Abstract
TDP-43 aggregation in the cytoplasm or nucleus is a key feature of the pathology of amyotrophic lateral sclerosis and frontotemporal dementia and is observed in numerous other neurodegenerative diseases, including Alzheimer's disease. Despite this fact, the inciting events leading to TDP-43 aggregation remain unclear. We observed that endogenous TDP-43 undergoes reversible aggregation in the nucleus after the heat shock and that this behavior is mediated by the C-terminal prion domain. Substitution of the prion domain from TIA-1 or an authentic yeast prion domain from RNQ1 into TDP-43 can completely recapitulate heat shock-induced aggregation. TDP-43 is constitutively bound to members of the Hsp40/Hsp70 family, and we found that heat shock-induced TDP-43 aggregation is mediated by the availability of these chaperones interacting with the inherently disordered C-terminal prion domain. Finally, we observed that the aggregation of TDP-43 during heat shock led to decreased binding to hnRNPA1, and a change in TDP-43 RNA-binding partners suggesting that TDP-43 aggregation alters its function in response to misfolded protein stress. These findings indicate that TDP-43 shares properties with physiologic prions from yeast, in that self-aggregation is mediated by a Q/N-rich disordered domain, is modulated by chaperone proteins and leads to altered function of the protein. Furthermore, they indicate that TDP-43 aggregation is regulated by chaperone availability, explaining the recurrent observation of TDP-43 aggregates in degenerative diseases of both the brain and muscle where protein homeostasis is disrupted.
INTRODUCTION
Aberrant protein aggregation is the key feature of the pathology of most neurodegenerative diseases, including Alzheimer's disease (AD), Parkinson's disease, Huntington's disease and amyotrophic lateral sclerosis (ALS) (1–3). However, although the ubiquitous presence of protein inclusions in neurodegenerative diseases suggests that they play a role in pathophysiology, there is no universal agreement as to what role they play. Protein aggregates themselves have been proposed to be (i) key toxic species promoting neuronal dysfunction and death; (ii) epiphenomena which associated with disease but themselves are unimportant; or (iii) structures which protect cells from harm by sequestering abnormally misfolded toxic proteins. Their presence alone on pathology is consistent with any of these possibilities.
The RNA-binding protein TDP-43 is the key component of protein aggregates in ALS, a degenerative disease of spinal and cortical motor neurons, and the overlapping clinical syndrome of frontotemporal lobar degeneration (FTLD) (4). Dominant mutations in TDP-43 cause familial ALS, indicating that altered TDP-43 function can drive the pathogenesis of the disease (5–9). To date, the only effect that these mutations have shown is to increase the propensity of purified TDP-43 to aggregate in vitro (10). Interestingly, TDP-43 aggregation is also seen in a wide variety of other neurodegenerative diseases, indicating that defining the role of TDP-43 aggregation in cellular function could have a broad impact on our understanding of neurodegeneration (11).
TDP-43 is structurally similar to heterogeneous ribonucleoprotein (hnRNP) A/B family members with two RNA recognition motifs (RRMs) and a C-terminal domain necessary for protein–protein interactions with other hnRNP proteins to mediate alternative splicing (12–15). Recent evidence supports that the C-terminal domain, in addition to mediating protein–protein interactions with other splicing factors, has properties of a Q/N-rich ‘prion domain’ similar to those observed in yeast prions (16–18). Yeast prions contain low-complexity Q/N-rich domains which can adopt an aggregated conformation that recruits the native form of the protein into an inactive aggregate (19,20). These are proposed to be ‘functional’ prions, which allow the yeast cell to respond to stress. The key regulators of yeast prion aggregation are chaperone proteins, in particular the Hsp40/Hsp70 system and Hsp104 (21). Although a few proteins have been proposed to function as physiologic prions outside of yeast and the requirement of the C-terminal domain for TDP-43 aggregation is well described (21–23), physiologic stimuli that regulate the reversible aggregation of TDP-43 have not been defined.
Here, we investigate the role of the prion domain in regulating physiologic aggregation of TDP-43 during heat shock. We observed that heat shock induces reversible nuclear aggregation of TDP-43 which is mediated by interaction between Hsp40/Hsp70 chaperone proteins and the C-terminal prion domain. TDP-43 nuclear aggregates lose their interaction with hnRNPA1 and change their RNA-binding partners, suggesting that heat shock-induced TDP-43 aggregation alters the function of the protein, analogous to what is observed for physiologic prions in yeast.
RESULTS
Heat shock induces reversible nuclear TDP-43 aggregation in cultured cells
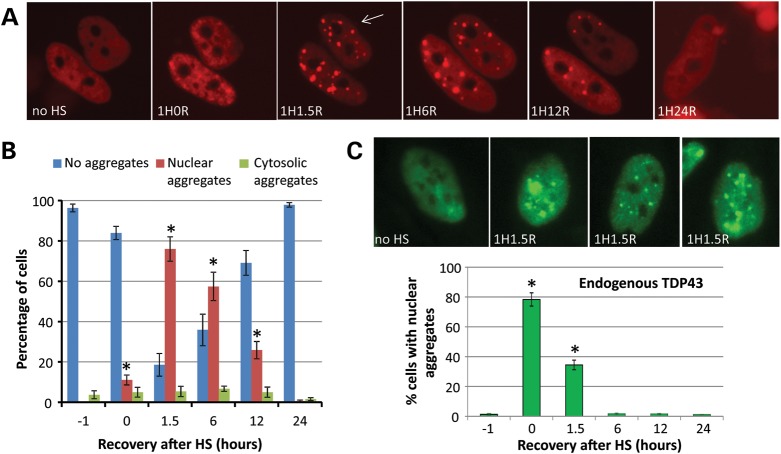
To investigate the effect of accumulating misfolded proteins on TDP-43 dynamics in live cells, we expressed Cherry-tagged TDP-43 in HeLa cells via lentivirus and subjected them to heat shock. Heat shock (1 h at 42°C, followed by recovery at 37°C) leads to accumulation of misfolded proteins in the cytoplasm, and a series of adaptive events including up-regulation of heat shock proteins and chaperones, which allow the cells to survive the stress (24). Ch-TDP43 normally has a grainy nuclear appearance; however, after heat shock Ch-TDP43 coalesced into distinct nuclear bodies (Fig. 1A and C). Time-lapse imaging of individual cells showed that these were maximal at ∼1.5 h after recovery at 37°C and resolved over the next 24 h. Although a small percentage of cells showed aggregates of Ch-TDP43 in the cytoplasm that stained for stress granule (SG) markers, these were much less common (<10% of cells) compared with those showing nuclear aggregates (>75% of cells; Fig. 1B). Nuclear aggregation after heat shock was not an artifact of the Ch-TDP43 fusion protein or lentiviral infection, as similar aggregation of endogenous TDP-43 was also observed after heat shock, with endogenous TDP-43 aggregates appearing and resolving slightly earlier than exogenously expressed Ch-TDP43 (Fig. 1C). Western blotting did not reveal decreased levels or fragmentation of TDP-43 during heat shock (Supplementary Material, Fig. S1), whereas HSP40 and HSP70 were up-regulated as expected (not shown). Comparison of heat shock-induced aggregation of wild-type TDP-43 to two different disease associated mutants (A315 T, M337 V) did not reveal a difference in the frequency or kinetics of aggregation (Supplementary Material, Fig. S2).
Figure 1.
Heat shock (HS) induces reversible, predominantly nuclear, TDP-43 aggregation in cell lines. (A) Live time-lapse imaging of Cherry-TDP43 in individual HeLa cells during HS. Normally, Cherry-TDP43 shows a grainy nuclear appearance (no HS). Immediately after 1 h at 42°C, TDP-43 showed minor nuclear condensation (1H0R indicates 1 h HS, 0 h recovery), which then coalesced to large nuclear bodies with additional hours of recovery at 37°C after HS (1H1.5R, 1H6R). TDP-43 began to redistribute to a grainy nuclear morphology after 12 h (1H12R) and returned to normal in all cells by 24 h after HS (1H24R). *P < 0.01 t-test indicated time versus no HS. (B) Although cytoplasmic aggregates (which stained with markers of SGs) were observed in a small number of cells (<5%), nuclear aggregates were far more common, appearing in ∼75% of Ch-TDP43 expressing cells at 1.5 h after HS. (C) Immunocytochemistry of TDP-43 in HeLa cells after HS, showing that endogenous TDP-43 forms nuclear bodies after HS indistinguishable from those formed by Ch-TDP43. The graph shows a time course and quantitation of HS-induced aggregation of endogenous TDP-43 in HeLa cells. *P < 0.001 t-test indicated time versus no HS.
Heat shock-induced nuclear TDP-43 aggregates stain with markers of nuclear stress bodies
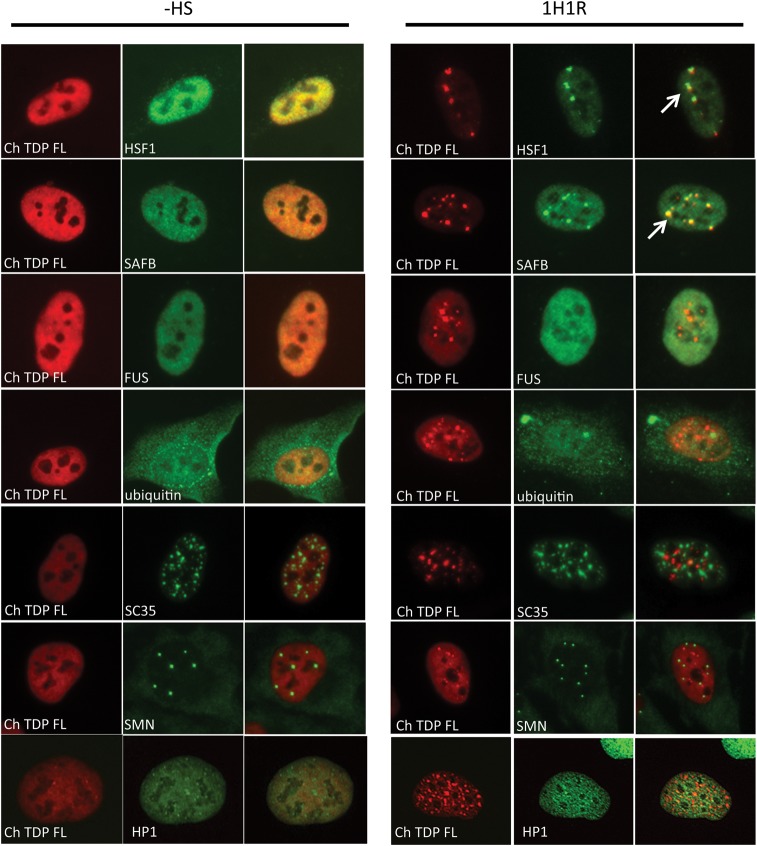
The nuclear aggregation of TDP-43 after heat shock was reminiscent of other stress regulated proteins, including transcription factors and RNA-binding proteins, which organize into nuclear stress bodies (25,26). Therefore, we co-stained Ch-TDP43 expressing cells before and after heat shock with markers for various nuclear bodies (Fig. 2). After heat shock Ch-TDP43 transiently co-localized with heat shock factor 1 (HSF1), the key transcriptional regulator of the heat shock response and scaffold attachment factor B (SAFB), a RNA-binding protein that localizes to nuclear stress bodies (27). The dynamics of TDP-43 association with nuclear stress bodies was similar to SAFB, in that it appeared later and persisted longer than HSF1 (Supplementary Material, Fig. S3) (28). The nuclear aggregates of TDP-43 did not stain with ubiquitin, supporting that they represent reversible functional aggregation, rather than formation of a pathologic inclusion. FUS/TLS, another RNA-binding protein mutated in ALS patients, did not co-localize with TDP-43 in nuclear stress bodies. Furthermore, markers of other nuclear bodies including nuclear speckles (SC35), gemini bodies (SMN) or heterochromatin (HP1) also did not co-localize TDP-43 either before or after heat shock. These results indicate that in heat shocked cells where misfolded proteins accumulate, TDP-43 reversibly aggregates in the nucleus in bodies that overlap with other proteins involved in the heat shock response.
Figure 2.
Nuclear TDP-43 aggregates co-localize with markers of nuclear stress bodies, but not FUS or other nuclear bodies. Cherry-TDP43 (Ch-TDP) was introduced into HeLa cells using lentivirus that were fixed and analyzed by immunocytochemistry either before heat shock (−HS) or 1 h after recovery from heat shock at 37°C (1H1R) at a time of peak TDP-43 aggregate formation. Nuclear aggregates of TDP-43 co-localized with the transcription factor HSF1 and the RNA-binding protein SAFB, previously described markers of nuclear stress bodies induced by heat shock. Heat shock-induced nuclear aggregates of TDP-43 did not co-localize with ubiquitin, suggesting that they are not marked for proteasomal degradation. Accumulation of large ubiquitin positive inclusions was observed after heat shock due to that of misfolded proteins. TDP-43 also did not co-localize with FUS, nuclear speckles (SC35), gemini bodies (SMN) or a heterochromatin marker (HP1) either before or after heat shock.
Determinants in the C-terminal prion domain of TDP-43 are critical for normal subnuclear localization, pathologic inclusion formation and heat shock-induced aggregation
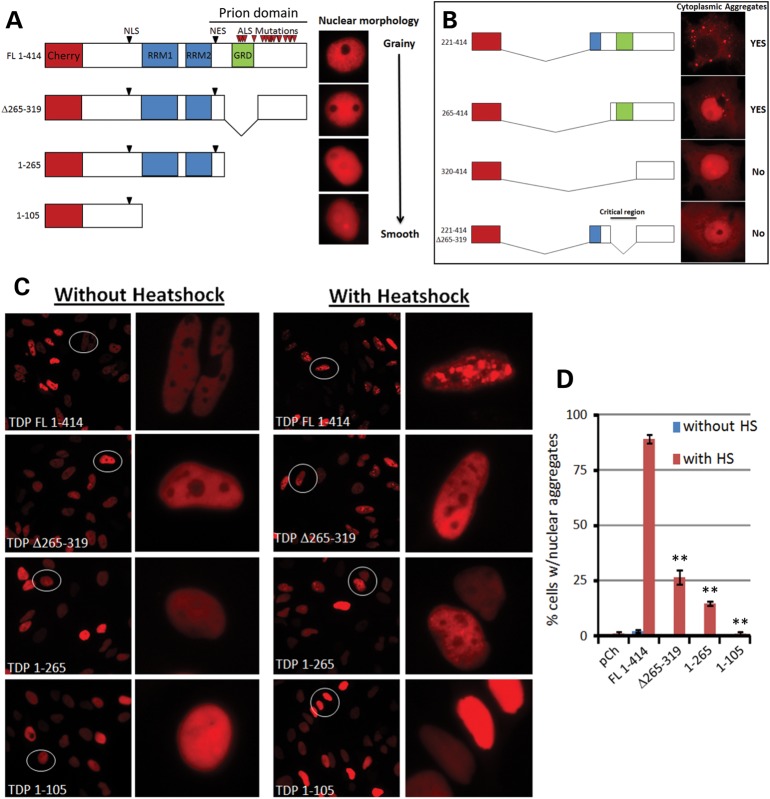
TDP-43 is an RNA-binding protein with two RNA-binding domains (RRM1 and RRM2), and a C-terminal domain that is critical for protein–protein interactions with other RNA-binding proteins (29) and for self-aggregation (22). We and others previously reported that the C-terminal region has properties similar to those of a prion-related Q/N-rich domain in yeast (16,17). These highly disordered domains are found in a variety of proteins which undergo functional aggregation and are particularly common in RNA-binding proteins involved in RNA granule formation such as SGs and P-bodies (30). To determine the role of the prion domain in TDP-43 for heat shock-induced nuclear aggregation, we generated a series of C-terminal deletions in Ch-TDP43 and examined their morphology in live cells (Fig. 3A). Deletion of the prion domain of TDP-43 led to smoothening of its normally grainy nuclear distribution. Interestingly, deletion of the particularly glycine-rich domain (GRD) with the prion domain (Δ265–319) had a similar effect. Deletion of the GRD region also markedly suppressed cytoplasmic aggregation of the C-terminal 25 kDa fragment of TDP-43 (Fig. 3B), which is observed in ALS and FTLD patient tissues and is enriched in pathologic inclusions (31).
Figure 3.
Determinants in the C-terminal prion domain are critical for normal subnuclear localization, cytoplasmic inclusion formation and heat shock-induced aggregation of TDP-43. (A) Schematic diagram of C-terminal deletion constructs (left) with preserved nuclear localization signal (NLS), with images of the corresponding nuclear appearance (right). Full-length TDP-43 showed a grainy nuclear appearance (FL 1-414), supporting that TDP-43 normally exists in subnuclear granular structures or microaggregates. Deletion of the C-terminal prion domain (1–265) led to a smooth nuclear appearance of TDP-43, with further deletion of the RNA-binding domains (RRM1 and RRM2) having no additional effect (1–105). Deletion of the GRD alone within the prion domain markedly attenuated the normal grainy nuclear appearance. (B) Schematic diagram of N-terminal deletions, where the NLS is removed. The nuclear export signal is also absent in constructs 265–414 and 320–414, but because they are missing the NLS they are also localized to the cytoplasm. As previously reported, a construct corresponding to the ∼25 kDa C-terminal fragment of TDP-43 (221–414), which mainly contains the prion domain, formed prominent cytoplasmic inclusions. Deletion of the GRD (320–414 or 221–414:Δ265–319) in the context of the cytoplasmic 25 kDa fragment completely blocked TDP-43 aggregation. (C) Analysis of heat shock-induced nuclear aggregation of TDP-43 constructs from (A). Deletion of the entire prion domain (1–265) or just the GRD (Δ265–319) markedly attenuated heat shock-induced aggregation of TDP-43. (D) Quantitation of the percentage of transfected cells with nuclear aggregates before and after heat shock (1 h at 42°C, 1.5 h recovery at 37°C). **P < 0.01 Student's t-test for indicated construct versus FL 1–414.
We next investigated the effect of deletions in the prion domain on the reversible nuclear aggregation of TDP-43 after heat shock (Fig. 3C). Deletion of either the entire prion domain (1–265) or just the GRD (Δ265–319) within the prion domain dramatically diminished heat shock-induced nuclear aggregation of TDP-43, indicating that the prion domain in TDP-43 mediates reversible stress-induced aggregation of the protein (Fig. 3D). This is reminiscent of regulated aggregation observed in other prion domain containing proteins, including TIA-1 in SG formation (32). Although determinants in the prion domain also mediate protein–protein interactions (33), the fact that deletion of this region blocks both stress-induced nuclear aggregation and pathologic cytoplasmic inclusion formation suggests that this region is critical for self-aggregation, regardless of the cellular location (nucleus or cytoplasm) or compartment-specific protein-binding partners.
Substitution of the prion domain from the yeast protein RNQ1 recapitulates heat shock-induced aggregation, but not TDP-43 splicing function
Prion domains are disordered regions characterized only by enrichment of glutamine (Q), asparagine (N), glycine (G) and tyrosine (Y) residues (34) and have a strong propensity to self-aggregate (35). We reasoned that if the role of the C-terminal prion domain of TDP-43 was to mediate heat shock-induced aggregation, substitution of the prion domain from another protein should recapitulate this phenomenon. Therefore, we generated a construct substituting the prion domain of the yeast protein RNQ1 for the prion domain of TDP-43. These domains share no primary sequence homology (Fig. 4A), and it is unlikely that determinants mediating interactions with other mammalian proteins are conserved between TDP-43 and RNQ1. Remarkably, the TDP-RNQ1 fusion protein formed heat shock-induced nuclear aggregates in a time course similar to normal full-length TDP-43 (Fig. 4B). To further investigate whether the prion domain of RNQ1 could substitute for the prion domain of TDP-43, we investigated the ability of the TDP-RNQ1 fusion protein to mediate splicing of CFTR exon 9, the most well-characterized alternative splicing target of TDP-43 (36). Transfection of full-length TDP-43 strongly promoted CFTR exon 9 exclusion, an effect that was blocked by deletion of the C-terminal prion domain (1–265) similar to previous reports. However, substitution of the prion domain from RNQ1 (TDP-RNQ1) was not able to recapitulate normal TDP-43 mediated exon 9 skipping (Fig. 4C). This is presumably because the CFTR exon skipping assay requires co-operation between TDP-43 and other RNA-binding proteins including hnRNPA1 and hnRNPA2/B1 (13), interactions which may not supported by the prion domain of RNQ1. Therefore, these experiments support that the prion domain of TDP-43 is the key mediator of heat shock-induced aggregation, a function that can be recapitulated by substitution with the prion domain from the yeast protein RNQ1. However, they also indicate that the TDP-43 prion domain is multifunctional, consistent with prior studies showing that the C-terminal region is critical for protein–protein interactions with other splicing factors.
Figure 4.
Substitution of the prion-related domain from the yeast protein RNQ1 recapitulates heat shock-induced aggregation, but not TDP-43 splicing function. (A) Sequence comparison of RNQ1 and TDP-43 Q/N-rich prion domains. For the construct YFP TDP-RNQ1, amino acids 266–414 from TDP-43 were substituted with amino acids 133–405 from RNQ1. (B) Time-lapse imaging of individual cells showing nuclear aggregation of full-length TDP-43 or TDP-RNQ1 after heat shock. The Q/N-rich prion domain of RNQ1 was able to substitute for the prion domain of TDP-43 to promote nuclear aggregate formation after heat shock, with a time course and reversibility similar to full-length TDP-43 (TDP FL). The graph below shows quantitation of the percentage of transfected cells with nuclear aggregates from three independent experiments. *P < 0.05 Student's t-test for construct at time indicated versus baseline without heat shock. (C) To assay the ability of TDP-43 and TDP-RNQ1 to alter splicing of a TDP-43 target gene, the CFTR splicing reporter construct GT13T5 was transfected into 293T cells with the indicated constructs. Full-length TDP-43 (TDP FL) promotes exclusion of exon 9 in the reporter, with an increase in the ratio of the bottom band (exon 9 excluded) to the top band (exon 9 included). Deletion of the C-terminal prion domain blocked the ability of TDP-43 to mediate alternative splicing of exon 9 (TDP 1–265). Substitution of with the prion domain from RNQ1 was not able to restore the ability of TDP-43 to promote the exclusion of exon 9 (TDP-RNQ1). The graph below shows the ratio of exon 9 exclusion/inclusion from three independent experiments for the indicated constructs.
The prion domain of TDP-43 is able to functionally replace the prion domain of TIA-1 to induce cytoplasmic SG formation
TIA-1 is an RNA-binding protein with a similar domain structure to TDP-43 and was one of the first mammalian proteins recognized to have a prion-like Q/N-rich domain (32). TIA-1 organizes the formation of cytoplasmic SGs, a process that requires the prion domain to mediate self-aggregation. Although TDP-43 is present in SGs under some conditions, unlike TIA-1, it cannot seed SG formation in the absence of a stressor (37). Therefore, to determine if the prion domain of TDP-43 can mediate self-aggregation in the context of another mammalian protein, we substituted the prion domain of TDP-43 for the prion domain of TIA-1 and measured SG induction (Fig. 5). Transfection of cells with TIA-1-induced SG formation (visualized by staining for the SG marker G3BP) in ∼26% of cells, whereas TDP-43 did not significantly induce SGs above transfection with YFP alone. Likewise, construct TDP-TIA (with the RNA-binding domains of TDP-43 and the prion domain of TIA-1) did not induce SG formation. Interestingly, TIA-TDP (the RNA-binding domains of TIA-1 and the prion domain of TDP-43) induced SG formation in ∼50% of cells, even more effectively than TIA-1 itself. Furthermore, when cells were treated with the potent SG inducer arsenite, TIA-TDP localized with SGs in 100% of cells, identical to full-length TIA-1, whereas TDP-43 did not form either nuclear aggregates or localize to SGs under these conditions. These data indicate that the prion domain of TDP-43 is able to functionally replace the prion domain of TIA-1 in both SG induction and localization.
Figure 5.
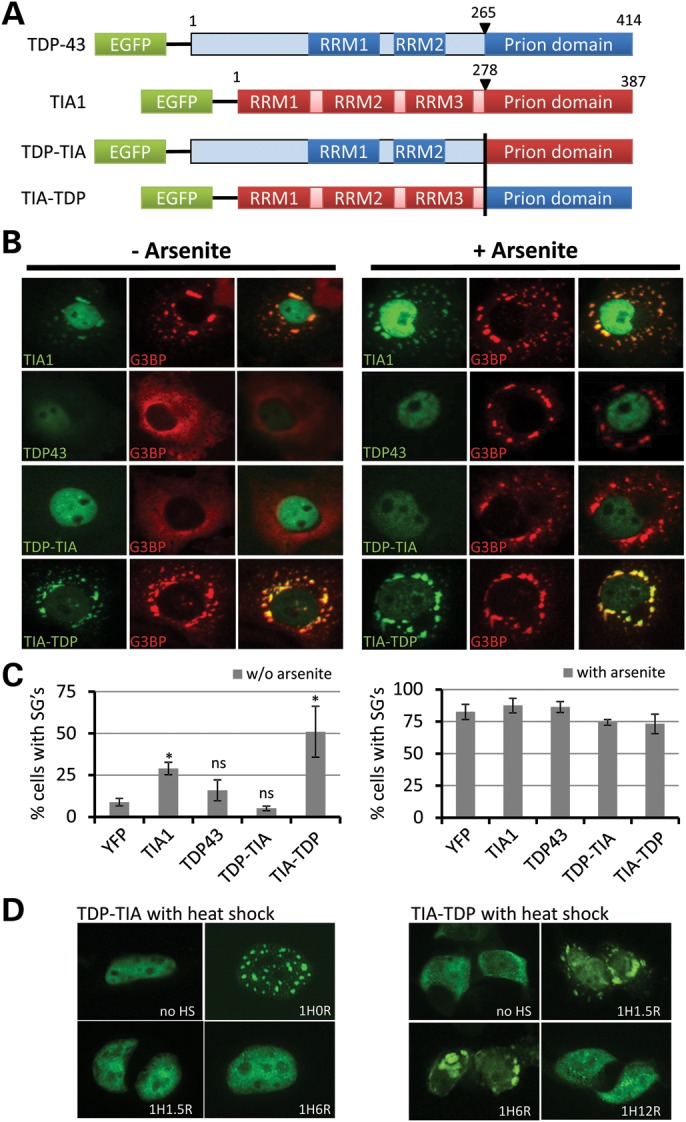
The C-terminal domain of TDP-43 is able to functionally replace the prion domain of TIA-1. Cells were transfected with YFP-tagged TIA-1, TDP-43 or one of two chimeric constructs (shown in A): ‘TDP-TIA’, N-terminal region (RRM domains) from TDP-43 with the C-terminal prion domain of TIA-1; ‘TIA-TDP’, N-terminal region (RNA-binding domains) from TDP-43 with the C-terminal prion domain of TIA-1. (B) SG formation was analyzed by immunocytochemistry for the SG marker G3BP in the absence (left) or the presence (right) of arsenite, which induces SGs in the majority of cells. The percentage of transfected cells with G3BP-positive SGs is shown in (C). Transfection of YFP-TIA-1 (TIA1) induced SG formation above the baseline caused by transfection alone (YFP) and YFP-TIA1 co-localized with G3BP. YFP-TDP43 transfection showed a minor increase in cells with SGs above YFP alone, and TDP-43 did not co-localize with G3BP-positive SGs or form nuclear aggregates, even when cells were treated with arsenite. TDP-TIA also did not induce SG formation, indicating that the neither the RNA binding domains of TDP-43 nor the prion domain of TIA-1 was sufficient to induce SGs. In contrast, TIA-TDP strongly induced SG formation, indicating that the prion domain of TDP-43 can functionally replace the prion domain of TIA-1 to promote SG induction. *P < 0.05 Student's t-test for construct versus. YFP control. ns, not significant. (D) After heat shock, TDP-TIA formed nuclear aggregates identical to full-length TDP-43, whereas TIA-TDP localized to cytoplasmic SGs identical full-length TIA-1, showing that the prion domains of these proteins are interchangeable, with their stress induced localization mediated by the N-terminal regions which contain the nuclear localization signal and RNA-binding domains.
Given that TIA1 and TDP-43 aggregate into different granules after heat shock (cytoplasmic SGs and nuclear stress bodies, respectively), we investigated the localization of the TIA1-TDP and TDP-TIA1 after heat shock (Fig. 5D). As expected, the TDP-TIA1 fusion formed nuclear aggregates like full-length TDP-43, whereas the TIA1-TDP fusion localized to cytoplasmic SGs after heat shock similar to full-length TIA-1. These data strongly support that the prion domains of TDP-43 and TIA-1 are functionally interchangeable, and the localization to different cellular compartments after heat shock-induced aggregation is determined by the N-terminal region containing the NLS, NES and RRM domains.
Chaperones modulate heat shock-induced aggregation of TDP-43 and binding to the splicing co-factor hnRNPA1
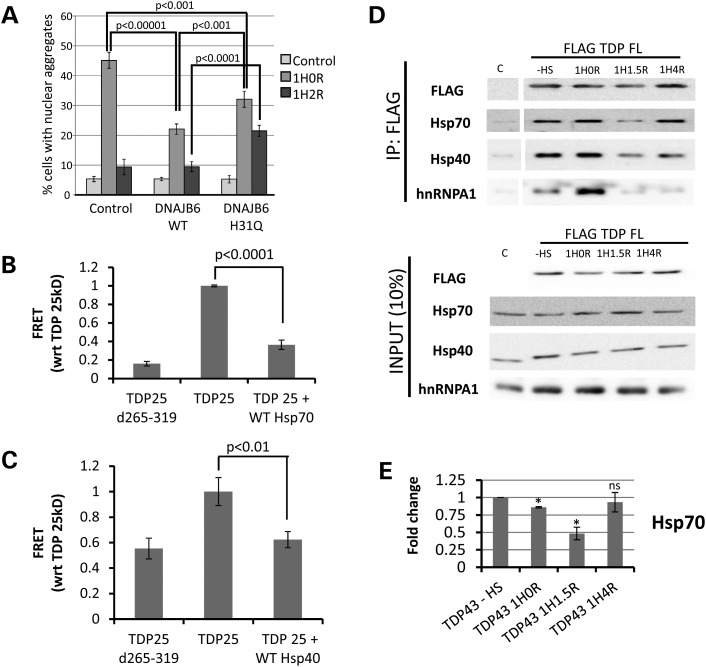
Prion domains are disordered low complexity domains, with an innate tendency to self-aggregate (35). Previous proteomics work found that TDP-43 is constitutively bound to HSP40 and HSP70 chaperone proteins (38), which are key modulators of yeast prion propagation (39). We also recently observed that point mutations in the Hsp40 family member DNAJB6 cause an inherited limb girdle muscular dystrophy in humans, characterized by aberrant protein aggregation, including TDP-43 aggregation (40). Using co-immunoprecipitation for endogenous TDP-43, we confirmed that TDP-43 is constitutively bound to HSP40 and HSP70 chaperones in HeLa cells (Supplementary Material, Fig. S4). Based on these findings, we hypothesized that chaperones are constitutively bound to TDP-43, but become titrated away to bind to misfolded proteins after heat shock, leading to TDP-43 aggregation. To test this hypothesis, we examined whether increased expression of the HSP40 family member DNAJB6 could suppress heat shock-induced aggregation of TDP-43. Indeed, we observed that overexpression of wild-type DNAJB6 markedly suppressed the formation of heat shock-induced TDP-43 nuclear aggregates (Fig. 6A). The suppression of TDP-43 aggregation was attenuated by a point mutant in DNAJB6 (H31Q), which acts as a dominant negative by failing to activate HSP70 ATPase activity (41). The dominant negative H31Q mutant also led to longer persistence of the TDP-43 nuclear aggregates after heat shock, supporting that endogenous chaperones regulate the normal dynamics of heat shock-induced TDP-43 aggregation. Finally, knockdown of DNAJB6 also led to an increase in heat shock-induced aggregation of endogenous TDP-43 in the nucleus, strongly supporting that the availability of this HSP40 co-chaperone regulates TDP-43 aggregation (Supplementary Material, Fig. S5).
Figure 6.
Heat shock (HS)-mediated aggregation of TDP-43 is mediated by chaperone interaction with the prion domain and is accompanied by loss of binding to hnRNPA1. (A) 293T cells were transfected with either wild-type (WT) or mutant (H31Q) versions of the Hsp40 family member DNAJB6, and nuclear aggregate formation of endogenous TDP-43 was determined by immunostaining. WT DNAJB6 markedly decreased nuclear TDP-43 aggregation after HS. This was partially diminished by the H31Q mutation which fails to activate HSP70 ATPase activity. P-values shown for Student's t-test. (B and C) FRET-based aggregation measurements of the prion domain-containing 25 kDa fragment of TDP-43 (TDP25). Overexpression of HSP70 (B) or HSP40 (C) markedly reduced the aggregation of the TDP-43 25 kDa C-terminal fragment, similar to the effect of deleting the GRD which is critical for aggregation (Fig. 3) (TDP25 d265–319). (D) Co-immunoprecipitation experiments examining binding partners of TDP-43 during HS-induced nuclear aggregation. HeLa cells were infected with lentivirus encoding Flag-tagged TDP-43 (FLAG-TDP FL) and underwent HS for 1 h at 42C and collected before (−HS) or 0 h (1H0R), 1.5 h (1H1.5R) or 4 h (1H4R) after recovery at 37°C. Both HSP70 and HSP40 showed loss of interaction with TDP-43 at the time of maximum aggregation (∼1.5 h; Fig. 1) and began to re-associate with TDP-43 by 4 h of recovery. Binding to hnRNPA1 initially increased immediately after HS (at a time when nuclear aggregation of TDP-43 is not yet present), but was severely diminished at 1.5 h and did not recover back to baseline as rapidly as HSP40 and HSP70. (E) Quantitation of densitometry measurements from three independent experiments of HSP70 pulled down with Flag-TDP43 during HS-induced TDP-43 aggregation. *P < 0.05 indicated time point versus no HS control. ns, not significant.
To confirm that suppression of TDP-43 aggregation by DNAJB6 was through binding to the prion domain, we developed a fluorescence resonance energy transfer (FRET)-based assay to quantitatively measure the co-aggregation of the TDP-43 prion domain fused to YFP and cyan fluorescent protein (CFP) (Y-TDP43PRD and C-TDP43PRD), similar to assays developed previously for measuring aggregation of polyglutamine containing proteins (42) (Supplementary Material, Fig. S6). Formation of insoluble cytoplasmic aggregates by Y-TDP43PRD and C-TDP43PRD correlated with FRET signal, which similar to heat shock-induced aggregation of TDP-43 (Fig. 3) was markedly suppressed by deletion of the critical GRD (265–319; Fig. 6B and C). Overexpression of either the Hsp40 (DNAJB6) or HSP70 (HSPA1A) significantly attenuated the FRET signal, supporting that these chaperones modulate TDP-43 aggregation through binding to the C-terminal prion domain.
Finally, we examined the dynamics of the interaction between TDP-43 and endogenous chaperones during heat shock, using co-immunoprecipitation. 293T cells expressing Flag-tagged TDP-43 were heat shocked as before, immunoprecipitated for Flag and blotted for HSP40, HSP70 and the mRNA splicing co-factor hnRNPA1 (Fig. 6D and E). Consistent with previous reports and as shown above with endogenous protein (Supplementary Material, Fig. S4), we observed a constitutive interaction of TDP-43 with HSP40, HSP70 and hnRNPA1 (38). One hour after recovery from heat shock, at a time of maximal TDP-43 nuclear aggregate formation, the binding of TDP-43 to HSP40 and HSP70 was decreased from baseline. Binding was partially restored after further recovery (4 h), at the time when TDP-43 has largely disaggregated. Interestingly, at the time of peak nuclear aggregate formation (1H1.5R), TDP-43 also lost its interaction with the splicing co-factor hnRNPA1, suggesting that TDP-43 aggregation may alter its function by disrupting its binding to other protein interactors.
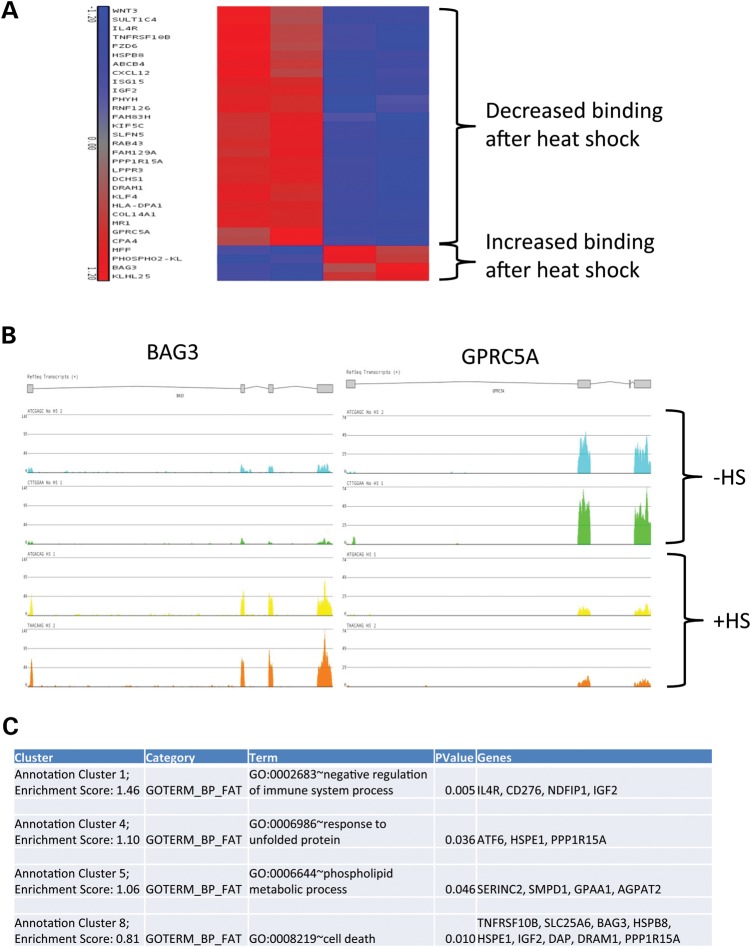
To further investigate whether heat shock-induced aggregation of TDP-43 alters the function of the protein, we immunoprecipitated TDP-43 before and after heat shock-induced nuclear aggregation similar to above and performed next-generation sequencing to identify RNAs bound in a complex with the protein (RIP-seq; Fig. 7). We observed that of the TDP-43 binding partners that changed after heat shock (>2-fold, P < 0.05), the large majority showed decreased binding to TDP-43 (27/31 transcripts = 87%), with only a small number showing increased binding to TDP-43 (4 of 31 transcripts = 13%). To determine whether this simply reflected a change in the overall abundance of these transcripts after heat shock, we compared our results with two previous whole transcriptome analyses after heat shock (43,44). Remarkably, of the 27 transcripts which showed decreased binding to TDP-43 after heat shock on RIP-seq, 87% (23 of 27 transcripts, HeLa cells) and 81% (22 of 27 transcripts, 293T cells) either showed no change in overall abundance or changed in the opposite direction (i.e. decreased binding to TDP-43 but increased overall abundance after heat shock; Supplementary Material, Fig. S8). This strongly supports that they reflect a true change in the cadre of mature mRNAs bound to TDP-43 after heat shock, with loss of binding observed with the majority of these transcripts. Pathway analysis revealed significant enrichment in genes involved in the unfolded protein response, phospholipid metabolism and cell death (Fig. 7C). These data indicate that TDP-43 aggregation during heat shock directly influences its mRNA-binding partners, which could have direct functional consequences on the cellular response to heat shock.
Figure 7.
RIP-seq analysis of TDP-43 RNA-binding partners after heat shock. RIP followed by next generation sequencing (RIP-seq) was performed in 293T cells expressing Flag-TDP43 before or after (1 h 42°C, 1.5 h 37°C) heat shock at the time of maximal nuclear TDP-43 aggregation. (A) Transcripts which were differentially bound to TDP-43 after heat shock (P < 0.05, fold >2) are shown on heat map analysis. 87% (27/31 transcripts) of transcripts showed decreased binding to TDP-43, with 13% (4 of 31 transcripts) showing increased binding to TDP-43. These changes did not correlate with overall levels of these transcripts induced by heat shock, as the majority of transcripts that decreased binding to TDP-43 showed an increase in overall abundance after heat shock (see text). (B) Representative images of RIP-seq genomic alignments, showing examples of transcripts which increased (BAG3) and decreased (GPRC5A) in binding to TDP-43 after heat shock induced (+HS). (C) Pathway analysis of transcripts differentially bound to TDP-43 before and after heat shock (P < 0.05) generated by DAVID functional annotation using gene ontology biological pathway terms. The second highest enrichment score was for genes involved in the unfolded protein response.
These findings support a model whereby the loss of binding to chaperones during proteotoxic stress (heat shock) induces to the reversible aggregation of TDP-43. Furthermore, they support that heat shock-induced TDP-43 aggregation is physiologic in that it alters its function, leading to changes in binding to both protein and RNA-binding partners. A schematic model showing that physiologic aggregation of TDP-43 during heat shock is shown in Figure 8.
Figure 8.
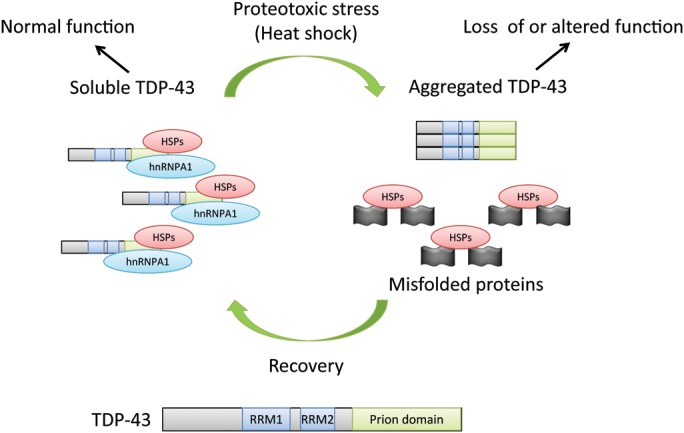
Schematic model of the TDP-43 prion domain as a sensor of misfolded protein stress. TDP-43 is constitutively bound to chaperones (HSPs) to maintain the disordered Q/N-rich prion-related domain in a conformation which is soluble and able to interact with its normal binding partners (hnRNPs). In the setting of heat shock, chaperones are titrated away by increased amounts of misfolded proteins in the cell. Loss of interaction between the prion domain and chaperones promotes self-aggregation of TDP-43, loss of interaction with its normal binding partners (hnRNPA1 shown) and hence a loss of TDP-43 function. This could in part explain the interruption of RNA splicing seen during heat shock. As cells recover from heat shock and new chaperone proteins are translated, they disaggregate TDP-43 and return it to a normal state. Although based on observations of heat shock-induced nuclear aggregation here, a similar mechanism could explain nuclear or cytoplasmic aggregation in the context of other types of proteotoxic stress, including other neurodegenerative diseases (i.e. 50% of Alzheimer's cases) where TDP-43 aggregation is observed in the context of additional misfolded proteins.
DISCUSSION
Here, we report that TDP-43 undergoes reversible nuclear aggregation due to the accumulation of misfolded proteins during heat shock, a behavior that is regulated by the interaction between the Hsp40/Hsp70 co-chaperone system and the C-terminal prion domain of TDP-43. This suggests that TDP-43 exhibits the behavior of a physiologic prion, or functional protein aggregate, analogous to those reported in yeast and invertebrates.
Proteins with prion-like behavior are best characterized in yeast (19,20). The Sup35 protein is normally required for translational termination, but under certain conditions, it forms a self-propagating aggregated conformation transmitted to offspring, which is dependent on the intrinsically disordered Q/N-rich prion domain. Sup35 aggregation is primarily induced by environmental stress and leads to loss of Sup35 function, and widespread read through of stop codons, allowing the rapid emergence of novel phenotypes (24). Therefore, rather than representing a disease, prion domain-mediated aggregation of Sup35 is proposed to be adaptive strategy to resist stressful conditions. Interestingly, aggregation of yeast prions is primarily modulated by chaperone proteins of the Hsp104 and Hsp70/Hsp40 systems, similar to what we observed for aggregation of the prion domain of TDP-43 (39,45).
There has also been evidence of prion-like behavior of a Q/N-rich protein in regulating synaptic activity in Aplysia, and more recently in Drosophila (46–50). CPEB (or its ortholog Orb2) is a RNA-binding protein involved in regulating local synaptic protein synthesis. Synaptic activity appears to induce aggregation of CPEB, dependent on the Q/N-rich prion domain, to regulate local mRNA translation and maintain synaptic facilitation. Although we focused here on heat shock-induced TDP-43 aggregation in a tissue culture model system, TDP-43 has been observed in both axons and dendrites of neurons (51,52), and prion domain regulated TDP-43 aggregation could also play a similar role in regulating synaptic function in mammalian neurons. It is important to note that while yeast prions show template-based propagation and amyloid formation, aggregation of proteins like TDP-43 and TIA-1 with Q/N-rich prion domains in higher organisms is reversible and whether aggregation is based on template propagation or another mechanism has not yet been clearly demonstrated.
What is the functional consequence of heat shock-induced nuclear aggregation of TDP-43? Our data indicate that aggregated TDP-43 loses its interaction with the binding partner hnRNPA1 and alters the mRNA species bound in RNA–protein complexes with TDP-43, suggesting that TDP-43 aggregation alters its function. Interestingly, it has been known for some time that heat shock interrupts RNA splicing, a block which persists for hours after recovery, similar to the time course of TDP-43 aggregation (53). Furthermore, reactivation of mRNA splicing after heat shock is promoted by the Hsp40/Hsp70 co-chaperone system (54). Given that aggregated TDP-43 may be non-functional and overexpression of Hsp40/Hsp70 co-chaperones suppresses TDP-43 aggregation, it is tempting to speculate that heat shock-induced aggregation of TDP-43 could contribute to this arrest in mRNA splicing.
In addition to heat shock-induced aggregation of TDP-43 correlating to loss of binding to a protein-binding partner, we observed using RIP-seq that the cadre of mature mRNA species bound to TDP-43 is also altered, with an enrichment in those involved with the unfolded protein response. In addition to intronic sequences, TDP-43 is known to bind to the 3′UTR of many transcripts and may regulate RNA stability, transport and/or translation after splicing (29,52,55,56). Therefore, it is possible that release of these mature mRNA transcripts from a complex with TDP-43 after heat shock may also play a role in regulating the cellular response to heat shock.
Although we observed that the C-terminal prion domain of TDP-43 could be replaced by the Q/N-rich region of the yeast prion RNQ1 to allow heat shock-induced aggregation, the TDP-RNQ1 fusion construct did not mediate splicing of a TDP-43 target, exon 9 of the CFTR pre-mRNA (29). This is consistent with the idea that the C-terminal domain of TDP-43 is multifunctional, i.e. the elements mediating chaperone interaction and self-aggregation (a property common to all prion domains) is shared with RNQ1, but specific structural elements for interacting with other hnRNPs are not conserved between TDP-43 and RNQ1. This is in keeping with the observations of others that an overlapping region of the C-terminal Q/N-rich domain is necessary for both interacting with hnRNPA2 and the formation of insoluble protein aggregates (57).
One aspect that was initially surprising about our findings was that while TDP-43 occasionally translocated to the cytoplasm and co-localized with markers of SGs, this was quite rare (<5% of cells) compared with the formation of nuclear aggregates (∼80% of cells). TDP-43 has variously been reported to be present in subnuclear bodies in neurons (58), cytoplasmic SGs in non-neuronal cells under a variety of conditions (37,59,60), acute response granules with markers of P-bodies in neuronal dendrites (51) and in RNPs transported along developing axons (52). We believe that this is indicative of the fact that TDP-43 aggregates and localizes to different RNA–protein complexes depending on the cell type and cellular context. Interestingly, the nuclear aggregates we observed co-localized with markers of nuclear stress bodies, including the key transcriptional regulator of the heat shock response, HSF1. While the organization of these structures is mediated in part by SatIII non-coding RNAs (25), the formation of nuclear TDP-43 aggregates was dependent on the C-terminal prion domain (and could be recapitulated with the prion domain of the yeast protein RNQ1), strongly indicating that self-binding of the prion domain, rather than binding to RNA elements, mediated the aggregation of TDP-43 into nuclear bodies after heat shock.
Implications for TDP-43 aggregation in disease pathology
TDP-43 pathology was initially proposed to be specific for ALS and a subset of FTLD cases (i.e. FTLD–TDP); however, abnormal TDP-43 staining has been reported in many neurodegenerative and myodegenerative diseases, including AD, Parkinson's disease and inclusion body myopathies (reviewed in 11). The fact that TDP-43 aggregation is so widespread suggests that it could be part of a normal cellular stress response, in particular the accumulation of misfolded proteins. This is in keeping with the observation that TDP-43 aggregation can be induced by proteasome inhibition (16,61,62) or accumulation of proteins in the secretory pathway by tunicamycin (63). Our data suggest that the mechanism of TDP-43 aggregation in these cases, and perhaps in diseases where misfolded proteins accumulate, is due to ‘sensing’ of the altered protein homeostasis state of the cell by the C-terminal prion domain, which self-aggregates in response to the availability of chaperone proteins. This is in keeping with the idea that the protein homeostasis network is delicately balanced (particularly in the brain) and that the aggregation of one misfolded protein will lead to the aggregation of others (64,65). It is important to note that while chaperone availability appear to be a key factor, other protein-binding partners or post-translational modifications may also play a role in heat shock-induced TDP-43 aggregation and remain to be explored.
The nuclear aggregates of TDP-43 we observed were reversible and did not stain with ubiquitin and do not appear to represent inclusions. It is more likely they represent a form of functional subnuclear structure, consistent with the fact that they transiently overlap with markers of nuclear stress bodies. It is notable that while cytoplasmic aggregates of TDP-43 are frequently pointed out, nuclear aggregates are also a common feature of TDP-43 pathology in ALS and FTLD (31), and in VCP-related FTLD, nuclear aggregation is the most common pathology (66). Therefore, much like SG-like structures have been proposed to represent ‘pre-inclusions’ of TDP-43 in the cytoplasm (67,68), it is possible that the nuclear aggregation behavior we observed could represent the initial stages of nuclear inclusion pathology observed in disease states. What causes the initially reversible aggregation of TDP-43 (in either the cytoplasm or the nucleus) to form an irreversible inclusion body remains unclear, but may involve post-translational modification such as cross-linking or phosphorylation (69), or a decreased degradation capacity of aggregated TDP-43 (23).
We recently reported that mutations in the Hsp40 family member DNAJB6 lead to dominantly inherited myopathy with protein aggregates, and TDP-43 aggregation is a prominent feature in diseased muscle (40). It remains to be determined whether these DNAJB6 mutations disrupt a specific interaction between the co-chaperone and TDP-43 or rather reflect a global alteration in protein homeostasis induced by DNAJB6 mutations in patient myofibers. However, the fact that overexpression of DNAJB6 can suppress physiologic aggregation of TDP-43 after heat shock and either knocking down or using dominant negative DNAJB6 led to increased TDP-43 aggregation suggests that TDP-43 is a direct client protein of DNAJB6, and as such may be a key player in mediating TDP-43 prion-like aggregation in muscle.
MATERIALS AND METHODS
Plasmids and constructs
Flag-tagged human TDP-43 fused to pCherry was described previously (16). TDP-43 deletion constructs and fusion constructs (TIA-TDP, TDP-TIA, TDP-RNQ1) were generated using polymerase chain reaction mutagenesis, cloned into the pCherry-C1, pYFP-C1 or pCFP-C1 vector and sequenced in their entirety and are available on request.
Cell culture and transfection
COS-7, HeLa, 293T and immortalized mouse motor neuron-like NSC-34 cells were cultured in high glucose formulation of Dubelcco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% (vol/vol) fetal bovine serum, 2 mm l-glutamine and penicillin/streptomycin. For transfection, DNA was mixed with TransitLT1 (Mirus) at a ratio of 1 µg DNA/ 3 ml of TransitLT1. Cells were transfected for 48 h, unless otherwise indicated.
Lentivirus production and infection
Lentiviruses were produced as described previously (70). Briefly, 293T cells were plated onto six-well plates and transfected with a packaging vector (Δ8.9), envelope vector (vesicular stomatitis virus-glycoprotein) and transfer vector encoding Ch TDP-43, Ch TDP 43 Δ265–319, Ch TDP 1-265 or Ch TDP 1-105. Media was changed once at 12 h and collected at 72 h and applied directly to HeLa cells resulting in ∼95% infection efficiency.
Heat shock, live cell imaging and immunocytochemistry
Imaging of cells was performed in a climate-controlled chamber (In Vivo Scientific) at 37°C and 5% CO2, and images were acquired with a Cool Snap HQ2 CCD camera (Photometrics) mounted on a Nikon Eclipse Ti-U microscope, controlled using MetaMorph software (Molecular Devices). For heat shock experiment, HeLa cells were transfected with Ch-TDP43 constructs for 48 h. After transfection, cells were subjected to heat shock (42°C, 5% CO2) for 1 h. After heat shock, cells were allowed to recover at 37°C (with 5% CO2) for indicated times (0–24 h). Immunocytochemistry was performed as described previously (16). Antibodies used were as follows: rabbit anti-TDP-43 raised to amino acids 1–260 (Proteintech, 1:250), rabbit anti-HSF (Cell Signaling Technology, 1:250), mouse anti-SAFB (Abcam, 1:250), mouse anti-ubiquitin (Millipore, 1:250), rabbit anti-FUS (MBL International, 1:250), mouse monoclonal anti-SC35 (Abcam, 1:250), goat anti-rabbit Alexa 488 or Alexa 594 (Invitrogen, 1:500) and goat anti-mouse Alexa 488 or Alexa 594 (Invitrogen, 1:500).
FRET assay
FRET assay using 293T cells was performed and data analyzed as described previously (16). Briefly, 250 000 cells/cm2 were seeded in 24-multiwell plates and grown for 24 h in growth media. Cells were transfected with TransIT-LT1 Transfection Reagent (Mirus Bio Corporation) in a 1:3 (µg/ml) ratio according to the manufacturer's protocol using the following amounts of plasmids per well: 125 ng CFP-tagged and 375 ng YFP-tagged test plasmid for FRET determinations; 125 ng CFP alone, for CFP beed through determination from the sample FRET; 375 ng YFP alone, for YFP crossover activation determinations; and no DNA, for background determination. After 24 h, the cells were trypsinized and plated in 96-well plate in quadruplicate and allowed to adhere for additional 24 h. Cells were fixed with 4% paraformaldehyde, washed with phosphate buffered saline (PBS) and read in an Infinite M1000 plate reader (Tecan Group Ltd, Männedorf, Switzerland). Data were represented as FRET per donor.
Protein extraction, immunoblotting and immunoprecipitation
For the study of TDP-43 solubility, sequential fractionation was performed as described (71). Briefly, 293T cells were washed twice with cold PBS, lysed in cold radio immunoprecipitation assay (RIPA) buffer for 15 min, sonicated and centrifuged at 100 000g at 4°C for 30 min. Supernatant was collected for RIPA-soluble fraction. Pellets were washed, sonicated and centrifuged twice to completely remove RIPA-soluble fraction. Pellets were resuspended in urea buffer for additional 15 min, sonicated, centrifuged at 100 000g at 22°C for 30 min and RIPA-insoluble supernatant was collected. 1 mm phenylmethylsulfonyl fluoride and protease inhibitors were added to all buffers prior to use. Protein concentration was determined using Micro BCA protein assay kit (Thermo Scientific), and equal amounts of proteins (35 µg/sample) were resolved on 10% sodium dodecyl sulfate polyacrylamide gel electropheresis (SDS–PAGE). For immunoprecipitation of Flag-tagged TDP-43, 293T cells were transfected with FLAG-TDP43 FL for 48 h and were subjected to heat shock, as described. Cell pellets were collected, washed twice with cold PBS, lysed in co-IP buffer (1× tris buffered saline, 1 mm NaCl, 1% Triton X-100, 10% glycerol supplemented with phosphatase and protease inhibitors) for 15 min, and spun at 12 000g for 25 min. Lysate was collected and protein concentration was determined using Micro BCA protein assay kit. 1000 µg of total proteins from whole cell extracts were immunoprecipitated overnight at 4°C using Flag-conjugated magnetic beads (Sigma-Aldrich), then beads were washed three times with immunoprecipitation (IP) wash buffer (20 mm Tris, pH 7.5, 1 mm ethylenediaminetetraacetic acid, 150 mm NaCl, 10% glycerol). Flag-conjugated magnetic beads were resuspended in 40 µl of 2× SDS loading buffer with β-mercaptoethanol and heated at 70°C for 5 min to elute the samples. Equal amounts of total proteins (35 µg/sample) and 20 µl of eluted IP samples were resolved in 10% SDS–PAGE and analyzed in western blotting according to standard procedures. Antibody incubation was done overnight at 4°C and probed with horse radish peroxidase (HRP)-conjugated secondary antibody for 2 h. Antibodies used were rabbit polyclonal anti-TDP-43 antibody raised to amino acids 1–260 (Proteintech, 1:1000), rabbit polyclonal anti-TDP-43 antibody raised to amino acids 350–414 (Novus Biologicals, 1:500), mouse monoclonal anti-FLAG (Sigma-Aldrich, 1:1000), rabbit anti-Hsp70/Hsp72, rabbit anti-Hsp40 (Enzo Life Sciences, 1:1000). Secondary antibodies used were goat anti-mouse HRP and goat anti-rabbit HRP (Jackson ImmunoResearch Laboratories, 1:10 000).
RNA-binding protein immunoprecipitation (RIP) was performed using the RiboCluster Profiler RIP-Assay Kit (MBL International, catalog # RN1001) as per the manufacturer's instructions. 293T cells were infected with lentivirus encoding wild-type Flag-TDP43. Two independently performed biologic replicates were collected for RIP (i) without heat shock or (ii) after 1 h 42°C followed by 1.5 h recovery at 37°C, at the time of maximal TDP-43 aggregation. The resulting RNA was analyzed and quantified by spectrophotometry. Uninfected 293T cells were included as a control for pull downs. The resulting RNA was submitted for library preparation and single-end sequencing on an Illumina Hi-Seq. The resulting reads were aligned to the human genome (hg19 build) using TOPHAT, and bam files imported into Partek software (Partek Incorporated, St Louis, MO, USA) for analysis. Transcripts were annotated to RefSeq build 1-4-2013, and analysis of variance (ANOVA) performed to identify transcripts bound to TDP-43 which were significantly increased or decreased either before or after heat shock. Pathway analysis was performed using database for annotation, visualization and integrated discovery bioinformatics resource (http://david.abcc.ncifcrf.gov/). For comparison of genes which are differentially expressed after heat shock, independent data sets (Affymetrix Human Exon 1.0 ST Arrays in HeLa cells before and after heat shock (43); RNA-seq before and after heat shock in 293T cells (44)) were obtained from the Gene Expression Omnibus (GSM669693, GSM669696, GSM670092, GSM670093, GSM1063575, GSM1063576). Data sets were imported into Partek for differential expression analysis, and transcripts which showed increased or decreased binding to TDP-43 after heat shock were analyzed for a change in overall abundance after heat shock. For the RNA-seq data set from Shalgi et al. (44), replicates were not available so only genes with a fold change greater than 2 were included as an ANOVA P-value could not be calculated.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institutes of Health (NIH) (NS055980 and NS069669 to R.H.B.) and (AG031867 and AG042095 to C.C.W.), the Neuroscience Blueprint Core (NS057105) to Washington University, the Hope Center for Neurological Disorders, the Children's Discovery Institute, and the Muscular Dystrophy Association to R.H.B. and C.C.W. R.H.B. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul Anderson for providing TIA-1 plasmids and Francisco Baralle for CFTR reporter plasmids.
REFERENCES
- 1.Skovronsky D.M., Lee V.M., Trojanowski J.Q. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. doi:10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 2.Winklhofer K.F., Tatzelt J., Haass C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. doi: 10.1038/sj.emboj.7601930. doi:10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voisine C., Pedersen J.S., Morimoto R.I. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol. Dis. 2010;40:12–20. doi: 10.1016/j.nbd.2010.05.007. doi:10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen-Plotkin A.S., Lee V.M., Trojanowski J.Q. TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. doi:10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L., 3rd, Bigio E.H., Caselli R., et al. TDP-43 A315 T mutation in familial motor neuron disease. Ann. Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. doi:10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoseki A., Shiga A., Tan C.F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol. 2008;63:538–542. doi: 10.1002/ana.21392. doi:10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 7.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. TARDBP Mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. doi:10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 8.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. doi:10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Deerlin V.M., Leverenz J.B., Bekris L.M., Bird T.D., Yuan W., Elman L.B., Clay D., Wood E.M., Chen-Plotkin A.S., Martinez-Lage M., et al. TARDBP Mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. doi:10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson B.S., Snead D., Lee J.J., McCaffery J.M., Shorter J., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. doi:10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baloh R.H. TDP-43: the relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS J. 2011;278:3539–3549. doi: 10.1111/j.1742-4658.2011.08256.x. doi:10.1111/j.1742-4658.2011.08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou S.H., Wu F., Harrich D., Garcia-Martinez L.F., Gaynor R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratti E., Dork T., Zuccato E., Pagani F., Romano M., Baralle F.E. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. doi:10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H.Y., Wang I.F., Bose J., Shen C.K. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. doi:10.1016/S0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 15.He Y., Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. doi:10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuentealba R.A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M.I., Weihl C.C., Baloh R.H. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 2010;285:26304–26314. doi: 10.1074/jbc.M110.125039. doi:10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cushman M., Johnson B.S., King O.D., Gitler A.D., Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J. Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. doi:10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udan M., Baloh R.H. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. doi:10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickner R.B., Edskes H.K., Roberts B.T., Baxa U., Pierce M.M., Ross E.D., Brachmann A. Prions: proteins as genes and infectious entities. Genes Dev. 2004;18:470–485. doi: 10.1101/gad.1177104. doi:10.1101/gad.1177104. [DOI] [PubMed] [Google Scholar]
- 20.Halfmann R., Alberti S., Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. doi:10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler J., Tyedmers J., Bukau B., Mogk A. Chaperone networks in protein disaggregation and prion propagation. J. Struct. Biol. 2012;179:152–160. doi: 10.1016/j.jsb.2012.05.002. doi:10.1016/j.jsb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Budini M., Buratti E., Stuani C., Guarnaccia C., Romano V., De Conti L., Baralle F.E. Cellular model of TAR DNA-binding protein 43 (TDP-43) aggregation based on its C-terminal Gln/Asn-rich region. J. Biol. Chem. 2012;287:7512–7525. doi: 10.1074/jbc.M111.288720. doi:10.1074/jbc.M111.288720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang I.F., Chang H.Y., Hou S.C., Liou G.G., Way T.D., James Shen C.K. The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat. Commun. 2012;3:766. doi: 10.1038/ncomms1766. doi:10.1038/ncomms1766. [DOI] [PubMed] [Google Scholar]
- 24.Tyedmers J., Madariaga M.L., Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. doi:10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biamonti G., Vourc'h C. Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. doi:10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Y.S., Zhang B., Spector D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. doi:10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denegri M., Chiodi I., Corioni M., Cobianchi F., Riva S., Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. doi:10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weighardt F., Cobianchi F., Cartegni L., Chiodi I., Villa A., Riva S., Biamonti G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 1999;112:1465–1476. doi: 10.1242/jcs.112.10.1465. [DOI] [PubMed] [Google Scholar]
- 29.Buratti E., Baralle F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 2008;13:867–878. doi: 10.2741/2727. doi:10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 30.Thomas M.G., Loschi M., Desbats M.A., Boccaccio G.L. RNA Granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. doi:10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. doi:10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 32.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. doi:10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y.M., Baralle F.E. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. doi:10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberti S., Halfmann R., King O., Kapila A., Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. doi:10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. doi:10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buratti E., Brindisi A., Pagani F., Baralle F.E. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 2004;74:1322–1325. doi: 10.1086/420978. doi:10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. doi:10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 38.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. doi:10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reidy M., Masison D.C. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion. 2011;5:245–249. doi: 10.4161/pri.5.4.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harms M.B., Sommerville R.B., Allred P., Bell S., Ma D., Cooper P., Lopate G., Pestronk A., Weihl C.C., Baloh R.H. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol. 2012;71:407–416. doi: 10.1002/ana.22683. doi:10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hageman J., Rujano M.A., van Waarde M.A., Kakkar V., Dirks R.P., Govorukhina N., Oosterveld-Hut H.M., Lubsen N.H., Kampinga H.H. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. doi:10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Pollitt S.K., Pallos J., Shao J., Desai U.A., Ma A.A., Thompson L.M., Marsh J.L., Diamond M.I. A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron. 2003;40:685–694. doi: 10.1016/s0896-6273(03)00697-4. doi:10.1016/S0896-6273(03)00697-4. [DOI] [PubMed] [Google Scholar]
- 43.Pandey R., Mandal A.K., Jha V., Mukerji M. Heat shock factor binding in Alu repeats expands its involvement in stress through an antisense mechanism. Genome Biol. 2011;12:R117. doi: 10.1186/gb-2011-12-11-r117. doi:10.1186/gb-2011-12-11-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. doi:10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardill J.P., Dulle J.E., Fisher J.R., True H.L. Requirements of Hsp104p activity and Sis1p binding for propagation of the [RNQ(+)] prion. Prion. 2009;3:151–160. doi: 10.4161/pri.3.3.9662. doi:10.4161/pri.3.3.9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Si K., Lindquist S., Kandel E.R. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. doi:10.1016/S0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 47.Si K., Choi Y.B., White-Grindley E., Majumdar A., Kandel E.R. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. doi:10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Salazar A.M., Silverman E.J., Menon K.P., Zinn K. Regulation of synaptic Pumilio function by an aggregation-prone domain. J. Neurosci. 2010;30:515–522. doi: 10.1523/JNEUROSCI.2523-09.2010. doi:10.1523/JNEUROSCI.2523-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruttner S., Stepien B., Noordermeer J.N., Mommaas M.A., Mechtler K., Dickson B.J., Keleman K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron. 2012;76:383–395. doi: 10.1016/j.neuron.2012.08.028. doi:10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majumdar A., Cesario W.C., White-Grindley E., Jiang H., Ren F., Khan M.R., Li L., Choi E.M., Kannan K., Guo F., et al. Critical role of amyloid-like oligomers of drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. doi:10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Wang I.F., Wu L.S., Chang H.Y., Shen C.K. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. doi:10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 52.Fallini C., Bassell G.J., Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet. 2012;16:3703–3718. doi: 10.1093/hmg/dds205. doi:10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yost H.J., Lindquist S. RNA Splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-x. doi:10.1016/0092-8674(86)90382-X. [DOI] [PubMed] [Google Scholar]
- 54.Vogel J.L., Parsell D.A., Lindquist S. Heat-shock proteins Hsp104 and Hsp70 reactivate mRNA splicing after heat inactivation. Curr. Biol. 1995;5:306–317. doi: 10.1016/s0960-9822(95)00061-3. doi:10.1016/S0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 55.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C., et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. doi:10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. doi:10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Budini M., Romano V., Avendano-Vazquez S.E., Bembich S., Buratti E., Baralle F.E. Role of selected mutations in the Q/N rich region of TDP-43 in EGFP-12xQ/N-induced aggregate formation. Brain Res. 2012;1462:139–150. doi: 10.1016/j.brainres.2012.02.031. doi:10.1016/j.brainres.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 58.Wang I.F., Reddy N.M., Shen C.K. Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl Acad. Sci. USA. 2002;99:13583–13588. doi: 10.1073/pnas.212483099. doi:10.1073/pnas.212483099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderwyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. doi:10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aulas A., Stabile S., Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegen. 2012;7:54. doi: 10.1186/1750-1326-7-54. doi:10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ju J.S., Miller S.E., Hanson P.I., Weihl C.C. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J. Biol. Chem. 2008;283:30289–30299. doi: 10.1074/jbc.M805517200. doi:10.1074/jbc.M805517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Eersel J., Ke Y.D., Gladbach A., Bi M., Gotz J., Kril J.J., Ittner L.M. Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS One. 2011;6:e22850. doi: 10.1371/journal.pone.0022850. doi:10.1371/journal.pone.0022850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leggett C., McGehee D.S., Mastrianni J., Yang W., Bai T., Brorson J.R. Tunicamycin produces TDP-43 cytoplasmic inclusions in cultured brain organotypic slices. J. Neurol. Sci. 2012;317:66–73. doi: 10.1016/j.jns.2012.02.027. doi:10.1016/j.jns.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. doi:10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 65.Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. doi:10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neumann M., Mackenzie I.R., Cairns N.J., Boyer P.J., Markesbery W.R., Smith C.D., Taylor J.P., Kretzschmar H.A., Kimonis V.E., Forman M.S. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J. Neuropathol. Exp. Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. doi:10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 67.Parker S.J., Meyerowitz J., James J.L., Liddell J.R., Crouch P.J., Kanninen K.M., White A.R. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem. Internat. 2012;60:415–424. doi: 10.1016/j.neuint.2012.01.019. doi:10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 68.Dewey C.M., Cenik B., Sephton C.F., Johnson B.A., Herz J., Yu G. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. doi:10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cohen T.J., Hwang A.W., Unger T., Trojanowski J.Q., Lee V.M. Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 2012;31:1241–1252. doi: 10.1038/emboj.2011.471. doi:10.1038/emboj.2011.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baloh R.H., Schmidt R.E., Pestronk A., Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J. Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winton M.J., Igaz L.M., Wong M.M., Kwong L.K., Trojanowski J.Q., Lee V.M. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.