Abstract
Gap-repair assays have been an important tool for studying the genetic control of homologous recombination in yeast. Sequence analysis of recombination products derived when a gapped plasmid is diverged relative to the chromosomal repair template additionally has been used to infer structures of strand-exchange intermediates. In the absence of the canonical mismatch repair pathway, mismatches present in these intermediates are expected to persist and segregate at the next round of DNA replication. In a mismatch repair defective (mlh1Δ) background, however, we have observed that recombination-generated mismatches are often corrected to generate gene conversion or restoration events. In the analyses reported here, the source of the aberrant mismatch removal during gap repair was examined. We find that most mismatch removal is linked to the methylation status of the plasmid used in the gap-repair assay. Whereas more than half of Dam-methylated plasmids had patches of gene conversion and/or restoration interspersed with unrepaired mismatches, mismatch removal was observed in less than 10% of products obtained when un-methylated plasmids were used in transformation experiments. The methylation-linked removal of mismatches in recombination intermediates was due specifically to the nucleotide excision repair pathway, with such mismatch removal being partially counteracted by glycosylases of the base excision repair pathway. These data demonstrate that nucleotide excision repair activity is not limited to bulky, helix-distorting DNA lesions, but also targets removal of very modest perturbations in DNA structure. In addition to its effects on mismatch removal, methylation reduced the overall gap-repair efficiency, but this reduction was not affected by the status of excision repair pathways. Finally, gel purification of DNA prior to transformation reduced gap-repair efficiency four-fold in a nucleotide excision repair-defective background, indicating that the cillateral introduction of UV damage can potentially compromise genetic interpretations.
Keywords: transformation, mismatch repair, recombination, methylation, nucleotide excision repair, base excision repair
1. Introduction
Bacterial DNA is methylated at discrete sites on adenine and cytosine as part of endogenous restriction-modification systems. In Escherichia coli, an additional methylation mark is added by the Dam methyltransferase, which methylates the N6 position of adenine (6meA) in GATC sequences. Dam methylation is exploited as a strand-discrimination signal during mismatch repair (MMR), where it is the transiently un-methylated state of newly synthesized DNA strands that initiates their selective removal [1]. Though eukaryotic DNA also can be methylated (e.g., the C5 position of cytosine), methylation primarily alters chromatin structure to affect gene expression, and notably is not used as a strand discrimination signal during MMR.
In contrast to the DNA of many eukaryotes, the DNA of Saccharomyces cerevisiae does not appear to be intentionally methylated. The lack of methylation prompted an examination by Hoeskstra and Malone of the effects of 6meA on genetic stability in yeast, which was accomplished through over-expression of the E. coli Dam methyltransferase. In contrast to the very strong effects that chemical methylating agents have on genome stability, Dam-directed methylation had only minor effects on recombination and mutagenesis [2]. Though other types of methylated bases are efficiently removed by the base excision repair (BER) pathway (reviewed in [3]), it was found that yeast DNA was methylated to a greater extent in strains that were defective in components of the nucleotide excision repair (NER) pathway [4]. This result was unexpected because the NER machinery is generally assumed to remove only those lesions that distort the DNA helix or that block RNA polymerase [5]. The yeast data not only suggested NER-directed removal of subtle base modifications, it raised the possibility that introducing E. coli DNA into eukaryotic cells might trigger unwanted repair reactions. The potential ability of NER to target base modifications that do not distort the DNA helix has been supported by in vitro studies using purified human NER components [6].
Methylated DNA isolated from E. coli is commonly used for yeast genome modification and as a tool to study homologous recombination. In one type of recombination assay, a chromosomal sequence is used as a template to repair a gap present on an introduced plasmid. We have used such gap-repair assays to study the effect of sequence divergence on homologous recombination [7] and to characterize strand-exchange intermediates formed during recombination [8, 9]. Strand-exchange intermediates can be inferred by sequence analysis of recombination products generated in an MMR-defective background, where mismatches formed during recombination are expected to persist. Such analyses have been complicated, however, by residual removal of recombination-generated mismatches [8]. Here, a gap-repair assay was used to examine the basis of MMR-independent mismatch removal in yeast. We find that Dam methylation is responsible for most of the mismatch correction observed in strains lacking the canonical MMR system and that the aberrant repair is mediated by the NER pathway. In addition, we document a general inhibitory effect of methylation on gap-repair efficiency as well as the introduction of substantial UV damage into DNA that is gel-purified prior to its use in transformation.
2. Materials and methods
2.1. Strain construction
All strains were derived from SJR3409 (W303 derivative; MATa can1Δ∷his3Δ3′,18 leu2-3,112 his3Δ ura3-1 trp1 mlh1Δ∷kan RAD5), which contains a diverged, chromosomal his3 allele. The MLH1 parent of this strain (HK2210) was constructed by Hannah Klein. DNA-repair genes (RAD14, MAG1, OGG1, NTG1, NTG2 or UNG1) were deleted by one-step gene replacement using PCR-generated deletion cassettes. The BER-deficient strain contained deletions of the five DNA N-glycosylase genes and was generated by crossing haploid strains SJR3612 (SJR3409 MATa ung1Δ∷loxP ogg1Δ∷loxP-hyg-loxP mag1Δ∷loxP) and SJR3898 (SJR3409 MATα mag1Δ∷loxP-TRP1-loxP ntg1Δ∷loxP-hyg-loxP ntg2Δ∷loxP-URA3Kl-loxP).
2.2. Media and growth conditions
Cells were grown nonselectively in YPD (1% Bacto-yeast extract, 2% Bacto peptone, 2% dextrose; 1.5% agar for plates) supplemented with 500 μg/mL adenine hemisulfate. His+ or Leu+ colonies were selected on synthetic complete (SC) medium deficient in histidine (SC-his) or leucine (SC-leu), respectively. Ura- segregants were selected on SC medium containing 0.1% 5-fluoroorotic acid (5-FOA; [10]). All growth was at 30°C.
2.3. Plasmids
A URA3/ARS-containing plasmid (pSR987; [9]) was used to determine efficiency of repairing a gap within HIS3 as well as the distribution of crossover and noncrossover (CO and NCO, respectively) events among His+ recombinants. To isolate independent NCO events for sequence analyses, cells were transformed with a CEN plasmid containing the same gapped HIS3 substrate (pSR1015; [8]). The methylation status of plasmids was determined by the E. coli strain used as the host. Plasmids isolated from the dam+dcm+ strain DH5α [F−Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 phoA supE44 thi-1 gyrA96 relA1] were designated as fully methylated, while those isolated from a dam- dcm strain [ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 R(zgb210∷Tn10) TetS endA1 rspL136 (StrR) dam13∷Tn9 (CamR) xylA-5 mtl-1 thi-1 mcrB1 hsdR2; New England Biolabs] were considered to be un-methylated. An in vitro methylated version of the un-methylated plasmid was generated using purified Dam methyltransferase (New England Biolabs) and methylation was confirmed by insensitivity to digestion with MboI.
2.4. Gap-repair experiments
Gap-repair efficiency was measured as previously described using a mix of BssHII-digested pSR987 [8] and a circular LEU2-containing plasmid (pRS315; [11]) in a 10:1 weight ratio. Unless otherwise specified, linearized DNA was purified from an agarose gel prior to use in transformation experiments. Following co-transformation, His+ or Leu+ transformants were independently selected on SC-his or SC-leu plates, respectively, and repair efficiency was calculated as the ratio of His+ to Leu+ colonies. The stability of the URA3 marker on repaired plasmids was used to determine whether a NCO or CO event occurred. Stability was determined by growing His+ colonies for three days in SC-his medium, diluting cells 1:100 in YPD and growing overnight to allow plasmid loss, and finally spotting onto 5-FOA plates. An autonomous plasmid produced by a NCO event was unstable and the corresponding transformant grew on 5-FOA medium. By contrast, plasmid integration at the chromosomal donor locus was indicated by a failure to grow on 5-FOA.
For DNA sequence analysis of NCO products, His+ transformants were obtained using pSR1015. Entire colonies (without prior purification) were inoculated into 96-well microtiter plates containing SC-his medium and grown for 3 days. Following DNA extraction, the repaired plasmid allele (or chromosomal donor allele) was amplified by PCR and sequenced by the Duke University DNA Analysis Facility. As reported previously, heteroduplex DNA was only observed in the recipient allele [8, 9].
2.5. Statistical analysis
Mean His+:Leu+ ratios obtained in different strain backgrounds or using plasmids with different methylation states were compared using Student's t-test. The relative numbers of CO versus NCO events, as well as the distributions of NCOs among the classes defined in the text, were compared using a two-tailed Fisher Exact Test. p<0.05 was considered significant.
3. Results
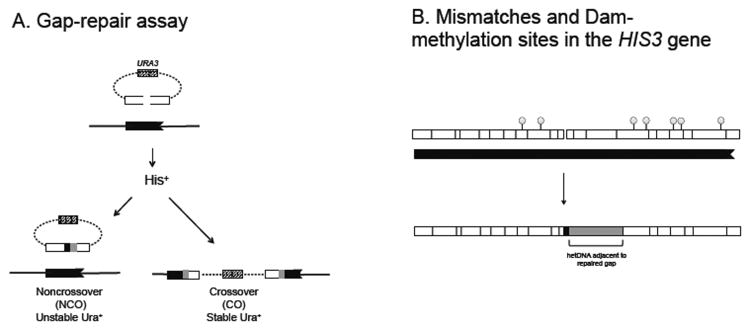
We previously described a plasmid-based gap-repair assay that allows characterization of the molecular structures of individual repair events resulting from homologous recombination [9]. As illustrated in Fig. 1A, the plasmid contains a full-length HIS3 gene with a centrally located 8-bp gap created by restriction digest. Upon transformation, the gapped plasmid uses a truncated his3 allele on chromosome V as a donor, repair template. The donor sequence contains ∼800 bp of total homology with the gapped plasmid allele and encodes a mutant protein lacking 11 amino acids at the C-terminus. In order to monitor the transfer of DNA strands between recombining molecules, 19 silent single-nucleotide polymorphisms (SNPs) were engineered into the chromosomal his3 allele, giving rise to 2% sequence divergence between the chromosomal donor and the recipient plasmid alleles (Fig. 1B). Heteroduplex DNA (hetDNA) formed by strand exchange contains mismatches whose repair usually leads to conversion to donor sequence or, less often, to restoration to plasmid sequence. In the absence of MMR, however, mismatches in hetDNA are expected to persist and can be detected by sequence analysis of the His+ colonies derived from gap repair. In sequence chromatograms, a dual peak at the position of a given SNP corresponds to hetDNA. All experiments reported here were performed in mlh1Δ, MMR-deficient strain backgrounds in order to allow detection of hetDNA.
Fig. 1.
Gap-repair assay. (A) The white box indicates the gapped, full-length his3 allele on the plasmid; the black box corresponds to the his3Δ3′ repair template on chromosome V; and the hatched box represents the plasmid-encoded URA3 gene. hetDNA in repaired plasmids is indicated by gray boxes. (B) Positions of SNPs (black lines) and Dam-methylation sites (lollipops) relative to the gap in the plasmid allele are indicated to scale. An example of a hetDNA (gray) adjacent to the repaired gap is illustrated.
In addition to monitoring hetDNA formation, two other parameters were measured in gap-repair experiments: the overall efficiency of gap repair and the distribution of repair products into crossover and noncrossover (CO and NCO, respectively) events. The former was determined as the frequency of His+ transformants derived from the gapped plasmid relative to the frequency of Leu+ colonies obtained with an uncut plasmid. CO and NCO transformants were distinguished based on the stability of a URA3 marker present on the repaired plasmid (see Materials and Methods for details). Here, we focus specifically on the hetDNA detected in NCO products.
3.1 Mismatches in hetDNA are repaired in the absence of MMR
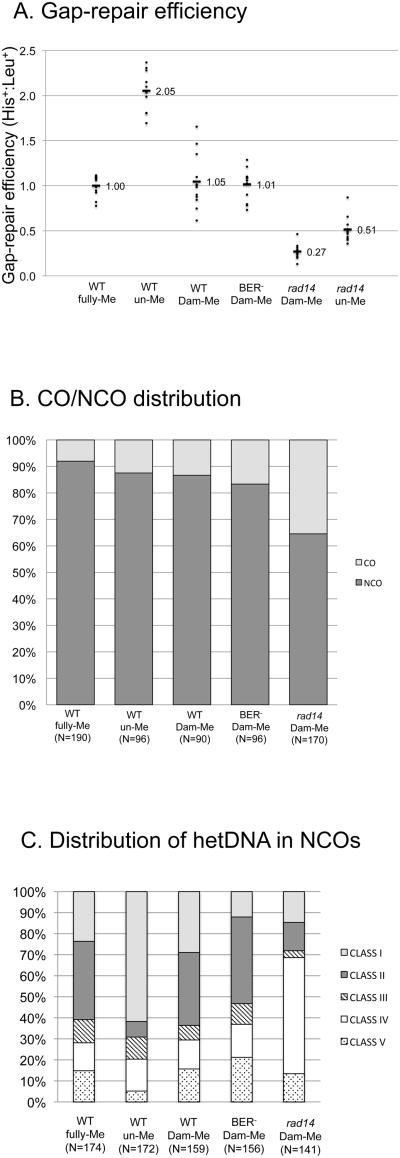
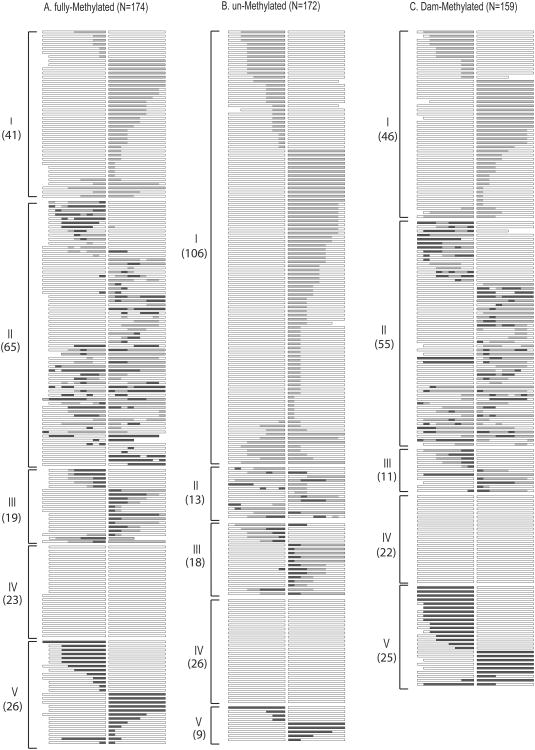
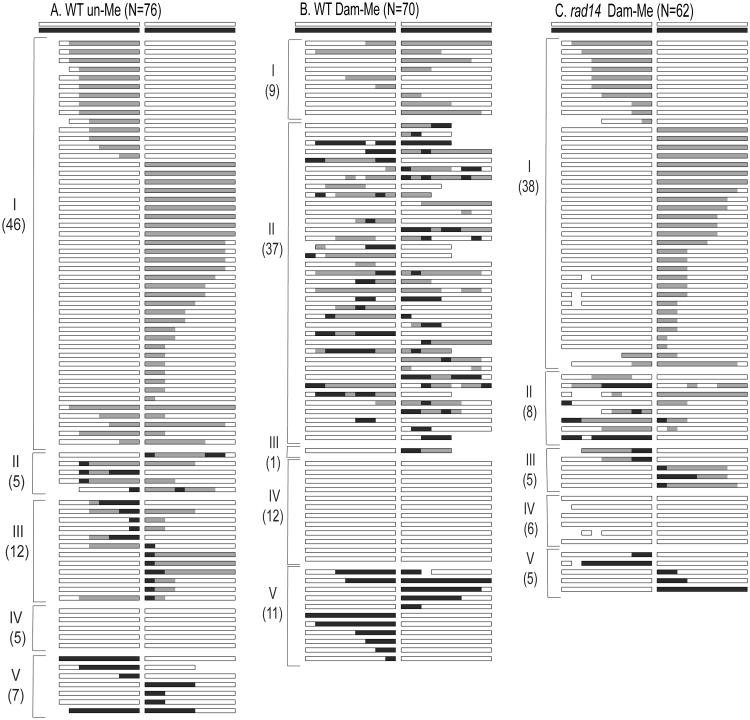
The ratio of His+:Leu+ colonies obtained when DNA isolated from a dam+dcm+ strain (DH5α; see Materials and Methods) was used to transform an MMR-defective (mlh1 Δ) yeast strain was set to 1.00 to facilitate comparisons of gap-repair efficiency when the methylation status and strain background were varied (Fig. 2A). We designate the mlh1Δ strain that is proficient in other repair pathways as WT, and refer to DH5α-derived plasmid DNA as being fully-methylated (fully-Me). As reported in previous analyses [7], approximately 90% of His+ colonies obtained with fully-Me DNA reflected NCO events (Fig. 2B). Among the 174 NCOs sequenced, five classes of strand-transfer events were identified, only the first three of which contained hetDNA (Fig. 3A). Class I events had the “continuous” hetDNA tracts predicted to be produced in the absence of MMR. By contrast, Class II events had “discontinuous” tracts of hetDNA interrupted by patches of restoration and/or gene conversion (GC), indicating intermittent repair of mismatches. Approximately equal numbers of restoration and GC tracts were observed, suggesting no bias in terms of which strand was removed during the repair reaction. The median length of individual repair tracts was ∼70 bp, and multiple tracts were often observed in a single recombinant. Finally, Class III events had a GC tract immediately adjacent to the repaired gap, followed by continuous hetDNA. Because the GC tracts in Class III events could reflect either repair events or gap expansion prior to hetDNA formation, only the first two classes of hetDNA-containing events were considered to be informative in terms of hetDNA analysis. Among the 106 NCO events that had informative hetDNA, the Class II events with discontinuous hetDNA tracts outnumbered the Class I events with continuous, uninterrupted hetDNA; there were 65 Class II but only 41 Class I events. Because MMR was disabled in the strain used to monitor gap repair, the frequent presence of restoration/GC tracts interspersed with hetDNA indicates an alternative and robust mechanism that removes mismatches in recombination intermediates.
Fig. 2.
Gap-repair efficiency, CO-NCO distribution and hetDNA classes. (A) Each data point corresponds to the His+:Leu+ ratio obtained in an independent transformation experiment. The mean ratio for each strain relative to WT transformed with fully-Me DNA is given, with the WT ratio set to 1.00. (B) Percentages of CO and NCO events are indicated by light and dark shading, respectively. (C) Distributions of sequenced NCO products into five classes (I-V; see text for description) based on the presence and position of hetDNA in the repaired plasmid allele. The total number of events analyzed (N) in panels B and C is given below the strain genotype and plasmid-methylation status. All DNAs were gel-purified prior to use for transformation and all strains were mlh1Δ.
Fig. 3.
Sequence analysis of NCO products. DNA was isolated from individual His+ colonies following transformation of an mlh1Δ strain with (A) fully-Me, (B) un-Me or (C) Dam-Me plasmid that had been gel purified. Each horizontal line represents an independent repair event, with a box representing homology on each side of the gap. White areas correspond to plasmid sequence, black areas to chromosomal sequence and gray areas to hetDNA. Gaps in individual boxes correspond to an absence of sequence information. Classes I-V are described in the text and the number of products in each category is indicated.
The two remaining classes of gap-repair events were Class IV events, in which there was no detected sequence transfer from the chromosomal donor to the plasmid, and Class V events with a continuous GC tract that extended uninterrupted from the gap. The numbers of Class IV and Class V events are usually comparable (23 and 26 events in the present analysis), leading us to previously suggest that these events most likely reflect the detection of only one strand of segregated hetDNA [9].
3.2 Methylation triggers co-repair of mismatches in hetDNA intermediates
The plasmid substrate used in the transformation experiments described above was isolated from E. coli strain DH5α, which contains the Dam and Dcm methylation systems. Dam generates 6meA at GATC sequences, while Dcm methylates the C5 position of the internal cytosine in CCAGG and CCTGG sequences. Because yeast DNA lacks methylation, we hypothesized that the trigger for MMR-independent mismatch removal might be the presence of plasmid-derived methylated bases in hetDNA intermediates. To investigate this possibility, we used plasmid DNA isolated from a dam−dcm−E. coli host strain in yeast transformation experiments, and refer to this DNA as being unmethylated (un-Me). Though the overall gap-repair efficiency was approximately 2-fold higher when the un-Me plasmid was used (Fig. 2A; p<0.001), the proportion of CO events among repair products was not altered (Fig. 2B; p=0.30). The molecular profile of NCO events obtained with the un-Me plasmid, however, was strikingly different from that observed with the fully-Me plasmid. In contrast to only 39% continuous hetDNA tracts with the fully-Me plasmid substrate, the proportion of continuous tracts among informative hDNA-containing products increased to 89% when the un-Me plasmid was used in transformation experiments (Fig. 2C and Fig. 3; p<0.0001). The most dramatic change associated with plasmid methylation was in the relative proportions of Class I versus Class II events, but we also note a proportional increase in Class IV relative to Class V events when the un-Me plasmid was used (p=0.015). This reflected a loss in Class V events (26/174 obtained with fully-Me DNA compared to 9/172 with un-Me DNA; p=0.004), suggesting that some may correspond to methylation-triggered GC tracts that repair all hetDNA.
To confirm the role of methylation in mismatch removal, the un-Me plasmid DNA was methylated in vitro using purified Dam methyltransferase. Use of Dam-methylated (Dam-Me) DNA reduced the overall transformation efficiency 2-fold, down to a level indistinguishable from that observed with the fully-Me plasmid (Fig. 2A; p=0.60). Significantly, use of Dam-Me DNA was associated with a decrease in the proportion of continuous hetDNA-containing events to 46% (Fig. 2C and Fig. 3C), which is indistinguishable from the level obtained using fully-Me DNA (p=0.33). With regard to the specific effect of Dam-Me on mismatch removal, the his3 sequences on the gapped plasmid contain multiple Dam sites (indicated as lollipops in Fig. 1B), but are devoid of Dcm sites. Together, these results demonstrate that presence of 6meA in recombination intermediates is sufficient to trigger most of the MMR-independent removal of mismatches that occurs in the gap-repair assay used here.
3.3. Elimination of DNA N-glycosylases does not affect gap-repair efficiency or reduce mismatch removal
Given that Dam-mediated methylation can trigger coincident mismatch removal in yeast, we hypothesized that one or more of the DNA N-glycosylases, at least some of which have specificity for methylated bases, might be responsible for reducing the repair efficiency and/or in generating discontinuous hetDNA tracts. To examine these possibilities, we eliminated the five DNA N-glycosylases that can initiate base excision repair (BER) in yeast: Mag1, Ogg1, Ntg1, Ntg2 and Ung1 [3]. For simplicity, we refer to this quintuple-mutant strain as BER-defective. When the Dam-Me DNA was used, the overall gap-repair efficiency in the BER-defective strain was indistinguishable from that in WT (p=0.74; Fig. 2A), as was the distribution of repair events into NCO and CO products (p=0.55; Fig. 2B). Among the NCO events isolated in the BER-defective strain, the proportion that contained informative hetDNA also was similar to that in WT (p=0.08; Fig. 2C and Fig. S1B). Among the hetDNA-containing events, however, the proportion of continuous relative to discontinuous tracts was reduced in the BER-deficient background. Whereas 46% of hetDNA-containing tracts in WT were continuous, only 23% were continuous in the BER-defective strain (p=0.0002). The observed increase in the proportion of discontinuous hetDNA tracts is the opposite of what would be expected if BER were responsible for triggering coincident mismatch removal. Instead, these data suggest that BER-initiated removal of 6MeA antagonizes the coincident repair of mismatches.
3.4 NER is required for efficient gap repair of gel-purified plasmid
The nucleotide excision repair (NER) pathway was previously implicated in removing 6meA generated by overexpressing the E. coli dam gene in yeast [4]. To examine whether NER activity is relevant to the generation of discontinuous hetDNA tracts in NCO events, we deleted the RAD14 gene, which is required for damage recognition during NER [3]. When the Dam-Me plasmid was used in the gap-repair assay, there was an unexpected, four-fold reduction in overall repair efficiency in the absence of RAD14 (Fig. 2A; p<0.0001). This reduction, however, was related neither to the methylation status of the plasmid (a similar 4-fold reduction relative to WT was observed when un-Me plasmid was used; Fig 2A) nor to the level of sequence identity between the recombination substrates (data not shown). In addition to the reduced repair efficiency associated with NER loss, the proportion of COs among His+ colonies obtained using the Dam-Me plasmid was elevated from 13% in wild type (WT) to 35% in the rad14 Δ background (Fig. 2B; p=0.0009). These data suggest that, at least in the system used here, there is a strong and unexpected pro-recombination role for Rad14 that differentially affects CO and NCO formation.
In addition to the reduction in overall gap-repair efficiency and enrichment in CO events associated with Rad14 loss, there was a dramatic decrease in the proportion of NCO events that contained informative hetDNA (i.e., Class I and II events; Fig. 2C and Fig. S1A). In WT transformed with the Dam-Me plasmid, ∼65% of NCO events had hetDNA tracts; in the rad14 Δ background, only ∼30% of NCOs had detectable hetDNA (Fig. 3; p<0.001). There was no change, however, in the proportional of Class I relative to Class II events in the rad14 Δ background (46/106 Class I events in WT versus 23/46 in rad14 Δ; p=0.72).
3.5 The role of NER in promoting gap-repair reflects the use of gel-purified plasmid
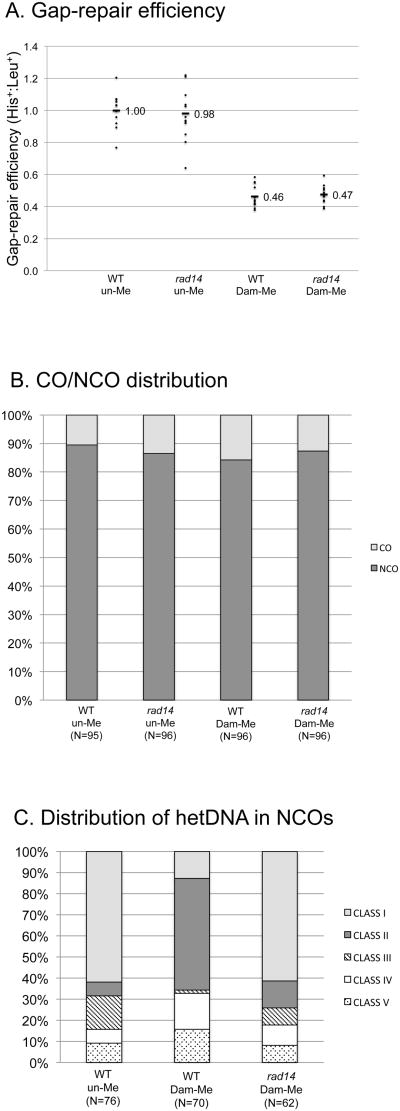
In gap-repair assays, linearized plasmid DNA is typically gel-purified prior to transformation in order to eliminate any background transformants derived from uncut plasmid DNA (e.g., see [12, 13]). We similarly purified the DNA used in the experiments described above. Gel-purification requires visualization of DNA with UV light, however, raising the possibility that the perceived requirement for NER during gap-repair might reflect a need to remove UV-induced DNA damage. To examine this possibility, we transformed linearized un-Me or Dam-Me plasmids that had not been gel-purified into the WT and rad14 Δ strains. It should be noted that presence of uncut plasmid in the system used here does not compromise transformation results, as uncut plasmid contains a small insertion that inactivates the HIS3 gene [9]. Strikingly, the use of un-purified DNA completely eliminated the requirement of Rad14 for efficient gap repair (Fig. 4A; p=0.62 and p=0.66 for un-Me and Dam-Me plasmids, respectively). In addition, the increased proportion of CO products observed when gel-purified, Dam-Me DNA was introduced into the rad14 Δ background was eliminated when un-purified DNA was used (Fig. 4B). By contrast, there was no detectable effect of fragment purification on transformation of the WT strain, suggesting that functional NER is sufficient to remove any UV damage that is introduced during gel purification. These results demonstrate that NER is not required for the gap-repair process per se, and only becomes necessary when the transformed plasmid contains UV damage. It is important to note that, as observed with gel-purified DNA, presence of 6meA in the un-purified plasmid reduced repair efficiency approximately 2-fold in both WT and NER-defective backgrounds.
Fig 4.
Gap-repair efficiency, CO-NCO distribution and hetDNA classes obtained using un-purified plasmid DNA. (A) Each data point corresponds to the His+:Leu+ ratio obtained in an independent transformation experiment. The mean ratio for each strain is relative to that obtained when the WT strain was transformed with un-Me DNA. (B) Percentages of CO and NCO events are indicated by light and dark shading, respectively. (C) Distributions of sequenced NCO products into five classes (I-V; see text for description) based on the presence and position of hetDNA in the repaired plasmid allele. The total number of events analyzed (N) in panels B and C is given below the strain genotype and plasmid-methylation status. All strains were mlh1Δ.
3.6 NER is responsible for the methylation-associated removal of mismatches in hetDNA
The dramatic effect of gel purification on gap repair in the rad14 Δ strain prompted us to re-examine whether the presence of UV damage might have masked a role for NER in the methylation-dependent removal of mismatches in hetDNA. We thus sequenced NCO products obtained when un-purified DNA was transformed into the WT and rad14 Δ strains. For the WT strain, His+ transformants isolated using un-Me or Dam-Me DNA were sequenced; for the rad14 Δ strain, only transformants obtained using Dam-Me DNA were examined. The results of molecular analyses of the products obtained using un-purified DNA are presented in Fig. 5 and are summarized in Fig. 4C. As observed when WT was transformed with gel-purified DNA, the relative numbers of NCO events with continuous and discontinuous hetDNA tracts (Class I and Class II events, respectively) obtained when the WT strain was transformed with un-purified DNA were dramatically affected by the methylation status of the DNA. That is, Class I events greatly outnumbered Class II events (47 and 5 events, respectively) when un-Me DNA was used, but there was an excess of Class II events (37 Class II versus 9 Class I events) when the DNA was Dam-methylated (p<0.0001). When NER was defective, however, the use of un-purified, Dam-Me DNA yielded a distribution of events that was indistinguishable from that obtained when the WT strain was transformed with un-Me DNA (p=0.22). These data demonstrate that it is the NER pathway that is responsible for generating discontinuous tracts of hetDNA when the transforming plasmid contains 6meA.
Fig. 5.
Sequence analysis of NCO products derived using linearized plasmid that was not gel purified. DNA was isolated from individual His+ colonies following transformation of an mlh1Δ strain with (A) un-Me or (B) Dam-Me plasmid, or (C) transformation of an mlh1Δ rad14 Δ strain with Dam-Me DNA. Each horizontal line represents an independent repair event, with a box representing homology on each side of the gap. White areas correspond to plasmid sequence, black areas to chromosomal sequence and gray areas to hetDNA. Gaps in individual boxes correspond to an absence of sequence information. Classes I-V are described in the text and the number of products in each category is indicated.
4. Discussion
In the gap-repair experiments reported here, a linearized plasmid was transformed into MMR-defective (mlh1 Δ) strains containing a diverged repair template, allowing mismatches formed in strand-exchange intermediates to be monitored. Plasmid DNA isolated from E. coli strains containing the Dam methyltransferase is standardly used in such gap-repair assays. The analyses presented here demonstrate that the subtle modification to DNA structure conferred by 6meA can lower the overall gap-repair efficiency and is sufficient to trigger the sporadic removal of mismatches present in recombination intermediates, resulting in conversion of continuous to discontinuous hetDNA. These effects were eliminated when the transforming DNA was isolated from a dam‒ dcm‒ E. coli strain, and were fully restored when the un-Me DNA was treated in vitro with purified Dam methyltranferase. Though Dam methylation clearly is sufficient to alter gap-repair efficiency and to generate discontinuous hetDNA, we cannot formally exclude a similar role for Dcm methylation or other types of non-yeast base modifications. In addition to defining the major source of MMR-independent mismatch removal, the data also provide an explanation for why discontinuous hetDNA tracts were only rarely observed in our initial molecular analyses of gap repair [9]. In early experiments, the transforming DNA was prepared by PCR amplification and hence would have been un-Me; in later analyses, restriction-digested plasmid DNA was used in transformation experiments [8]. That Dam methylation interferes with the overall efficiency of gap repair suggests that the presence of 6MeA either interferes with plasmid maintenance or with one or more steps during recombination. In terms of recombination, a non-yeast base modification could perturb 5′-end resection; it could reduce the formation of Rad51 nucleoproteins filaments and/or the efficiency of subsequent strand exchange; or it could interfere with the loading of replication proteins required for extending an invading or captured 3′ end.
The primary goal of the studies reported here was to determine whether methylation of transforming DNA triggers the coincident removal of recombination-generated mismatches and if so, whether the removal reflects NER and/or BER activity. Experiments reported by Hoekstra and Malone almost 25 years ago suggested that NER removes 6meA from yeast DNA, but did not examine whether BER might also be involved [4]. In their experiments, the E. coli Dam methyltransferase was over-expressed and modification of yeast genomic DNA was monitored using methylation-sensitive restriction enzymes. Our initial examination of whether NER activity directed against Dam-Me plasmid DNA triggers coincident mismatch removal was compromised by an unexpected requirement of NER for efficient gap repair. In addition, loss of NER inexplicably increased the proportion of CO products and reduced the proportion of NCO intermediates that contained hetDNA. We found, however, that the profound effects of NER status were strictly limited to plasmid that had been gel-purified prior to use in transformation experiments. We thus assume that it is UV damage inadvertently introduced during fragment purification that affects gap repair in an NER-defective background. Unrepaired UV damage, for example, would be expected to block plasmid replication and thereby decrease gap-repair efficiency. Similarly, the reduction in detected hetDNA could reflect replication of only one strand of the plasmid, with the other strand containing a replication-blocking UV lesion(s). With regard to the proportional increase in CO events in the rad14Δ background, it is possible that the Rad1-Rad10 complex, which processes intermediates during recombination [14] as well as during NER, is more available for recombination reactions when NER is disabled. CO events in particular are promoted by Rad1-Rad10 in gap-repair assays [7, 13, 15] and in assays of double-strand break induced recombination between ectopic chromosomal repeats [16]. It is important to emphasize that when NER was functional, gel purification had no detectable effect on the recombination parameters examined. The NER system is thus able to efficiently remove UV damage in plasmid DNA that otherwise affects gap repair.
When un-purified DNA was used in gap-repair experiments, the hetDNA profile obtained with Dam-Me DNA in an NER-defective (rad14Δ) background was indistinguishable from that obtained with un-Me DNA in a WT (NER-proficient) background. This demonstrates that it is indeed the NER pathway that is responsible for generating most of the discontinuous hetDNA tracts observed when Dam-Me DNA was used in gap-repair experiments. It should be noted that the mismatch-containing DNA targeted by the NER machinery in this assay is only hemi- methylated, with the relevant hetDNA intermediates being comprised of one plasmid and one chromosomal DNA strand. Interestingly, the NER system did not appear to discriminate between the Dam-Me and un-Me strands when initiating repair, as we observed similar numbers of restoration and gene conversion events. If the Dam-Me strand contributed by the plasmid were preferentially targeted for removal, then gene conversion would have predominated over restoration; if the un-Me, chromosomal strand were preferentially targeted, then restoration should have predominated. In addition, based on the lengths of gene conversion and restoration tracts, the NER machinery often removed more than the canonical 25-30 nucleotides. Either the initial incision reactions were perturbed, or the initial gap was enlarged, perhaps by the 5′ to 3′ activity of Exo1 [17].
Given the lack of significant helical distortion associated with Dam methylation of adenine, it seemed likely to us that 6meA might be recognized and the modified base excised by one or more of the yeast DNA-N-glycosylases. Removal of the resulting apurinic/apyrimidinic site by BER would involve either excision of a single nucleotide or the excision of an oligonucleotide as part of flap generated by strand-displacement DNA synthesis (short- and long-patch BER, respectively; [18]). In principle, long-patch BER could be responsible for coincident mismatch removal. Mismatch correction during gap repair was not only still observed in a yeast strain with all five glycosylase genes deleted, the proportion of hetDNA tracts that were discontinuous actually increased. This increase can be explained if BER and NER compete for the removal of 6meA. Loss of BER thus would be accompanied by an increase in NER efficiency and a concomitant shift towards proportionally more discontinuous hetDNA tracts.
Even though isolation of plasmid DNA from a dam- dcm- E. coli host eliminated most mismatch removal in yeast, small numbers of gene conversion and restoration events persisted. This suggests the existence of an additional pathway of MMR-independent mismatch removal that is not triggered by DNA methylation. MMR-independent removal of mismatches in mitotic and meiotic recombination intermediates was previously reported in budding yeast [19] and, as observed here, the residual removal could not be attributed to the NER machinery. Though only small numbers of events were analyzed in the earlier study, they were similar to those reported here in that repair tracts were much shorter than those typically generated by the MMR machinery and both restoration and gene conversion events were observed. This is in stark contrast to the very biased activity of the MMR system in removing mismatches in meiotic recombination intermediates, which results mostly in gene conversion events [20]. In addition, it is possible that it is the minor pathway observed here, rather than gap expansion, that is responsible for the residual meiotic gene conversion that persists in the absence of MMR [21]. Finally, it should be noted that MMR-independent removal of mismatches in meiotic recombination intermediates also has been observed in Schizosaccharomyces pombe[22] and in Drosophila melanogaster[23], suggesting evolutionary conservation of an alternative pathway(s) of mismatch processing. At least in S. pombe, however, it is the NER pathway that is responsible for the removal of mismatches in MMR-defective backgrounds [22].
In summary, the results presented here demonstrate that the use of bacterially methylated DNA in yeast gap-repair experiments reduces the overall efficiency of repair and triggers the coincident removal of mismatches in recombination intermediates, resulting in discontinuous hetDNA tracts. We demonstrate that 6meA in particular can initiate NER in vivo, but without obvious strand discrimination, resulting in approximately equal numbers of gene conversion and restoration tracts in NCO products. The versatility of yeast NER in terms of recognizing a subtle helical distortion caused by 6meA is unexpected and likely extends to other types of non-distorting base modifications. Whether the detection and processing of 6meA is unique to budding yeast or extends to other eukaryotes is presently unclear. It should be noted, however, that a very low level of 6MeA has been detected in eukaryotic genomes [24]. Whether this specific modification is intentionally generated or is pathological is not known, but the latter seems more likely given the ability of 6meA to be targeted by the NER machinery. In addition to the methylation-specific effects observed, our data also support the existence of an additional MMR-independent pathway(s) that removes mismatches present in recombination intermediates. Finally, results raise a cautionary note with regard to using bacterial DNA for directed genome modifications and to interpreting genetic results obtained when gel-purified DNA fragments are introduced into an NER-compromised background.
Supplementary Material
Highlights.
Mismatches formed during mitotic gap repair are removed in the absence of the mismatch repair (MMR) pathway
Bacterial methylation of plasmid DNA triggers MMR- independent mismatch removal
Nucleotide excision repair is responsible for methylation- triggered mismatch removal
Methylation of plasmid DNA reduces gap-repair efficiency
Acknowledgments
The authors thank Nayun Kim and Kevin Lehner for intellectual contributions made throughout the course of this work. We also acknowledge other lab members and Tom Petes for helpful comments on the manuscript. This work was supported by NIH grant GM38464 to SJ-R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Ann Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 2.Hoekstra MF, Malone RE. Expression of the Escherichia coli dam methylase in Saccharomyces cerevisiae: effect of in vivo adenine methylation on genetic recombination and mutation. Mol Cell Biol. 1985;5:610–618. doi: 10.1128/mcb.5.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boiteux S, Jinks-Robertson S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013;193:1025–1064. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoekstra MF, Malone RE. Excision repair functions in Saccharomyces cerevisiae recognize and repair methylation of adenine by the Escherichia coli dam gene. Mol Cell Biol. 1986;6:3555–3558. doi: 10.1128/mcb.6.10.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 6.Huang JC, Hsu DS, Kazantsev A, Sancar A. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc Natl Acad Sci USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welz-Voegele C, Jinks-Robertson S. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics. 2008;179:1251–1262. doi: 10.1534/genetics.108.090233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchel K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 2013;9:e1003340. doi: 10.1371/journal.pgen.1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchel K, Zhang H, Welz-Voegele C, Jinks-Robertson S. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell. 2010;38:211–222. doi: 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-fluoroorotic acid as a selective agent in yeast molecular genetics. Meth Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 11.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haghnazari E, Heyer WD. The DNA damage checkpoint pathways exert multiple controls on the efficiency and outcome of the repair of a double-stranded DNA gap. Nucleic Acids Res. 2004;32:4257–4268. doi: 10.1093/nar/gkh717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Symington LS, Kang LE, Moreau S. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:4649–4656. doi: 10.1093/nar/28.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 15.Schiestl RH, Prakash S. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol Cell Biol. 1990;10:2485–2491. doi: 10.1128/mcb.10.6.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazon G, Lam AF, Ho CK, Kupiec M, Symington LS. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat Struct Mol Biol. 2012;19:964–971. doi: 10.1038/nsmb.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 19.Coic E, Gluck L, Fabre F. Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 2000;19:3408–3417. doi: 10.1093/emboj/19.13.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Detloff P, White MA, Petes TD. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone JE, Petes TD. Analysis of the proteins involved in the in vivo repair of base- base mismatches and four-base loops formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics. 2006;173:1223–1239. doi: 10.1534/genetics.106.055616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleck O, Lehmann E, Schar P, Kohli J. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat Genet. 1999;21:314–317. doi: 10.1038/6838. [DOI] [PubMed] [Google Scholar]
- 23.Radford SJ, Sabourin MM, McMahan S, Sekelsky J. Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics. 2007;176:53–62. doi: 10.1534/genetics.107.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.