Abstract
Autosomal recessive parkinsonism genes contribute to maintenance of mitochondrial function. Two of these, PINK1 and parkin, act in a pathway promoting autophagic removal of depolarized mitochondria. Although recruitment of parkin to mitochondria is PINK1-dependent, additional components necessary for signaling are unclear. We performed a screen for endogenous modifiers of parkin recruitment to depolarized mitochondria and identified hexokinase 2 (HK2) as a novel modifier of depolarization-induced parkin recruitment. Hexose kinase activity was required for parkin relocalization, suggesting the effects are shared among hexokinases including the brain-expressed hexokinase 1 (HK1). Knockdown of both HK1 and HK2 led to a stronger block in parkin relocalization than either isoform alone, and expression of HK2 in primary neurons promoted YFP-parkin recruitment to depolarized mitochondria. Mitochondrial parkin recruitment was attenuated with AKT inhibition, which is known to modulate HK2 activity and mitochondrial localization. We, therefore, propose that Akt-dependent recruitment of hexokinases is a required step in the recruitment of parkin prior to mitophagy.
INTRODUCTION
Mutations in several different genes cause autosomal recessive parkinsonism in humans (1). Several lines of evidence suggest that two of these, parkin and PINK1, share a common genetic pathway (2–4) whose critical function is to remove damaged mitochondria from the cell (3).
PINK1 is a mitochondrial kinase whose protein levels are regulated through voltage-dependent proteolysis (5,6). Parkin, an E3 protein-ubiquitin ligase (7), is typically cytosolic or nuclear but under conditions of mitochondrial depolarization is relocalized to the outer mitochondrial membrane (OMM) (3). Once recruited, parkin ubiquitylates several OMM proteins, leading to the removal of the ubiquitin-tagged depolarized mitochondria (3,8,9).
Parkin relocalization is dependent upon the accumulation of PINK1 on the OMM and its kinase activity (10–12). Whether additional pathways are required for these events is unclear. Therefore, we undertook a genome-wide shRNA knockdown screen in an unbiased evaluation of the requirement of each gene for parkin recruitment. We recovered PINK1 and identified hexokinase 2 (HK2), which catalyzes the conversion of glucose to glucose-6-phosphate, as a novel modifier of parkin recruitment to mitochondria. We show that hexokinase activity is critical for parkin recruitment and acts downstream of Akt, known to control recruitment of hexokinases to mitochondria (13–16), but upstream of PINK1. These results suggest that cellular signaling plays an important role in the activity of PINK1/parkin.
RESULTS
High-content screen for modifiers of carbonyl cyanide m-chlorophenylhydrazone-induced parkin relocalization to depolarized mitochondria
As parkin is localized to the cytosol under basal conditions but PINK1 accumulates on the OMM, we hypothesized that there would be additional proteins necessary for parkin recruitment to depolarized mitochondria. To investigate this in an unbiased manner, we used genome-wide shRNA interference to screen for modifiers of parkin recruitment to depolarized mitochondria in cells treated with CCCP.
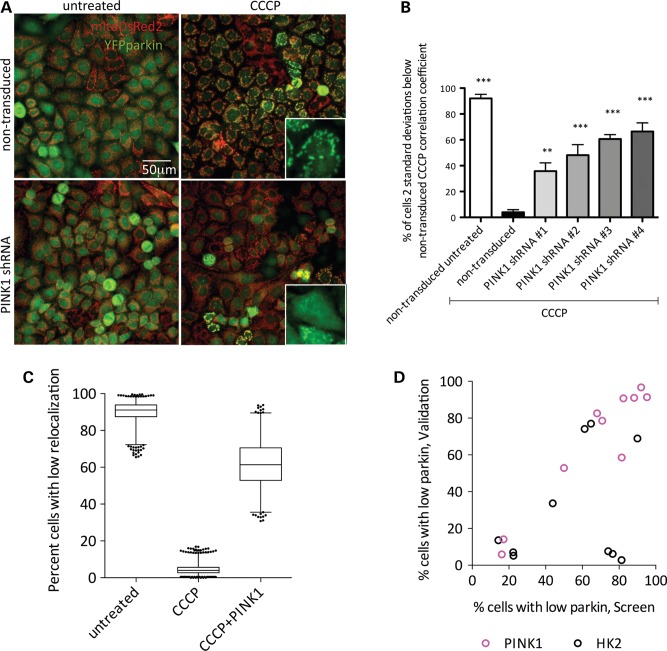
We transfected Flp-In HeLa cells expressing YFP-tagged Parkin (3) with mitochondrially targeted DsRed2. For initial assay optimization, we confirmed that these cells rapidly recruited parkin to mitochondria. As an automated measure of relocalization, we used Pearson's correlation coefficient for the signal intensity between areas of MitoDsRed2 fluorescence and regions of YFP-parkin fluorescence. We also calculated the number of cells that had a correlation coefficient more than 2 standard deviations from the population mean (i.e. did not have parkin relocalization) (Supplementary Material, Fig. S1A). Parkin recruitment using either measure was substantially complete at concentrations of CCCP >1 μm within 1–2 h of exposure (Supplementary Material, Fig. S1A and B). We next transduced this cell line with four different lentiviral shRNA constructs targeting endogenous PINK1 and treated with CCCP (2 μm, 1 h, 37°C). As reported, shRNA-mediated decrease of PINK1 expression in HeLa cells decreased YFP-parkin translocation under depolarizing conditions (Fig. 1A) (17,18). Under control conditions, >90% of cells had low correlation between parkin and MitoDsRed2, and after CCCP treatment <10% of cells had low correlation coefficients. This effect was reversed with 40–60% of cells not responding to CCCP by application of any of four independent shRNA sequences against PINK1 (Fig. 1B). These results show that the HeLa-derived cell line exhibited PINK1-dependent CCCP-induced recruitment of parkin to mitochondria as expected (17,18).
Figure 1.
High-content screen for modifiers of CCCP-induced parkin relocalization to depolarized mitochondria. (A) HeLa cells expressing YFP-parkin and MitoDsRed2 were transduced with PINK1 shRNA lentiviral particles, treated with 2 μm CCCP for 1 h and imaged on a Cellomics VTI arrayscan. Scale bar: 50 μm, insets show higher magnification of YFP-parkin. (B) Cultures were treated as in (A) using four independent shRNA clones directed against PINK1 and imaged. Bars show the percentage of cells from 200 counted for which the correlation coefficient between YFP-parkin and MitoDsRed2 was more than 2 standard deviations below the mean of CCCP-treated controls. Error bars denote the SEM, n = 3 wells per condition. One-way ANOVA with Dunnett's post hoc test; **P < 0.01; ***P < 0.001 versus CCCP-treated control. (C) Percentage of cells with low correlation coefficients in the high-content screen. Central line shows the median, the box denotes 1–3 quartiles and range bars denote 1–99 percentiles. (D) Comparison of percent cells with low correlation coefficients from the primary screen (x-axis) versus validation (y-axis). Each point represents a different PINK1 (magenta circles) or HK2 (black circles) shRNA.
We next undertook a genome-wide shRNA screen to find additional modifiers of parkin recruitment. We utilized a library of ∼75 000 independent clones targeting 15 000 genes. We transduced each construct independently and measured parkin relocalization following treatment with 2 μm CCCP. On each plate, we included controls of non-transduced untreated cells, non-transduced CCCP-treated cells and PINK1 shRNA-transduced, CCCP-treated cells (Fig. 1C). These controls allowed us to count the percentage of cells without relocalization, as well as ensured that lentiviral transduction was efficient for each plate. Throughout the screen, these different control conditions were effectively separated and we calculated a z-score between untreated and CCCP-treated controls of 0.706 (Fig. 1C). For the screen, we used a slightly longer timepoint (2 h) than the initial shRNA experiments (1 h; Fig. 1A and B) to minimize the number of genes that might marginally slow parkin recruitment rather than block it. Furthermore, because a PINK1 shRNA control was used on each plate, we were able to show that the block in parkin recruitment mediated by PINK1-deficiency was maintained at 2 h. (Fig. 1C).
Screen data were ranked by false discovery rate (FDR)-adjusted P-values generated from two-proportion z-tests of the percentage of cells for each shRNA construct without parkin relocalization. The mean percentage of cells without parkin recruitment across all shRNA-transduced and CCCP-treated conditions was 14.36%. Eighteen genes met the criteria of having at least three clones arrayed on multiple plates with significantly different recruitment. After a secondary screen, two genes were validated as modifying parkin recruitment. We recovered eight clones for PINK1, distinct from the positive control PINK1 clone included in the screen. We additionally found seven shRNA clones for HK2 and confirmed four of these in the validation step (Fig. 1D, Table 1). We, therefore, considered HK2 to be a novel candidate modifier of parkin recruitment to mitochondria.
Table 1.
HK2 and PINK1 results from the screen and validation
| Clone # | Gene | Screen percent of cells with low correlation coefficient | Validation percent of cells with low correlation coefficient |
|---|---|---|---|
| TRCN0000037671 | HK2 | 43.9 | 33.64 |
| TRCN0000037672 | HK2 | 14.22 | 13.59 |
| TRCN0000037673 | HK2 | 61.28 | 74.09 |
| TRCN0000197099 | HK2 | 76.53 | 6.13 |
| TRCN0000196260 | HK2 | 74.06 | 7.62 |
| TRCN0000195582 | HK2 | 81.36 | 2.78 |
| TRCN0000199344 | HK2 | 64.68 | 76.99 |
| TRCN0000195340 | HK2 | 89.95 | 68.93 |
| TRCN0000037669 | HK2 | 22.33 | 7.04 |
| TRCN0000037670 | HK2 | 22.46 | 5.16 |
| TRCN0000007097 | PINK1 | 16.14 | 5.85 |
| TRCN0000007098 | PINK1 | 50 | 52.86 |
| TRCN0000007099 | PINK1 | 70.87 | 78.57 |
| TRCN0000007100 | PINK1 | 68 | 82.61 |
| TRCN0000007101 | PINK1 | 95.19 | 91.43 |
| TRCN0000196758 | PINK1 | 17.03 | 14.07 |
| TRCN0000197083 | PINK1 | 92.04 | 96.72 |
| TRCN0000199193 | PINK1 | 88.24 | 91.04 |
| TRCN0000199255 | PINK1 | 81.42 | 58.54 |
Endogenous HK2 modifies parkin recruitment to mitochondria
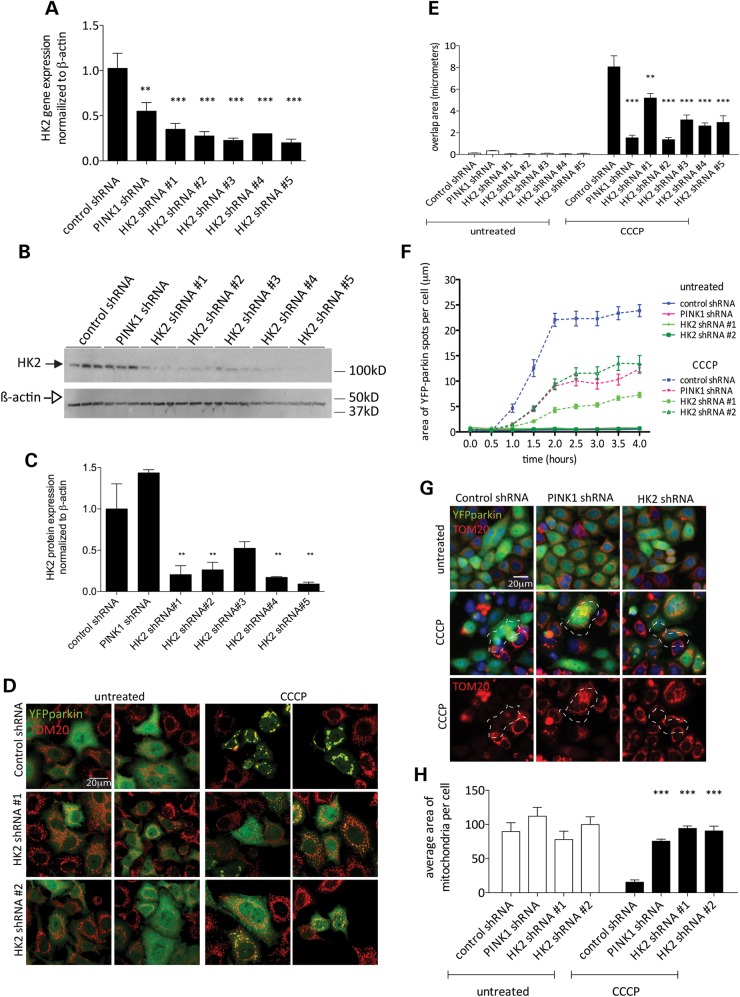
We next generated a series of HeLa clones with stable expression of five different lentiviral shRNA constructs targeting HK2 and a stable PINK1 shRNA line in parallel. Stable integrated clones each expressing one of the five different shRNA sequences had lower HK2 mRNA (Fig. 2A) and protein levels (Fig. 2B), with residual HK2 protein of ∼20–30% of the parental line (Fig. 2C). For all five independent HK2 sequences, the reduction in HK2 protein levels resulted in attenuated CCCP-dependent YFP-parkin mitochondrial recruitment (Fig. 2D and E). Having established that all five sequences were effective in blocking CCCP-induced parkin recruitment, we took two HK2 shRNA sequences forward for subsequent experiments, with some additional controls for off-target effects used in later experiments.
Figure 2.
HK2 deficiency decreases YFP-parkin recruitment to mitochondria. (A) Relative HK2 mRNA levels in YFP-parkin HeLa cell lines with stable expression of lentiviral shRNA constructs targeting HK2 or PINK1 normalized to β-actin. Error bars are SEM (n = 4). One-way ANOVA with Dunnett's post hoc test; **P < 0.01; ***P < 0.001 versus control shRNA. (B) Western blot of PINK1 and HK2 shRNA lines probed for HK2 (closed arrow) and β-actin (open arrow). (C) Quantitation of (B) with HK2 values normalized to β-actin relative to control shRNA; error bars are SEM (n = 3). One-way ANOVA with Dunnett's post hoc test; **P < 0.01 versus control shRNA. (D) Cell lines stably expressing YFP-parkin, and either control, PINK1 or HK2 shRNA-treated 2 μm CCCP (2 h), were fixed, stained with anti-TOM20 and imaged by confocal microscopy. Scale bar: 20 μm. (E) Stable HeLa lines expressing control, PINK1 or HK2 shRNA were treated with 2 μm CCCP (2 h in low-glucose media), fixed and imaged on the Cellomics VTI arrayscan. Bars show the mean overlap area between YFP-parkin and MitoDsRed2 structures (n = 6 wells per condition, error bars indicate the SEM). Data from CCCP-treated conditions were analyzed by one-way ANOVA with Dunnett's post hoc test; **P < 0.01; ***P < 0.001 versus CCCP-treated control shRNA. (F) Living cells labeled with Hoechst 33342 were imaged on the Cellomics VTI arrayscan every 30 min for 4 h after treatment with 2 μm CCCP. Data points are mean areas of discreet YFP-parkin ‘spots’ per cell and error bars are SEM. Data were analyzed by repeated measure two-way ANOVA with Bonferroni's post hoc test. All PINK1 and HK2 shRNA lines were significantly different from CCCP-treated control shRNA at all timepoints after 1h (P < 0.001). (G) Cell lines were treated for 24 h with 2 μm CCCP, fixed and stained with anti-TOM20. Scale bar: 20 μm. (H) Quantitation from (G), of the total TOM20 signal (n = 10 wells per condition, error bars indicate SEM). Data from CCCP-treated conditions were analyzed by one-way ANOVA with Dunnett's post hoc test, ***P < 0.001, significantly different from CCCP-treated control shRNA.
The above effects could represent either a slowing of parkin recruitment or a limit in the total amount of parkin recruited. To distinguish between these possibilities, we used live-cell assays to visualize the same cells over several hours after CCCP treatment, counting the area of discrete YFP-parkin-positive signal per cell. In control cells, the maximal recruitment signal occurred by ∼2 h (Fig. 2F). Although the amount of recruitment was diminished in the PINK1 shRNA line and HK2 cell lines, the time course appeared similar (Fig. 2F). We, therefore, infer that deficiency in PINK1 or HK2 limits total parkin recruitment rather than slowing the kinetics of recruitment.
HeLa cells in culture can be induced to remove all mitochondria by autophagy after extended exposure to CCCP (3). To determine whether HK2 deficiency blocks mitophagy, we assayed total mitochondrial volume by staining for TOM20. Similar to recruitment of parkin, we found that either PINK1 or two independent HK2 shRNA sequences could completely block the removal of mitochondria after exposure to 2 μm CCCP for 24 h (Fig. 2G and H).
These results suggested that in both transient assays (Fig. 1D) and in stable cell lines (Fig. 2D and E), endogenous HK2 is required for recruitment of parkin to mitochondria and subsequent mitophagy. Two sets of experiments were performed to exclude trivial explanations for these effects unrelated to HK2 knockdown. We were first concerned that the shRNA sequences might have off-target effects directed against PINK1. However, PINK1 mRNA levels were unchanged with HK2 knockdown even though we could reliably detect a decrement in the PINK1 shRNA stable cell lines (Supplementary Material, Fig. S2A). Second, we investigated whether cell lines with HK2 knockdown displayed any differences in mitochondrial membrane potential. Using tetramethylrhodamine ethyl ester (TMRE) to label active mitochondria in the PINK1 and HK2 cell lines, we observed no difference in basal TMRE fluorescence among cell lines (Supplementary Material, Fig. S2B) and no differences in the rate of CCCP-induced loss of TMRE signal (Supplementary Material, Fig. S2C and D).
Hexokinase activity is genetically upstream of PINK1 and crucial for parkin recruitment in HeLa cells and in primary cortical neurons
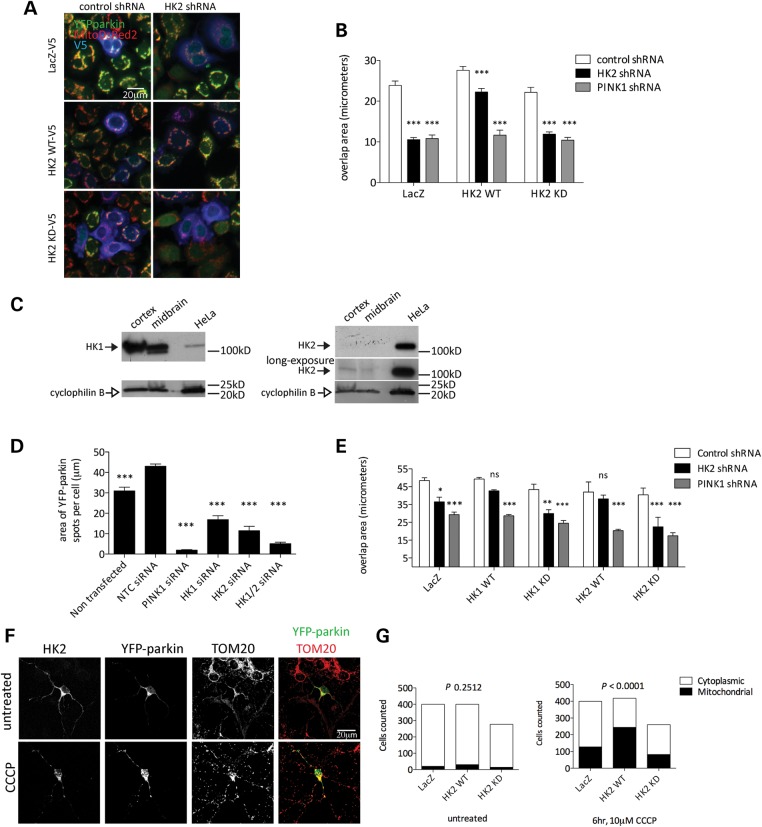
To further exclude that off-target effects of shRNA sequences were responsible for diminished YFP-parkin recruitment, we performed rescue experiments by reintroducing HK2 expression plasmids into the stable HK2-deficient cell lines. We also tested whether hexokinase activity is necessary for HK2 to support parkin recruitment by comparing transfection with either a wild-type (WT) or D209A/D657A HK2 construct that cannot phosphorylate glucose (19). Both variants of HK2 were expressed at similar steady-state levels as estimated by western blotting (data not shown). We found that WT, but not catalytically inactive HK2, could rescue parkin relocalization in HK2-deficient cell lines (Fig. 3A and B). Interestingly, HK2 expression had no effect on parkin localization following PINK1 knockdown. These results suggest, first, that it is hexokinase activity that controls parkin recruitment and, second, that PINK1 is required for the effects of HK2 in HeLa cells.
Figure 3.
Kinase active HK2 promotes depolarization-induced parkin recruitment in HeLa cells and primary cortical neurons. (A) HeLa lines stably expressing YFP-parkin, MitoDsRed2 and either control (left panels) or HK2 (right panels) shRNA transfected with V5-tagged LacZ (upper panels), HK2 WT (middle panels) or HK2 KD (lower panels) constructs and treated with 2 μm CCCP (2 h, low-glucose media). Scale bar: 20 μm. (B) Quantitation from (A) of the mean overlap area between YFP-parkin and MitoDsRed2 in cells co-expressing V5-tagged constructs (n = 6 wells per condition, error bars are SEM). Data from CCCP-treated conditions were analyzed by two-way ANOVA with Bonferroni's post hoc test; ***P < 0.001, versus CCCP-treated control shRNA for each construct. (C) Immunoblots of HK1 and HK2 (filled arrows) in the human frontal cortex, human midbrain and HeLa lysates with cyclophilin B (open arrows) as a loading control. (D) Effects of knockdown of both HK1 and HK2 on parkin recruitment. HeLa cells stably expressing YFP-parkin were transfected with siRNA against PINK1, HK1, HK2 or both HK1 and HK2 and were treated with 2 μm CCCP (2 h, low-glucose media) for 2 h, fixed and analyzed for the mean area of discreet YFP-parkin ‘spots’ per cell (n = 5 wells per condition, error bars are SEM). Data from CCCP-treated conditions were analyzed by one-way ANOVA with Dunnett's post hoc test; ***P < 0.001, significantly different from non-targeting control siRNA condition. (E) HeLa lines stably expressing YFP-parkin, MitoDsRed2 and either control, HK2 or PINK1 shRNA were transfected with V5-tagged LacZ, HK1 WT, HK1 KD, HK2 WT or HK2 KD constructs and treated with 2 μm CCCP (2 h, low-glucose media). Data graphed are the mean overlap area between YFP-parkin and MitoDsRed2 in cells co-expressing V5-tagged constructs (n = 3 wells per condition, error bars are SEM). Data from CCCP-treated conditions were analyzed by two-way ANOVA with Bonferroni's post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001, versus CCCP-treated control shRNA for each construct. (F) Primary cortical neurons transfected with YFP-parkin and V5-tagged LacZ, HK2 WT or HK2 KD constructs were treated with 10 μm CCCP for 6 h, fixed and labeled for TOM20 and V5. Scale bar: 20 μm. (G) Quantification of the proportion of neurons with discreet areas of mitochondria-localized YFP-parkin assessed under blinded conditions. P-values are by chi-squared test.
There are two active hexokinases in humans, HK2 and HK1. Based on the above results, we inferred that either HK1 or HK2 should be able to support parkin relocalization. However, HK1 was not recovered in the initial screen in HeLa cells (Supplementary Material, Table S1). To resolve whether HK1 could substitute for HK2, we first examined relative HK1 and HK2 protein levels in HeLa cells compared with brain (Fig. 3C). We found that HK2 was expressed at higher levels in HeLa cells than in the brain and, conversely, HK1 was expressed at high levels in the human cerebral cortex or the midbrain, with only modest expression in HeLa. These results suggest that hexokinase activity might be related predominantly to HK2 in HeLa cells. However, there is some HK1 expression and it was, therefore, possible that the residual parkin recruitment seen with HK2 knockdown might represent HK1. To address this, we performed dual knockdown of both isoforms and found that there was an additive effect of knocking down HK1 on an HK2 background (Fig. 3D). Furthermore, parkin recruitment in HK2 knockdown HeLa cells was rescued by overexpression of kinase active, but not kinase-dead (D657A) HK1 (Fig. 3E). These results suggest that both HK1 and HK2 contribute to parkin relocalization but that in HeLa cells, HK2 activity predominates.
It has been controversial whether parkin can be recruited to depolarized mitochondria in neurons, with both positive (3,18,20,21) and negative results reported (22). Potentially relevant in the context of glucose metabolism, the metabolic state of neurons has been suggested to regulate parkin recruitment (22). We, therefore, decided to repeat the above experiments in neurons.
Based on expression patterns described above (Fig. 3C), we reasoned that we would be unlikely to be able to knockdown HK2 in neurons, and so we used overexpression of HK2 with the kinase-dead variant as a negative control. In our hands, without exogenous expression of hexokinase (i.e. with endogenous HK1/2 only), ∼25% of neurons showed clear parkin relocalization in neurons although with longer exposure to CCCP (Fig. 3F and G). In agreement with experiments in HeLa cells, we found that expression of WT, but not kinase-inactive HK2 promoted increased CCCP-dependent mitochondrial YFP-parkin localization in neurons (Fig. 3F and G). These results suggest that hexokinase activity promotes parkin recruitment in multiple cell types, including neurons.
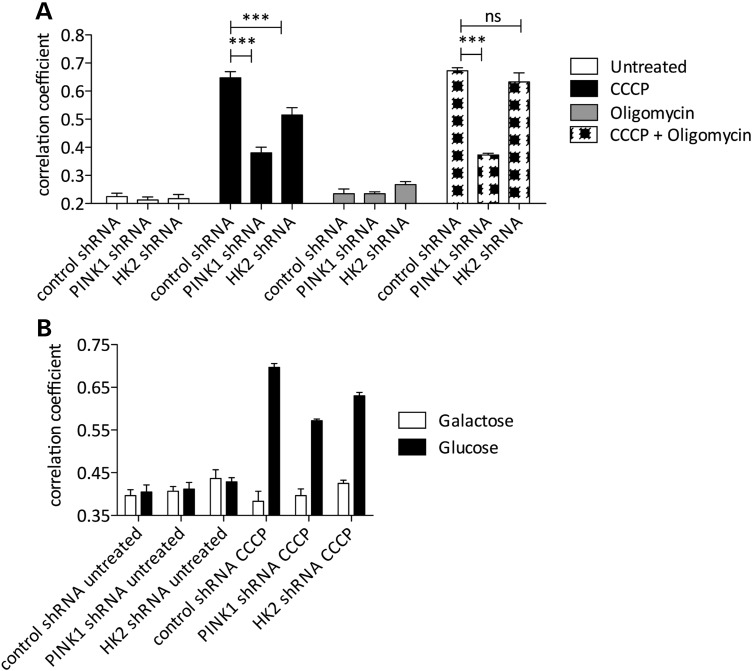
One proposed explanation for previous negative results for parkin relocalization in neurons is that CCCP-induced relocalization of parkin to mitochondria does not occur in cells reliant on oxidative phosphorylation (22). Given that hexokinase activity is thought to be critical for a shift from oxidative phosphorylation to glycolysis (23), we decided to examine how metabolic status would affect these results. First, we used oligomycin to inhibit mitochondrial H+ ATP synthase and block glycolysis, and found that the effects of HK2 knockdown, but not PINK1 knockdown, were reverted (Fig. 4A). Second, we confirmed previous results that growth of HeLa cells on galactose rather than glucose prevented CCCP-induced parkin relocalization (22), and further showed that this was seen in both HK2- and PINK1- deficient cells (Fig. 4B). These results suggest that parkin relocalization under conditions of mitochondrial depolarization requires both ATP synthesis and active PINK1 signaling.
Figure 4.
Parkin recruitment requires both oxidative phosphorylation and PINK1 signaling. (A) HeLa lines stably expressing YFP-parkin/MitoDsRed2 and either control, PINK1 or HK2 shRNA were treated with 2 μm CCCP and 10 μm oligomycin as indicated (2 h, low-glucose media), and the mean overlap area between YFP-parkin and MitoDsRed2 was analyzed (n = 4 wells per condition, error bars are SEM). Data from CCCP-treated conditions were analyzed by two-way ANOVA with Bonferroni's post hoc test; ***P < 0.001, versus control shRNA for each treatment condition. (B) HeLa lines stably expressing YFP-parkin/MitoDsRed2 and either control, PINK1 or HK2 shRNA were cultured in glucose or galactose complete media and treated with 2 μm CCCP (2 h, low-glucose media or galactose-containing media), and the mean overlap area between YFP-parkin and MitoDsRed2 was analyzed (n = 6 wells per condition, error bars are SEM).
Akt-dependent HK2 recruitment to mitochondria contributes to parkin relocalization
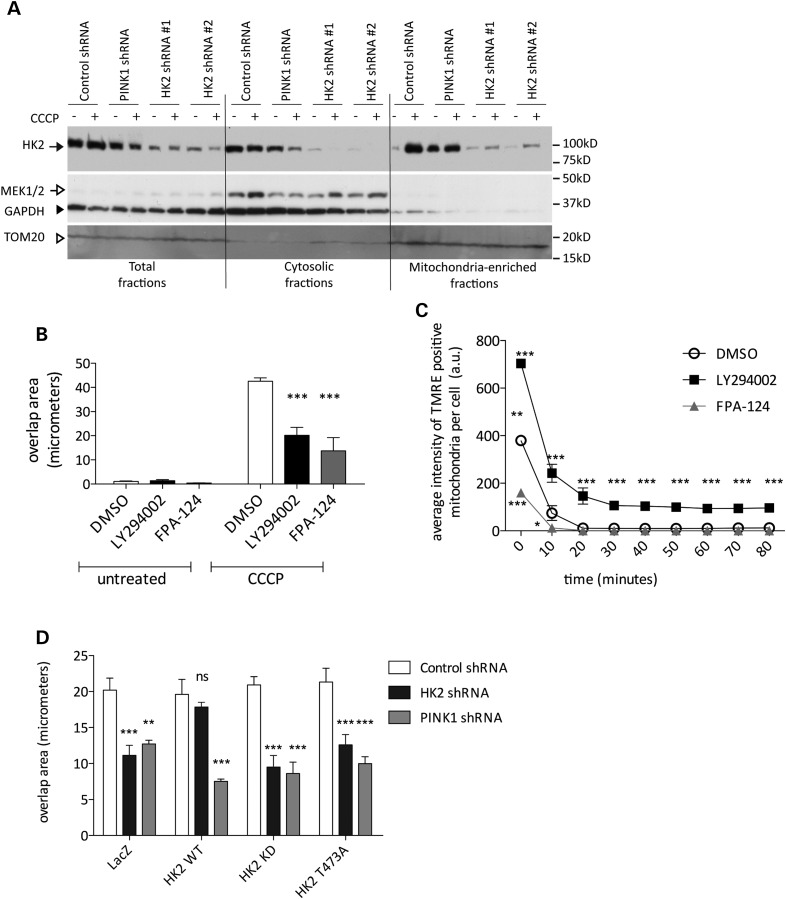
In the above experiments with neurons, we noted that HK2 was redistributed to mitochondria after CCCP treatment (Fig. 3F). It has been shown previously that both glucose conversion to glucose-6-phosphate and HK-2-dependent cell survival are increased upon binding of HK2 with VDAC at the OMM (14,24,25). We confirmed that we could recruit endogenous HK2 to mitochondria after CCCP treatment (Fig. 5A). Interestingly, the recruitment of HK2 occurred both in control cell lines and in PINK1-deficient lines, suggesting that the effects of CCCP on HK2 recruitment are not signaled by PINK1.
Figure 5.
Akt promotes parkin relocalization by HK2. (A) YFP-parkin stable HeLas were treated with 2 μm CCCP for 2 h and equal amounts of cellular fractions per lane were loaded. The closed arrow indicates HK2, the open arrow indicates MEK1/2 for cytosol, the closed arrowhead indicates GAPDH as a loading control and the open arrowhead indicates TOM20 as a marker of mitochondria. (B) YFP-parkin stable HeLas were pretreated with Akt inhibitors (50 μm each, 1 h) prior to treatment with 2 μm CCCP (2 h) and the mean overlap area between YFP-parkin and MitoDsRed2 was measured using the co-localization bioapplication on the Cellomics VTI arrayscan (n = 6 wells per condition). Values are mean overlap area, error bars are SEM. Data were analyzed by two-way ANOVA with Bonferroni's post hoc test comparing with DMSO control; ***P < 0.001. (C) YFP-parkin stable HeLa cells were labeled with TMRE prior to pretreatment with AKT inhibitors and treatment with 2 μm CCCP. TMRE was imaged on the Cellomics VTI arrayscan (n = 6 wells per condition). Points show TMRE intensities normalized to the 0 timepoint for each condition, error bars are SEM. Data were analyzed by repeated measures two-way ANOVA with Bonferroni's post hoc test comparing with CCCP-treated DMSO control; *P < 0.05, **P < 0.01, ***P < 0.001 (D) HeLa lines were transfected with V5-tagged LacZ, HK2 WT, HK2 KD or HK2 T473A constructs and treated with 2 μm CCCP (2 h, low-glucose media). Bars show mean overlap area between YFP-parkin and MitoDsRed2 structures in cells co-expressing V5-tagged constructs; error bars are SEM (n = 4 wells per condition). Data were analyzed by two-way ANOVA with Bonferroni's post hoc test, **P < 0.01, ***P < 0.001, significantly different from CCCP-treated control shRNA.
The recruitment of HK2 to the OMM is enhanced by phosphorylation of HK2 at T473 by Akt (14,26,27). To further understand the mechanism by which HK2 recruits parkin to mitochondria, we used pharmacological and genetic approaches. We first inhibited Akt signaling with two different Akt inhibitors, LY294002 and FPA-124 (50 μm each). Both compounds significantly decreased the amount of parkin recruitment after CCCP treatment (Fig. 5B). The same effects were seen in both control cell lines and in HK2-deficient cells (data not shown). These effects were not explained by protection against mitochondrial depolarization, as TMRE staining was diminished after CCCP treatment in the presence of the two inhibitors, and LY294002 increased TMRE staining in the cells indicating hyperpolarization (Fig. 5C).
Second, we investigated the ability of T473A mutant HK2 to support parkin relocalization in HK2-deficient HeLa lines (Fig. 5D). We found that T473A or kinase-dead HK2 was unable to rescue parkin relocalization. Collectively, these results show Akt-mediated phosphorylation of HK2 at T473 is required for subsequent PINK1- and HK2-dependent parkin recruitment.
DISCUSSION
Here, we identified HK2 as a modifier of parkin recruitment to mitochondria prior to mitophagy in both transformed cells and neurons. Mechanistically, we show that the hexokinase activity of HK2 is critical and PINK1 must be expressed. Furthermore, we establish that Akt contributes to recruitment of parkin via phosphorylation of T473, resulting in recruitment of HK2 to the OMM.
Unbiased screens have been used recently to identify several aspects of parkin signaling, including interaction with (28) and ubiquitylation of HK1 and HK2 (9). Here, the identification of multiple shRNA sequences for PINK1 demonstrated that the screen had worked, as this was an expected result (12,29,30). Several approaches were used to authenticate HK2 as a modifier for parkin recruitment. First, we recovered multiple independent sequences from the initial screen. Second, we were able to replicate the effects with both transient knockdown and generation of stable lines. Third, in the stable situation, we used five independent sequences. Fourth, we were able to rescue the effects by re-expression of HK2 cDNA. Fifth, inactive mutants of HK2 could not rescue the shRNA, showing sequence specify. Collectively, these data show that HK2 is a true modifier of parkin recruitment.
We note that the relative expression of HK1 compared with HK2 is higher in the brain. However, the observation that hexokinase activity itself is required for parkin recruitment suggests that HK1 or HK2 should be equally effective in this pathway. Although we show that HK2 can promote parkin recruitment in neurons, it is likely that HK1 would be more important in the brain. Our results also show that Akt-dependent recruitment of HK2 to mitochondria is important. The recruitment of hexokinase to mitochondria supports glycolysis, allowing for energy metabolism in the absence of oxygen (23). This has implications for both aging (31) and tolerance to oxidative stress (32), both processes implicated in Parkinson's disease. We speculate that diminishment of Akt signaling would be of interest to examine in the context of parkinsonism, and there is some evidence that Akt signaling is altered in sporadic PD (33,34).
Our results show that HK2 expression will not rescue PINK1 deficiency, placing it downstream of PINK1 in a pathway. Interestingly, there is evidence of disruption of Akt signaling in PINK1-deficient cells (35). Therefore, one interpretation of our data is that loss of PINK1 limits hexokinase activity and recruitment because of altered Akt signaling. Furthermore, mTor, a downstream target of Akt, plays a role in PINK1 phenotypes both in vitro and in vivo (36). We, therefore, consider these observations to be consistent with a signaling pathway that involves control of recruitment of cytosolic proteins to the OMM. In this context, it is interesting that phosphorylation of T473 of HK2 by Akt has been recently shown to diminish the effects of oxidative stress in cardiomyocytes (37). Therefore, we propose that Akt-mediated recruitment of hexokinase to mitochondria is a general cell biological pathway that is of benefit to protect against oxidative stress, which is also a function ascribed to PINK1 both in neurons (38–40) and in other tissues (41).
Parkin has been shown to be recruited to the OMM in many studies (3,8,12,17,18,20,29,30,42,43). Whether this occurs in neurons has been controversial, although we note that it was robust under the conditions we tested. Our data would be consistent with the proposal that the PINK1/parkin pathway is dependent on bioenergetic status (22), but suggest that part of the apparent discrepancy between studies may be related to the amount of hexokinase activity in neurons. Although it has been suggested that neurons are exclusively reliant on oxidative phosphorylation and glial cells alone are capable of glycolysis, there is evidence in the human brain that both pathways may be present in both cell types (44). Specific conditions, including oxygen–glucose deprivation (24), can induce HK2 in neurons, suggesting that PINK1-dependent parkin translocation could be active within neurons at least under some circumstances. Therefore, mitochondrial turnover by PINK1 may not be a constitutive pathway in all cells.
Once recruited to the OMM, parkin then ubiquitylates several target proteins (8,9). Of interest is that HK1 and HK2 are targets for parkin. As parkin binds to hexokinases, this may be part of the mechanism by which HK2 is required for parkin recruitment. This observation also suggests that parkin activity may be terminated by turnover of HK1/2 after ubiquitylation. If hexokinase activity were then lost, the PINK1/parkin pathway would not then be active.
The above data highlight that the signaling pathways important for PINK1- and parkin-mediated mitophagy, and by extension for recessive parkinsonism, have multiple additional aspects that are active in cells. Additional studies are required to identify whether there are further controls of the PINK1/parkin pathway.
MATERIALS AND METHODS
Plasmids, cell lines and human tissue
The YFP-parkin HeLa cell line and the MitoDsRed2 plasmid were obtained from Dr Richard Youle (NINDS, Bethesda, MD, USA). The HK1 and HK2 constructs (45) were obtained from Addgene (Addgene plasmids 23730 and 23854) and transferred into pLenti6/V5-DEST using the Gateway recombination technology (Invitrogen). D209A/D657A and T473A mutations were introduced by the QuikChange II site-directed mutagenesis kit (Stratagene) and verified by sequencing. Frozen samples from the human frontal cortex (male, 45 years) and ventral midbrain (male, 48 years) were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland Baltimore, MD, USA.
Cell culture and transfections
YFP-parkin stable HeLas were maintained in DMEM (Lonza) containing 4.5 g/l glucose, 2 mm l-glutamine and 10% fetal bovine serum (Lonza) with 200 μg/ml Hygromycin B (Invitrogen) for selection at 37°C in 5% CO2. Transient transfection of HeLa cells was performed using PEI (polyethyeleneimine) (Polysciences) as described (46) or Lipofectamine 2000 (Invitrogen). Stable YFP-parkin/MitoDsRed2 HeLas were generated by Lipofectamine 2000 transfection followed by selection in 400 μg/ml G418 (Invitrogen) and subcloning in the presence of 200 μg/ml Hygromycin B and 400 μg/ml G418. YFP-parkin HeLa and YFP-parkin/MitoDsRed2 cells expressing lentiviral shRNA constructs were generated by transduction with lentiviral particles at a multiplicity of infection (MOI) of ∼8 in the presence of 8 µg/ml polybrene (Sigma), and selected with 2 μg/ml puramycin (Invitrogen). To further reduce HK2 levels, YFP-parkin/MitoDsRed2-co-expressing HeLa cells were transduced twice more with lentiviral particles at 8 MOI on sequential days. All shRNA lines were maintained in the presence of 200 μg/ml Hygromycin B and 1 μg/ml puramycin. LY294002 (Calbiochem), FPA124 (Santa Cruz Biotechnology), oligomycin (Sigma) and CCCP (Sigma) were used as indicated. Transient siRNA transfections were performed using Dharmafect reagent 1 according to the manufacturer's protocol with each siRNA at 25 nm and cells were treated and harvested 3 days after transfection (Thermo Scientific). To measure parkin relocalization in the absence of glucose, cells were cultured for three consecutive passages in glucose-free DMEM (Invitrogen) supplemented with 10 mm galactose (Sigma) and 10% FBS before plating.
Primary cortical neuron culture and transfections
Primary neurons from cortices were prepared from C57BL6/J newborn (P0) pups. After tissue dissection, neuronal cells were dissociated by papain (Worthington), plated onto coverslips precoated with poly-d-lysine (Neuvitro) at 0.7 × 106 cells/well in Basal Medium Eagle (Sigma) supplemented with B27, N2, glutaMAX-I, (Invitrogen) and 0.45% glucose (Sigma-Aldrich). Three micromolar cytosine arabinoside (Sigma-Aldrich) was added the day following plating. Half of media was replaced at 1, 4 and 6 days after plating (DIV). Neurons were transfected at 7 DIV using a modified CaPO4 method (47) and treated with CCCP 48 h following transfection. Animal studies were in accordance with the guidelines approved by the Institutional Animal Care and Use Committees of the National Institute of Child Health and Human Development, NIH.
Lentiviral shRNA screen
Transduction particles were purchased as the Mission Human Virus Library LIBHVI (Sigma). YFP-parkin/MitoDsRed2 stable HeLas were plated at 1200 cells per well in 96 well plates in 100 μl of DMEM (Invitrogen) and 10% FBS (Gibco). The following day, media was replaced with 95 μl of fresh media containing 8.4 μg/ml polybrene and cells were transduced with 5 μl of lentiviral transduction particles. On each plate, five wells were left untransduced and one well was transduced with a PINK1 shRNA (see Supplementary Material). The day after transduction, media was changed to 120 μl DMEM/10% FBS per well, and 72 h post-transduction, cells were treated for 2 h with 2 μm CCCP (Sigma) in serum-free DMEM. Two wells per plate were not CCCP-treated as negative controls for parkin recruitment. Cells were fixed in 50 μl of 4% paraformaldehyde (PFA) containing 1 μg/ml Hoechst. The Cellomics VTI ArrayScan co-localization bioapplication was used to quantify YFP-parkin mitochondrial localization by measurement of Pearson's correlation coefficient between areas of MitoDsRed2 fluorescence and regions of YFP-parkin fluorescence in the cytoplasm in 200 cells per condition. Raw data were filtered to exclude genes outside a 2-sigma cut-off or those that did not have three independent shRNAs. Two-proportion z-tests were performed for the percentage of cells for each shRNA construct with correlation coefficients >2 standard deviations below the mean of CCCP-treated non-transduced controls and calculated P-values were adjusted for multiple test correction using an FDR procedure. A series of genes with the lowest P-values and >3 shRNA clones, as well as 18 genes with at least 3 significantly different clones arrayed on multiple plates of the screen were validated under the same conditions used in the initial screen.
TMRE labeling
YFP-parkin HeLa cells were loaded with 80 nm TMRE and 1 μg/ml Hoechst in serum-free low-glucose DMEM (Invitrogen) for 15 min. Media containing 40 nm TMRE was added before imaging, and CCCP was added in the presence of 40 nm TMRE. All image capture and analysis was performed on the Cellomics VTI arrayscan using a live-cell chamber (37°C, 5% CO2). Analysis was performed using the spot detection bioapplication for TMRE-positive mitochondria within the cytosol, and mitochondria were measured for total intensity in a minimum of five fields at ×20 magnification.
SDS–PAGE and western blotting
Cells for western blot were lysed in 1× cell lysis buffer (Cell Signaling) with complete protease inhibitors (Roche), and lysates were clarified by centrifugation at 16 000 g for 10 min. Proteins were resolved on 4–20% Criterion TGX pre-cast gels (Bio-Rad) in SDS/Tris–glycine running buffer, and transferred to polyvinylidenedifluoride membranes. Membranes were blocked with 5% nonfat milk prepared in Tris-buffered saline containing 0.1% Tween 20 and incubated for 1–2 h at RT or overnight at 4°C with the indicated primary antibody. Membranes were washed (3 × 5 min at RT), incubated with horseradish peroxidase-conjugated anti-mouse or rabbit IgG (Jackson ImmunoResearch Laboratories), washed again and developed using enhanced chemiluminescence plus reagent (Thermo Scientific). Primary antibodies used for western blot were rabbit anti-HK2 (Cell Signaling, 1:1000), rabbit anti-HK1 (cell signaling 1:1000), rabbit anti-cyclophilin B (Abcam 1:5000), rabbit anti-TOM20 (Santa Cruz Biotechnology, 1:5000), rabbit anti-GAPDH (Abcam, 1:3000), mouse anti-β-actin (Sigma, 1:5000), rabbit anti-MEK1/2 (Cell Signaling 1:1000). Human brain samples were processed using dounce homogenization in cell lysis buffer at 10% (w/v) on ice.
Immunofluorescence
Following fixation in 4% PFA, neurons or HeLas were blocked with 5% FBS in PBS with 0.1% Triton-X100. Primary antibodies were incubated for 2 h at RT or overnight at 4°C. After washing with PBS, secondary fluorescently labeled antibodies (Invitrogen) were diluted in blocking buffer and incubated for 1 h at RT followed by washing and then mounting with ProLong Gold antifade reagent (Invitrogen) or stored in PBS for analysis on the Cellomics VTI arrayscan. Primary antibodies used for ICC experiments were rabbit anti-TOM20 (Santa Cruz Biotechnology, 1:1000), mouse anti-V5 (1:500, Invitrogen) and rabbit anti-V5 (1:1000, Abcam). All Alexa fluor secondary antibodies used for ICC were from Invitrogen and used at 1:500.
qRT-PCR analysis
cDNA was synthesized from Trizol-extracted RNA using SuperScript III according to the manufacturer's instructions (Invitrogen). We measured cDNA abundance using Sybr Green on an Applied BioSystems HT-7900 qRT-PCR system with β-actin as a normalization gene. We performed serial dilutions of cDNA to find primer pairs with 100% efficiency that produced a single product. Primers are listed in Supplementary Material.
Cellular fractionation
After treatment with CCCP (2 h, 2 μm), cells were harvested by scraping and centrifuged at 260 g for 4 min and resuspended in 0.5 ml of mitochondrial isolation (225 mm mannitol, 50 mm sucrose, 5 mm HEPES, pH 7.3, and complete protease inhibitor). Samples were pipetted to resuspend and then passed through an insulin syringe for 10 strokes to lyse cells. Nuclei and unbroken cells were pelleted at 1000 g for 10 min at 4°C and resuspended in 250 μl of mitochondrial isolation buffer, lysed and pelleted as above. Supernatants were combined and centrifuged at 12 000 g for 10 min. Supernatants from this step were taken as the cytosolic fraction, and pellets washed twice with isolation buffer and pelleted at 12 000 g for 10 min. Mitochondrial pellets were resuspended in 50 µl of 1× cell lysis buffer, 10× cell lysis buffer was added to total fractions for a final concentration of 1× and fractions lysed on ice for 20 min. Lysates were clarified by centrifugation at 16 000 g for 10 min and total protein for each fraction was measured by Pierce 660 nm Protein Assay (Thermo).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Richard Youle, NINDS, for the gift of the HeLa YFP-parkin cell line.
REFERENCES
- 1.Bonifati V. Autosomal recessive parkinsonism. Parkinsonism Relat. Disord. 2012;18(Suppl. 1):S4–S6. doi: 10.1016/S1353-8020(11)70004-9. doi:10.1016/S1353-8020(11)70004-9. [DOI] [PubMed] [Google Scholar]
- 2.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. doi:10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 3.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. doi:10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. doi:10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 5.Lin W., Kang U.J. Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. doi:10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takatori S., Ito G., Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci. Lett. 2008;430:13–17. doi: 10.1016/j.neulet.2007.10.019. doi:10.1016/j.neulet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Trempe J.-F., Sauvé V., Grenier K., Seirafi M., Tang M.Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G., et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013 doi: 10.1126/science.1237908. 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 8.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L.J., Hess S., Chan D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. doi:10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., Harper J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013 doi: 10.1038/nature12043. 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S.H.Y., Renton A.E.M., Harvey R.J., Whitworth A.J., Martins L.M., et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. doi:10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. doi:10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narendra D.P., Jin S.M., Tanaka A., Suen D.-F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azoulay-Zohar H., Israelson A., Abu-Hamad S., Shoshan-Barmatz V. In self-defence: hexokinase promotes voltage-dependent anion channel closure and prevents mitochondria-mediated apoptotic cell death. Biochem. J. 2004;377:347. doi: 10.1042/BJ20031465. doi:10.1042/BJ20031465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto S., Murphy A.N., Brown J.H. Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ. 2008;15:521–529. doi: 10.1038/sj.cdd.4402285. doi:10.1038/sj.cdd.4402285. [DOI] [PubMed] [Google Scholar]
- 15.Pastorino J.G., Shulga N., Hoek J.B. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. doi:10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 16.Sun L., Shukair S., Naik T.J., Moazed F., Ardehali H. Glucose phosphorylation and mitochondrial binding are required for the protective effects of hexokinases I and II. Mol. Cell. Biol. 2008;28:1007–1017. doi: 10.1128/MCB.00224-07. doi:10.1128/MCB.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. doi:10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 18.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L.A., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. doi:10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardehali H., Yano Y., Printz R.L., Koch S., Whitesell R.R., May J.M., Granner D.K. Functional organization of mammalian hexokinase II retention of catalytic and regulatory functions in both the NH2- and COOH-terminal halves. J. Biol. Chem. 1996;271:1849–1852. doi: 10.1074/jbc.271.4.1849. doi:10.1074/jbc.271.4.1849. [DOI] [PubMed] [Google Scholar]
- 20.Cai Q., Zakaria H.M., Simone A., Sheng Z.-H. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. doi:10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joselin A.P., Hewitt S.J., Callaghan S.M., Kim R.H., Chung Y.-H., Mak T.W., Shen J., Slack R.S., Park D.S. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum. Mol. Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. doi:10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- 22.Van Laar V.S., Arnold B., Cassady S.J., Chu C.T., Burton E.A., Berman S.B. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. doi:10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen P.L. Mitochondrial matters of the heart: a plethora of regulatory modes to maintain function for a long lifetime. J. Bioenerg. Biomembr. 2009;41:95–98. doi: 10.1007/s10863-009-9219-5. doi:10.1007/s10863-009-9219-5. [DOI] [PubMed] [Google Scholar]
- 24.Cheung E.C., Ludwig R.L., Vousden K.H. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc. Natl Acad. Sci. USA. 2012;109:20491–20496. doi: 10.1073/pnas.1206530109. doi:10.1073/pnas.1206530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf A., Agnihotri S., Micallef J., Mukherjee J., Sabha N., Cairns R., Hawkins C., Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 2011;208:313–326. doi: 10.1084/jem.20101470. doi:10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimenez-Cassina A., Lim F., Cerrato T., Palomo G.M., Diaz-Nido J. Mitochondrial hexokinase II promotes neuronal survival and acts downstream of glycogen synthase kinase-3. J. Biol. Chem. 2009;284:3001–3011. doi: 10.1074/jbc.M808698200. doi:10.1074/jbc.M808698200. [DOI] [PubMed] [Google Scholar]
- 27.Pastorino J.G., Hoek J.B., Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer Res. 2005;65:10545–10554. doi: 10.1158/0008-5472.CAN-05-1925. doi:10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- 28.Okatsu K., Iemura S.-I., Koyano F., Go E., Kimura M., Natsume T., Tanaka K., Matsuda N. Mitochondrial hexokinase HKI is a novel substrate of the Parkin ubiquitin ligase. Biochem. Biophys. Res. Commun. 2012;428:197–202. doi: 10.1016/j.bbrc.2012.10.041. doi:10.1016/j.bbrc.2012.10.041. [DOI] [PubMed] [Google Scholar]
- 29.Geisler S., Holmström K.M., Treis A., Skujat D., Weber S.S., Fiesel F.C., Kahle P.J., Springer W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–878. doi: 10.4161/auto.6.7.13286. doi:10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. doi:10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M., Wang B., Fei J., Santanam N., Blough E.R. Important roles of Akt/PKB signaling in the aging process. Front. Biosci. (Schol. Ed.) 2010;2:1169–1188. doi: 10.2741/s125. doi:10.2741/S125. [DOI] [PubMed] [Google Scholar]
- 32.Kerner J., Lee K., Hoppel C.L. Post-translational modifications of mitochondrial outer membrane proteins. Free Radic. Res. 2011;45:16–28. doi: 10.3109/10715762.2010.515218. doi:10.3109/10715762.2010.515218. [DOI] [PubMed] [Google Scholar]
- 33.Malagelada C., Jin Z.H., Greene L.A. RTP801 is induced in Parkinson's disease and mediates neuron death by inhibiting Akt phosphorylation/activation. J. Neurosci. 2008;28:14363–14371. doi: 10.1523/JNEUROSCI.3928-08.2008. doi:10.1523/JNEUROSCI.3928-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timmons S., Coakley M.F., Moloney A.M., O'Neill C. Akt signal transduction dysfunction in Parkinson's disease. Neurosci. Lett. 2009;467:30–35. doi: 10.1016/j.neulet.2009.09.055. doi:10.1016/j.neulet.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 35.Akundi R.S., Zhi L., Büeler H. PINK1 enhances insulin-like growth factor-1-dependent Akt signaling and protection against apoptosis. Neurobiol. Dis. 2012;45:469–478. doi: 10.1016/j.nbd.2011.08.034. doi:10.1016/j.nbd.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tain L.S., Mortiboys H., Tao R.N., Ziviani E., Bandmann O., Whitworth A.J. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. doi:10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts D.J., Tan-Sah V.P., Smith J.M., Miyamoto S. Akt phosphorylates HK-II at Thr473 and increases mitochondrial HK-II association to protect cardiomyocytes. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.482026. 10.1074/jbc.M113.482026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akundi R.S., Zhi L., Sullivan P.G., Büeler H. Shared and cell type-specific mitochondrial defects and metabolic adaptations in primary cells from PINK1-deficient mice. Neurodegener. Dis. 2012 doi: 10.1159/000345689. 10.1159/000345689. [DOI] [PubMed] [Google Scholar]
- 39.Cooper O., Seo H., Andrabi S., Guardia-Laguarta C., Graziotto J., Sundberg M., McLean J.R., Carrillo-Reid L., Xie Z., Osborn T., et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003985. 141ra90 doi:10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao Z., Gandhi S., Burchell V.S., Plun-Favreau H., Wood N.W., Abramov A.Y. Cell metabolism affects selective vulnerability in PINK1-associated Parkinson's disease. J. Cell Sci. 2011;124:4194–4202. doi: 10.1242/jcs.088260. doi:10.1242/jcs.088260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddall H.K., Yellon D.M., Ong S.-B., Mukherjee U.A., Burke N., Hall A.R., Angelova P.R., Ludtmann M.H.R., Deas E., Davidson S.M., et al. Loss of PINK1 increases the heart's vulnerability to ischemia-reperfusion injury. PLoS One. 2013;8:e62400. doi: 10.1371/journal.pone.0062400. doi:10.1371/journal.pone.0062400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gegg M.E., Cooper J.M., Chau K.-Y., Rojo M., Schapira A.H.V., Taanman J.-W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. doi:10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziviani E., Tao R.N., Whitworth A.J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. doi:10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gjedde A., Marrett S. Glycolysis in neurons, not astrocytes, delays oxidative metabolism of human visual cortex during sustained checkerboard stimulation in vivo. J. Cereb. Blood Flow Metab. 2001;21:1384–1392. doi: 10.1097/00004647-200112000-00002. doi:10.1097/00004647-200112000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Johannessen C.M., Boehm J.S., Kim S.Y., Thomas S.R., Wardwell L., Johnson L.A., Emery C.M., Stransky N., Cogdill A.P., Barretina J., et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. doi:10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A., Greggio E., Beilina A., Kaganovich A., Chan D., Taymans J.-M., Wolozin B., Cookson M.R. The Parkinson's disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. doi:10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang M., Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. doi:10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.