Abstract
A functionally important proline residue is highly conserved in the cytosolic TIR signalling domains of human Toll-like receptors (TLRs). The anti-viral Toll, TLR3, is unusual as it has alanine instead of proline at this position and is the only human TLR that associates directly with the adaptor molecule TRIF rather than MyD88. Here we report that a mutant TLR3 that substitutes the BB-loop alanine for proline (A795P) enhances NF-κB activation but is incapable of mediating TRIF dependent IRF3 responses. Wild-type and A795P TLR3 associate constitutively with both TRIF and MyD88 and activation induces additional binding of TRIF to the wild-type and of MyD88 to the A795P mutant receptors respectively. In addition activation of A795P but not wild-type TLR3 leads to the recruitment TRAF6, a downstream signal transducer of the MyD88 dependent pathway. These results show that adaptor specificity can be conferred by minimal determinants of the TIR domain.
Keywords: Innate-immunity, Toll-like receptors, signalling adaptor proteins
Introduction
Toll-Like Receptors (TLRs) play an important role in innate immunity by recognising specific molecular patterns expressed by microbes. TLR signalling requires dimerization of the receptor extracellular domains, mediated by microbial products, leading to the dimerisation of the receptor cytoplasmic Toll/interleukin-1 receptor (TIR) domains (1, 2). The activated conformation of the receptor provides a scaffold for the recruitment of the downstream signalling molecules that activate transcription factors including NF-κB and interferon response factors (IRFs), leading to the production of pro-inflammatory cytokines and type 1 interferons.
TLRs use four signalling adaptor proteins (3). Of these TRIF and MyD88 adaptor molecules are required for the activation of IRF3/7 and NF-κB respectively. TRIF is the largest member of the adaptor family and is required for TLR3-induced signalling and, together with TRAM, the MyD88-independent signalling pathway of TLR4 whereas MyD88 couples to all the human TLRs except TLR3. Acting as a platform, TRIF accumulates other signalling molecules to the ‘TRIF signalosome’ and activates a range of cellular responses mediated by other modules in TRIF as well as the TIR (4). TRAF3 binds to the N-terminus of TRIF leading to phosphorylation of IRF-3 and 7 by the TANK binding kinase-1 (TBK-1). On the other hand Receptor-interacting protein-1 (RIP-1) can bind to the C-terminus of TRIF causing both FADD (Fas associated death domain) dependent apoptosis and activation of NF-κB by the IκB kinase complex. The role of TRAF6 in TRIF signalling is controversial. While some studies found that TRAF6 functions exclusively in MyD88 directed signalling others suggest a secondary role in TRIF mediated activation of NF-κB (5-7).
The TIR domains of the TLRs and their cognate adaptors are approximately 200 amino acids and fold into a characteristic α–β structure (8). The loops connecting these secondary structure elements mediate receptor-adaptor interactions. Of particular interest is the BB-loop that links the second β-sheet to the second α-helix of the TIR. A naturally occurring mutation of the conserved proline residue in the BB loop of TLR4 was identified as the cause of LPS hypo-responsiveness in C3H/HeJ mice. This missense mutation substituted proline with a histidine residue (P712H) and displayed dominant negative properties (9, 10). The importance of the BB-loop in TLR and adaptor function is further emphasised by the inhibitory properties of cell permeable peptide mimics (11, 12).
The BB-loop proline is conserved in all human TLRs except for TLR3 which has alanine at this position and previous studies suggest that this residue plays an important role in recruiting TRIF into the signalling complex (13). To test this idea further we have studied the signalling characteristics of a mutant TLR3 that replaces the BB-loop alanine for proline. We show that this single amino acid change switches TLR3 from a TRIF to a MyD88 directed signalling pathway.
Materials and methods
Plasmids and reagents
Full length pCMV-Myc-Mal, pCMV-MyD88-Myc and pcDNA3.1-TLR7 were a kind gift from Dr Alexander Weber (University of Tübingen, Germany); pcDNA3.1-Myc-TRIF was a kind gift from Dr Rongtuan Lin (McGill University, Canada); pcDNA3.1-Flag-TLR2,-Flag-TLR3, -Flag-Mal and -HA-Mal were a kind gift from Dr Ashley Mansell, (Monash Institute of Medical Research, Australia); pcDNA3.1-TBK1-Myc was a kind gift from Dr Ivan Dikic (Institute of Biochemistry, Frankfurt Medical School, Germany); pcDNA3-Myc-TRAF6 was a kind gift from Dr William Dalton, Moffit Cancer Centre, USA); pcDNA.I-RIP1-Myc was a kind gift from Dr Michelle Kelliher (University of Massacheusetts Medical School, USA) and Gal4-IRF-3,-5, -7 and Gal4-luciferase constructs were a kind gift from Dr Kate Fitzgerald (University of Massacheusetts Medical School, USA). Vector pEYFPN1-TLR5-YFP was obtained from Clontech and reporter gene constructs NF-κB-, IFN-β-luciferase and pRL-TK were a kind gift from Dr Clare Bryant, University of Cambridge, UK. Myc-TRAM was cloned into the pCMV-Myc vector at sites HindIII and KpnI and Flag-TRAF6 into the pEFBOS vector at sites XhoI and HindIII. All BB-loop mutants were generated using the QuikChange II site-directed mutagenesis kit with pfu-Turbo (Stratagene) using the above mentioned plasmids.
Cell culture and reagents
Human embryonic kidney (HEK) 293, HEK293T and HEK293 -derived MyD88-deficient (I3A) (14) cells were cultured in DMEM medium supplemented with 10% FCS (Invitrogen) and 100 U/ml Penicillin/Streptomycin and maintained in a 37°C humidified atmosphere. Pam3Cys4K was obtained from EMC microcollections and CL097 Imidazoquinoline compound, FLA-ST Ultrapure Flagellin from S. typhimurium and Poly (I:C) HMW synthetic analog of dsRNA; were all sourced from InvivoGen.
Luciferase reporter assay
HEK293 or HEK293 MyD88-deficient (I3A) cells were seeded at 1 × 105cells/well in a 96-well plate 36 h prior to transfection with jetPEI (Polyplus). NF-κB-, IFN-β-, or UAS-Gal4- dependent gene expression was determined using luciferase reporter constructs concomitantly with indicated vectors. Gal4-fusions IRF3, IRF5 and IRF7, were used to analyse activation of IRF activity. The Renilla luciferase-thymidine kinase encoding plasmid (pRL-TK) was used to normalize for transfection efficiency, and pcDNA3.1 empty vector was used to maintain constant DNA. Transfected cells were lysed using Passive lysis buffer (Promega) and assayed for luciferase and Renilla activity using luciferase assay reagent (Promega). Luminescence readings were corrected for Renilla activity and expressed as fold increase over non-stimulated control values. Data is presented as mean +/− SEM of one of three independent experiments. Statistical analysis was performed using two-way ANOVA where significance is relative to wild-type TLR plasmid transfection: *P<0.05, **P<0.01, ***P<0.001.
Immunoprecipitation and immunoblot analysis
HEK293T cells were seeded at 1.5 × 105 cells/well in a 6-well plate and were transfected using JetPEI with the indicated plasmids where the total amount of DNA (3 μg per well) was kept constant. Twenty-four hours post transfection, cells were lysed in KalB buffer as described (21). EZview™ Red Anti-Flag M2 affinity beads (25 μl, 50% slurry, Sigma) were incubated with the cell lysates for 2 h at 4°C. The immune complexes were precipitated, washed and eluted by addition of sample buffer for resolution by SDS-PAGE and immunoblotting with either anti-Myc (Abcam), anti-HA (Abcam) or anti-Flag antibody (Sigma). Immunocomplexes were visualized by using SuperSignal West Pico chemiluminescent substrate solution (Pierce) followed by exposure to X-ray film (Hyperfilm ECL, Amersham Pharmacia) to detect Chemiluminescence.
Densitometry analysis for IP western band intensities was calculated using the Totallab software (Nonlinear Dynamics), verifying for non-saturation and subtracting background. Values were expressed as a percentage of the total area of each band normalized to the strongest band.
Small interfering RNA (siRNA) knockdown of TRIF, RNA extraction, qPCR
The 25-bp duplex of targeting stealth siRNA or non-targeting (NT) siRNA (Invitrogen) was transfected into sub-confluent HEK293/293T cells, using siRNA jetPRIME® (Polyplus). Double-stranded siRNAs containing equal parts of the following antisense sequences was used to knock down TRIF: siRNA2, 5′-CCCAUUGACGGUGUUUCGGACUGGA-3′; siRNA3, 5′-CCAUCACUUCCUAGCGCCUUCGACA-3′. The NT siRNA were low GC and medium GC stealth RNAi negative control duplexes (Invitrogen). Forty eight hours after transfection the cells were either transfected with the relevant constructs or analysed by real-time quantitative PCR (qPCR) or western blot using anti-TRIF (Cell Signaling) and β-actin (Ambion).
Total RNA from cells were purified using RNeasy Plus kit and QiaShredder (QIAgen). First strand cDNA was synthesised from the total RNA using SuperScript II reverse transcriptase (Invitrogen) with random hexamer (Promega) as described in the protocol from Untegasser lab (http://www.untergasser.de/lab/protocols/cdna_synthesis_superscript_ii_v1_0.htm).
The cDNA served as template for real-time qPCR analysis to determine the level of gene knockdown using Maxima SYBR Green/ROX qPCR SuperMix (Thermo Scientific). Relative gene levels of TRIF was determined and normalised against internal control of GAPDH to determine the relative gene knockdown using pfaffl method. The gene-specific primers for TRIF: 5′-TCCTCCCTGTTCCCTTCC-3′, 5′-CCTGGAAATCCTCGCAGA-3′; and GAPDH: 5′-GAAGGTGAAGGTCGGAGTC-3′; 5′-GAAGATGGTGATGGGATTTC were designed to span the exon-exon junction to eliminate amplification of genomic DNA.
Results
The BB-loop alanine residue of TLR3 confers specificity for TRIF directed activation of IRF-3 and IFN-β
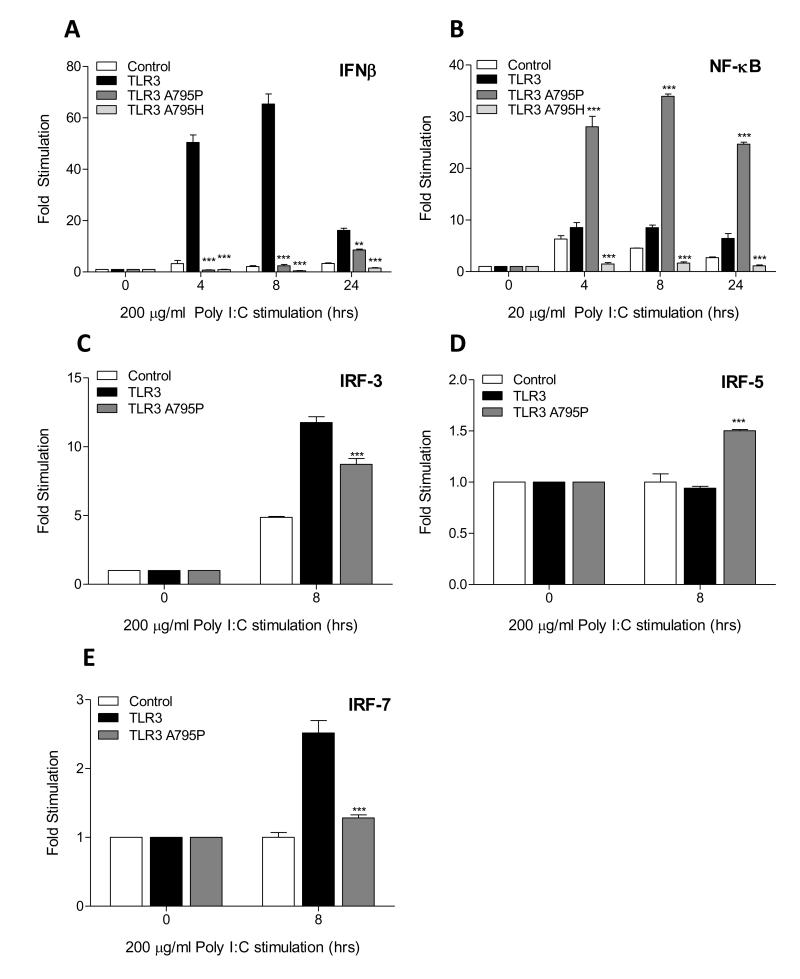
The ten human TLRs all have a proline-glycine dipeptide in the BB-loop except for TLR3 which has alanine-glycine (Supplemental Figure S1). In view of this we asked whether the BB-loop alanine and proline residues played a role in receptor selectivity for TRIF and MyD88 respectively. To this end, we mutated A795 of TLR3 to a proline and also to histidine, a mutation previously shown to disrupt TRIF recruitment and IRF3 directed signalling (13). HEK293 cells were transfected with either: wild-type TLR3, TLR3 A795P or TLR3 A795H, and downstream activation of the IFN-β and NF-κB reporter genes was assayed. Following poly (I:C) stimulation, wild-type TLR3 strongly activated the IFN-β promoter whereas mutants A795P and A795H were not responsive (Fig. 1A). As expected wild-type TLR3 supports a small but sustained induction of NF-κB but surprisingly the A795P, potentiated this response by almost 20 fold (Fig. 1B). A795H was unable to activate the NF-κB-driven reporter and exerted a dominant negative effect on the endogenous response to poly (I:C).
Figure 1. BB-loop alanine-proline mutant of TLR3 is defective for TRIF signaling but enhances NF-κB activation.
(A-B) HEK293 cells were transfected with empty vector or expression plasmid for TLR3, TLR3 A795P or TLR3 A795H mutants (10 ng) with the IFN-β (25ng) or NF-κB (10ng) promoter reporter. After 24 h, cells were stimulated with poly (I:C) (200 μg/ml) (A) or (20 μg/ml) (B) for 4, 8 and 24 h. (C-E) TLR3 or TLR3 A795P mutant (5 ng) was transfected with either 15 ng of Gal4-IRF3, Gal4-IRF5, or Gal4-IRF7 along with UAS-Gal4-luc reporter (30 ng). Following stimulation cell lysates were harvested for assessment of luciferase reporter gene activity. All results were comparative to a constitutively expressed ‘reference’ reporter gene, TK-Renilla and fold induction relative to unstimulated cells. Data is presented as mean +/− SEM of one of three independent experiments. Statistical analysis was performed using two-way ANOVA where significance is relative to WT TLR3 plasmid transfection: *P<0.05, **P<0.01, ***P<0.001.
We next sought to investigate the effect of the TLR3 A795P mutant on downstream IRF activity. As TLR3 signalling activates IRF-3 and IRF-7, but not IRF-5 (15), we measured activation of these transcription factors by wild-type and A795P mutant. For these experiments we used an in vivo reporter assay that employs hybrid proteins consisting of the yeast Gal4 DNA binding domain (16). UAS(Gal)-driven luciferase reporter gene was used to measure activation of the IRF fusion proteins. As expected wild-type TLR3 activated IRF-3 and IRF-7 but not IRF-5. In contrast, the TLR3 A795P mutant inhibited poly (I:C)-induced activation of IRF-3 and IRF-7 but activated IRF-5 when compared to wild-type TLR3 (Fig. 1C-E).
Constitutive and inducible binding of MyD88 and TRIF to wild-type and TLR3 A795P mutant
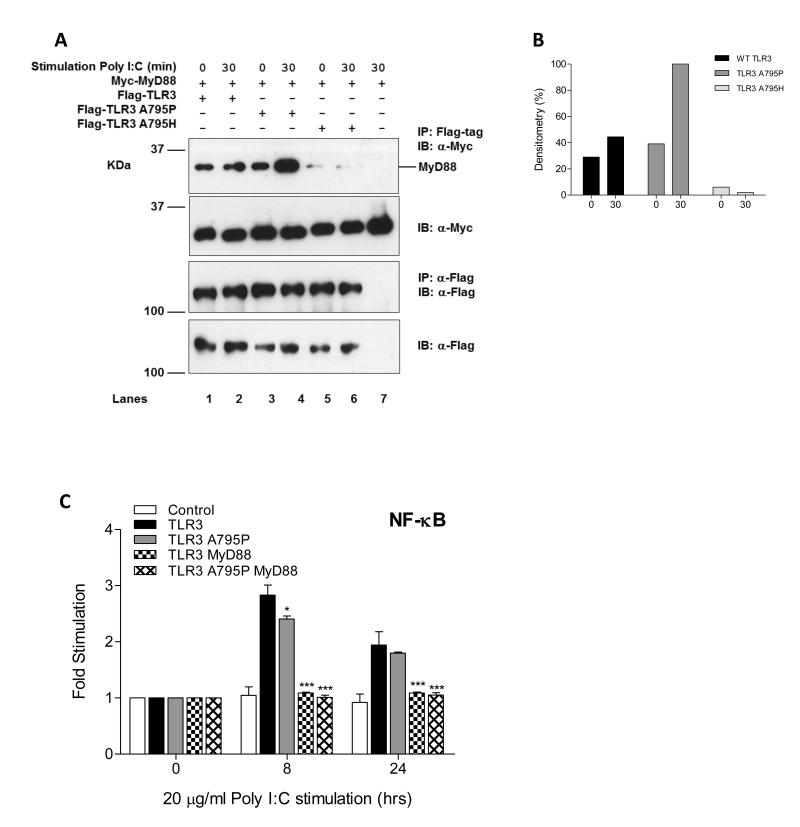
We next asked whether a switch in adaptor specificity from TRIF to MyD88 accounts for the observed enhancement of NF-κB activation by TLR3 A795P. Both wild-type and A795P but not A795H TLR3 associate with MyD88 in the absence of stimulation. However, treatment with poly (I:C) induces a 2 – fold increase in MyD88 binding to A795P but not to wild-type TLR3 (Fig. 2A-B). To investigate the adaptor specificity of the A795P receptor further we used a derivative of the HEK293 cell line (I3A) that does not express MyD88. As shown in Fig. 2C A795P mutant induces a 2.5 fold increase in NF-κB activity but the 20-30 fold enhancement seen in Fig. 1B is not observed. Interestingly reconstitution of MyD88 into the HEK293 I3A cells caused a suppression of this low level response by both the wild-type and A795P TLR3. This suggests that overexpression of MyD88 exerts a dominant negative effect on TRIF mediated activation of NF-κB, an effect that was also demonstrated in another study in which MyD88 negatively regulated TLR3/TRIF-induced corneal inflammation (17).
Figure 2. Constitutive and inducible binding of MyD88 to the wild-type and A795P TLR3.
(A) HEK293T cells were transfected with 3 μg of total DNA containing either: 0.5 μg of full-length Myc-tagged MyD88 with either 2.5 μg of Flag-TLR3 (lanes 1-2), Flag-TLR3 A795P (lanes 3-4) or Flag-TLR3 A795H (lanes 5-6) in the presence or absence of poly (I:C) (100 μg/ml) for 30 minutes. Lysates were immunoprecipitated with α-M2 Flag agarose beads (top panel) and detected by Western blot. Results are representative of three independent experiments. (B) Densitometry analysis for IP western band intensities. (C) HEK293 MyD88-deficient (I3A) cells were co-transfected with empty vector or expression plasmid for wild-type TLR3 or TLR3 A795P mutant (10 ng) and wild-type MyD88 (5 ng) with the NF-κB promoter reporter (10 ng). After 24 h, cells were stimulated with poly (I:C) (20 μg/ml) for 8 and 24 h followed by harvesting of the cell lysates and assessment of luciferase reporter gene activity.
To further explore the mechanism by which the A795P TLR3 mutant up-regulates NF-κB activation, we asked whether the adaptor molecule TRIF can still associate with this mutant receptor. These experiments show that TRIF can bind constitutively to both wild-type and A795P TLR3. However, after stimulation TRIF binding to the wild-type TLR3 is enhanced 2.5 fold compared to the mutant A795P with only a 1.5 fold increase. This is the reciprocal pattern to that observed with MyD88 (Fig. 3A-B, compared to Fig 2A, B). To determine whether the enhanced signalling to NF-κB by A795P TLR3 is TRIF-dependent, activation of NF-κB and IFN-β was examined in the presence and absence of targeted functional gene silencing of TRIF. In HEK293 cells, endogenous TRIF protein was constitutively expressed in the presence of a NT siRNA control, whereas TRIF protein levels were markedly reduced after treatment with two TRIF-targeted siRNAs (Fig. 3C). Expression of TRIF mRNA as measured by qPCR also demonstrated a significant reduction when compared to NT siRNA control levels (Fig. 3D). As shown in Fig 3E and F, silencing of TRIF abolished signalling to both IFN-β and NF-κB by wild-type TLR3 in response to treatment with ligand. By contrast A795P TLR3 retained the ability to activate NF-κB in the absence of TRIF protein, a finding which supports the conclusion that the mutant receptor signals to NF-κB by a MyD88 dependent pathway.
Figure 3. TLR3 A795P signaling response is not TRIF-dependent.
(A) HEK293T cells were transfected with 3 μg of total DNA containing either: 1 μg of full-length Myc-tagged TRIF with either 2.0 μg of Flag-TLR3 (lanes 1-2), Flag-TLR3 A795P (lanes 3-4) or Flag-TLR3 A795H (lanes 5-6) in the presence or absence of poly (I:C) (100 μg/ml) for 30 minutes. Results are representative of three independent experiments. (B) Densitometry analysis for IP western band intensities. (C-F) Two different TRIF-targeted siRNAs or non-targeting (NT) siRNA at 10 nM, were transfected into HEK293 cells and left for 48 h. (C) Following siRNA transfection, cell lysates were probed with anti-TRIF antibody for endogenous TRIF detection. (D) Total RNA was extracted and relative expression levels of TRIF were determined by qPCR. (E-F) HEK293 cells were transfected with empty vector or expression plasmid for TLR3, TLR3 A795P or TLR3 A795H mutants (10 ng) with the NF-κB (10ng) or IFN-β (25ng) promoter reporter. After 24 h, cells were stimulated with poly (I:C) (20 μg/ml) (E) or (200 μg/ml) (F) for 8 h followed by harvesting of the cell lysates and assessment of luciferase reporter gene activity.
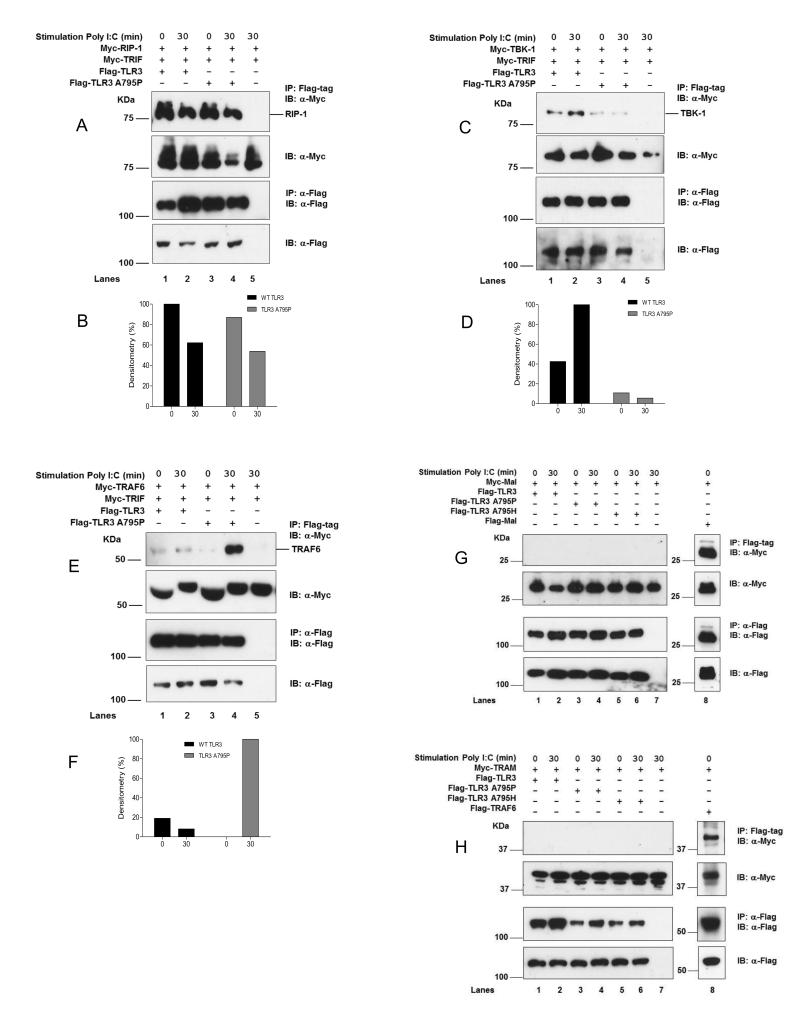
We also examined the association of other signal transducers that are involved in the TRIF and MyD88 directed pathways. We found that the death-domain kinase RIP-1 that is required for TRIF dependent activation of NF-κB binds constitutively to wild-type and A795P TLR3 (Fig 4A-B). On the other hand tank-binding kinase 1 (TBK-1), required for TRIF dependent IRF3 activation, is present at much lower levels in complexes with the A795P mutant (Fig 4C-D). Interestingly, TRAF6, an E3-ubiquitin ligase recruited to the myddosome complex in MyD88 directed signalling (5), formed a complex with the A795P receptor upon poly (I:C) stimulation whereas wild-type TLR3 did not (Fig 4EF). The bridging adaptors Mal and TRAM were unable to bind either wild-type TLR3 or the A795P/H mutants (Fig 4G-H). Taken together these results indicate that the mutant A795P shifts the adaptor specificity of TLR3 towards MyD88.
Figure 4. TLR3 A795P suppresses TBK-1 but enhances TRAF6 recruitment.
HEK293T were transfected with 3 μg of total DNA containing either: (A) 0.5 μg of full-length Myc-tagged RIP-1 and Myc-TRIF, (C) 0.5 μg of Myc-tagged TBK-1 and Myc-TRIF, (E) 0.5 μg of Myc-tagged TRAF6 and Myc-TRIF, (G) 0.2 μg of Myc-tagged Mal, (H) 1 μg of Myc-tagged TRAM; with either 2.0 μg of Flag-TLR3 (lanes 1-2), Flag-TLR3 A795P (lanes 3-4) or Flag-TLR3 A795H (lanes 5-6, for Mal and TRAM (G-H) only) in the presence or absence of poly (I:C) (100 μg/ml) for 30 minutes. Results are representative of three independent experiments. (B, D, and F): quantitation by densitometry.
BB-loop mutations within the TIR domain of other TLRs alters signalling
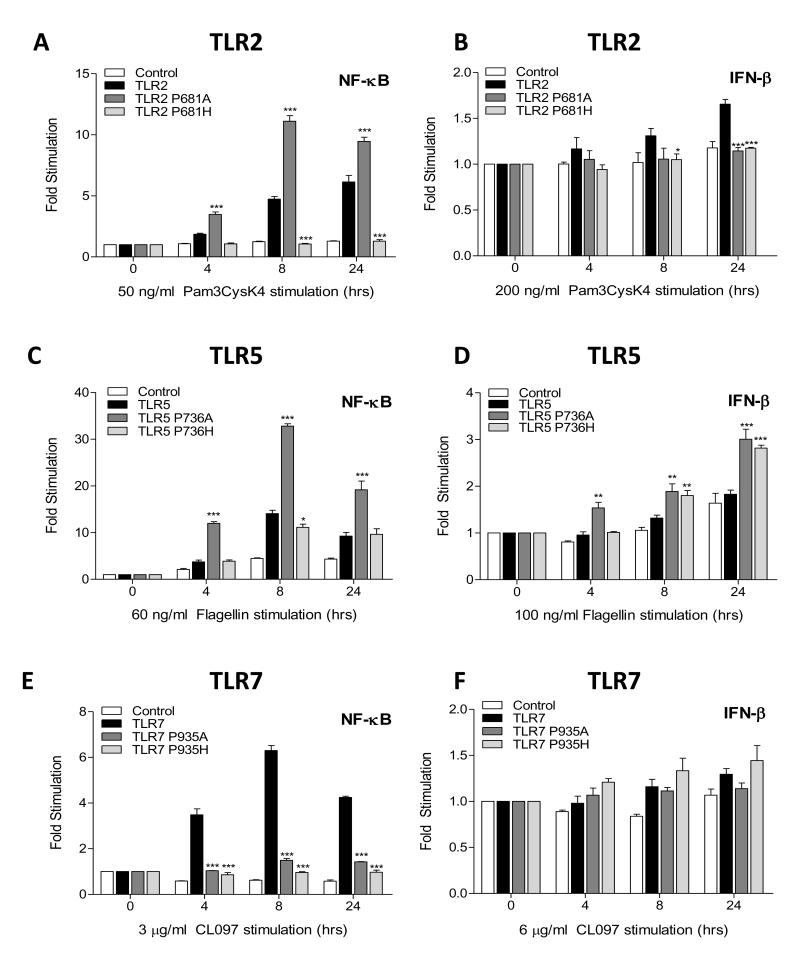
In view of the striking properties of the TLR3 A795P mutant and a recent finding that TRIF can mediate activation of NF-κB and mitogen-activated protein kinases (MAPKs) by TLR5 (18) we investigated the effect of mutating the BB-loop proline residue in other TLRs. As expected, upon treatment of HEK293 cells with the TLR2 agonist Pam3CSK4, transfected wild-type TLR2 induced strong NF-κB reporter gene activity over time, whilst the BB-loop TLR2 P681H mutant inhibited this response (Fig. 5A). Interestingly, the TLR2 P681A mutant significantly enhanced NF-κB activity by 3 fold at early time points when compared to wild-type TLR2. By contrast, IFN-β-driven reporter gene activity for both mutant TLR2 constructs showed no response following Pam3CSK4 stimulation, whereas a minor increase over time was observed for wild-type TLR2 (Fig. 5B). For TLR5, an expected NF-κB response was induced by flagellin, and like TLR2 P681A mutant, TLR5 P736A mutant also significantly enhanced NF-κB reporter gene activity, whereas mutant TLR5 P736H was at levels similar to that of wild-type TLR5 response (Fig. 5C). Both TLR5 mutants induced a small IFN-β signal but wild-type TLR5 remained unresponsive (Fig. 5D). In the case of TLR7 neither BB-loop mutant drove NF-κB or IFN-β activity in response to imidazoquinolines whereas wild-type receptor signalled normally (Fig. 5E-F). These results demonstrate that a BB-loop alanine is not sufficient to confer TRIF directed signalling to MyD88 dependent TLRs.
Figure 5. BB-loop proline-alanine mutant is not sufficient to confer TRIF directed signaling.
HEK293 cells were transfected with empty vector (control) or expression plasmid for wild-type TLR2, TLR2 P681A or TLR2 P681H mutants (10 ng) (A-B); wild-type TLR5, TLR5 736A or TLR5 P736H mutants (10 ng) (C-D); wild-type TLR7, TLR7 P935A or TLR7 P935H mutants (10 ng) (E-F); with either the NF-κB promoter reporter (10 ng) or IFN-β promoter reporter (25 ng). After 24 h, cells were stimulated with either Pam3CysK4 (50 ng/ml) (A) or (200 ng/ml) (B); Flagellin (60 ng/ml) (C) or (100 ng/ml) (D); CL097 (3 μg/ml) (E) or (6 μg/ml) (F); for 4, 8 and 24 h and the cell lysates harvested for assessment of luciferase reporter gene activity. Statistical analysis was performed using two-way ANOVA where significance is relative to each WT TLR plasmid transfection: *P<0.05, **P<0.01, ***P<0.001.
Discussion
The activity of human TLR3 is required for protection against encephalitis caused by herpes simplex virus 1 and although not essential probably participates in host defence against viral infection in other tissues (19, 20). Activation of TLR3 by viral nucleic acid leads to the production of interferon β and λ which inhibit viral replication. The anti-viral functions of TLR3 are mediated by interferon response factors (IRF) that require TRIF for activation. Here we report the remarkable finding that the specificity of TLR3 for TRIF depends critically on a single residue A795 in the TIR domain ‘BB-loop’. Interestingly, IRF-5, which is not a target of the TLR3 signalling pathway (21), was activated by the A795P mutant, a response that is also observed in MyD88-dependent signalling by TLR7 and 8. We also show that the mutant receptor associates with TRIF and MyD88 constitutively, either directly or perhaps as part of detergent resistant membrane microdomains (22). Treatment with the ligand poly (I:C) enhances the binding of TRIF to wild-type TLR3, and MyD88 to the A795P mutant receptor, providing further evidence that this mutation alters adaptor specificity. Furthermore, the activation of NF-κB by the A795P receptor was found to be dependent upon MyD88 and largely independent of TRIF. The partial inhibition observed in NF-κB activity induced by the A795P mutant in cells targeted with TRIF siRNAs (Fig 3E) suggests that this receptor may retain the ability to activate NF-κB by the TRIF pathway or that the presence of TRIF enhances the MyD88 directed response. The inability of TLR3 A795H to complex with TRIF confirms another study in which this mutant was unable to recruit TRIF, highlighting the BB-loop of TLR3 as a critical determinant for TRIF-TIR interaction (13). In addition to alterations in TIR adaptor specificity the A795P receptor, but not the wild-type, recruits the downstream signal transducer TRAF6, further evidence of a switch to MyD88 directed signalling. Previous studies showed that TRAF6 is essential for MyD88 signalling in murine macrophages but not for TLR3 signalling. On the other hand the related signal transducer TRAF3 but not TRAF6 is essential for the induction of type 1 interferons and the anti-inflammatory cytokine IL-10 (5, 7).
Our finding that MyD88 binds constitutively to TLR3 indicates that adaptor usage by the TLRs is more promiscuous than previously thought. In this regard recent reports have provided evidence that Mal and MyD88 have regulatory roles in normal TLR3 signalling (23, 24). In cells lacking MyD88, TRIF-TLR3 activation of IFN-β is significantly enhanced although TRAM-TRIF signalling by TLR4 is suppressed (23). Thus the repressive function of MyD88 is specific for TLR3 and our finding that MyD88 is constitutively associated with TLR3 suggests that this dominant negative effect acts at the level of adaptor recruitment. Another study revealed that the absence of Mal also enhances signalling by TLR3, consistent with Mal serving a bridging function to bring MyD88 to the membrane (25) although in this study we could not detect a direct interaction of Mal with TLR3. A possible mediator of TLR3 regulation by Mal/MyD88 is IRAK-2. IRAK-2 can associate with MyD88 and TRAF6 to activate the transcription factor NF-κB (26, 27) and also indirectly associates with TLR3 (28). Thus formation of TLR3/TRIF complexes may allow for recruitment of TRAF6 which then forms a complex with IRAK-2 and MyD88 and signals to NF-κB. In the case of the A795P mutant, a conformational change in TRIF might inhibit TBK-1 binding and allow for enhanced TRAF6 recruitment, which in turn recruits more IRAK-2 and MyD88 resulting in a strong up-regulation of NF-κB activity. However our finding that TRIF is not required for NFκB activation by the A795P receptor argues against such an indirect model.
Different stoichiometric combinations of receptors and adaptors cluster into membrane microdomains and regulate a specific signalling response against a particular pathogen (22). Therefore, our results may suggest that in the absence of stimulus TLR3 is primed in microdomains with MyD88, TRIF or both adaptors, and that binding of ligand leads to the formation of an active signalosome with TRIF and wild-type TLR3 on the one hand or with MyD88 in the case of the A795P mutant receptor.
At present the molecular mechanisms by which post receptor complexes form during TLR signalling are unclear. The variability of different receptor and adaptor TIR surfaces and potential electrostatic complementarity may underlie the functional specificity of different modules in the TLR signalling response (29). In this regard we describe BB-loop mutants of TLR2 and TLR5 that exhibit similar up-regulation of NF-κB to that seen with the TLR3 A795P mutant. It may be that the adaptor molecules bound to these mutants undergo a conformational change that favours enhanced recruitment of downstream molecules. Conversely, the mutation may also inhibit or restrict further recruitment of other adaptor molecules that negatively regulate NF-κB. As both TLR2 and TLR5 do not signal from endosomes it may be that altering their BB-loop to an alanine is not sufficient for it to switch adaptor specificity with TRIF. By contrast in the case of endosomal TLR7, both mutations within the BB-loop disrupted NF-κB signalling. However the P935A mutant also inhibited IFN-β activity, suggesting that switching to TRIF may require more than just a single amino acid substitution. Crystal structures of several isolated TIR domains are known but there are no structures of receptor/adaptor TIR complexes (30). However modelling studies suggest that ligand binding to the receptor ectodomain causes dimerization of the TIR domains in a symmetrical arrangement (31-33). The receptor BB-loops are predicted to form a critical part of this homodimerization interface, a conclusion that is supported by site-directed mutagenesis and studies with cell-permeable peptide inhibitors. Comparison of the crystal structures of TLR2 TIR and the inactive mutant P681H (34) shows that although the BB-loops adopt a similar overall conformation (Supplemental Figure S2) the histidine residue protrudes from the loop in a way that the proline cannot due to geometrical constraints. It seems likely that this either prevents dimerization of the mutant or forms a scaffold that is unable to support the binding of adaptors. By contrast it is likely that substitution of proline for alanine would not cause such a steric hindrance of dimerization or binding interfaces but could confer a different adaptor specificity or preference. It is interesting to note that the Drosophila Toll receptors also have a hydrophobic residue at this position, usually valine.
In view of the functional consequences of the proline substitution in the human TLR3 we carried out a phylogenetic analysis to discover whether Ala795 is conserved across species. As shown in Supplemental Figure S3 alanine is found at this position in TLR3 of all vertebrates with the exception of fish which have proline. Fish TLR3 is also activated by double stranded RNA and this leads to the production of interferons and the activation of NF-κB (35). In contrast to mammals, the BB-loop of the fish TRIF adaptor has alanine and mutation of this residue to proline inactivates signalling to both IFNβ and NF-κB suggesting that reciprocal mutations in the receptor and adaptor TIRs can complement each other (36). Fishes have a paralogue of TLR3, TLR22, which is also involved in anti-viral responses mediated through TRIF (37). TLR22 has proline in the BB-loop and like TLR3 detects dsRNA (38). It may be that TLR22 is specialised to recognise dsRNA at the cell surface. Mammals may have lost this TLR when they began to live on land and it became dispensable due to redundancy (39).
In conclusion this study shows that a single residue in the BB-loops of TIR domains plays a critical role in adaptor selectivity. Structural studies of complex formation by TLR3 with TRIF and other TLR/adaptor interactions, will provide a detailed understanding of the molecular topology that governs downstream signalling responses. The study emphasises the importance of TIR-TIR interactions for inflammatory signalling processes and that BB-loop mediated protein-protein interactions are important targets for the development of new anti-inflammatory drugs.
Supplementary Material
Acknowledgements
We thank Drs K. Fitzgerald, A. Weber, A. Mansell, R. Lin, I. Dikic, W. Dalton and M. Kelliher for the kind gift of expression plasmids and cell lines and Drs C. Bryant, J. Stack, M. Gangloff and M. Moncrieffe for helpful discussions during the course of this work. A final thank you to Mr A. Liaunardy for help with the qPCR and siRNA analysis.
This work was supported by a UK Medical Research Council programme grant G1000133-E01 to NJG
References
- 1.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 2.Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 3.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 4.Han KJ, Yang Y, Xu LG, Shu HB. Analysis of a TIR-less splice variant of TRIF reveals an unexpected mechanism of TLR3-mediated signaling. J. Biol. Chem. 2010;285:12543–12550. doi: 10.1074/jbc.M109.072231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, Mann M, Karin M. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 6.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 7.Gohda J, Matsumura T, Inoue J. Cutting edge: TNFR-associated factor (TRAF) 6 is essential for MyD88-dependent pathway but not toll/IL-1 receptor domain-containing adaptor-inducing IFN-beta (TRIF)-dependent pathway in TLR signaling. J. Immunol. 2004;173:2913–2917. doi: 10.4049/jimmunol.173.5.2913. [DOI] [PubMed] [Google Scholar]
- 8.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. U S A. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 10.Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J. Exp. Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toshchakov VY, Fenton MJ, Vogel SN. Cutting Edge: Differential inhibition of TLR signaling pathways by cell-permeable peptides representing BB loops of TLRs. J. Immunol. 2007;178:2655–2660. doi: 10.4049/jimmunol.178.5.2655. [DOI] [PubMed] [Google Scholar]
- 12.Couture LA, Piao W, Ru LW, Vogel SN, Toshchakov VY. Targeting Toll-like receptor (TLR) signaling by Toll/interleukin-1 receptor (TIR) domain-containing adapter protein/MyD88 adapter-like (TIRAP/Mal)-derived decoy peptides. J. Biol. Chem. 2012;287:24641–24648. doi: 10.1074/jbc.M112.360925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nature Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Commane M, Jiang Z, Stark GR. IL-1-induced NFkappa B and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK) Proc. Natl. Acad. Sci. U S A. 2001;98:4461–4465. doi: 10.1073/pnas.071054198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 16.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 17.Johnson AC, Li X, Pearlman E. MyD88 functions as a negative regulator of TLR3/TRIF-induced corneal inflammation by inhibiting activation of c-Jun N-terminal kinase. J. Biol. Chem. 2008;283:3988–3996. doi: 10.1074/jbc.M707264200. [DOI] [PubMed] [Google Scholar]
- 18.Choi YJ, Im E, Chung HK, Pothoulakis C, Rhee SH. TRIF mediates Toll-like receptor 5-induced signaling in intestinal epithelial cells. J. Biol. Chem. 2010;285:37570–37578. doi: 10.1074/jbc.M110.158394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Audry M, Ciancanelli M, Alsina L, Azevedo J, Herman M, Anguiano E, Sancho-Shimizu V, Lorenzo L, Pauwels E, Philippe PB, Perez de Diego R, Cardon A, Vogt G, Picard C, Andrianirina ZZ, Rozenberg F, Lebon P, Plancoulaine S, Tardieu M, Valerie D, Jouanguy E, Chaussabel D, Geissmann F, Abel L, Casanova JL, Zhang SY. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J. Exp. Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, Sancho-Shimizu V, Lorenzo L, Puel A, Picard C, Chapgier A, Plancoulaine S, Titeux M, Cognet C, von Bernuth H, Ku CL, Casrouge A, Zhang XX, Barreiro L, Leonard J, Hamilton C, Lebon P, Heron B, Vallee L, Quintana-Murci L, Hovnanian A, Rozenberg F, Vivier E, Geissmann F, Tardieu M, Abel L, Casanova JL. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 21.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 22.Triantafilou M, Brandenburg K, Kusumoto S, Fukase K, Mackie A, Seydel U, Triantafilou K. Combinational clustering of receptors following stimulation by bacterial products determines lipopolysaccharide responses. Biochem. J. 2004;381:527–536. doi: 10.1042/BJ20040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siednienko J, Gajanayake T, Fitzgerald KA, Moynagh P, Miggin SM. Absence of MyD88 results in enhanced TLR3-dependent phosphorylation of IRF3 and increased IFN-beta and RANTES production. J. Immunol. 2011;186:2514–2522. doi: 10.4049/jimmunol.1003093. [DOI] [PubMed] [Google Scholar]
- 24.Johnson AC, Li X, Pearlman E. MyD88 functions as a negative regulator of TLR3/TRIF-induced corneal inflammation by inhibiting activation of c-Jun N-terminal kinase. J. Biol. Chem. 2008;283:3988–3996. doi: 10.1074/jbc.M707264200. [DOI] [PubMed] [Google Scholar]
- 25.Kenny EF, Talbot S, Gong M, Golenbock DT, Bryant CE, O’Neill LA. MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J. Immunol. 2009;183:3642–3651. doi: 10.4049/jimmunol.0901140. [DOI] [PubMed] [Google Scholar]
- 26.Wesche H, Gao X, Li X, Kirschning CJ, Stark GR, Cao Z. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J. Biol. Chem. 1999;274:19403–19410. doi: 10.1074/jbc.274.27.19403. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 28.Keating SE, Maloney GM, Moran EM, Bowie AG. IRAK-2 participates in multiple toll-like receptor signaling pathways to NFkappaB via activation of TRAF6 ubiquitination. J. Biol. Chem. 2007;282:33435–33443. doi: 10.1074/jbc.M705266200. [DOI] [PubMed] [Google Scholar]
- 29.Dunne A, Ejdeback M, Ludidi P, O’Neill LAJ, Gay NJ. Structural complementarity of Toll/Interleukin1 Receptor Identity Regions in Toll-like receptors and the adaptors Mal and MyD88. J. Biol. Chem. 2003;278:41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 30.Ve T, Gay NJ, Mansell A, Kobe B, Kellie S. Adaptors in Toll-Like Receptor Signaling and their Potential as Therapeutic Targets. Curr. Drug Targets. 2012;13:1360–1374. doi: 10.2174/138945012803530260. [DOI] [PubMed] [Google Scholar]
- 31.Nunez Miguel R, Wong J, Westoll JF, Brooks HJ, O’Neill LA, Gay NJ, Bryant CE, Monie TP. A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signalling adaptor proteins. PLoS ONE. 2007;2:e788. doi: 10.1371/journal.pone.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bovijn C, Ulrichts P, De Smet AS, Catteeuw D, Beyaert R, Tavernier J, Peelman F. Identification of interaction sites for dimerization and adapter recruitment in Toll/interleukin-1 receptor (TIR) domain of Toll-like receptor 4. J. Biol. Chem. 2012;287:4088–4098. doi: 10.1074/jbc.M111.282350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toshchakov VY, Szmacinski H, Couture LA, Lakowicz JR, Vogel SN. Targeting TLR4 signaling by TLR4 Toll/IL-1 receptor domain-derived decoy peptides: identification of the TLR4 Toll/IL-1 receptor domain dimerization interface. J. Immunol. 2011;186:4819–4827. doi: 10.4049/jimmunol.1002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu YW, Tao X, Shen BH, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 35.Phelan PE, Mellon MT, Kim CH. Functional characterization of full-length TLR3, IRAK-4, and TRAF6 in zebrafish (Danio rerio) Mol. Immunol. 2005;42:1057–1071. doi: 10.1016/j.molimm.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Fan S, Chen S, Liu Y, Lin Y, Liu H, Guo L, Lin B, Huang S, Xu A. Zebrafish TRIF, a Golgi-localized protein, participates in IFN induction and NF-kappaB activation. J. Immunol. 2008;180:5373–5383. doi: 10.4049/jimmunol.180.8.5373. [DOI] [PubMed] [Google Scholar]
- 37.Matsuo A, Oshiumi H, Tsujita T, Mitani H, Kasai H, Yoshimizu M, Matsumoto M, Seya T. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 2008;181:3474–3485. doi: 10.4049/jimmunol.181.5.3474. [DOI] [PubMed] [Google Scholar]
- 38.Xiao X, Qin Q, Chen X. Molecular characterization of a Toll-like receptor 22 homologue in large yellow croaker (Pseudosciaena crocea) and promoter activity analysis of its 5′-flanking sequence. Fish Shellfish Immunol. 2011;30:224–233. doi: 10.1016/j.fsi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Oshiumi H, Matsuo A, Matsumoto M, Seya T. Pan-vertebrate toll-like receptors during evolution. Curr. Genomics. 2008;9:488–493. doi: 10.2174/138920208786241234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.