The development and availability of genomic applications for use in clinical care is accelerating rapidly. the routine use of genomic information, however, is beyond most health-care providers’ formal training, and the challenges of understanding and interpreting genomic data are compounded by the demands of clinical practice. nearly all physicians, for example, agree that genetic variations may influence drug response, but only a small fraction feel adequately informed about pharmacogenomic testing.1 Clinical decision support (CDS) embedded into clinical information systems, such as the electronic health record (EHR) and the personal health record (PHR), is recognized as being necessary to facilitate the appropriate use of genomic applications.2–4
CDS provides clinical knowledge and patient-specific information, filtered or presented at particular times to enhance clinical care.5 CDS solutions can assist clinical-care providers with personalizing care and can incorporate the preferences of health-care consumers. EHRs and PHRs theoretically may support access to and storage of genetic data. These systems may also support data exchange between repositories and enable CDS embedment and linkage. The use of EHRs and PHRs in this manner depends on characteristics of the underlying health information technology (IT) infrastructure. This article seeks to provide a common ground for discussing CDS for genetic testing and for data access processes among heterogeneous health IT infrastructures.
There are many lessons learned from more than five decades of experience with CDS that can be applied to CDS implementation in the era of genomic data. Indeed, existing CDS technologies already play a role in supporting genetic testing and data access processes. In the following sections, we provide an overview of existing frameworks for local evaluation of health IT infrastructures for CDS, processes for genetic testing and data access, and the rationale behind the Electronic Medical Records and Genomics (eMERGE) Network’s6 work on establishing a common ground for discussing CDS solutions among heterogeneous IT infrastructures. We also provide examples from eMERGE to illustrate that we can characterize genomic CDS using frameworks from the pregenomic CDS era, and outline lessons learned from implementing pregenomic CDS that can account for variation in health IT infrastructure. Finally, we propose a framework to describe opportunities for genomic CDS that can support provider- and consumer-initiated genetic testing and data access processes.
The work in this article is complementary to that of the Clinical Sequence Exploratory Research Electronic Records Working Group, also in this special issue.7 The Working Group’s manuscript surveys the six current Clinical Sequence Exploratory Research sites on the processes used for variant annotation, curation, report generation, and integration into the EHR, in order to determine commonalities, determine gaps, and to suggest future directions. This article takes a more top–down approach to system desiderata.
BACKGROUND
CDS and preimplementation evaluation frameworks
CDS interventions among heterogeneous IT infrastructures may vary in terms of the technologies on which they are built, their available features, and their configurations. There are existing frameworks for characterizing features and configurations and for assisting with assessing challenges before implementation. CDS technologies include internal (e.g., internally developed and off-the-shelf technologies), proprietary external, and open standards. Off-the-shelf technologies are those provided, as designed, by a vendor. If a clinical system can integrate with external CDS (e.g., via an application programming interface), such technologies might provide additional functionality. External CDS technologies may include open-standards software and add-ons for purchase by clinical system vendors. For example, an external CDS technology may support the “ infobutton” standard8 that facilitates context-specific links to websites.
CDS technologies are designed with different CDS features and implemented using different configurations. Features of CDS interventions might include data entry for patient information through forms and templates, or reminders, alerts, and order sets, which can provide actionable content. Less often described, although still relevant CDS features, are those for data visualization or summarization. There are more granular descriptions for CDS features, for example, taxonomies describing backend capabilities9 and the types of support that can be provided (i.e., front-end tools).10 Such taxonomies should be referenced for a more detailed overview of possible CDS features available through clinical systems and can be leveraged to evaluate local systems. Individual CDS features may or may not be available, or they may be configured differently across institutions.
CDS configurations may be characterized as passive, semiactive, and active. These terms are synonymous with definitions for knowledge resources, information retrieval tools, and classic CDS described by the Agency for Healthcare Research and Quality.11 We can describe configurations by the method in which patient-specific information is submitted and how patient-specific recommendations are generated. With passive CDS/knowledge resources, information submission and generation of recommendations are both manual processes. For semiactive CDS/ information retrieval, information submission is automated, and generation of recommendations is manual. Active CDS/ classic CDS involves automated information submission and automatically generated recommendation processes. CDS configurations and examples are summarized in Table 1.
Table 1.
Clinical decision support configurations
| CDS configurations | Description (adapted from ref. 11) | Personalized medicine examples |
|---|---|---|
| Passive CDS/knowledge resources | Manual submission of patient data and manual retrieval of patient-specific knowledge | Warfarin dose schedule (a stand-alone external CDS technology, such as that at warfarindosing. org) |
| Semiactive CDS/information retrieval tool | Automated submission of patient data and manual retrieval of patient-specific knowledge | GeneInfo sheets23 (a knowledge resource capable of adopting the “infobutton” standard)8 |
| Active CDS/classic CDS | Automated submission of patient data and automated retrieval of patient-specific knowledge | PREDICT project19 (leverages home-grown system CDS functionalities); CLIPMERGE program24 (leverages commercial system off-the-shelf functionalities) |
| Other forms of CDS | Visualization and summary data | SMART Genomics Advisor (an application that leverages the SMART architecture)48 |
CDS, clinical decision support; CLIPMERGE, Clinical Implementation of Personalized Medicine through Electronic Health Records and Genomics; PREDICT, Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment; SMART, Substitutable Medical Apps Reusable Technologies.
Frameworks, such as the ten key considerations for successful implementation proposed by Cresswell et al.,12 and the eight-dimension conceptual model proposed by Sittig and Singh,13 might be useful for assessing implementation challenges and guiding local approaches to CDS implementation. In this perspective, we illustrate the application of Sittig and Singh’s model to assess genomic CDS interventions.
The authors of this article represent participants within the eMERGE Network who are uniquely suited to provide insight into common routes for delivering genomic CDS interventions, particularly to support genetic testing and data access processes within clinical information systems. We therefore focus on CDS solutions in the context of the workflow and communication dimension of the model. This dimension includes processes to assure patient care tasks are carried out efficiently. The following section provides an overview of genetic testing and data access processes.
Phases of genetic testing and previous efforts to conceptualize the process of genetic testing
Three phases of genetic testing, including the preanalytic, analytic, and postanalytic phases, are defined according to the US Centers for Disease Control and Prevention Notice of Intent published in the Federal Register, Vol. 65, No. 87, 5/4/2000 25928. The preanalytic phase involves determining when and what genetic tests are appropriate to answer a clinical question. It also includes the collection and transportation of the appropriate biospecimen to the testing laboratory. The analytic phase involves steps to perform interrogative analyses of genetic material. The postanalytic phase involves reporting and interpreting genetic test results.
In 2008, the American Health Information Community’s Personalized Health Care Workgroup and the Office of the National Coordinator for Health Information Technology published a “Personalized Healthcare Detailed Use Case,” describing the process of performing a genetic test with focuses on the exchange of personal health history, family health history, and genetic testing information between clinical providers and health-care consumers.14,15 Distinct from that work, we emphasize approaches to deliver genomic data using CDS embedded in the EHR/PHR during genetic testing and data access processes. The following section provides insight into our rationale for establishing a common ground for discussing CDS solutions among heterogeneous IT infrastructures.
eMERGE Network
More than 20 relevant groups were recently brought together by the US National Institutes of Health National Human Genome Research Institute to share lessons from implementing genomic applications.16 One important consensus reached by the group was that collaborative projects can help maximize the generalizability and usefulness of genomic medicine approaches. There are, however, heterogeneous IT infrastructures among collaborating institutions, which can pose challenges for replicating implementation strategies.
We believe a common ground for discussing genomic data delivery, which also encompasses variability among institutions, needs to be established in order to account for the heterogeneity of IT infrastructures. Motivated by the range of approaches used to report genomic data, members of the eMERGE Network have, for example, summarized potential genomic data result-handling pathways.17 The eMERGE Network EHR integration workgroup plans to develop consensus and concepts for providing access to genomic applications, including EHR/PHR-linked decision support. Several sites in eMERGE and elsewhere have already developed genomic CDS, operating on structured genetic test results.18–22 In the following section, we describe five examples from members of the eMERGE Network and illustrate that common CDS configurations and technologies used to describe pregenomic CDS can also be used to characterize genomic CDS.
EXAMPLES OF GENOMIC CDS FROM THE eMERGE NETWORK
Five illustrative examples and their CDS configurations are summarized in Table 1. Examples from the institutions of the authors of this article include the warfarin dose schedule Web application (WarfarinDosing.org), infobutton-linked GeneInfo sheets,23 the PREDICT (Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment) project,19 the CLIPMERGE (Clinical Implementation of Personalized Medicine through Electronic Health Records and Genomics) program,24 and the SMART (Substitutable Medical Apps Reusable Technologies) Genomics Advisor (http://smartplatforms.org/smart-app-gallery/genomics-advisor/). Three of these examples use external CDS technologies, two of which utilize open standards (the “infobutton” standard8 and SMART architecture standards) for integrating into existing clinical systems. Websites such as WarfarinDosing.org provide a standalone external technology for passive CDS that allows users to input clinical and genomic variables to recommend a starting daily dose for warfarin. Such tools, however, require users to correctly transcribe and match variables and values within their EHR with those found on the website. GeneInfo sheets were created to provide actionable genetic information at the point of care. The sheets were created as a knowledge resource for a delimited set of use cases and are accessible through the Intermountain EHR system’s infobutton architecture presented in the problem list and prescription order entry system.23 This could be configured for OpenInfobutton,8 which can be leveraged to deliver semiactive CDS across more EHR systems. The SMART Genomics Advisor is built using the SMART platform, which allows the insertion of nonvendor applications into otherwise-monolithic EHR products and which can be used to facilitate other forms of CDS. The SMART Genomics Advisor-augmented SMART Diabetes Monograph App, for example, provides a mashup of data and the calculation and visualization of single-nucleotide polymorphism data relevant for clinical care of diabetes. Genomics Advisor is available as a stand-alone external CDS technology or it can be integrated with other applications. Application developers have achieved integration with patient genetic data from an instance of open- SNP (http://opensnp.org/). Currently, the SMART architecture enables “apps” to run against several EHRs, PHRs, and clinical data repositories that support the SMART application programming interface.
In addition to external CDS technologies, some institutions leverage CDS functionality provided within their local clinical systems. The PREDICT project, for example, utilizes the CDS functionality of its homegrown EHR, StarPanel, to provide active CDS. PREDICT was first launched in September 2010 and is currently designed to include both preemptive testing and “just in time,” indication-based testing.19 To date, >11,000 individuals have been tested in PREDICT using the Illumina ADME platform, which includes 34 genes and 184 variants. Genomic CDS is currently available for five “drug–genome interactions”: clopidogrel and CYP2C19 variants; simvastatin and SLCO1B1 variants; thiopurines and TPMT variants; tacrolimus and CYP3A5 variants; and warfarin and VKORC1, CYP2C9 variants. In contrast to the example provided above for passive warfarin CDS, by linking with the EHR, the PREDICT warfarin CDS can query the EHR for demographic variables, interacting medications, and genetic variants, and then calculate a recommended dose schedule.
Commercial clinical systems often also have off-the-shelf CDS functionality that can be leveraged. The CLIPMERGE project, for example, uses Epic Systems Best Practice Alert technology to deliver genome-informed dosing guidelines and guidelines for medication selection. Initially, the CLIPMERGE program focuses on delivering pharmacogenomic CDS and targets patients who are likely to have drug–genome interactions, including clopidogrel and CYP2C19 variants; warfarin and VKORC1, CYP2C9 variants; simvastatin and SLCO1B1 variants; tricyclic antidepressants and CYP2D6, CYP2C19 variants; and selective serotonin reuptake inhibitors and CYP2D6 variants.24 Other universities are also developing similar systems with commercial EHRs.7,18,25 Moreover, some clinical system vendors provide support for customized interface development that can be leveraged for other forms of CDS (e.g., Cerner MPages technology). This type of external CDS system is also the basis for the GeneInsight system21 from Brigham and Women’s Hospital, which has been integrated into local EHRs of Brigham and Women’s Hospital and the Dana-Farber Cancer Institute via single sign-on and context passing.7
In addition to characterizing genomic CDS using common CDS frameworks for configurations and technologies, there are several lessons learned from implementing pregenomic CDS that can account for variation in health IT infrastructures and that are relevant for genomic CDS. We draw from the experiences of others to overcome challenges to CDS implementation.
LESSONS FROM PREGENOMIC CDS AND CHALLENGES FOR GENOMIC CDS
Managing shared knowledge for CDS interventions
Early adopters of genomic medicine generally indicate a reliance on expert committees to survey the available evidence locally and further state that this process often leads to similar conclusions among different institutions.16 Looking forward, we can learn from previous experiences with knowledge management for CDS interventions. Efforts such as the GuideLines Into DEcision Support26 and the Clinical Decision Support Consortium27 projects were established to circumvent duplicated efforts and should be leveraged to create sharable genomic CDS content. We can also benefit from the previous finding that collaborators did not accept knowledge management solutions provided by these projects without customization. 28 Given that the acceptance of knowledge management solutions will vary by institution, the Clinical Decision Support Consortium now provides editing tools and a legal framework to make the process more accessible.29 The evolving nature of genomic knowledge also exacerbates the challenge of making knowledge management solutions accessible. For example, although commercial systems often provide off-the-shelf CDS functionality,30 they currently require manual creation of CDS rules, which creates challenges to scaling up and maintaining the EHR CDS rule base. Scalable knowledge management solutions will require automated approaches to access, update, assess the quality, and document the provenance of knowledge.
Improving the effectiveness of CDS interventions
Another challenge recognized by those with early experiences with genomic applications is a lack of institutional and clinician acceptance of the supporting evidence.16 For this reason, in the PREDICT program, two of the early drug–genome interactions implemented (clopidogrel and warfarin) were validated within the EHR-linked DNA biorepository before implementation in the clinical workflow.31,32 The reason for doing this was twofold: (i) to counter potential local criticism of the “real-world applicability” of the genetic effect, and (ii) to locally train a multiethnic regression model for warfarin.
Apart from demonstrating the value of interventions, previous work with CDS suggests that user interface characteristics, information content, and integration with workflow and clinical decision-making processes may influence acceptance by health-care providers and should be explored in early validation efforts.33–35 A report by the Agency for Healthcare Research and Quality examining fourteen factors associated with successful CDS systems34 confirms and identifies new factors that favorably affect health-care processes. These include “clinician– system interaction features,” such as provision of decision support as part of clinician workflow, and “content features,” such as provision of a recommendation, rather than an assessment.11 Although further understanding of how these features translate to acceptance of CDS is needed, we can still draw from these experiences when designing and conducting studies to validate genomic CDS. The avoidance of design issues such as information overload and “alert fatigue,” for example, will be particularly important to consider in the design of genomic CDS interventions. In addition, new CDS capabilities are needed.
With the possibility of a genetic test result and recommended action today being outdated tomorrow, CDS interventions can be leveraged to provide clinical users with educational materials that may support evidence-based practice, revised interpretations of genetic test results, and appropriate actions to take given these updates. Overby et al.36 found that an existing taxonomy for assessing CDS system functional requirements may not cover a sufficient range of CDS capabilities when considering pharmacogenomics data. The taxonomy for example did not cover semiactive CDS capabilities. Although previous experiences with semiactive CDS indicate its underuse, semiactive CDS may provide a venue for clinical provider and health-care consumers to learn about new genomics evidence on an ongoing basis. Many suggestions for improving the effectiveness of CDS interventions proposed by Sittig et al.38 should also be considered in the genomic CDS era. Suggestions for facilitating the use of complex patient data include the following: automatically summarizing patient data into a brief synopses of pertinent information; prioritizing and filtering recommendations to the user given both patient- and provider-specific data; and considering comorbidities to provide the most appropriate recommendations.
Decision support architecture and standard approaches for CDS interventions
Variation in decision support architecture also makes transferring successful interventions across institutions challenging. A review by Wright and Sittig indicates four architectures for CDS linked with clinical systems that track well chronologically. 39 The earliest architectures were stand-alone decision support systems, followed by decision support integrated into clinical systems, standards for sharing decision support content, and, most recently, service models. Even though the state of the art has evolved, all of these approaches continue to be built and used today. For all approaches, major difficulties exist in transferring successful interventions across institutions. We have learned from experiences with pregenomic CDS that standard terminologies, standard approaches to representing knowledge, and standard approaches to leverage knowledge are needed for effective scaling.40 Use of standard terminologies such as the National Library of Medicine’s Unified Medical Language System (http://www.nlm.nih.gov/research/umls) and the Logical Observation Identifiers Names and Codes (http://loinc.org), for example, provide a language for communicating health-care concepts among systems. Standard approaches to represent knowledge, such as the Health Level 7 Arden Syntax standard for representing rules,41 are needed to support translation into a computable format that can be used for CDS. Standard approaches to leveraging knowledge in a computable format to generate CDS, such as the Health Level 7 Context- Aware Knowledge Retrieval (“Infobutton”) standard,8 also improve transportability and adaptability. One of the major challenges for genomic CDS is that the common approach used by laboratory medicine professionals—interpretation of genetic tests during the analytic phase, followed by generation of text reports—will not be sufficient to link genomic data with CDS technologies. Standards cannot be leveraged until genetic observations are captured in a computable, structured format.
Standard terminologies for the exchange of CDS interventions
There are limitations to using standard terminologies for pregenomic CDS that also apply to genomic CDS. Wright and Sittig highlighted the two main limitations in using standards:39 (i) there are too many to choose from, and (ii) standards constrain what a user can encode to what was included in the scope of the standard. Similar to our previous experiences in the pregenomic era, there will also be many nonstandard approaches in implementing genomic CDS. We can learn from these experiences by leveraging CDS technologies that support existing terminologies to facilitate data exchange but that do not rely on emerging and incomplete standards. There is also a balance between structuring data and inclusion of contextual information. There exist standard terminologies such as those within the Logical Observation Identifiers Names and Codes for structuring genetics data, including family history. Even so, it is relatively rare for family history information to be captured as structured text, due in part to difficulties in entering details as structured data and clinician perceptions of structured data as being difficult to interpret.42 PHRs provide another venue for entering, reporting, retrieving, and displaying genealogy and genetic test data. Technologies such as GenePING,43 developed at Harvard–Massachusetts Institute of Technology, provide support for secure storage and sharing of these data.
It is clear that we can learn much from pregenomic CDS experiences and that we are able to leverage existing frameworks to characterize health IT infrastructures for CDS locally. Existing frameworks, however, do not appear to be sufficient to characterize implementation from the perspective of end-user workflow and communication processes considered in this commentary. In the following sections, we propose a framework for discussing opportunities for genomic CDS that support health-care provider- and consumer-initiated genetic testing and data access processes.
OPPORTUNITIES FOR GENOMIC CDS
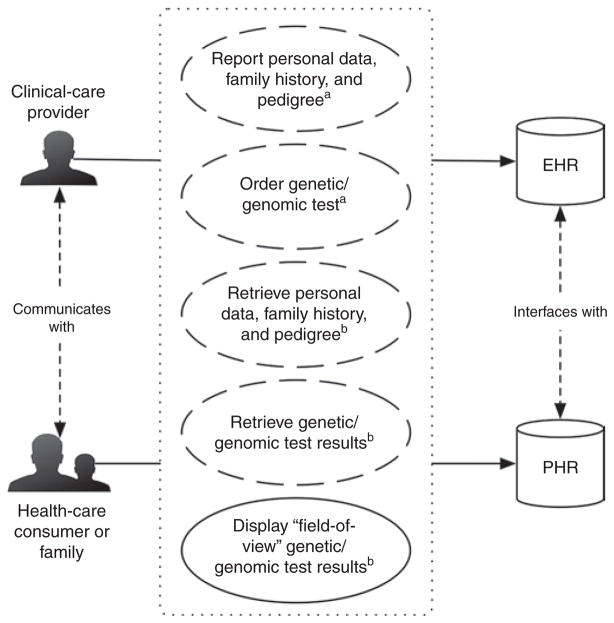
We can conceptualize genetic testing and data access processes within health IT infrastructures by considering two axes: (i) clinical system transaction initiation and (ii) stakeholder-driven clinical system interaction points. These axes are considered in Figure 1, which illustrates opportunities for CDS interventions during genetic testing and data access processes.
Figure 1. Clinical system transactions for clinical decision support (CDS) interventions during genetic testing and data access processes.
Dashed and solid ovals indicate human-initiated and computer-initiated clinical system transactions, respectively. Superscript letters indicate the phase of genetic testing in which clinical system transactions occur (i.e., apreanalytic phase and bpostanalytic phase). Clinical system interactions are illustrated by solid arrows pointing from stakeholders (i.e., clinical-care provider and health-care consumer or family) to clinical systems (i.e., the electronic health record and personal health record systems). EHR, electronic health record; PHR, personal health record.
Human- and computer-initiated clinical system transactions
Genetic testing during the preanalytic phase is most often human initiated (e.g., a clinical-care provider orders a genetic test). Test results from the analytic phase, including data interpretations, are then returned to the requestor during the postanalytic phase. The diseases and traits for which there are commercially available genetic tests have more than tripled in the past ten years,44 and there are several institutions that currently or will soon have whole-genome or whole-exome sequencing data available from Clinical Laboratory Improvement Amendments–certified laboratories.
When either large-scale genotyping (e.g., from genome-wide association studies or focused drug metabolism genotyping/ sequencing platforms) or whole-genome/whole-exome sequencing are made available within EHRs, human-initiated steps during the preanalytic phase can be replaced by computer-initiated data retrieval. Subsequently, the analytic phase can be simplified to involve primarily data reanalysis and reinterpretation. During the postanalytic phase, filtered or “field-of-view” results can then be displayed. To illustrate this concept, SMART Genomics Advisor application developers demonstrate “field-of-view” data visualization by displaying only the subset of known genetic factors associated with diabetes that are filtered from the full set of single-nucleotide polymorphisms captured in openSNP using their SMART Genomics Advisor-augmented SMART Diabetes Monograph app. Although we foresee a move to support this new model, we do not see traditional approaches for ordering genetic testing disappearing anytime soon. Stakeholders involved in human- and computer-initiated data retrieval include clinical-care providers and health-care consumers. These stakeholders have different points of interaction with clinical systems.
Stakeholder-driven clinical system interaction points and information needs
Clinical-care providers may interact with data through an EHR, whereas health-care consumers may do the same through a PHR. The goal of a PHR is to make health information accessible to patients; however, PHRs vary quite a bit in their design, functionality, and implementation.45 For example, electronic PHRs may be “tethered” to facilitate data exchange with systems such as EHRs. Alternatively, PHRs may be untethered and installed on isolated personal computers. PHRs are relevant for this discussion given the fact that health-care consumers can initiate genetic testing through direct-to-consumer genetic testing services, and it is likely that use of direct-to-consumer testing will influence clinical-care provider decisions.46,47 Clinical-care providers and health-care consumers both need information to assess genetic testing options during the preanalytic phase of genetic testing. Needs specific to clinical-care providers include knowledge for selecting a genetic test, for assessing risk, and for interpreting risk for a genetic condition using existing patient data. Needs of health-care consumers might be educational in nature and may also include understanding what the results will tell them and whether they can be acted upon to improve health. In addition, health-care consumers may have practical information needs such as understanding how test results might affect their insurance and whether a direct-to-consumer laboratory is Clinical Laboratory Improvement Amendments certified.
During the postanalytic phase of genetic testing, clinical-care providers and health-care consumers need support for performing interpretations and care-planning activities. Specific needs for both include knowledge of the time when the results become available, interpretations of results in the context of a clinical action or health-care consumer goal, and knowledge of when updates to interpretations are made and applied to past data.
CONCLUSION
Although there is heterogeneity among institutions integrating genomic CDS, there are common stakeholders, common genetic testing and data access processes, and common clinical system transactions for linking CDS. The eMERGE Network EHR integration workgroup plans to develop consensus and concepts for providing access to genomic applications, including EHR/PHR-linked decision support. We provide examples of genomics CDS and illustrate that they can be characterized using common frameworks for CDS configurations and technologies. We also describe several lessons learned from implementing pregenomic CDS that can account for variation in health IT infrastructures and that are relevant for genomic CDS. Existing frameworks, however, do not appear to be sufficient to characterize implementation from the perspective of end-user workflow and communication processes.
Acknowledgments
C.L.O. and G.H. were supported by the US National Institutes of Health (NIH) National Library of Medicine (NLM) grant 5T15LM007079. I.K. was supported by NIH National Human Genome Research Institute (NHGRI) grant 3 U01 HG006828 and NIH NLM grant U54 LM008748. M.S.W. was supported by NIH NHGRI grant 3 U01 HG006382. J.S. was supported by NIH NHGRI grant 5 U01 HG006388. E.B., O.G., and C.W. were supported by NIH NHGRI grant U01 HG006380. C.W. was supported by NIH NLM grant R01 LM009886. J.C.D. was supported by NIH NHGRI grant U01 HG006378. P.T.-H. was supported by NIH National Center for Research Resources (NCRR) grant 1 UL1 RR025014, the Washington State Life Sciences Discovery Fund grant “Northwest Institute for Genetic Medicine,” and NIH NHGRI grant U01 HG006507. G.H. was supported by NIH NLM grant R01 LM006910. The authors declare no competing financial interests.
References
- 1.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91:450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 2.Brinner KA, Downing GJ American Health Information Community Personalized Health Care Workgroup. Advancing patient-centered pediatric care through health information exchange: update from the American Health Information Community Personalized Health Care Workgroup. Pediatrics. 2009;123(suppl 2):S122–S124. doi: 10.1542/peds.2008-1755N. [DOI] [PubMed] [Google Scholar]
- 3.Kawamoto K, Lobach DF, Willard HF, Ginsburg GS. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med Inform Decis Mak. 2009;9:17. doi: 10.1186/1472-6947-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman MA, Williams MS. Electronic medical records and personalized medicine. Hum Genet. 2011;130:33–39. doi: 10.1007/s00439-011-0992-y. [DOI] [PubMed] [Google Scholar]
- 5.Teich JM, Osheroff JA, Pifer EA, Sittig DF, Jenders RA CDS Expert Review Panel. Clinical decision support in electronic prescribing: recommendations and an action plan: report of the joint clinical decision support workgroup. J Am Med Inform Assoc. 2005;12:365–376. doi: 10.1197/jamia.M1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarczy-Hornoch P, Amendola L, Aronson SJ, et al. A survey of informatics approaches to whole-exome and whole-genome clinical reporting in the electronic medical record. Genet Med. 2013;15:824–832. doi: 10.1038/gim.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Context-aware knowledge retrieval (infobutton) product brief. [Accessed 16 May 2011.];HL7 International Wiki Site. http://wiki.hl7.org/index.php?title=Infobutton_project.
- 9.Wright A, Goldberg H, Hongsermeier T, Middleton B. A description and functional taxonomy of rule-based decision support content at a large integrated delivery network. J Am Med Inform Assoc. 2007;14:489–496. doi: 10.1197/jamia.M2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright A, Sittig DF, Ash JS, et al. Development and evaluation of a comprehensive clinical decision support taxonomy: comparison of front-end tools in commercial and internally developed electronic health record systems. J Am Med Inform Assoc. 2011;18:232–242. doi: 10.1136/amiajnl-2011-000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobach DF, Sanders GD, Bright TJ, et al. Enabling Health Care Decisionmaking Through Clinical Decision Support and Knowledge Management. Agency for Healthcare Research and Quality; Rockville, MD: 2012. [PMC free article] [PubMed] [Google Scholar]
- 12.Cresswell KM, Bates DW, Sheikh A. Ten key considerations for the successful implementation and adoption of large-scale health information technology. J Am Med Inform Assoc. 2013;20(e1):e9–e13. doi: 10.1136/amiajnl-2013-001684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sittig DF, Singh H. A new sociotechnical model for studying health information technology in complex adaptive healthcare systems. Qual Saf Health Care. 2010;19(suppl 3):i68–i74. doi: 10.1136/qshc.2010.042085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.United States Department of Health and Human Services, Office of the National Coordinator for Health Information Technology. Personalized Healthcare Detailed Use Case. Rockville, MD: 2008. [Google Scholar]
- 15.Glaser J, Henley DE, Downing G, Brinner KM Personalized Health Care Workgroup of the American Health Information Community. Advancing personalized health care through health information technology: an update from the American Health Information Community’s Personalized Health Care Workgroup. J Am Med Inform Assoc. 2008;15:391–396. doi: 10.1197/jamia.M2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15:258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starren J, Williams MS, Bottinger EP. Crossing the Omic Chasm: A Time for Omic Ancillary Systems. JAMA. 2013;14:1–2. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther. 2012;92:563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stack CB, Gharani N, Gordon ES, Schmidlen T, Christman MF, Keller MA. Genetic risk estimation in the Coriell Personalized Medicine Collaborative. Genet Med. 2011;13:131–139. doi: 10.1097/GIM.0b013e318201164c. [DOI] [PubMed] [Google Scholar]
- 21.Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011;32:532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J. 2011;17:528–536. doi: 10.1097/PPO.0b013e318238216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Fiol G, Williams MS, Maram N, Rocha RA, Wood GM, Mitchell JA. Integrating genetic information resources with an EHR. AMIA Annu Symp Proc. 2006:904. [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx Program: Clinical Implementation of Personalized Medicine Through Electronic Health Records and Genomics-Pharmacogenomics. Clin Pharmacol Ther. 2013;94:214–217. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson DR, Conlon M, Baralt C, Johnson JA, Clare-Salzler MJ, Rawley-Payne M. University of Florida Clinical and Translational Science Institute: transformation and translation in personalized medicine. Clin Transl Sci. 2011;4:400–402. doi: 10.1111/j.1752-8062.2011.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomotan EA, Hoeksema LJ, Edmonds DE, Ramírez-Garnica G, Shiffman RN, Horwitz LI. Evaluating the use of a computerized clinical decision support system for asthma by pediatric pulmonologists. Int J Med Inform. 2012;81:157–165. doi: 10.1016/j.ijmedinf.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon BE, Simonaitis L, Goldberg HS, et al. A pilot study of distributed knowledge management and clinical decision support in the cloud. Artif Intell Med. 2013 Mar 30; doi: 10.1016/j.artmed.2013.03.004. in press e-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28.Eichner J, Das M. Challenges and Barriers to Clinical Decision Support (CDS) Design and Implementation Experienced in the Agency for Healthcare Research and Quality CDS Demonstrations. Agency for Healthcare Research and Quality; Rockville, MD: 2010. Technology ANRCfHI. [Google Scholar]
- 29.Hongsermeier T, Maviglia S, Tsurikova L, et al. A legal framework to enable sharing of Clinical Decision Support knowledge and services across institutional boundaries. AMIA Annu Symp Proc. 2011;2011:925–933. [PMC free article] [PubMed] [Google Scholar]
- 30.Sittig DF, Wright A, Meltzer S, et al. Comparison of clinical knowledge management capabilities of commercially-available and leading internallydeveloped electronic health records. BMC Med Inform Decis Mak. 2011;11:13. doi: 10.1186/1472-6947-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaney JT, Ramirez AH, Bowton E, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther. 2012;91:257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez AH, Shi Y, Schildcrout JS, et al. Predicting warfarin dosage in European- Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13:407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krall MA, Sittig DF. Clinician’s assessments of outpatient electronic medical record alert and reminder usability and usefulness requirements. Proc AMIA Symp. 2002:400–404. [PMC free article] [PubMed] [Google Scholar]
- 34.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005;330:765. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kannry JL, Kushniruk AW, Denny JC, Hoffman M, Levy H, Williams MS. Integrating Genomic Test Results and Decision Support at the Point of Care: Genomics and the EHR. In: Babyatsky M, Giovanni M, Murray M, editors. Clinical Genomics: Practical Applications in Adult Patient Care. McGraw Hill; New York: 2013. [Google Scholar]
- 36.Overby CL, Tarczy-Hornoch P, Kalet IJ, et al. Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice. J Pers Med. 2012;2:241–256. doi: 10.3390/jpm2040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates DW, Kuperman GJ, Wang S, et al. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003;10:523–530. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sittig DF, Wright A, Osheroff JA, et al. Grand challenges in clinical decision support. J Biomed Inform. 2008;41:387–392. doi: 10.1016/j.jbi.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright A, Sittig DF. A four-phase model of the evolution of clinical decision support architectures. Int J Med Inform. 2008;77:641–649. doi: 10.1016/j.ijmedinf.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawamoto K, Del Fiol G, Lobach DF, Jenders RA. Standards for scalable clinical decision support: need, current and emerging standards, gaps, and proposal for progress. Open Med Inform J. 2010;4:235–244. doi: 10.2174/1874431101004010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pryor TA, Hripcsak G. Sharing MLM’s: an experiment between Columbia- Presbyterian and LDS Hospital. Proc Annu Symp Comput Appl Med Care. 1993:399–403. [PMC free article] [PubMed] [Google Scholar]
- 42.Patel VL, Arocha JF, Kushniruk AW. Patients’ and physicians’ understanding of health and biomedical concepts: relationship to the design of EMR systems. J Biomed Inform. 2002;35:8–16. doi: 10.1016/s1532-0464(02)00002-3. [DOI] [PubMed] [Google Scholar]
- 43.Adida B, Kohane IS. GenePING: secure, scalable management of personal genomic data. BMC Genomics. 2006;7:93. doi: 10.1186/1471-2164-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. [Accessed 15 March 2013.];GeneTests/GeneReviews. http://www.genetests.org.
- 45.Archer N, Fevrier-Thomas U, Lokker C, McKibbon KA, Straus SE. Personal health records: a scoping review. J Am Med Inform Assoc. 2011;18:515–522. doi: 10.1136/amiajnl-2011-000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaufman DJ, Bollinger JM, Dvoskin RL, Scott JA. Risky business: risk perception and the use of medical services among customers of DTC personal genetic testing. J Genet Couns. 2012;21:413–422. doi: 10.1007/s10897-012-9483-0. [DOI] [PubMed] [Google Scholar]
- 47.Gollust SE, Gordon ES, Zayac C, et al. Motivations and perceptions of early adopters of personalized genomics: perspectives from research participants. Public Health Genomics. 2012;15:22–30. doi: 10.1159/000327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandl KD, Mandel JC, Murphy SN, et al. The SMART Platform: early experience enabling substitutable applications for electronic health records. J Am Med Inform Assoc. 2012;19:597–603. doi: 10.1136/amiajnl-2011-000622. [DOI] [PMC free article] [PubMed] [Google Scholar]