Abstract
Death is a vital developmental cell fate. In Caenorhabditis elegans, programmed death of the linker cell, which leads gonadal elongation, proceeds independently of caspases and apoptotic effectors. To identify genes promoting linker-cell death, we performed a genome-wide RNA interference screen. We show that linker-cell death requires the gene pqn-41, encoding an endogenous polyglutamine-repeat protein. pqn-41 functions cell autonomously, and is expressed at the onset of linker-cell death. pqn-41 expression is controlled by the MAP kinase kinase SEK-1, which functions in parallel to the Zn-finger protein LIN-29 to promote cellular demise. Linker-cell death is morphologically similar to cell death associated with normal vertebrate development and polyglutamine-induced neurodegeneration. Our results may, therefore, provide molecular in-roads to understanding non-apoptotic cell death in metazoan development and disease.
Programmed cell death is essential for metazoan development and is required to sculpt organs, eliminate harmful cells, and counter cell division (1, 2). Apoptosis, an extensively studied cell death process, requires caspase activation, and is accompanied by a stereotypical morphological signature including chromatin compaction, cytoplasmic shrinkage, and no gross disruption of organelles (3). Mice lacking key apoptotic effectors such as caspase-3, caspase-9, Apaf-1, or Bax and Bak, have mild defects, and can survive to adulthood (4). Given the prevalence of cell death during murine development (5), these observations raise the possibility that a non-apoptotic cell death pathway plays key roles in animal development. Although genes promoting necrotic cell death have been identified (6), they are not required for normal development (7). Indeed, genes dedicated to non-apoptotic developmental cell death have not been described.
We previously described the programmed death of the C. elegans linker cell during male reproductive system development (8). The linker cell leads the migration of the male gonad and dies between the fourth larval stage (L4) and adulthood (9). Linker cell demise is orchestrated by a cell-autonomous process independent of all C. elegans caspases and other known cell death genes (8, 10). Electron microscopy (EM) of the dying linker cell reveals non-apoptotic features, including crenellation (indentation) of the nuclear envelope, uncondensed chromatin, and organelle swelling (8) (Fig. 1C). However, linker cell death does not require the unfolded protein response or other stress responses (tables S1 and S2). Similar morphological features are seen during normal developmental death of neurons in the vertebrate spinal cord and ciliary ganglion (11–13), suggesting that linker cell-type death is morphologically conserved.
Fig. 1.
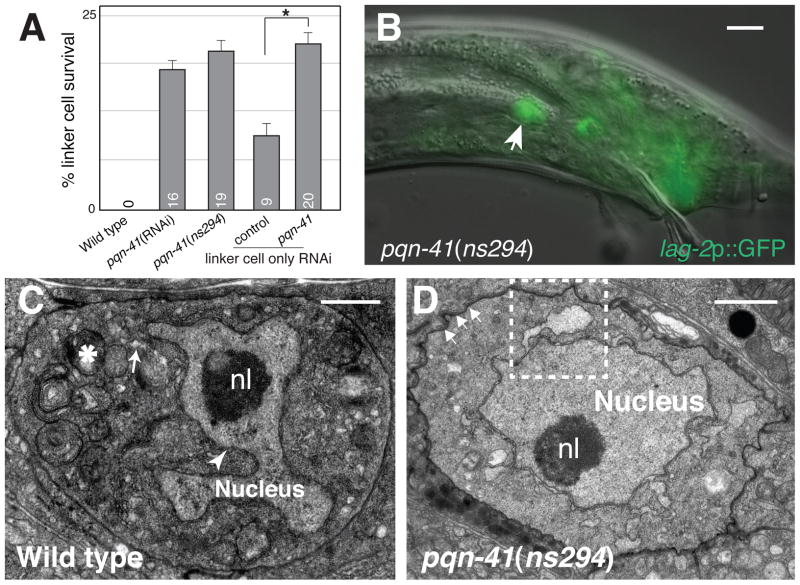
PQN-41 is required for linker cell death. (A) Linker cell survival 2–4 hours after the L4 molt. Numbers, percentages. Error bars, SEM. Asterisk, p<0.04. n≥50. Increased survival in rde-1 may reflect reduced let-7 miRNA function (8). (B) Adult pqn-41(ns294) male with surviving linker cell (arrow). Scale bar, 10 μm. (C) Electron micrograph of dying linker cell in a wild-type animal. nl, nucleolus. Swollen ER and mitochondria, arrow, asterisk. Arrowhead, nuclear envelope crenellation. (D) Electron micrograph of linker cell in (B). Arrows, adhesion junctions. Dashed box, enlarged in fig. S2A. Scale bars in (C, D), 2 μm.
To uncover the molecular mechanisms leading to linker cell death, we performed a genome-wide RNA interference (RNAi) screen to identify genes whose inactivation prevents linker cell death. 18,132 bacterial clones, expressing double-stranded RNA (dsRNA) corresponding to 89% of predicted protein-coding C. elegans genes, were fed individually to RNAi-sensitized males expressing a lag-2 promoter::GFP linker cell reporter (14). The screen strategy was validated by recovery of the lin-29 gene, previously identified as required for linker cell death (8). We recovered five additional RNAi clones leading to linker cell survival. Three clones also affect linker cell migration, and may thus affect multiple aspects of linker cell fate. One of the remaining clones, derived from the gene pqn-41 (Fig. 2A), caused linker cell survival in 20% of animals (Fig. 1A) with no obvious pleiotropic effects (fig. S1), and was further studied.
Fig. 2.
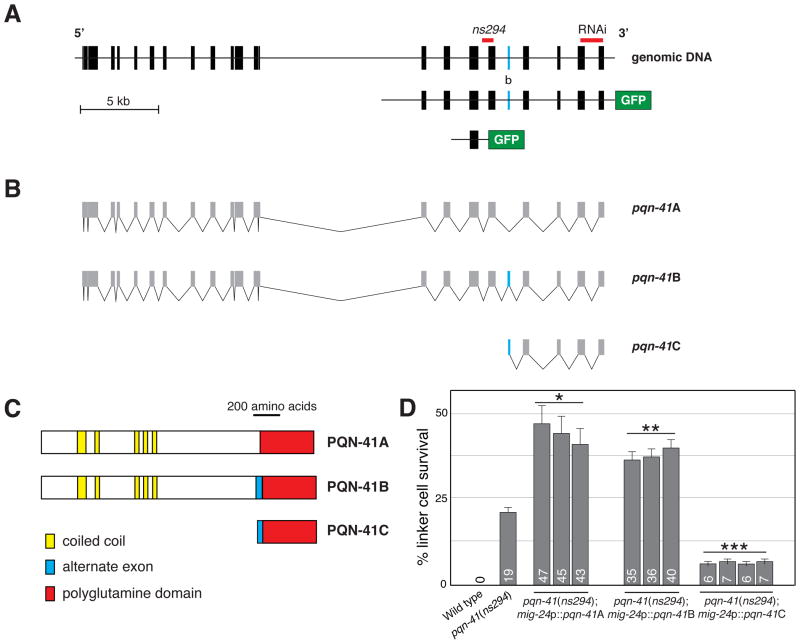
PQN-41C promotes linker cell death. (A) pqn-41 genomic region. Alternative exon b is labeled with the letter “b”. GFP not to scale. (B) Three mRNAs generated by the pqn-41 region. (C) Predicted protein structures for mRNAs in (B). (D) Linker cell survival in pqn-41(ns294) mutants expressing the indicated cDNAs. Numbers, percentages. Error bars, SEM. *, different from pqn-41(ns294), p<0.0009, n=138. **, p<0.0007, n=236. ***, p<0.004, n=229.
To confirm that the clone we identified inactivates pqn-41, we generated a deletion allele, ns294, in the gene. pqn-41(ns294) animals lack 337 nucleotides, removing portions of intron 17 and exon 18 of the predicted genomic structure (Fig. 2A). Consistent with our RNAi results, pqn-41(ns294) adult males possess an inappropriately surviving linker cell (Fig. 1A,B). Furthermore, in pqn-41(ns294) mutants, the linker cell persists at least 24 hours after the L4-adult transition (19% survival, n=79), suggesting that reducing pqn-41 function not only delays, but may block linker cell death. RNAi against pqn-41 in pqn-41(ns294) animals does not increase linker cell survival (16% survival, n=108), suggesting that ns294 may be a strong loss-of-function allele. To test whether pqn-41(ns294) mutants have defects in developmental apoptosis, we scored for surviving pharyngeal cells that normally die apoptotically (15), and observed none (n=10). Thus, the non-apoptotic nature of linker cell death may reflect distinct molecular machineries.
None of the pqn-41(ns294) surviving cells exhibit nuclear crenellation (n=19), as observed by light microscopy. To examine surviving linker cell morphology at higher resolution, we performed serial-section EM. We found that the cell forms adherens junctions to surrounding epithelia (8), and that nuclear envelope crenellation is mild (Fig. 1D; n=2). In both animals, however, organelle swelling was evident (Fig. 1D; fig. S2A,B), suggesting that pqn-41 may be required for nuclear crenellation but not organelle swelling.
To determine if pqn-41 functions cell-autonomously, we examined rde-1(ne219) RNAi deficient mutants (16), containing a mig-24 promoter::rde-1 cDNA transgene restoring RNAi only in the linker cell (17). Feeding pqn-41 RNAi bacteria to these mutants resulted in linker cell death defects similar to systemic RNAi (Fig. 1A), suggesting that pqn-41 functions cell-autonomously. To further examine this issue, we assessed whether expression of pqn-41 within the linker cell can restore linker cell death to pqn-41(ns294) mutants. The pqn-41 locus encodes at least three alternate transcripts we designated pqn-41A, B, and C (Fig. 2). pqn-41A/B span the locus, differing by alternative in-frame exon b. pqn-41C mRNA initiates immediately upstream of exon b (Fig. 2B). We found that expression of pqn-41C is sufficient to promote linker cell demise in pqn-41(ns294) mutants (Fig. 2D), strongly suggesting that pqn-41 functions cell autonomously to promote linker cell death.
pqn-41C encodes a protein composed of runs of glutamine residues with one to eight residues per run (Fig. 2C; fig. S3A). 151 of the 427 pqn-41C codons encode glutamine. The average number of non-glutamine amino acids interrupting adjacent glutamine residues in PQN-41C is 1.8 and is the second smallest in the C. elegans proteome (fig. S4). Glutamine-rich domains are hallmarks of some neurodegenerative disease (ND) proteins and of Q/N-rich prions (18). Both domain classes can adopt coiled-coil structures (18). Similarly, PQN-41C is predicted to contain six coiled-coil motifs demarcated by flanking prolines (CC1-6; fig. S5). Three sequences outside the coiled-coil motifs are conserved among nematodes (CD1-3; fig. S3B), as is the overall proportion of glutamines (38%, 37%, and 37% in C. remanei, C. brenneri and C. briggsae, respectively).
ND and Q/N-rich polyglutamine proteins tend to aggregate in cells (18). Similarly, we found that a PQN-41C::GFP protein forms cytoplasmic aggregates in the linker cell (Fig. 3G), suggesting that PQN-41C shares structural features with these proteins. To understand the importance of the coiled-coil and conserved regions to PQN-41C function, we examined the effects of protein truncations on the ability of PQN-41C to rescue pqn-41(ns294) mutants (fig. S6A). Truncation of consecutive pairs of coiled-coil domains abolishes PQN-41C rescuing activity. Coil-breaking prolines in coiled-coil domains 2, 4, and 6, also reduce rescue efficiency. Thus, sequences encoding the coiled-coil regions are important for PQN-41C function. Truncation of conserved domain CD3 also abolishes rescue, whereas deletion of the CD1/2 domains has only modest effects.
Fig. 3.
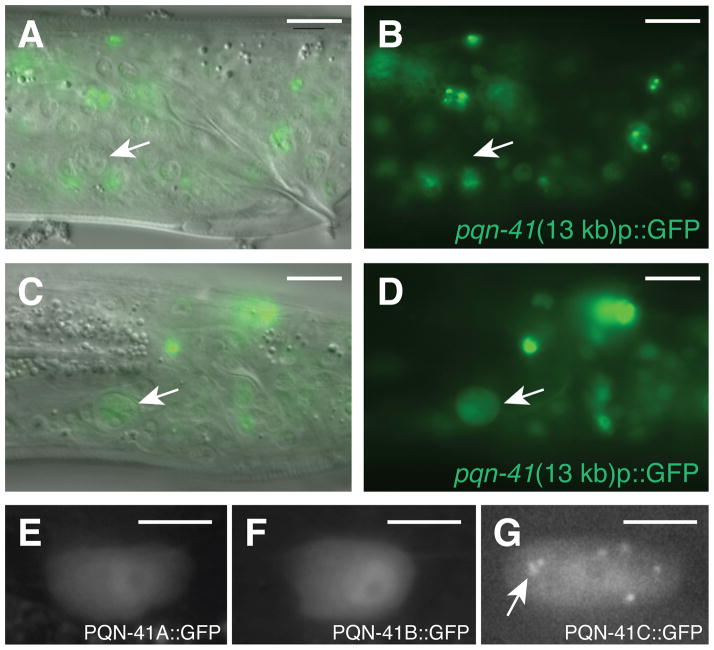
PQN-41 is expressed as the linker cell dies. (A) DIC and fluorescence image of late L4 male expressing the 13 kb pqn-41::GFP reporter in Fig. 2A. Arrow, linker cell. (B) Fluorescence image only. (C) Same as (A) except in an older L4 animal. (D) Fluorescence image only. (E–G) Expression of mig-24 promoter::pqn-41A, B, and C cDNA GFP translational fusions, respectively. Arrow, cytoplasmic puncta. Scale bars, 10 μm (A–D), 5 μm (E–G).
Expression of polyglutamine repeats is often toxic. However, PQN-41C does not exhibit non-specific cellular toxicity. Expression of PQN-41C in the linker cell of pqn-41(ns294) males starting in L2 larvae did not cause precocious cell death (n=227). Rather, cell death was initiated appropriately at the L4-adult transition. Likewise, expression of PQN-41C in the hermaphrodite distal tip cells or the anchor cell did not kill these cells (n>38). Thus, PQN-41C requires the appropriate cellular context to promote death.
To investigate the functions of the pqn-41A/B transcripts, we tested whether they could restore linker cell death to pqn-41(ns294) mutants. We found that both transcripts enhance linker cell survival in pqn-41(ns294) (Fig. 2D) and wild-type animals (fig. S6B). While other polyglutamine proteins can protect cells from polyglutamine toxicity (19), it is puzzling that PQN-41A/B protect the linker cell, given that both proteins contain the glutamine-rich sequences of PQN-41C. It is possible that the N-terminus of PQN-41A/B overrides the cell death-promoting activity of the glutamine-rich domain. Supporting this idea, the N-terminus is sufficient to block linker cell death (fig. S6B). Furthermore, PQN-41A/B::GFP proteins expressed in the linker cell fail to aggregate (Fig. 3E, F).
ND and Q/N-rich proteins often contain coiled-coil motifs outside the glutamine-rich domain, or associate with proteins containing such domains (18). Similarly, PQN-41A and B N-termini are predicted to contain at least five coiled-coil motifs (fig. S7). We found, however, that deletion of sequences encoding different coiled-coil domains did not abolish ectopic survival induced by pqn-41B (fig. S6B).
We next sought to characterize pqn-41 expression. A 13 kb DNA fragment derived from the pqn-41 locus and fused to gfp sequences is broadly expressed in transgenic animals in most cells starting in the embryo (Figs. 2A, 3). However, expression is only switched on in the linker cell as the cell begins to die (Fig. 3A–D). A 2.5 kb DNA sub-fragment derived from this reporter promotes gfp expression nearly exclusively in the linker cell, and only upon cell death initiation (Fig. 2A; fig. S8). The sequences driving linker cell expression of pqn-41 overlap with those deleted in pqn-41(ns294) mutants, and lie upstream of the pqn-41C start (Fig. 2A), suggesting that they may control pqn-41C expression.
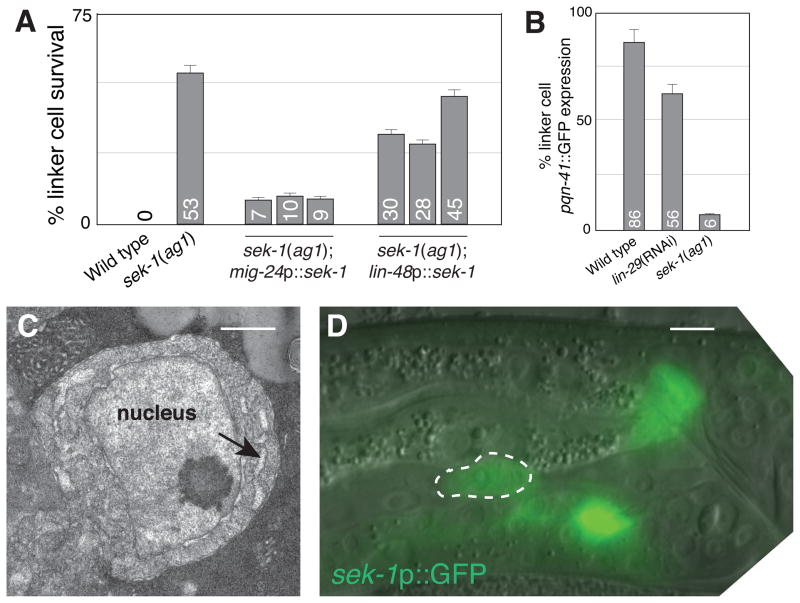
We previously demonstrated a role for the LIN-29 Zn-finger transcription factor in linker cell death (8)(fig. S9B). lin-29 is expressed in the linker cell before the cell begins to die, suggesting that lin-29 might control pqn-41 expression. However, pqn-41 expression is only modestly disturbed in lin-29(RNAi) males (Fig. 4B; fig. S10A). To identify strong regulators of pqn-41 expression, we tested clones identified in our RNAi screen. A clone directed against the gene tir-1, encoding a p38 MAP kinase scaffolding protein important for C. elegans innate immunity, neuronal differentiation and stress responses (20, 21), blocks linker cell death, as does the tir-1(qd4) genetic lesion (fig. S9B). We tested whether other genes involved in innate immunity and stress affect linker cell death. Two independent alleles of the p38 cascade MAPKK gene sek-1 strongly block linker cell death (Fig. 4A; fig. S9; table S2); however, none of the other genes we tested had an effect (table S2). Thus, sek-1 promotes linker cell death independently of innate immunity, stress response, and neuronal differentiation pathways. Unlike in lin-29(RNAi) animals, pqn-41 expression is often not detected in sek-1 mutants (Fig. 4B), suggesting that sek-1 functions upstream of pqn-41. Consistent with this idea, double mutants carrying a strong loss-of-function lesion in sek-1 and the pqn-41(ns294) allele have a survival defect similar to that of sek-1 mutants alone (fig. S9B). Furthermore, EM of surviving linker cells in sek-1(ag1) mutants reveals no nuclear crenellation, but some ER swelling (n=2; Fig. 4C), consistent with regulation of pqn-41 by sek-1. The differential effects of lin-29 and sek-1 on pqn-41 expression suggest these genes function independently. Indeed, we found that sek-1 expression does not require lin-29 function and vice versa (fig. S10B,C). Furthermore, lin-29 mutants do not exhibit the extent of ER swelling of sek-1 mutants (fig. S11; n=3). Finally, strong loss-of-function mutations in lin-29 and sek-1 interact additively (fig. S9B), suggesting these genes may indeed function in parallel.
Fig. 4.
SEK-1 MAPKK is required for linker cell death. (A) Linker cell survival in sek-1(ag1) mutants expressing the indicated promoter::cDNA constructs. Numbers, percentages. Error bars, SEM. n≥70. (B) Expression of the 2.5 kb pqn-41::GFP reporter in late L4 (wild-type) or young adult (lin-29, sek-1) animals. n≥50. (C) EM of linker cell in fig. S9A. Arrow, swollen ER. Scale bar, 2 μm. (D) Late L4 male expressing a sek-1 promoter::GFP reporter. Scale bar, 5 μm.
To determine where sek-1 functions, we examined its expression using a sek-1 genomic region::GFP reporter. This transgene was expressed in the linker cell throughout the cell’s development (Fig. 4D). Expression of a sek-1 cDNA using the mig-24 linker cell-specific promoter restored cell death in sek-1 mutants to a greater extent than expression of sek-1 using the lin-48 promoter, active in surrounding cells (22) (Fig. 4A). These results support a cell-autonomous role for sek-1 in linker cell death.
The studies described here, as well as the morphological similarities between linker cell death and vertebrate developmental cell death, raise the possibility that PQN-41-like proteins might mediate non-apoptotic developmental cell death in vertebrates. The vertebrate proteins MED12 and p400 may be good candidates for such proteins. They are the most similar in primary sequence structure to PQN-41, contain glutamine-rich C-termini (fig. S12), are nuclearly localized, and are important in tumor formation (23, 24). Intriguingly, EM studies reveal that nuclear envelope crenellation is strongly associated with several polyQ expansion diseases (fig. S13). Our studies, thus, raise the possibility that these disease proteins might promote neurodegeneration by inappropriately activating a linker cell death-type process.
Supplementary Material
Acknowledgments
We thank J. Darnell and Shaham lab members for discussions, N. Tishbi for technical assistance, D. Kim, C. Bargmann, M. Kinet, M. Kato, P. Sternberg, K. Nishiwaki, and K. Matsumoto for reagents. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, funded by the National Center for Research Resources (NCRR). ESB is supported in part by the Rockefeller Women & Science Fellowship Program and NIH training grant CA09673. S.S. is supported by NIH grant R01HD042680.
References
- 1.Abraham MC, Shaham S. Trends Cell Biol. 2004;14:184. doi: 10.1016/j.tcb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs Y, Steller H. Cell. 2011;147:742. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr JF, Wyllie AH, Currie AR. Br J Cancer. 1972;26:239. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honarpour N, et al. Dev Biol. 2000;218:248. doi: 10.1006/dbio.1999.9585. [DOI] [PubMed] [Google Scholar]
- 5.Coucouvanis E, Martin GR. Cell. 1995;83:279. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 6.Declercq W, Vanden Berghe T, Vandenabeele P. Cell. 2009;138:229. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Newton K, Sun X, Dixit VM. Mol Cell Biol. 2004;24:1464. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham MC, Lu Y, Shaham S. Dev Cell. 2007;12:73. doi: 10.1016/j.devcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Sulston JE, Albertson DG, Thomson JN. Dev Biol. 1980;78:542. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- 10.Ellis HM, Horvitz HR. Cell. 1986;44:817. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 11.Pilar G, Landmesser L. J Cell Biol. 1976;68:339. doi: 10.1083/jcb.68.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oppenheim RW, et al. J Neurosci. 2001;21:4752. doi: 10.1523/JNEUROSCI.21-13-04752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borsello T, Mottier V, Castagne V, Clarke PG. J Comp Neurol. 2002;453:361. doi: 10.1002/cne.10411. [DOI] [PubMed] [Google Scholar]
- 14.Simmer F, et al. Curr Biol. 2002;12:1317. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 15.Ellis RE, Horvitz HR. Development. 1991;112:591. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]
- 16.Tabara H, et al. Cell. 1999;99:123. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 17.Tamai KK, Nishiwaki K. Dev Biol. 2007;308:562. doi: 10.1016/j.ydbio.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 18.Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Cell. 2010;143:1121. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Proc Natl Acad Sci U S A. 2002;99:17131. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberati NT, et al. Proc Natl Acad Sci U S A. 2004;101:6593. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang CF, Bargmann CI. Genes Dev. 2005;19:270. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson AD, Fitzsimmons D, Hagman J, Chamberlin HM. Development. 2001;128:2857. doi: 10.1242/dev.128.15.2857. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs M, et al. Cell. 2001;106:297. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 24.Makinen N, et al. Science. 2011;334:252. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.