/−) and Cnt mice (Fig. 6A–C). The tsGFP-VSVG was not retained at the Golgi in HdhQ150(+/−)/Cav1(+/−) neurons, and trafficking with time resembled that of neurons from Cnt animals (Fig. 6A and C). Specifically, movement out of the Golgi was nearly complete within 45 min at the permissive temperature, as judged by the loss of overlap of the GFP signal and the GM130 antibody staining (Fig. 6C, 45 min) and the disappearance of the vesicles with time (Fig. 6C, 90 min). Quantification of the trafficking data revealed that the percent of perinuclear tsGFP-VSVG was similar in neurons from both HdhQ150(+/−)/Cav1(+/−) and Cnt relative to HdhQ150(+/−) mice (Fig. 6E, Cnt (black); HdhQ150(+/−) (gray); HdhQ150(+/−)/Cav1(+/−) (blue)). Thus, loss of the Cav1–mhtt interaction restored vesicle trafficking and allowed Cav1-dependent cholesterol exit from the Golgi. Reduction in Cav1 levels was sufficient to rescue the post-Golgi trafficking defect.
Loss or reduction of Cav1 suppresses motor decline in HdhQ150(+/−) and HdhQ150(+/+) animals
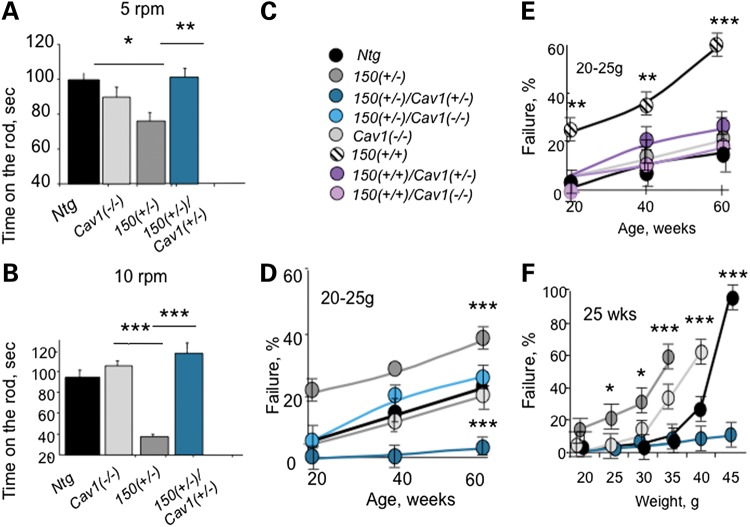
To test whether observed Cav1-dependent trafficking defects were relevant for HD pathophysiology in vivo, we evaluated whether loss or reduction of Cav1 expression in HdhQ150(+/−) and HdhQ150(+/+) mice altered the onset of motor decline in these animals (Fig. 7). If impaired caveolin-related trafficking caused or contributed to pathophysiology, then loss or reduction of Cav1 in the progeny of the HdhQ150(+/−)/Cav1(+/−) crosses should suppress the progression of these phenotypes. Motor decline was examined by two standard tests (28). The rotorod test measured the ability of animals to maneuver a rotating rod (Fig. 7A–C). Testing was performed on age- and gender-matched mice once a day for the total of 10 days to ensure that the results reflected motor function and to avoid learning bias. All animals at 7 weeks of age displayed robust and equivalent performance. However, at 25 weeks of age, we observed substantial decline in rotorod performance at 5–10 rpm in HdhQ150(+/−) animals relative to Cav1 (−/−) and Ntg littermates (Fig. 7A and B). Remarkably, reduction of Cav1 in HdhQ150(+/−)/Cav1(+/−) animals was sufficient to prevent motor decline (Fig. 7A and B). On the average, rotorod performance in the crosses was equivalent to that of their Ntg and Cav1 (−/−) littermates, neither of which expresses mhtt. Thus, the HdhQ150(+/−)/Cav1(+/−) animals inherited the same complement of the disease allele, as did their HdhQ150I(+/−) littermates, but reduction of Cav1 in the crosses suppressed or delayed the motor decline.
Figure 7.
Loss or reduction of Cav1 in HD mice improves motor performance. 150 denotes HdhQ150. Ntg denotes non-transgenic littermates. (A and B) Rotarod performance. Presented is the time on the rod rotating with the fixed speed of 5 (A) or 10 (B) rpm. Five female mice 55 weeks old from indicated genotypes were examined. Mice were allowed three attempts to stay on the rod for 2 min. Mice were tested for 10 days and the best time of three attempts was averaged and plotted. Significant deficiency was observed in 150 (+/−) mice. 150(+/−)/Cav1(+/−) mice performed better than 150(+/−) mice under both conditions. (C–E) Hanging bar performance. The mouse was allowed to hang on the thin metal bar using its front paws (C, animal key). A healthy mouse usually engages its hindpaws and tail to maneuver on the bar and move along the bar towards the post. HD mice typically do not engage hindpaws and cannot hang on the bar >20–30 s. HD mice crosses with reduced Cav1 expression (HdhQ150(+/−)/Cav1(+/−)) or (HdhQ150(+/+)/Cav1(+/−)) perform substantially better than age- and gender-matched HD (HdhQ150(+/−)) or (HdhQ150(+/+)). The results of the hanging bar test demonstrate that 150(+/−) (D) or 150(+/+) (E) mice crosses with loss or reduction of Cav1 expression perform on the bar significantly better relative to age- and gender-matched HD littermates. 30–40 mice were tested in every age group. Only mice with weight between 20 and 25 g were taken into analysis. (F) 150(+/−)/Cav1(+/−) mice perform better on the bar test than other mice in respect to increased body weight. All mice were around 25 weeks of age. Heavier mice performed poorly independent of the genotype. However, 150(+/−)/Cav1(+/−) (blue) performed better than control Ntg (black) or 150(+/+) (white) and 150(+/−) (gray) mice. Ten female mice were tested for every point. Results are expressed as mean ± SEM. *P < 0.05; **P < 0.01 and ***P < 0.001.
As a second measure of motor function, animals were tested for grip strength (28) (Fig. 7C–F). We selected animals of approximately the same weight (20–25 g) in the indicated age groups (Fig. 7C–E). The mice were allowed to grab with their forelimbs a narrow wire rod (D < 0.25 cm) suspended 30 cm above a padded surface (Fig. 7C–F). Each mouse was released and observed for 1 min. Mice scoring positive for this test fell within 30 s. The entire group of animals was tested together, and the results were expressed as a percent of failure (Fig. 7C–F). The performance of HdhQ150(+/−) (Fig. 7C and D) and HdhQ150(+/+) (Fig. 7C and E) relative to controls is shown. The Ntg (black) and Cav1 (−/−) animals (light gray) perform comparably well at all ages tested, with an approximate 20% drop in success rate when animals reached a year of age (Fig. 7D and E). Compared with controls, HdhQ150(+/+) (Fig. 7E, striped) or HdhQ150(+/−) (Fig. 7D, dark gray) mice performed poorly on the bar starting as early as 10 weeks of age. At all ages tested, HdhQ150(+/+) performed worse than HdhQ150(+/−), and in each mouse line, the percent of failure progressively increased with age. By 60 weeks of age, 65% of HdhQ150(+/+) (Fig. 7E, striped) and 40% of HdhQ150(+/−) (Fig. 7D, dark gray) animals failed the test. In contrast, loss or reduction of Cav1 in the crosses conferred a remarkable improvement and substantial delay in the onset of motor decline relative to their HdhQ150(+/−) and HdhQ150(+/+) littermates (Fig. 7D and E). HdhQ150(+/+)/Cav1(+/−) (Fig. 7C and E), HdhQ150(+/+)/Cav1 (−/−) (Fig. 7C and E), HdhQ150 (+/−)/Cav1 (−/−) (Fig. 7D) and HdhQ150 (+/−)/Cav1(+/−) (Fig. 7D) animals were indistinguishable from Ntg and Cav1 (−/−) littermates (Fig. 7D and E). The performance on the bar test for all animals declined with weight (Fig. 7F). The HdhQ150(+/−)/Cav1(+/−) animals consistently outperformed the other lines. Taken together, the results indicated that mice-expressing mhtt had impaired motor function, but loss or reduction of Cav1 in the HdhQ150(+/−) or HdhQ150(+/+) crosses rescued the motor deficit.
Loss or reduction of Cav1 inhibits the formation of inclusions in HdhQ150(+/+) and HdhQ150(+/−) animals
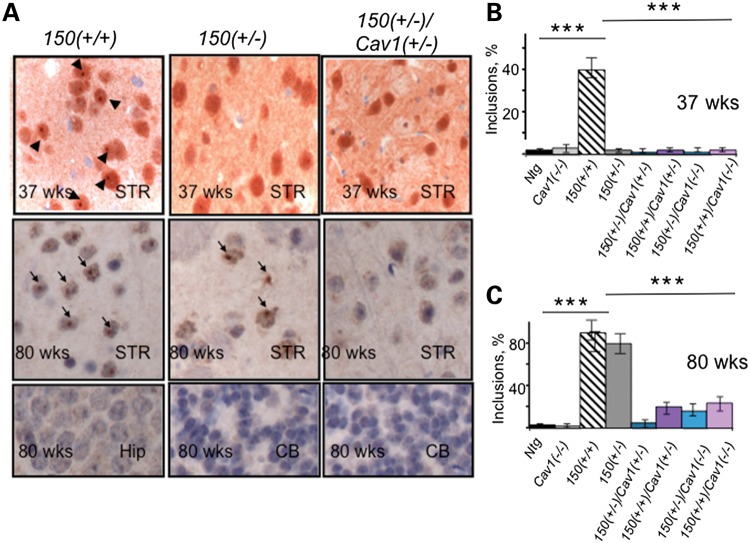
We next tested whether reduction or loss of Cav1 in mhtt-expressing animals might also suppress formation of inclusion bodies that mark disease progression (29,30). We quantified the incidence of inclusions visualized using ubiquitin specific antibody in two brain regions susceptible to cell death in HD (cortex and striatum) and in two that were resistant (hippocampus and cerebellum). The inclusions were measured in HdhQ150(+/+) and in HdhQ150(+/−) animals, and compared with those observed in HdhQ150(+/−)/Cav1 (+/−), HdhQ150(+/−)/Cav1 (−/−), HdhQ150(+/+)/Cav1 (+/−) and HdhQ150(+/+)/Cav1 (−/−) crosses at 37 and 80 weeks of age (Fig. 8A–C). For all regions of the brain, H&E staining revealed no obvious signs of neurodegeneration in control Ntg or Cav1 (−/−) animals. The number of neurons detected at 37 and 80 weeks of age were similar in the control animals, and no inclusions were detected with the ubiquitin antibody at any age in any brain region (Fig. 8A and B). In contrast, as judged by histological examination, the HdhQ150(+/+) mice form distinct inclusions by 37 weeks and the number increased with age (Fig. 8A–C). By 80 weeks, inclusions in HdhQ150(+/+) mice were observed in the majority of neurons (Fig. 8A and C). The HdhQ150(+/−) animals displayed few observable inclusions at 37 weeks, but they were prevalent (in about 50% of neurons) at 80 weeks (Fig. 8A and C). Thus, development of inclusions in HdhQ150(+/+) and HdhQ150(+/−) animals with age depended on the gene complement of expressed mhtt.
Figure 8.
Loss or reduction in Cav1 in HD animals significantly delays the appearance of inclusions in striatum. (A) Inclusions identified with ubiquitin staining were readily seen only in the striatum of 150(+/+) mouse but not in 150(+/−) or 150(+/−)/Cav1(+/−) at 37 weeks (top panel). Scale bar, 10 µm. At 80 weeks of age, inclusions were found in striatal neurons from both 150(+/+) and 150(+/−) mice but not in 150(+/−)/Cav1(+/−) mice (middle panel). Bottom panel represents regions of the brain unaffected in HD, hippocampus (Hip) and cerebellum (CB) of corresponding mice 80 weeks of age. No inclusions were observed in these brain regions. (B and C) Quantification of striatal inclusions in HD mice of all genotypes at 37 (B) and 80 (C) weeks of age. Appearance of nuclear inclusions is reduced in 80-week-old mice-expressing mhtt and reduced levels of Cav1 compared with HD mice. Data in (A–C) represent quantification of six randomly selected fields from three mice of each genotype. Results are expressed as mean ± SEM; ***P < 0.001.
In the same animals, we examined hippocampus and cerebellum, two regions relatively spared in HD. In these brain regions, we observed no significant ubiquitin staining in either HdhQ150(+/+) or HdhQ150(+/−) animals at either 37 or 80 weeks of age (Fig. 8A). In marked contrast, neither HdhQ150(+/−)/Cav1 (+/−) (Fig. 8A–C) nor HdhQ150(+/+)/Cav1 (+/−) animals developed inclusion bodies at any age tested (Fig. 8A–C). Few to no inclusions were observed at 80 weeks in the striatum, and no inclusions were formed in hippocampus or cerebellum of the same animals (Fig. 8A–C). Thus, loss of reduction of the mhtt/Cav1 interaction in HdhQ150(+/−)/Cav1(+/−) or HdhQ150(+/+)/Cav1(+/−) animals was sufficient to reduce or eliminate formation of inclusions, a hallmark associated with HD pathophysiology.
DISCUSSION
Expression of mhtt results in widespread toxicity manifested in trafficking defect, loss of cholesterol homeostasis, motor decline and inclusion formation (31). Here, we demonstrate that loss or reduction of Cav1 in HdhQ150(+/−) or HdhQ150(+/+) animals suppresses these phenotypes. Remarkably, loss or reduction of Cav1 restores post-Golgi caveolar-related trafficking, suppresses cholesterol accumulation in the ER/Golgi region, reestablishes exocytosis from the Golgi to the plasma membrane, inhibits or significantly delays the onset of motor decline, reduces weight loss and reduces inclusion formation associated with the disease. These findings imply that a deleterious interaction of mhtt and Cav1 promotes trafficking defects and toxicity in vivo. Indeed, three independent methods, FRET (Fig. 1), IP and GST pull-down (11), indicate that Cav1 directly interacts with mhtt. We propose that an aberrant mhtt interaction with Cav1 is a causative factor of pathophysiology in vivo and provides a direct link between lipid dysfunction (12–15) and post-Golgi trafficking defects (8–11), both of which are implicated in cellular and animal models of HD. The trafficking defect we observe occurs only when mhtt and Cav1 are coexpressed and confers a toxic ‘gain of function’. Loss of the normal htt–Cav1 interaction in Cav1 (−/−) animals is not beneficial, and likely contributes to the mild motor (32) and cognitive impairment (33) that develops in these animals. However, there is a sufficient back-up mechanism for cholesterol transport in Cav1 (−/−) animals to support their survival.
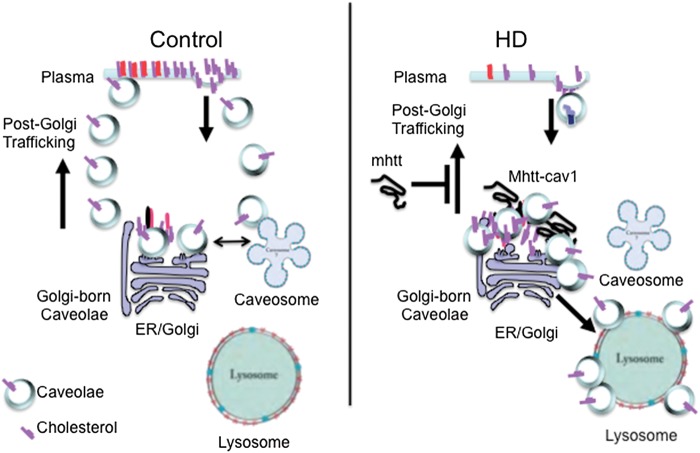
The detailed mechanism for the ‘gain-of-function’ phenotype is as yet unknown. However, caveolar post-Golgi exit is driven by caveolin oligomerization, binding of cholesterol in the cis-Golgi, and transport of cholesterol to the plasma membrane. Endosomal exit from the ER-Golgi occurs in a complex with domain-containing protein 1 (UBXD1) and valosin-containing protein (VCP) (34) (Fig. 9). Mutants of VCP or UBXD1 block cholesterol transport, and Cav1 accumulates on late endosome/lysosome membranes (35,36). Since a similar phenotype arises in mhtt-expressing cells, mhtt might interact with the Cav1–VCP–UBXD1 complex, and misdirect Cav1 to the lysosomes, resulting in abnormal compartmentalization of cholesterol and its accumulation in the Golgi and/or lysosomes (Fig. 9). Indeed, accumulation of cholesterol at the Golgi will independently suppress post-Golgi trafficking to the plasma membrane (37). Accumulation of Cav1 on the surface of the lysosomes is not observed normally. Thus, mhtt-dependent diversion of caveolin to lysosomes provides a plausible mechanism for a new phenotype that is distinct from the normal htt–Cav1 interaction (Fig. 9).
Figure 9.
Proposed model for cholesterol accumulation in HD caused by aberrant Cav1–mhtt interaction. Under normal conditions, Cav1 mediates post-Golgi trafficking of cholesterol from the Golgi to the cell surface (Golgi-born caveolae), and cycling of cholesterol from the cell surface. The intracellular trafficking is directed to and from the Golgi and plasma membrane via association in the caveosomes. In HD, mhtt binds Cav1 and impairs cholesterol exit from the Golgi. Instead, Golgi buildup of cholesterol results in aberrant shunting of Cav1 and cholesterol to the lysosome.
Indeed, the mislocalization of cholesterol and Cav1 during post-Golgi trafficking has emerged as a common theme in Alzheimer's disease (AD) (38), Prion disease (39), Parkinson's disease (40), lysosomal diseases (41) and HD (42). In Niemann–Pick Type C (NPC), lysosomal enzymes are transported from the Golgi to the endosomes via phosphomannosyl receptor interactions (43). Mutations in NPC1 or NPC2 proteins block endosomal lipid trafficking and divert cholesterol to the lysosomes, where it becomes a repository for free cholesterol and sphingolipids (44). Similar to what we have observed for HD, lipid storage diseases are characterized by neurodegeneration and accumulation of cholesterol and sphingolipids by a caveolar-related trafficking mechanism. Moreover, abnormal cholesterol trafficking could contribute to the inhibition of the lysosomal degradation of mhtt aggregates. Our data demonstrate that, indeed, normalization of cholesterol homeostasis results in reduction of the aggregation in HdhQ150(+/−)/Cav1(+/−) or HdhQ150(+/+)/Cav1(+/−) crosses. In AD, production of β-amyloid is also cholesterol dependent (45) and is found in a membrane fraction with caveolae (46). Faulty caveolar-related trafficking and secretion of lipids is a plausible source of the extracellular plaques, which are rich in cholesterol (46). Caveolar-related cholesterol accumulation in the endosomal–lysosomal compartment is also observed in prion disease (47). Glycosylphosphatidyl inositol anchoring of prion protein (PrP) in the ER directs the PrP protein to the Golgi and into cholesterol-rich caveolae (47). In caveolae, PrP is slowly degraded, and degradation appears to be cholesterol sensitive (48). Prusiner and colleagues have raised the possibility that caveolae are the sites of PrP aggregation (47). In fact, PrP can provoke an HD ‘phenocopy’ (49). Affected individuals are heterozygous for a 192-nucleotide insertion within the PrP coding region, which encodes an expanded PrP with eight extra octapeptide repeats. The PrP repeat expansions are well characterized and provoke early-onset, slowly progressive, atypical prion diseases (49). The autosomal dominant pattern of inheritance of PrP results in a wide range of clinical features, many of which overlap with those of HD (49). Although prion and lipid storage diseases arise from distinct mutations or misfolded proteins relative to HD, the common occurrence of lipid accumulation suggest that mhtt affects at least some of the same steps of cholesterol trafficking.
We demonstrate here using new mouse models that features of in vivo pathophysiology are remarkably suppressed in the HdhQ150 (+/−)/Cav1 (+/−) or HdhQ150 (+/+)/Cav1 (+/−) crosses. The improvement we observe in motor function of these crosses is not explained by differences in genetic background. The HdhQ150(+/−) (19) and Cav1 (+/−) (50) mice were backcrossed to generate isogenic strains, and each experiment was controlled using the appropriate littermates. Moreover, our HdhQ150(+/+) knock-in animals bred from the cross retain properties similar to the parental line. We find in our animals that grip strength is impaired early (Fig. 7D–F), and inclusions are widespread at 80 weeks (20 months) (Fig. 8A–C). Similar impairment in grip strength and widespread aggregate deposition is present in another strain derived from the HdhQ150 knock-in mice (51). The early decline in rotorod performance we observe is similar to the original HdhQ150(+/+) knock-in strain (19), from which our mice were bred. Thus, the beneficial effects we observed in crossing HdhQ150(+/−) and Cav1(+/−) animals appear to arise from loss or reduction of Cav1.
Caveolar transport represents a mechanism by which cholesterol efflux can be modulated in response to changes in cellular cholesterol content (52,53). Cells transport newly synthesized cholesterol in a Cav1-dependent manner, and blocking movement from ER to caveolae results in cholesterol accumulation in the lumen of the ER in most cell types (11). Furthermore, inhibitors of cholesterol biosynthesis cause of lipid raft disruption (52,53), which is reversed upon repletion of cholesterol (53). Collectively, these results indicate that caveolar cholesterol transport is inversely related to cholesterol biosynthesis.
Our findings provide a direct demonstration of this relationship. Brown and Goldstein established that a delicate balance of intracellular cholesterol is regulated by a feedback loop in which the level of cholesterol determines whether it is synthesized or suppressed. Since toxicity arises if cholesterol is too high or too low, the ER serves as both the cellular sensor of cholesterol level and the site of its synthesis (16,54). Thus, a caveolar-related trafficking block and cholesterol accumulation at the ER/Golgi region is predicted to result in a decrease in cholesterol biosynthesis. We propose that disruption of this sensitive equilibrium by altered caveolar trafficking contributes to toxicity and likely accounts for the fact that cholesterol accumulation (3,11,55,56) and suppressed biosynthesis (12–15) are both observed in HD postmortem brain, cell models and animal models of HD. The degree of cholesterol accumulation is likely to vary according to developmental stage and cell type (57). Collectively, the results from our novel animal models imply that mhtt–Cav1 interaction promotes toxicity in vitro and in vivo, and loss of reduction of Cav1 suppresses disease phenotypes. Cav1 (−/−) and Cav1(+/−) animals are viable, and reduction of Cav1 is sufficient to improve HD pathophysiology. Cav1, therefore, is a reasonable candidate for therapeutic targeting.
MATERIALS AND METHODS
Animals and breeding
The Institutional Animal Care and Use Committee approved all procedures involving animals. The following mouse models were used: homo- and heterozygous HD mice with 150 CAG repeats ‘knocked in’ to the endogenous mouse allele, HdhQ150 (19), and Cav1 null mice, Cav1 (−/−) (50). The HdhQ150 and Cav1 (−/−) mice were generated on the C57BL6 background HdhQ150 (19,50). Therefore, we used C57BL6 (Cnt) mice and non-transgenic littermates (Ntg) as controls.
Antibodies, immunofluorescence, and western analysis
Primary antibodies were: mouse monoclonal Cav1 (1:50, #610057, BD Transduction Laboratories, KY, USA), rabbit polyclonal Cav1 (1:100, #610059, BD Transduction Laboratories), rabbit polyclonal GM130 anti-cis-Golgi (1:50, Calbiochem, CA, USA), mouse monoclonal TGN38 anti-trans-Golgi (1:250, # 610898, BD Transduction Laboratories) and rabbit polyclonal anti-ubiquitin (1:200, Dako, Denmark, USA). Secondary antibodies were: goat anti-mouse Cy 5 (1:1500, Amersham, PA, USA), goat anti-mouse TMR (1:400), goat anti-rabbit Cy5 (1:1500, Amersham) and goat anti-rabbit TMR (1:50, Molecular Probes, Eugene, OR, USA). Hoechst dye was added to a final concentration of 1.0 µg/ml to visualize nuclei. Immunostaining and western blotting was performed as described previously (11).
Human skin fibroblasts
Age- and gender-matched control and HD human fibroblasts were from Coriell Cell Repository. The number of the CAG repeats expressed from each allele is provided in Table 2. Cells were cultured in Eagle's MEM on coverslips and were used for immunolabeling on Days 2–3 after plating. At least 30 cells were analyzed for each experiment.
Preparation of neuronal cell cultures
Preparation and culturing of primary striatal neurons has been previously reported (11). It is important to not that all experiments with estimation of free cholesterol levels using filipin were conducted in the glia-free cultures. That was achieved by culturing neurons in the Neurobasal medium starting 3 days after plating, and controlling glial cell proliferation with anti-mitotic reagents (11).
Inhibitors and fluorescent markers of endocytosis
C5-BODIPY-fatty acid labeled analog of LacCer was from Invitrogen, Alexa Fluor 594-labeled Tfn was from Molecular Probes, and DiI-LDL was from Biomedical Technologies Inc. (Stoughton, MA, USA). Endocytosis assays were preformed as described previously (11).
Confocal microscopy
Cells were imaged using confocal laser scanning microscope LSM 510 (Carl Zeiss) with ×100 or ×63 oil DIC objective (1.4 NA) with optical section set to ∼0.5 µm as described previously (11). For FRET donor dequenching experiments, a LSM 710 (Carl Zeiss) with ×40 water objective (1.1 NA) with optical section set to 2.5 µm was used. CFP signal was acquired using a 405-nm laser diode (5% power) excitation source; YFP signal was acquired using a 514-nm (2% power) Ar source. To dequench cerulean, 100 scans (pixel dwell ∼1.5 ms) were swept over a region of interest using the 514 nm (50% power) excitation source.
siRNA experiments
All reagents were from Dharmacon, Inc. (Chicago, IL, USA) as described previously (11).
Transfection of primary neurons and human skin fibroblasts with tsGFP-VSVG plasmid
The temperature-sensitive VSV-G (tsGFP-VSVG) plasmid was a gift from J. Lippincott-Schwartz (NIH, Bethesda, MD, USA) and M. McNiven (Mayo Clinic, Rochester, MN, USA) and is in pEGFP-N vector (Clontech). The tsGFP-VSVG protein is retained in the ER at a restrictive temperature of 40°C. Once cells are shifted to the permissive temperature (32°C), VSV-G protein is transported through the synthetic pathway to the Golgi and subsequently to the plasma membrane (58). Cells plated on coverslips were transfected using Lipofectamine™ 2000 reagent (Invitrogen, Carlsbad, CA, USA) and Nupherin™-neuron (BIOMOL, Plymouth Metting, PA, USA) as described previously (11). Neurons were transfected on Day 6 and fibroblasts on Day 2 after plating. Expression plasmid (4 µg) was mixed with 20 µl of Nupherin-neuron in 380 µl of serum-free and phenol red-free DMEM. After 30 min of incubation, DNA mixture was added to lipofectamine solution in DMEM at a ratio of 4:1 (liposome to DNA), incubated for another 45 min and added to the cells for 4 h. Fibroblasts were transfected using identical protocol but without Nupherin-neuron reagent. Cells were washed three times with DMEM and maintained in the incubator at restrictive temperature of 40°C and 5% CO2 for 16 h. Cells were shifted to the permissive temperature (32°C) and were fixed with 4% paraformaldehyde/PBS every 15 min following 2–3 h. Cells were costained with GM130 antibody to visualize the Golgi and Hoechst to identify nuclei. Experiments were performed at least three times in different cell plating. After fixation and immunostaining, cells were observed under LSM510 microscope using ×100 oil DIC lens.
Tissue culture and transfection of HEK293 T cells with fluorescently tagged fusion proteins
HEK293 T Cells (ATCC) were cultured in DMEM (ATCC 30-2002) supplemented with 10% fetal bovine serum, l-glutamine (Hyclone) and Pen/Strep (Hyclone). Cells were passaged every 3–4 days by treating with Trypsin and splitting 1:20. On Day 1, four-well glass-bottom dishes (LabTek 155382) were seeded at a density of 14 000 cells per well and allowed to adhere and spread overnight. On Day 2, cells were transfected with Lipofectamine 2000 (Invitrogen), as directed indicated by the manufacturer. Briefly, 1 µl of Lipofectamine (in 50 µl DMEM, no additives) was combined with 400 ng Cav–YFP and 2800 ng 23/51Qhtt–CFP plasmid DNA (in 50 µl DMEM, no additives). The mixture was allowed to incubate at room temperature for 20 min before adding to a single chamber-well. HEK293 T cells were incubated with the lipfectamine/DNA mixture for 18 h, then washed, fixed and mounted in media (Vectashield H-1000, no DAPI) for confocal imaging. The plasmids were constructed with a CMV promoter upstream of the expression cassette at the MacroLab Core Facility at UC Berkeley. Briefly, the gene for human Cav1 (full length, amino acid 1–178) was tagged with a C-terminal Citrine protein to generate the vector Cav1–YFP. The human htt gene with 23Q tract (full length, amino acid 1–3144) was tagged with a C-terminal Cerulean protein to generate the vector 23Q-CFP. Similarly, the 51Q mhtt gene was tagged with a C-terminal Cerulean protein to give the 51Q-CFP vector.
Filipin staining in neurons
Filipin staining has been previously described (11).
Quantitative image analysis
To measure filipin fluorescence, we used Metamorph software. A ROI was drawn around each cell to be measured. An identical ROI mask was placed in an area without fluorescent objects to measure and calculate background subtraction values. The net average fluorescence intensity of at least 30 cells was calculated for each genotype. Experiments were repeated in at least three independent platings.
Motor and neurological assessment
Rotating rod
Motor coordination and strength were assessed with a rotarod apparatus using the incremental fixed-speed rotarod protocol described previously (19) with minor modifications. Five female mice 25 weeks old from all nine genotypes (Fig. 2A) were allowed to stay on the rod 4.0 cm in diameter. Mice were trained to walk against the motion of a rotating drum. First, a training session of 3 min was done at a constant speed (3 rpm; rounds per minute). Then, 1 h later, mice were tested three times in 2-min trials at two different speeds: 5 and 10 rpm. The mean latency to fall off the Rotarod was recorded; mice remaining on the rod for >120 s were removed and their time scored as 120 s. The test was repeated for 10 days and the best time of three attempts was averaged over 10 days.
Hanging bar
Mice were lowered onto a parallel rod (D < 0.25 cm) placed 30 cm above a padded surface. Mice were allowed to grab the rod with their forelimbs, after which they were released and scored for their success (pass or failure) in holding onto the bar for 30 s. Three attempts were granted to pass the bar test. Any one successful attempt to hold onto the bar was scored as a pass. The percentage of animals that fell (and failed the test) was measured with age and recorded as a percent of the total number of animals tested. Thirty to 40 animals were measured in every group, for every time point.
Histology
Three mice from all nine genotypes (Fig. 2A) age 37 and 80 weeks were taken into analysis. Mice were terminally anesthetized by intraperitoneal injection of 10 µl of 8 mg/ml ketamine/1 mg/ml xylazine per gram of the body weight, after which they were perfused with 4% paraformaldehyde in phosphate buffered saline. Inclusions were detected using anti-ubiquitin antibody (rabbit polyclonal anti-Ubiquitin (1:200, Dako, Denmark). Hematoxylin & Eosin (H&E) staining was applied as described in (59). The sections were observed with the light microscope using 5, 20, 40 or ×100 lenses (Axiovert, C. Zeiss, Germany). Analysis was done by a blinded investigator who estimated the number of inclusions per neurons in five randomly selected fields in one brain section. Six sections per mouse were analyzed.
GC-TOF MS analysis for steroid compounds
Following decapitation, the brain regions (cortex, striatum, hippocampus, cerebellum) were isolated, snap-frozen with liquid nitrogen and freeze-dried until analysis (60). Lyophilized cells were then extracted by methanol:isopropanol:water (3:3:2) after disruption with stainless steel ball and supernatant were collected by centrifugation for 5 min at 16 100 × g. After complete drying, the extracts were derivatized with 10 μl of a solution containing 40 mg/ml of 98% methoxyamine hydrochloride (CAS No. 593-56-6, Sigma, St. Louis, MO, USA) in pyridine (silylation grade, Pierce, Rockford, IL, USA) at 30°C for 90 min followed by silylation with ninety microliters of MSTFA containing 1%TMCS (1 ml bottles, Pierce) at 37°C for 30 min. Metabolite identifications were made based on spectral similarity and retention time index using BinBase and were matched against the Fiehn mass spectral library of ∼1400 authentic metabolite spectra using retention index and mass spectrum information or the NIST05 commercial library (http://fiehnlab.ucdavis.edu/Metabolite-Library-2007/; (60)). The quantification was performed based on peak heights, which were calculated by unique m/z of each steroid compound.
Statistical analysis
Statistical analyses of means for three or more groups were performed using one-way analysis of variance (ANOVA) with the categories of genotype and age as independent factors followed by the Newman–Keuls post hoc test for multiple comparison. For analyses of means involving only two groups with a sample size n < 30, the F-test was used to determine whether the variances between the two groups were significantly different. For samples with a significant difference in variance, the Welch's t-test was applied. Student's t-test was applied for the samples (n ≥ 30) with an insignificant difference in variance. The significance level was set at 0.05 for all analyses. All statistical computations were carried out using Prism (Graphpad Software).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflicts of Interest statement. None declared.
FUNDING
This work was supported by the Mayo Foundation and the National Institutes of Health (NS40738, NS062384, NS069177 and ES020766 to C.T.M. and R01ES020715 to E.T.). We thank Ramandeep Takhter for help with TLC experiments.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr K. Johnson for help with mouse breeding, Mr Jordan Du Charme and Ms Maria Liang for help with animal testing, Dr Liang Zhang and Ms Courtney Hong for help with data analysis. We also acknowledge Dr Scott Gradia and Ms Abbey Hartland from UC Berkeley's MacroLab core facility for assistance in fluorescent protein vector cloning.
REFERENCES
- 1.McMurray C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010;11:786–799. doi: 10.1038/nrg2828. doi:10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trushina E., McMurray C.T. In: Protein Trafficking in Neurons. Bean A.J., editor. Elsevier: 2007. pp. 369–389. [Google Scholar]
- 3.Karasinska J.M., Hayden M.R. Cholesterol metabolism in Huntington disease. Nat. Rev. Neurol. 2011;7:561–572. doi: 10.1038/nrneurol.2011.132. doi:10.1038/nrneurol.2011.132. [DOI] [PubMed] [Google Scholar]
- 4.Costa V., Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO J. 2012;31:1853–1864. doi: 10.1038/emboj.2012.65. doi:10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koga H., Cuervo A.M. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol. Dis. 2011;43:29–37. doi: 10.1016/j.nbd.2010.07.006. doi:10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D., et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. doi:10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becanovic K., Pouladi M.A., Lim R.S., Kuhn A., Pavlidis P., Luthi-Carter R., Hayden M.R., Leavitt B.R. Transcriptional changes in Huntington disease identified using genome-wide expression profiling and cross-platform analysis. Hum. Mol. Genet. 2010;19:1438–1452. doi: 10.1093/hmg/ddq018. doi:10.1093/hmg/ddq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caviston J.P., Zajac A.L., Tokito M., Holzbaur E.L. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol. Biol. Cell. 2011;22:478–492. doi: 10.1091/mbc.E10-03-0233. doi:10.1091/mbc.E10-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Toro D., Alberch J., Lazaro-Dieguez F., Martin-Ibanez R., Xifro X., Egea G., Canals J.M. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. doi:10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 11.Trushina E., Singh R.D., Dyer R.B., Cao S., Shah V.H., Parton R.G., Pagano R.E., McMurray C.T. Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo. Hum. Mol. Genet. 2006;15:3578–3591. doi: 10.1093/hmg/ddl434. doi:10.1093/hmg/ddl434. [DOI] [PubMed] [Google Scholar]
- 12.Leoni V., Mariotti C., Nanetti L., Salvatore E., Squitieri F., Bentivoglio A.R., Bandettini di Poggio M., Piacentini S., Monza D., Valenza M., et al. Whole body cholesterol metabolism is impaired in Huntington's disease. Neurosci. Lett. 2011;494:245–249. doi: 10.1016/j.neulet.2011.03.025. doi:10.1016/j.neulet.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Valenza M., Carroll J.B., Leoni V., Bertram L.N., Bjorkhem I., Singaraja R.R., Di Donato S., Lutjohann D., Hayden M.R., Cattaneo E. Cholesterol biosynthesis pathway is disturbed in YAC128 mice and is modulated by huntingtin mutation. Hum. Mol. Genet. 2007;16:2187–2198. doi: 10.1093/hmg/ddm170. doi:10.1093/hmg/ddm170. [DOI] [PubMed] [Google Scholar]
- 14.Valenza M., Leoni V., Karasinska J.M., Petricca L., Fan J., Carroll J., Pouladi M.A., Fossale E., Nguyen H.P., Riess O., et al. Cholesterol defect is marked across multiple rodent models of Huntington's disease and is manifest in astrocytes. J. Neurosci. 2010;30:10844–10850. doi: 10.1523/JNEUROSCI.0917-10.2010. doi:10.1523/JNEUROSCI.0917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valenza M., Leoni V., Tarditi A., Mariotti C., Bjorkhem I., Di Donato S., Cattaneo E. Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington's disease. Neurobiol. Dis. 2007;28:133–142. doi: 10.1016/j.nbd.2007.07.004. doi:10.1016/j.nbd.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Brown M.S., Goldstein J.L. The LDL receptor and HMG-CoA reductase – two membrane molecules that regulate cholesterol homeostasis. Curr. Top. Cell Reg. 1985;26:3–15. doi: 10.1016/b978-0-12-152826-3.50008-5. [DOI] [PubMed] [Google Scholar]
- 17.Zou H., Stoppani E., Volonte D., Galbiati F. Caveolin-1, cellular senescence and age-related diseases. Mech. Ageing Dev. 2011;132:533–542. doi: 10.1016/j.mad.2011.11.001. doi:10.1016/j.mad.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowa G. Caveolae, caveolins, cavins, and endothelial cell function: new insights. Front. Physiol. 2012;2:120. doi: 10.3389/fphys.2011.00120. doi:10.3389/fphys.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C.H., Tallaksen-Greene S., Chien W.M., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X.J., Albin R.L., Detloff P.J. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. doi:10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 20.Stryer L., Haugland R.P. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. doi:10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simionescu N., Lupu F., Simionescu M. Rings of membrane sterols surround the openings of vesicles and fenestrae, in capillary endothelium. J. Cell. Biol. 1983;97:1592–1600. doi: 10.1083/jcb.97.5.1592. doi:10.1083/jcb.97.5.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauch D.H., Nagler K., Schumacher S., Goritz C., Muller E.C., Otto A., Pfrieger F.W. CNS Synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. doi:10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- 23.Funfschilling U., Saher G., Xiao L., Mobius W., Nave K.A. Survival of adult neurons lacking cholesterol synthesis in vivo. BMC Neurosci. 2007;8:1. doi: 10.1186/1471-2202-8-1. doi:10.1186/1471-2202-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein J.L., Brown M.S., Anderson R.G., Russell D.W., Schneider W.J. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu. Rev. Cell. Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. doi:10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 25.Murata M., Peranen J., Schreiner R., Wieland F., Kurzchalia T.V., Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc. Natl Acad. Sci. USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. doi:10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balch W.E., McCaffery J.M., Plutner H., Farquhar M.G. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. doi:10.1016/0092-8674(94)90359-X. [DOI] [PubMed] [Google Scholar]
- 27.Hirschberg K., Miller C.M., Ellenberg J., Presley J.F., Siggia E.D., Phair R.D., Lippincott-Schwartz J. Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J. Cell. Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. doi:10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks S.P., Dunnett S.B. Tests to assess motor phenotype in mice: a user's guide. Nat. Rev. Neurosci. 2009;10:519–529. doi: 10.1038/nrn2652. doi:10.1038/nrn2652. [DOI] [PubMed] [Google Scholar]
- 29.Hatters D.M. Protein misfolding inside cells: the case of huntingtin and Huntington's disease. IUBMB Life. 2008;60:724–728. doi: 10.1002/iub.111. doi:10.1002/iub.111. [DOI] [PubMed] [Google Scholar]
- 30.Kim T.W., Tanzi R.E. Neuronal intranuclear inclusions in polyglutamine diseases: nuclear weapons or nuclear fallout? Neuron. 1998;21:657–659. doi: 10.1016/s0896-6273(00)80581-4. doi:10.1016/S0896-6273(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Z., Diamond M.I. Huntington disease and the huntingtin protein. Prog. Mol. Biol. Transl. Sci. 2012;107:189–214. doi: 10.1016/B978-0-12-385883-2.00010-2. [DOI] [PubMed] [Google Scholar]
- 32.Trushina E., Du Charme J., Parisi J., McMurray C.T. Neurological abnormalities in caveolin-1 knock out mice. Behav. Brain Res. 2006;172:24–32. doi: 10.1016/j.bbr.2006.04.024. doi:10.1016/j.bbr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Gioiosa L., Raggi C., Ricceri L., Jasmin J.F., Frank P.G., Capozza F., Lisanti M.P., Alleva E., Sargiacomo M., Laviola G. Altered emotionality, spatial memory and cholinergic function in caveolin-1 knock-out mice. Behav. Brain Res. 2008;188:255–262. doi: 10.1016/j.bbr.2007.11.002. doi:10.1016/j.bbr.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Ritz D., Vuk M., Kirchner P., Bug M., Schutz S., Hayer A., Bremer S., Lusk C., Baloh R.H., Lee H., et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell. Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. doi:10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundy D.I., Li W.P., Luby-Phelps K., Anderson R.G. Caveolin targeting to late endosome/lysosomal membranes is induced by perturbations of lysosomal pH and cholesterol content. Mol. Biol. Cell. 2012;23:864–880. doi: 10.1091/mbc.E11-07-0598. doi:10.1091/mbc.E11-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayer A., Stoeber M., Ritz D., Engel S., Meyer H.H., Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J. Cell. Biol. 2010;191:615–629. doi: 10.1083/jcb.201003086. doi:10.1083/jcb.201003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying M., Grimmer S., Iversen T.G., Van Deurs B., Sandvig K. Cholesterol loading induces a block in the exit of VSVG from the TGN. Traffic. 2003;4:772–784. doi: 10.1034/j.1600-0854.2003.00134.x. doi:10.1034/j.1600-0854.2003.00134.x. [DOI] [PubMed] [Google Scholar]
- 38.Reiss A.B., Voloshyna I. Regulation of cerebral cholesterol metabolism in Alzheimer disease. J. Investig. Med. 2012;60:576–582. doi: 10.231/JIM.0b013e318246d973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis V., Hooper N.M. The role of lipid rafts in prion protein biology. Front. Biosci. (Landmark Ed.) 2011;16:151–168. doi: 10.2741/3681. doi:10.2741/3681. [DOI] [PubMed] [Google Scholar]
- 40.Liu J.P., Tang Y., Zhou S., Toh B.H., McLean C., Li H. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Mol. Cell. Neurosci. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. doi:10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Ballabio A., Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. doi:10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Valenza M., Cattaneo E. Emerging roles for cholesterol in Huntington's disease. Trends Neurosci. 2011;34:474–486. doi: 10.1016/j.tins.2011.06.005. doi:10.1016/j.tins.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Kim J.J., Olson L.J., Dahms N.M. Carbohydrate recognition by the mannose-6-phosphate receptors. Curr. Opin. Struct. Biol. 2009;19:534–542. doi: 10.1016/j.sbi.2009.09.002. doi:10.1016/j.sbi.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergo M.O., Young S.G. Extra Rabs unsnarl a lipid traffic jam. Nat. Med. 2002;8:662–664. doi: 10.1038/nm0702-662. doi:10.1038/nm0702-662. [DOI] [PubMed] [Google Scholar]
- 45.Guardia-Laguarta C., Coma M., Pera M., Clarimon J., Sereno L., Agullo J.M., Molina-Porcel L., Gallardo E., Deng A., Berezovska O., et al. Mild cholesterol depletion reduces amyloid-beta production by impairing APP trafficking to the cell surface. J. Neurochem. 2009;110:220–230. doi: 10.1111/j.1471-4159.2009.06126.x. doi:10.1111/j.1471-4159.2009.06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayashi H., Mizuno T., Michikawa M., Haass C., Yanagisawa K. Amyloid precursor protein in unique cholesterol-rich microdomains different from caveolae-like domains. Biochim. Biophys. Acta. 2000;1483:81–90. doi: 10.1016/s1388-1981(99)00174-2. doi:10.1016/S1388-1981(99)00174-2. [DOI] [PubMed] [Google Scholar]
- 47.Peters P.J., Mironov A., Jr, Peretz D., van Donselaar E., Leclerc E., Erpel S., DeArmond S.J., Burton D.R., Williamson R.A., Vey M., et al. Trafficking of prion proteins through a caveolae-mediated endosomal pathway. J. Cell. Biol. 2003;162:703–717. doi: 10.1083/jcb.200304140. doi:10.1083/jcb.200304140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilch S., Kehler C., Schatzl H.M. The prion protein requires cholesterol for cell surface localization. Mol. Cell. Neurosci. 2006;31:346–353. doi: 10.1016/j.mcn.2005.10.008. doi:10.1016/j.mcn.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Moore R.C., Xiang F., Monaghan J., Han D., Zhang Z., Edstrom L., Anvret M., Prusiner S.B. Huntington disease phenocopy is a familial prion disease. Am. J. Hum. Genet. 2001;69:1385–1388. doi: 10.1086/324414. doi:10.1086/324414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Razani B., Engelman J.A., Wang X.B., Schubert W., Zhang X.L., Marks C.B., Macaluso F., Russell R.G., Li M., Pestell R.G., et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. doi:10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- 51.Woodman B., Butler R., Landles C., Lupton M.K., Tse J., Hockly E., Moffitt H., Sathasivam K., Bates G.P. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res. Bull. 2007;72:83–97. doi: 10.1016/j.brainresbull.2006.11.004. doi:10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Smart E.J., Ying Y., Donzell W.C., Anderson R.G. A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J. Biol. Chem. 1996;271:29427–29435. doi: 10.1074/jbc.271.46.29427. doi:10.1074/jbc.271.11.6518. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Wandelmer J., Davalos A., Herrera E., Giera M., Cano S., de la Pena G., Lasuncion M.A., Busto R. Inhibition of cholesterol biosynthesis disrupts lipid raft/caveolae and affects insulin receptor activation in 3T3-L1 preadipocytes. Biochim. Biophys. Acta. 2009;1788:1731–1739. doi: 10.1016/j.bbamem.2009.05.002. doi:10.1016/j.bbamem.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. doi:10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 55.del Toro D., Xifro X., Pol A., Humbert S., Saudou F., Canals J.M., Alberch J. Altered cholesterol homeostasis contributes to enhanced excitotoxicity in Huntington's disease. J. Neurochem. 2010;115:153–167. doi: 10.1111/j.1471-4159.2010.06912.x. doi:10.1111/j.1471-4159.2010.06912.x. [DOI] [PubMed] [Google Scholar]
- 56.Luthi-Carter R., Taylor D.M., Pallos J., Lambert E., Amore A., Parker A., Moffitt H., Smith D.L., Runne H., Gokce O., et al. SIRT2 Inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc. Natl Acad. Sci. USA. 2010;107:7927–7932. doi: 10.1073/pnas.1002924107. doi:10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritch J.J., Valencia A., Alexander J., Sapp E., Gatune L., Sangrey G.R., Sinha S., Scherber C.M., Zeitlin S., Sadri-Vakili G., et al. Multiple phenotypes in Huntington disease mouse neural stem cells. Mol. Cell. Neurosci. 2012;50:70–81. doi: 10.1016/j.mcn.2012.03.011. doi:10.1016/j.mcn.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao H., Thompson H.M., Krueger E.W., McNiven M.A. Disruption of Golgi structure and function in mammalian cells expressing a mutant dynamin. J. Cell. Sci. 2000;113(Pt 11):1993–2002. doi: 10.1242/jcs.113.11.1993. [DOI] [PubMed] [Google Scholar]
- 59.Trenchev P., Konopka M. Combined haematoxylin, eosin and fluorescent stain for paraffin sections. Pathology. 1980;12:79–81. doi: 10.3109/00313028009060056. doi:10.3109/00313028009060056. [DOI] [PubMed] [Google Scholar]
- 60.Scholz M., Fiehn O. SetupX – a public study design database for metabolomic projects. Pac. Symp. Biocomput. 2007;12:169–180. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.