Summary
Tumor cells have an altered metabolic phenotype characterized by increased glycolysis and diminished oxidative phosphorylation. Despite the suspected importance of glycolysis in tumorigenesis, the temporal relationship between oncogene signaling, in vivo tumor formation and glycolytic pathway activity is poorly understood. Moreover, how glycolytic pathways are altered as tumors regress remains unknown. Here we use a switchable model of MYC-driven liver cancer, along with hyperpolarized 13C-pyruvate magnetic resonance spectroscopic imaging (MRSI) to visualize glycolysis in de novo tumor formation and regression. LDHA abundance and activity in tumors is tightly correlated to in vivo pyruvate conversion to lactate and is rapidly inhibited as tumors begin to regress, as are numerous glycolysis pathway genes. Conversion of pyruvate to alanine predominates in pre-cancerous tissues prior to observable morphologic or histological changes. These results demonstrate that metabolic changes precede tumor formation and regression and are directly linked to the activity of a single oncogene.
Introduction
Cellular transformation has been correlated with increased aerobic glycolysis and diminished oxidative phosphorylation (Warburg Effect) (Christofk et al., 2008; Dang and Semenza, 1999; Vander Heiden et al., 2009; Warburg, 1956). Increased glycolysis may contribute both to energy production and the generation of metabolic intermediates that sustain anabolic processes required for rapid tumor growth (Vander Heiden et al., 2009). Lactate dehydrogenase A (Ldha) is a key regulator of glycolysis and its inhibition can attenuate the growth of transplanted tumor cells (Fantin et al., 2006; Le et al., 2010). We sought to determine whether glycolytic changes could be detected prior to other pathological alterations. To visualize the temporal connection between oncogene signaling and glycolysis in de novo tumor formation we used a switchable transgenic model of MYC-driven liver cancer (Goga et al., 2007; Shachaf et al., 2004). Double transgenic LAP-tTA (LT2) × TRE-MYC (MYC) mice (hereafter called LT2/MYC) express the MYC oncogene specifically in hepatocytes in a doxycycline-regulated manner and give rise to tumors that have features of human hepatocellular carcinoma and hepatoblastoma (Cairo et al., 2008; Shachaf et al., 2004). This model allows us to examine in vivo glycolytic changes at different stages of liver tumor formation as well as the consequence of acutely inhibiting oncogene signaling in established tumors. Hyperpolarized 13C MRSI, an emerging metabolic imaging modality (Golman et al., 2006; Hu et al., 2010), was used to visualize glycolysis at different stages of de novo tumor formation and regression in LT2/MYC mice. We show that hyperpolarized 13C MRSI can detect metabolic changes during tumor formation as well as when tumors regress. We find that pyruvate to lactate conversion increased as tumors developed and was rapidly inhibited during regression. Numerous glycolysis pathway genes showed a similar expression pattern. Interestingly, conversion of pyruvate to alanine was predominant in very early stages of tumorigenesis, prior to any observable morphologic or histological changes. Changes in gene expression of glutamine and TCA-cycle pathways was also altered indicating that global metabolic changes occur as tumors form and regress. These results demonstrate that metabolic changes precede tumor formation and regression and are directly linked to the activity of a single oncogene such as MYC. Our study further highlights the potential of high alanine levels as an early biomarker for liver tumor development.
Results
Doxycycline-Regulated MYC Expression Precedes Liver Tumor Formation and is Inhibited During Regression
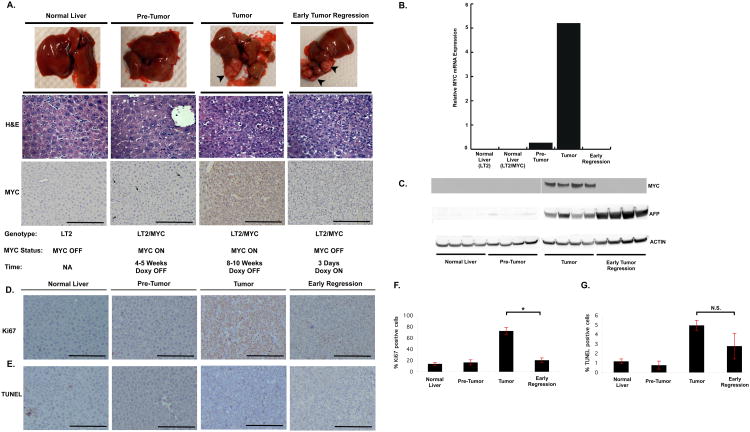
We first sought to define the different stages of tumorigenesis and regression. Control (LT2) mice and LT2/MYC mice kept continuously on doxycycline (doxy) do not express the MYC transgene and do not form tumors (Figure 1A-C). When doxycycline-containing food is withdrawn from LT2/MYC mice for 4-5 weeks the MYC transgene is modestly induced but there is no apparent phenotypic or histological changes to indicate tumor formation (Figure 1A-C). Prolonged removal of doxy for 8-10 weeks results in the formation of discrete tumor nodules, which have features of an aggressive malignancy. LT2/MYC tumors are comprised of small basophilic cells, with a high nuclear to cytoplasmic ratio, frequent mitotic figures indicating high proliferation and occasional apoptotic cells (Figure 1A and (Goga et al., 2007; Shachaf et al., 2004)). These tumor nodules are also associated with ∼ 20-fold increased MYC mRNA and protein expression compared to the pre-tumor liver tissues (Figure 1B-C). Immuno-histochemical staining for MYC shows a heterogeneous up-regulation in a small subset of pre-tumor liver cells, but a much more dramatic and uniform increased expression in tumor tissues (Figure 1A). This shows that the MYC transgene is not uniformly activated in all cells of the mouse liver. This resembles human tumor initiation and progression wherein activation of the driving oncogene may occur in subsets of cells within a tissue.
Figure 1. Switchable MYC expression in Primary Liver Tumor Formation and Early Regression.
A. Gross tissues, corresponding histology and MYC staining at different stages of tumor formation and early regression. No apparent histologic differences were noted between primary tumors and early-regression sections. MYC expression (indicated with arrowheads) is heterogeneous in pre-tumor tissue, is significantly increased in tumors, and is absent following 72 hrs of doxy treatment. B. Quantitative PCR and C. western blots show negligible MYC expression in early stage pre-tumor liver compared to elevated expression in established tumors. MYC mRNA and protein expression is switched off when doxy is administered for 3 days. C. The liver tumor marker, AFP is highly expressed in tumor tissue and persists once MYC is switched off. D. Comparison of staining for the Ki67 proliferation marker and E. TUNEL assay of liver samples at different stages of tumor development and regression. F. Graph depicting percentage of Ki67 positive cells. G. Graph depicting percentage of TUNEL positive cells. Error Bars represent S.E.M. * p < 0.004, N.S. = not significant.
This model system also allows for inhibition of MYC expression in primary tumors by adding doxycycline to their diet. Prolonged doxy administration can lead to complete regression of MYC-driven liver tumors, however, this takes several weeks ((Shachaf et al., 2004) and data not shown). Since we sought to examine the acute metabolic changes that occur once the expression of a driving oncogene such as MYC is inhibited, we treated mice with established tumors with doxy for only 72 hours. We found that at this time point MYC expression is completely inhibited (Figure 1A-C), however, tumors phenotypically and histologically appear nearly indistinguishable from primary tumors (Figure 1A). At the early stage of tumor regression we found that there was no substantial change in apoptosis, however, proliferation, as measured by Ki-67 staining, was significantly diminished (Figure 1D-G). These results indicate that acute inhibition of MYC precedes gross tumor regression.
In addition to following changes in tissue histology, we sought an independent marker of liver tumorigenesis that would allow us to examine different liver tumor stages in our models. Alpha-fetoprotein (AFP) is a well-established clinical biomarker of human HCC and hepatoblastoma, but is not detectable in normal liver. We therefore examined AFP expression in the different stages of liver tumor formation. We found that AFP is not detectable in non-tumor LT2 tissues and is negligible in the pre-tumor LT2/MYC tissues (Figure 1C). In contrast, primary liver tumors demonstrate abundant AFP expression (Figure 1C). Following 72 hours of doxy treatment, despite the complete inhibition of MYC expression, there is no discernible decrease in AFP expression (Figure 1B-C). Thus liver tissue histology and AFP expression indicate that MYC expression precedes tumor formation and liver tumor regression follows MYC inhibition.
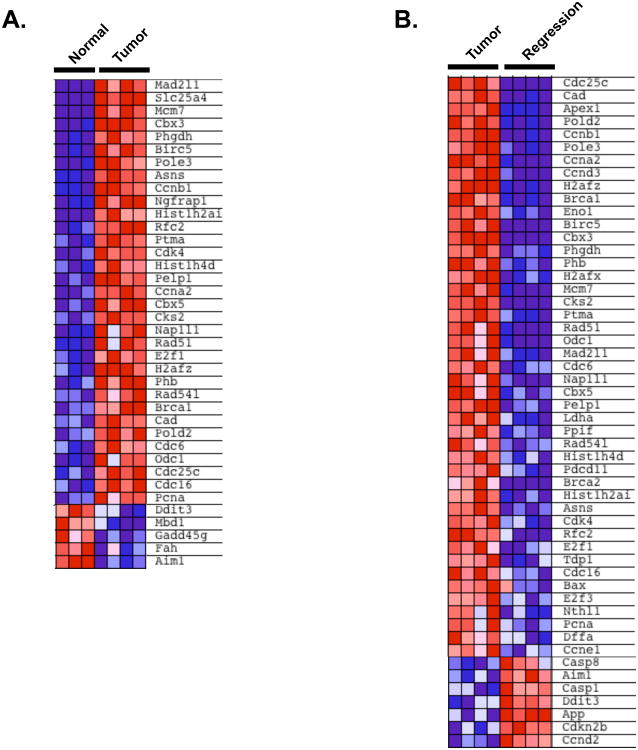
To better understand the gene expression changes that underlie MYC-driven tumor formation and regression, we performed mRNA profiling of liver samples at each stage. We selected a subset of 120 genes whose expression is altered in a MYC-dependent manner to determine if gene expression is changed in primary liver tumors as a consequence of inhibiting MYC (Table S1). Most of these genes were previously identified in studies looking at gene expression changes in tissue culture models. Genes in this test set are associated with diverse molecular functions including cell cycle, apoptosis, DNA damage, chromatin remodeling, and metabolic pathways such as glycolysis and amino acid metabolic pathways (annotated at www.myc-cancer-gene.org). We performed gene set enrichment analysis (GSEA), a computational method that determines whether there are concordant differences between two groups based on enrichment of the genes in one group compared with the other (Subramanian et al., 2005). This analysis confirmed that MYC target genes were significantly enriched in MYC tumors compared to normal livers (Figure 2A, p < 0.0001). Likewise, there was an enrichment of the MYC target genes in primary tumors compared to early regression samples (Figure 2B, p < 0.0001). Thus, MYC-driven tumorigenesis causes widespread changes in gene expression as primary tumors form and as they begin to regress.
Figure 2. Changes in Gene Expression During MYC-Driven Tumor Development and Regression.
GSEA analysis of 120 previously reported MYC-regulated genes. Shown are subsets of the 120 genes that are most abundantly enriched or down-regulated when comparing A. normal liver versus tumor tissues or B. tumor tissues versus early regression samples. See also Table S1.
Hyperpolarized 13C-Pyruvate Flux into Other Metabolites at Different Stages of MYC-Driven Liver Tumor Development and Regression
We next examined if dynamic changes in metabolic processes occur in vivo and if any metabolic changes precede overt tumorigenesis using the switchable MYC liver tumor model. We evaluated in vivo glycolysis at different stages of liver tumor formation and regression by examining the dynamic flux of pyruvate, a key product of glycolysis, into other metabolites. Hyperpolarized technology utilizing dynamic nuclear polarization (DNP) (Ardenkjaer-Larsen et al., 2003) can be used with the substrate [1-13C] pyruvate to specifically monitor LDH-catalyzed pyruvate to lactate flux in vivo. Hyperpolarized MRSI overcomes sensitivity limitations in traditional carbon MRSI caused by a low gyromagnetic ratio and low natural abundance of the MR detectable 13C isotope. Using a hyperpolarized approach, enhancement of sensitivity on the order of 50,000-fold can be achieved for in vivo metabolic imaging. [1-13C] pyruvate, which is converted to [1-13C] lactate, [1-13C] alanine, and [1-13C] bicarbonate in vivo (Figure 3A), has been recently used to visualize tumor tissues in a lymphoma transplant model as well as in a transgenic prostate cancer model (Albers et al., 2008; Day et al., 2007). In these prior instances, increased pyruvate conversion to lactate was observed in tumor versus adjacent tissue, however, in neither of these prior studies was the temporal association between oncogene signaling, LDH activity and in vivo glycolysis examined.
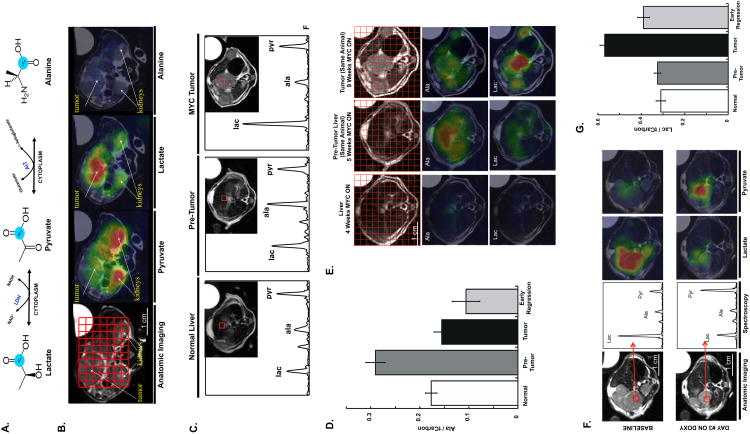
Figure 3. Hyperpolarized 13C metabolic Imaging of Liver Tumor Formation and Regression.
A. Diagram showing the conversion of 13C-pyruvate to lactate or alanine. B. Color overlay maps from spectroscopic grids show clear differences in metabolic profiles between liver tumors and other tissues. C. Hyperpolarized imaging reveals changes in metabolic profile during MYC-driven tumor formation. High pyruvate is detected in normal liver. Elevated alanine is associated with pre-tumor liver with modest MYC induction but no apparent phenotypic changes. A substantial increase in lactate is seen in developed tumor masses. D. A significant and reproducible pattern of high alanine in pre-tumor liver compared to controls (p = 0.003) and tumor (p = 0.0002) is observed. E. Heterogeneous elevation of alanine may correspond to areas of eventual tumor development. Color overlays generated from spectral grids are shown for a single animal over the course of disease progression after oncogene activation. The first column shows a healthy liver that has uniformly low alanine and lactate levels. The middle column shows a change in metabolic profile one week later, with substantially elevated alanine signal now observed. The alanine overlay highlights heterogeneity in the liver (bright red regions corresponding to highest signal intensity). The final column shows the liver 9 weeks after MYC activation, now consisting of a heterogeneous collection of distinct tumor nodules and regions of non-tumor. The lactate overlay shows the tumor nodules as the regions with the highest lactate signal intensity. Color intensities were normalized across the different time points according to metabolite. F. Imaging early tumor regression before and 72 hours after MYC inhibition. A dramatic decrease in lactate is detected in voxels at similar locations prior and after doxy treatment. Spectra for the indicated voxel and corresponding anatomic imaging is shown. G. Repeated studies demonstrated a significant increase in lactate during tumor formation compared to controls (p < 0.0001) and decrease upon early regression (p < 0.0002). Lactate and alanine are reported as normalized to total carbon (the sum of lactate, alanine, and pyruvate). Error Bars represent S.E.M. See also Figure S1.
In the cell, pyruvate may be converted to acetyl co-A, lactate or via transamination, into alanine. Conversion of pyruvate into lactate or alanine (Figure 3A) can be visualized by MRSI. We first examined the flux of pyruvate into other metabolites in established LT2/MYC liver tumors. MR anatomic imaging was used to visualize liver tumors, non-tumor tissues, and adjacent normal structures. Applying a compressed sensing technique, which allows rapid image acquisition across an entire liver, we performed hyperpolarized 13C 3D spectroscopic imaging with 0.034 cm3 per voxel resolution (Hu et al., 2010). Using this approach metabolic imaging of an entire mouse was achieved in ∼ 16 seconds (Figure 3B). We found that 13C-pyruvate could be readily detected in non-tumor tissues such as the kidneys (Figure 3B). In contrast, 13C-lactate was abundantly detected only in tumor and not in adjacent tissues (Figure 3B). 13C-alanine was not appreciably detected in established tumors (Figure 3B). Thus, consistent with prior studies, tumor tissues have increased flux of pyruvate to lactate consistent with increased LDH activity.
We next asked whether 13C-pyruvate flux changes during the course of primary liver tumor development. We imaged normal liver, pre-tumor liver tissues and established tumors using 13C-pyruvate 3D spectroscopic imaging (Figure 3C). We found that in pre-tumor tissues there is a significant increase in the conversion of 13C-pyruvate to 13C-alanine, which was not present in either non-tumor tissues or established tumors (Figure 3 C and D). Thus, conversion to alanine is the earliest metabolic change we detected and this preceded primary tumor formation. A sampling of spectra from various liver regions over the course of disease progression shows that while the average alanine signal is increased in the pre-tumor state, there is considerable heterogeneity between individual voxels in the pre-tumor liver (Figure 3E). We found that the regions in pre-tumor tissue with the most abundant alanine signal tended to be areas where tumor nodules would eventually develop. Thus, using this approach we can detect specific areas with elevated alanine levels in the pre-tumor state compared to the rest of the liver.
We next asked how 13C-pyruvate flux is altered when MYC is inhibited in established tumors. We imaged tumors at baseline and then again 3 days after the mice were fed a diet containing doxy to switch off MYC expression (Figure 1A-C). Following 3 days of doxy treatment liver tumors began to diminish in size, although substantial tumor mass was still present (Figure 3F) and was histologically indistinguishable from untreated tumors (Figure 1A). We examined 13C-pyruvate flux in the tumor tissue following MYC inhibition. We found a dramatic reversal of the 13C-pyruvate flux to lactate in early tumor regression (Figure 3F). Thus the ratio of 13C-lactate to total carbon rapidly returns to levels present in non-tumor tissue following MYC inhibition (Figure 3G).
In a series of control experiments, we found that constitutive expression of the tTA transcription factor in hepatocytes of LT2 transgenic mice compared to wild-type mice did not affect Lac/tCar or Ala/tCar ratios (Figure S1A). Likewise, doxy treatment of non-tumor bearing mice did not alter in vivo metabolic flux to either alanine or lactate (Figure S1B). This indicates that the results are not due to underlying toxicity from doxy treatment or transgene expression. We therefore conclude that inhibiting MYC in an established liver tumor inhibits glycolysis, resulting in diminished pyruvate to lactate flux.
We sought to understand the molecular basis for differences in 13C-pyruvate flux in livers at different stages of MYC-driven tumorigenesis. We first examined changes in gene expression of molecules that may impact 13C-pyruvate flux. The MCT1 transporter (SLC16A1) has been proposed to alter uptake of pyruvate into cells (Harris et al., 2009). We did not detect appreciable differences in MCT1 expression in tumor tissues or upon early tumor regression (Figure S1C). Moreover, the imaging studies were normalized to total 13C signal (tCar) and changes in MCT1 expression could be accounted for using this normalization.
Increased Alanine Metabolism Precedes Tumor Formation
We next examined gene expression changes in enzymes that regulate alanine and lactate production. The GPT1 (ALT) transaminase is a central regulator of pyruvate to alanine conversion. We found that GPT1 expression was moderately increased in pre-tumor tissues, consistent with the increased 13C-alanine observed in pre-tumor tissues (Figure 4A). In contrast, in tumor tissues or early regression samples GPT1 expression was dramatically decreased (Figure 4A). This is consistent with the absence of appreciable Ala/tCar found in imaging studies of established tumors.
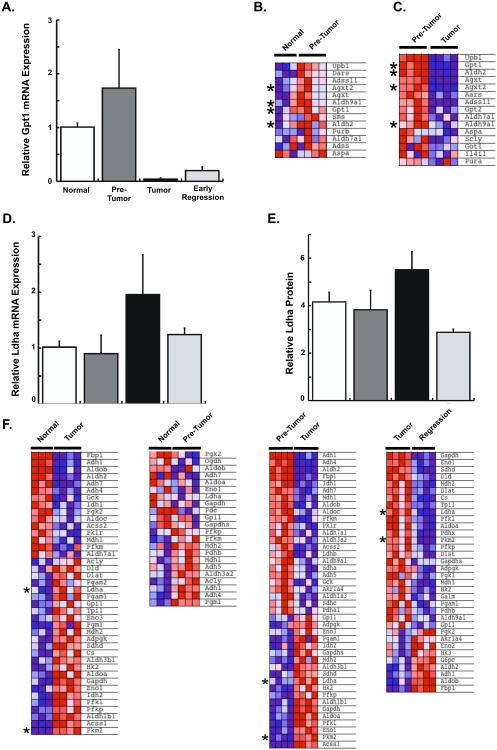
Figure 4. Expression of Key Regulators of the Alanine and Glycolysis Biosynthetic Pathways are Altered at Different Stages of MYC-driven Liver Tumor Development.
A. Gpt1 mRNA expression significantly increases during early stage pre-tumors and decreases in fully developed liver tumors. B. GSEA analysis of alanine-pathway genes reveals up-regulation of many genes in the pathway in pre-tumor samples compared to normal liver (p < 0.004). Enriched genes are shown. C. GSEA analysis of alanine-pathway genes comparing pre-tumor and tumor samples shows down-regulation of genes in this pathway (p < 0.005). Genes enriched in pre-tumors are shown. Alanine-pathway genes described in the text are indicated with an asterix. D. Ldha mRNA and E. protein expression show elevated Ldha expression only in primary tumors. F. GSEA analysis of glycolysis genes showing two-way comparisons of glycolytic pathway gene expression at different states of tumor formation and regression. Key glycolysis regulators, Ldha and Pkm2 are indicated with an asterix. Error Bars represent S.E.M. See also Figure S2, Table S2.
We next asked if in addition to GPT1 other genes in the alanine biosynthetic pathway are also up-regulated in pre-tumor tissues. Increased alanine production has been observed in association with cancer-related cachexia (Heber et al., 1985). Alanine can also be used for gluconeogenesis, generate cellular ATP, alter cellular re-dox potential and generate macromolecule intermediates useful for anabolism (Berg et al., 2002; Schindhelm et al., 2006), but to our knowledge this pathway has not been previously investigated as an early marker of tumor formation. Using the KEGG pathway database (www.genome.jp/kegg/) we examined 20 genes associated with alanine metabolism and found that most are up-regulated in early stage samples compared to normal livers (Figure 4B and Table S2, p < 0.004). In contrast, most of the alanine-pathway genes are down-regulated in the primary tumors compared to pre-tumor samples and remained low upon tumor regression (Figure 4C and Table S2, p < 0.005). Thus, increased expression of genes associated with alanine metabolism was associated with precancerous, early stages of tumor formation, but not in normal controls or primary and regressing tumors. Besides lactate, alanine is a major source of cytosolic pyruvate and transamination enables interconversion of alanine and pyruvate, which may then be used for gluconeogenesis or anabolic processes. Thus, our mRNA expression analysis shows that changes in genes associated with alanine metabolism precede primary tumor formation and raise the possibility that specific metabolic markers, such as alanine, may be useful to identify the earliest stages of tumor formation.
We next examined metabolic changes associated with primary tumors and early regression. Ldha has been shown to be the major mediator of tumor-associated glycolysis across various tumor types (Fantin et al., 2006; Vander Heiden et al., 2009) by converting pyruvate to lactate. The Ldha gene is also a direct transcriptional target of MYC in lymphoma and engineered fibroblast cells (Shim et al., 1997). We found that Ldha expression is similar in normal liver and pre-tumor tissues with low MYC expression (Figure 4D). In primary tumors, Ldha expression was increased substantially compared to normal liver, but expression returned to normal once MYC expression was inhibited in the early regression tumor samples (Figure 4D). Quantitative western blotting for Ldha also revealed similar results (Figure 4E).
While Ldha is a key regulator of glycolysis in tumor tissues (Dang and Semenza, 1999; Le et al., 2010), we asked if multiple glycolytic enzymes are coordinately up-regulated in primary tumors. We determined the expression of 72 genes associated with glycolysis (www.genome.jp/kegg/) and found that many glycolytic pathway genes, including Ldha and another key regulator Pkm2 (Christofk et al., 2008a; Vander Heiden et al.), were enriched in tumor samples compared to normal liver and pre-tumor samples (Figure 4F and Table S2). In contrast, once MYC was switched off, diminished expression of multiple glycolytic genes including Ldha and Pkm2 (Figure 4F and Table S2, p<0.007) was found in early regression samples. Thus, many, although not all, genes associated with the glycolysis-pathway are dynamically regulated as a consequence of MYC oncogene signaling as primary tumors form and regress.
Increased ALT and LDH Enzyme Activities Correlate with the Corresponding Metabolic Imaging Biomarkers
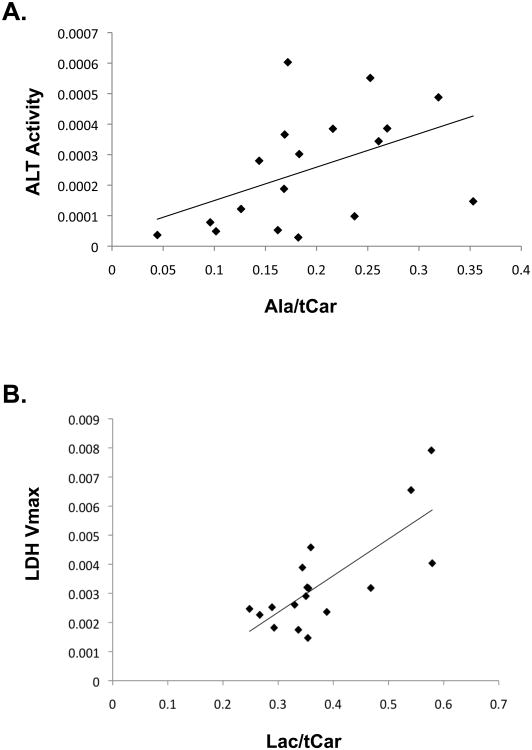
While the changes in Gpt1 and Ldha mRNA abundance observed in liver tumor formation and regression are consistent with the observed changes in metabolic imaging, additional translational or post-translational modifications could potentially also alter enzymatic activity that could not be accounted for entirely by differences in mRNA abundance. We therefore thought it was critical to determine the enzymatic activity of these molecules in liver tissues at different stages of tumorigenesis. To directly link changes of 13C-pyruvate flux observed during in vivo imaging to enzyme activity, we collected liver tissues immediately following imaging. Animals were sacrificed and tissues were collected for enzymatic activity assays. We found significantly elevated ALT enzyme activity in pre-tumor tissues compared to normal tissues or primary tumors and a correlation between ALT activity and Alanine/tCar ratio (Figure 5A, R = 0.47, p = 0.05). Finally, we also found a strong correlation between LDH Vmax and Lactate/tCar imaging (Figure 5B, R = 0.76, p < 0.0005).
Figure 5. ALT and LDH Enzyme Activities Correlate with the Corresponding Metabolic Imaging Biomarkers.
A. Graph of ALT activity versus alanine/tCar levels across various treatment conditions demonstrates a correlation between imaging and enzyme activity (p = 0.05). B. Graph of LDH activity versus lactate/tCar levels across various treatment conditions demonstrates a correlation between imaging and enzyme activity (p < 0.0005).
Expression of Glutamine and Tricarboxylic Acid Cycle (TCA) Pathway Genes Changes as Tumors Form and Regress
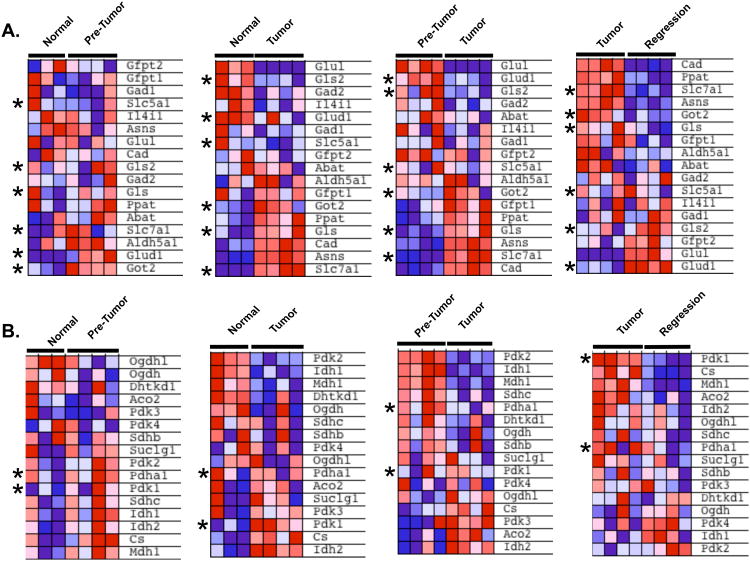
Several prior studies have identified increased glutamine uptake and catabolism as an important energy source in cells that over-express MYC (Dang, 2010; Wise et al., 2008; Yuneva et al., 2007), but the in vivo importance of this pathway is less clear. While our current studies with 13C-pyruvate imaging do not allow us to directly visualize glutamine catabolism, this pathway can feed into alanine production. We therefore examined if the expression of glutamine-pathway genes (Table S3) is altered in primary liver tissues as tumors form and regress. We find that a number of genes in this pathway are increased in the transition from normal to the pre-tumor state including glutamate dehydrogenase (Glud1) and the mitochondrial form of glutamic-oxaloacetic transaminase (Got2) (Figure 6A). The glutaminase genes (Gls and Gls2) were up-regulated in only a subset of the pre-tumor tissues (Figure 6A). Two glutamine transporters, Slc5a1 and Slc7a1, are direct transcriptional targets of MYC (Dang, 2010). We found Slc7a1 was modestly up-regulated in a subset of pre-tumor specimens compared to normal liver. With progression from the pre-tumor to tumor state we find that Slc7a1, Got2 and Gls are substantially further up-regulated, while Glud1 and Gls2 are markedly down-regulated (Figure 6A). Finally, there is a rapid reversal of these gene expression changes as tumors begin to regress, with Glud1 and Gls2 increasing and Slc7a1, Gls and Got2 being rapidly suppressed (Figure 6A). Our results demonstrate that gene expression of the glutamine pathway genes are altered during different stages of tumor formation, and are consistent with increased glutamine catabolism in MYC-driven liver tumors and rapid shut-off as tumors begin to regress.
Figure 6. Expression of Glutamine and TCA Cycle Pathway Genes as Tumors Form and Regress.
A. GSEA analyses of a subset of glutamine metabolism genes show an increase in expression as tumors develop and decrease in expression as they regress. Key regulators discussed in the text are indicated with an asterix. B. GSEA analysis of TCA cycle genes. Key regulators Pdha1 and Pdk1 are indicated with an asterix. See also Table S3.
The MYC oncogene in addition to promoting glycolysis has also been shown to increase mitochondrial biogenesis (Kim et al., 2008; Li et al., 2005). Thus it has been proposed that MYC may provide dual functions by driving both aerobic glycolysis and oxidative phosphorylation when sufficient oxygen is present (Dang, 2010). In addition to being converted to alanine or lactate, pyruvate can also be converted to CO2 and acetyl-CoA, which can enter the mitochondrial TCA-cycle. [1-13C]-pyruvate MRSI does not allow us to readily visualize the pyruvate that is converted to CO2 with high sensitivity. We therefore examined the gene expression changes of key regulators of the TCA cycle (Figure 6B, Table S3). Pyruvate dehydrogenase (Pdha1), the key regulator of pyruvate entry into the TCA cycle was elevated in the majority of pre-tumor tissues, but decreased as they progressed to tumors (Figure 6B). This is consistent with an increase in glycolysis in tumor tissues. Interestingly, however, Pdha1 was further down-regulated in the early regression samples compared to primary tumors. Thus the inability of Pdha1 to be up-regulated as tumors regress, may explain, at least in part, the rapid exit of tumor cells from the cell cycle (Figure 1D, F) if sufficient energy production cannot be sustained via the TCA-cycle and subsequent oxidative phosphorylation. We also examined the expression of Pyruvate dehydrogenase kinase enzymes responsible for inactivating the Pyruvate dehydrogenase complex and preventing glucose breakdown. The different isoenzymes (Pdk1-Pdk4) showed deregulated patterns of expression in each of the stages analyzed (Figure 6B).
Discussion
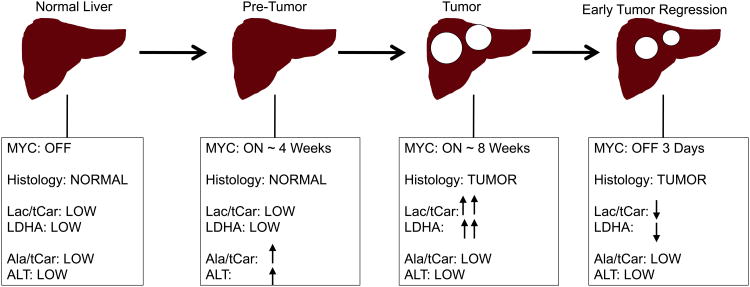
Increased conversion of glucose into lactate has been observed in tumor cells in culture, however, the ability to visualize dynamic changes in glycolysis as a function of oncogene signaling in vivo has been previously lacking. In this study, we sought to identify different stages of liver tumor formation and regression when the driving oncogene c-MYC was temporally regulated. We used hyperpolarized technology to assess metabolic consequences of MYC-driven liver tumor progression and regression. We found a strong correlation between LDH activity and in vivo conversion of pyruvate to lactate. Multiple glycolysis-pathway genes are up-regulated in tumors and their expression is rapidly repressed when MYC is switched off (Figure 4 and Figure 7). This MR-based system enables both rapid metabolic assessments and anatomical imaging. Using hyperpolarized 13C-pyruvate MRSI allows for non-invasive visualization of glycolytic changes within a de novo tumor beyond uptake and the first enzymatic step.
Figure 7. Changes in Metabolism Correlate with Stages of MYC-Driven Liver Tumor Development and Regression.
Schema summarizing the Lactate/tot Carbon, LDHA Vmax, Alanine/tot Carbon, ALT activity levels and metabolic gene expression changes in the different stages of MYC-induced liver tumor progression and regression.
Interestingly, we found that metabolic changes precede both tumor formation and regression. The earliest metabolic change observed was increased alanine in heterogeneous regions of the pre-tumor livers. Gpt1 gene expression and ALT enzymatic activity were elevated in this pre-tumor state and corresponded to the elevated Ala/tCar observed (Figure 7). Whether the elevated Ala/tCar spectra found is unique to livers in which MYC is the driving oncogene remains unknown. However, many MYC target genes potentially connect to increased alanine production. Multiple genes in the glutamine metabolic pathway are regulated by MYC signaling, including glutaminase (Gls), and the glutamine transporters (Slc5a1 and Slc7a1). Gls acts to convert glutamine to glutamate. Increased glutamate along with pyruvate are then converted by ALT to alanine and α-ketoglutarate, which in turn may enter the TCA cycle. Thus, elevated alanine production may be, in part, due to increased glutamine uptake and catabolism in pre-tumor tissues to glutamate (Figure S2).
We also examined other alanine pathway genes whose expression was altered in the pre-tumor state and found that two genes, Aldh2 and Aldh9a1, are known to be direct MYC-targets (www.myc-cancer-gene.org). Both genes are also substantially up-regulated in pre-tumor tissues compared to tumors (Figure 4B and Figure S2). Both Aldh2 and Aldh9a1 can participate in the production of beta-alanine from polyamine degradation. Beta-alanine plus pyruvate can in turn be converted into L-alanine and 3-oxopropanoate by alanine-glyoxylate aminotransferase 2 (Agxt2). Agxt2 is up-regulated in the pre-tumor state compared to normal liver and is diminished in primary tumors (Figure 4B-C). Ornithine decarboxylase (Odc1), a canonical MYC target gene, is rate-limiting for polyamine synthesis and is up-regulated in both pre-tumor and tumors. Thus, it is possible that MYC may regulate alanine production at the pre-tumor state both by up-regulating polyamine synthesis (via direct regulation of Odc1) and by their conversion into beta-alanine (via direct regulation of Aldh2 and Aldh9a1 in pre-tumor tissues). Why Odc1 expression remains high in tumors, while Aldh2 and Aldh9a1 expression diminishes is unclear, but it is well established that MYC may function as both a transcriptional activator and repressor depending on specific cellular contexts. Thus, there may be multiple MYC-driven pathways leading to increased alanine production in the pre-tumor state. However, it remains to be determined whether other oncogenes can also up-regulate the alanine pathway in pre-tumor tissues. As imaging and hyperpolarization techniques continue to improve, it should be possible to discern with increasing resolution if the pre-tumor tissues with the highest alanine production will ultimately develop into liver tumor nodules.
Finally, in the present study we employed a transgenic liver tumor model driven by the MYC oncogene. Other oncogenes including activated alleles of RAS, AKT, and PI3-kinase or loss of tumor suppressors (such as p53 and VHL) have also been shown to increase glycolysis (Dang, 2009). Thus, the deregulation of many glycolytic genes are not necessarily directly regulated by MYC, nor unique to MYC-driven tumors. Therefore, this approach should have utility beyond tumors driven by the MYC oncogene. Metabolic imaging via 13C-pyruvate is a rapid, non-radioactive approach to visualize glycolysis and alanine pathway changes in vivo and should provide new metabolic markers of cancer progression and regression. Clinical studies employing 13C-pyruvate MRSI imaging technology to visualize tumors in patients have recently begun. We propose that this approach may have utility to identify metabolic changes at the earliest steps of tumor formation and regression.
Experimental Procedures
Mouse Tumor Model
Tet-o-MYC/LAP-tTA (LT2/MYC) double transgenic mice were obtained by crossing two transgenic FVB strains(Goga et al., 2007). Under this system, the Tet-transactivator drove MYC expression only in the liver. MYC expression could be inactivated by doxy to bind and inactivate the Tet-transactivator. The Tet-o-MYC/LAP-tTA mice were weaned on doxy (200 mg/kg) until 4 weeks of age. Doxy was then removed from the diet to allow induction of expression of the human c-MYC transgene and tumorigenesis. Mice were monitored weekly for tumor development by inspection and palpating the abdomen. Tumors were confirmed by MR anatomic imaging. All animal studies were carried out under a protocol approved by the UCSF Institutional Animal Care and Use Committee.
Liver Histology and staining
Tissue sections from normal livers of control (LT2) mice, pre-tumor liver tissue and primary tumors from established or regressing LT2/MYC tumors were stained using a standard hematoxylin and eosin (H&E) protocol. Tumor morphology and tissue histology were examined. c-MYC (Epitomics, 1492) antibody was used to stain liver tissue sections for MYC. Ready-to-use Ki67 antibody (NeoMarkers) was used for Ki67 staining. TUNEL was preformed using ApopTag® Peroxidase In Situ Apoptosis Detection Kit (Millipore). All staining was performed on formalin-fixed, paraffin embedded, 5 micron tissue sections of mouse livers.
Real-time Quantitative PCR
Gene expression was determined by real-time quantitative PCR. Liver tissues from four representative samples from each group, normal LT2, early stage pre-tumors, established primary tumors and regressing tumors were analyzed. Total RNA from liver tissues was extracted (mirVana™ mirna isolation kit, Ambion), followed by Dnase treatment (Turbo DNA-free Dnase Treatment kit, Ambion) as per manufacturer's protocol. cDNA from one microgram of DNAse-treated total RNA was synthesized with the iScript™ cDNA synthesis Kit (Bio-rad). Real-time PCR was carried out using TaqMan probes (Applied Biosystems) for MYC, LDHA and GPT1 and SYBR green (Applied Biosystems) for MCT1 and compared to expression of the housekeeping gene, GAPDH.
Western blot analysis
Antibodies for c-MYC (Epitomics, 1492), AFP (NeoMarkers, RB-365-A1), and Actin (Sigma, A-1978) were purchased and used as indicated by the respective manufacturers.
mRNA Microarrays
Total RNA from LT2 controls (n=3), pre-tumor early stage samples (n=4), established LT2/MYC liver tumors (n=4) and early regression tumor samples (n=4) was extracted (mirVana™ mirna isolation kit, Ambion). RNA quality was assessed using the Nanodrop ND-8000 (Nanodrop Technologies, Inc.) and a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies) and run on Agilent mouse whole genome 4×44K Ink-jet arrays. Sample preparation, labeling, and array hybridizations were performed according to standard protocols from UCSF Shared Microarray Core Facilities and Agilent Technologies (http://www.arrays.ucsf.edu and http://www.agilent.com). RNA was amplified using the Sigma whole transcriptome amplification kit (Sigma-Aldrich), and subsequent Cy3-CTP labeling was performed using NimbleGen one-color labeling kits (Roche-NimbleGen Inc.). The labeled DNA was assessed using Nanodrop 8000, and equal amounts of Cy3 labeled target were hybridized to Agilent mouse whole genome 4×44K Ink-jet arrays. Hybridizations were performed for 14 hrs, according to the manufacturer's protocol (Agilent). Arrays were scanned using the Agilent microarray scanner and raw signal intensities were extracted with Feature Extraction v10.1 software.
Primary normalization and data extraction were done by the Microarray Core Facility. Briefly, raw log-intensities were normalized using the quantile normalization method (Bolstad et al., 2003). No background subtraction was performed, and the median feature pixel intensity was used as the raw signal before normalization. A one-way ANOVA model was used and specific contrasts were formulated to examine pair-wise comparisons of interest. Microarray data is available from the Gene Expression Omnibus: GSE28198.
Microarray data analyses
Effect of inactivation of the MYC oncogene in liver tumor models: Subsets of MYC target genes belonging to different pathways frequently reported to be targeted by MYC were selected from the MYC cancer gene database (http://www.myc-cancer-gene.org/ and Table S1). Genes selected are associated with various pathways including: cell cycle, apoptosis, DNA repair, chromatin remodeling, and glycolysis, alanine, and glutamine metabolic pathways. Hierarchical clustering was performed and the four study groups (LT2 controls, pre-tumor early stage mice, established LT2/MYC liver tumor and regressing LT2/MYC liver tumor mice) compared.
Comparison of specific metabolic pathways in developing and regressing MYC-driven liver tumors: Expression of genes associated with energy and anabolic metabolic pathways such as glycolysis and alanine metabolism was compared in the four study groups using gene set enrichment analysis (GSEA) (Subramanian et al., 2005). Gene lists for the glycolysis and alanine metabolic pathways were obtained from the KEGG database (http://www.genome.jp/kegg/pathway.html). Gene set enrichment analysis (GSEA) was used to determine the significance of differentially expressed MYC target genes and specific metabolic pathways in normal livers, early stage pre-tumors, primary developed tumors and tumors in early regression. Pre-defined gene lists including 120 MYC targets genes from multiple pathways regulated by MYC, 78 glycolysis genes and 20 alanine metabolism genes were used in GSEA. The gene set permutation was used instead of phenotype in order to avoid the potential problem of a small sample size.
Dynamic Nuclear Polarization and Injection of Hyperpolarized Pyruvate
The dynamic nuclear polarization and data acquisition protocols are described previously (Hu et al., 2010). An aliquot of 24 μL of [1-13C] pyruvic acid with 15 mM O×63 trityl radical and 1.5 mM Dotarem® gadolinium were polarized for 1 hour in a 3.35T magnetic field at 1.35 Kelvin using a Hypersense™ DNP polarizer (Oxford Instruments, Abingdon, UK). The power of the microwave irradiation was 20 mW. The solvent used to dissolve the frozen solid state sample was an aqueous mixture containing 40 mM Tris, 80 mM NaOH, and 0.3 mM Na2EDTA. The resulting pyruvate concentration was 80 mM, with a pH <7.5 and polarizations greater than 30% upon dissolution. A 350 μL volume of the dissolved pyruvate, followed by a 150 μL saline flush, were injected into mice through a tail vein catheter. A compressed sensing pulse sequence with 0.034 cm3 voxel resolution was used to acquire 3D MRSI data. The uptake of 13C-pyruvate and conversion to 13C-lactate and 13C-alanine were monitored with 3D spectroscopic imaging as described below. Mice were anesthetized with isoflurane and kept warm with a circulating-water heating pad.
Magnetic Resonance Imaging and Spectroscopic Imaging
All experiments were performed on a General Electric 3T (Waukesha, WI) MRI scanner equipped with 40 mT/m, 150 mT/m/ms gradients and a broadband RF amplifier. The RF coil was a custom built, dual-tuned 1H/13C transmit/receive design used previously (Albers et al., 2008; Hu et al., 2010). T2-weighted proton fast spin-echo (FSE) images were acquired and used as the anatomical reference on which 13C spectra were overlaid and co-registered. The axial FSE imaging parameters were: FOV = 8cm, 192×192 matrix, 2mm slice thickness, and NEX = 6. Carbon-13 hyperpolarized spectra were acquired as a volumetric grid using a compressed sensing 3D-MRSI sequence (Hu et al., 2010). Acquisition parameters were: variable flip angle, TE = 140ms, TR = 215ms, 16×16 in-plane phase encodes, center out phase encode order, 2.5mm × 2.5mm in-plane resolution, flyback readout in z with 16 points and 5.4mm resolution, 581 Hz spectral bandwidth, and 9.8 Hz spectral resolution. The 13C 3D-MRSI scan was started 30s after injection of the hyperpolarized 13C-pyruvate and lasted 16s. To quantify metabolism in the 13C spectra, the areas under the 13C-lactate, 13C-pyruvate-hydrate, 13C-alanine, and 13C-pyruvate peaks (total carbon-13 defined as the sum of all four) in magnitude spectra were calculated. The ratios lactate area to total carbon area (Lactate/tCar) and alanine area to total carbon (Alanine/tCar) were computed for each voxel and then averaged over all voxels of interest for each mouse to derive the final lac/tCar and ala/tCar values for statistical analysis.
Ex Vivo LDH Activity
Total LDH activity was measured through an NADH-linked spectrophotometric method by observing the decrease in absorbance of NADH at 339 nm after addition of varying concentrations of pyruvate. Around 0.2 grams of frozen tissue were thawed in lysis buffer containing 50 mM Tris (pH 8.2), 2mM DTT, 2mM EDTA, and 1% Triton x-100. The tissue was homogenized and centrifuged for 1 min at 7000 rpm at 4 °C. The supernatant was removed and diluted appropriately so that the reduction of NADH was linear over the first ten minutes of the assay. In microplate wells, 3 μl of the cell lysate were mixed with 147 μl of reaction buffer containing varying concentrations of pyruvate, 80 mM Tris (pH 7.2), 200 mM NaCl, and 200 μM NADH that was heated to 30°C. The assay was conducted immediately by monitoring the decrease in NADH absorbance, at 339 nm, for ten minutes using an Infinite M200 spectrophotometer (Tecan). Each pyruvate concentration was assayed in triplicate. The reaction rate was normalized to total protein concentration (Quick Start Bradford Protein Assay, BioRad). Each sample was assayed in triplicates, at 595 nm, after incubating at room temperature for at least 5 minutes. The absorbance of the sample was then recorded, and protein concentration calculated using a gamma-globulin standard curve. Once the sample's protein concentration had been acquired, it was used to calculate LDH reaction rate for the varying pyruvate concentrations. The LDH reaction rate, measured in μM NADH/min/[protein], was plotted against pyruvate concentrations according to Michaelis–Menten kinetics. The maximum velocity (Vmax) and Km values were then calculated using the Lineweaver–Burk plot.
Ex Vivo ALT Activity
ALT activity was measured with a commercially available enzymatic assay kit (MaxDiscovery™). A coupled reaction scheme was used in which the first step was alanine and alpha-ketoglutarate conversion to glutamate and pyruvate by ALT. Pyruvate and NADH were then converted to lactate and NAD+ by lactate dehydrogenase, and activity was detected spectrophotometrically by measuring absorbance at 339 nm.
Statistical Analysis
All statistical analysis was performed with the JMP® software package. For the 13C MRSI statistical analysis, experimental data were divided into four groups: Normal Liver (Tet-transactivator control mice [n=5]), Pre-Tumor (Tet-o-MYC/LAP-tTA mice with MYC on > 30 days and no tumor on anatomic imaging [n=5]), Tumor (Tet-o-MYC/LAP-tTA mice with MYC on and tumor confirmed by anatomic imaging [n=8]), and Early Regression (Tet-o-MYC/LAP-tTA tumor mice in which MYC has been off for 72 hours [n=4]). For animals with repeat acquisitions, e.g. two scans in a row to determine reproducibility, the average of the measurements was used. Normal Liver, Pre-tumor, Tumor, and Early Regression groups were compared with Tukey's test after a one-way analysis of variance (ANOVA) was performed. Note that 4 mice in the Tumor group had a baseline 13C MRSI imaging exam and a repeat 72 hours after MYC was kept on. The 4 mice in the Early Regression group received a 13C MRSI baseline study and then a follow-up 72 hours after inhibiting MYC. Thus, there are 8 total animals in the Tumor group (in which a baseline tumor 13C MRSI was performed) and 4 in the Early Regression group. Consequently, tissues were collected from 4 tumor group mice and 4 Early Regression group mice. For the expression comparisons, a student's t-test was used, and for enzyme activity correlations, the Pearson product-moment was computed, after which the student's t-test was used to test for statistical significance.
Supplementary Material
Highlights.
Metabolic changes precede tumor formation and regression.
13C-pyruvate MRSI imaging rapidly assess oncogene activity within a de novo tumor.
LDHA is increased substantially in tumors, but decreases in early regression.
Pyruvate to alanine conversion is a metabolic marker of early tumor formation.
Acknowledgments
We thank Dr. Noelle L'Etoile and members of the Goga and Vigneron labs for helpful discussions. We thank Mercedes Joaquin for mouse colony care and genotyping, Kristen Scott for her assistance in the animal imaging studies and Paniz Vafaei for her work on the LDH and ALT activity assays. We thank Drs. David Erle, Andrea Barczak, Yuanyuan Xiao, and Rebecca Barbeau from the Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility and NIH/NCRR UCSF-CTSI Grant Number UL1RR024131. We thank Linda Prentis for help with processing liver tissue samples. This work was supported by NIH R21CA137298 (to D.V. and A.G.), K08CA104032 and 1R01CA136717 (to A.G.), R01EB007588 (to D.V) and the UCSF Liver Center P30DK026743. A.G. is a V-Foundation Scholar. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen YF, Nelson SJ, Vigneron DB, Kurhanewicz J. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68:8607–8615. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L, Stryer L. Biochemistry. New York: W.H. Freeman; 2002. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, Goga A, Balakrishnan A, Semeraro M, Gresh L, Pontoglio M, Strick-Marchand H, Levillayer F, Nouet Y, Rickman D, Gauthier F, Branchereau S, Brugieres L, Laithier V, Bouvier R, Boman F, Basso G, Michiels JF, Hofman P, Arbez-Gindre F, Jouan H, Rousselet-Chapeau MC, Berrebi D, Marcellin L, Plenat F, Zachar D, Joubert M, Selves J, Pasquier D, Bioulac-Sage P, Grotzer M, Childs M, Fabre M, Buendia MA. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Dang CV. PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Sci Signal. 2009;2:pe75. doi: 10.1126/scisignal.297pe75. [DOI] [PubMed] [Google Scholar]
- Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13:820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66:10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- Harris T, Eliyahu G, Frydman L, Degani H. Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells. Proc Natl Acad Sci U S A. 2009;106:18131–18136. doi: 10.1073/pnas.0909049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber D, Byerly LO, Chlebowski RT. Metabolic abnormalities in the cancer patient. Cancer. 1985;55:225–229. doi: 10.1002/1097-0142(19850101)55:1+<225::aid-cncr2820551304>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Hu S, Lustig M, Balakrishnan A, Larson PE, Bok R, Kurhanewicz J, Nelson SJ, Goga A, Pauly JM, Vigneron DB. 3D compressed sensing for highly accelerated hyperpolarized (13)C MRSI with in vivo applications to transgenic mouse models of cancer. Magn Reson Med. 2010;63:312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindhelm RK, Diamant M, Dekker JM, Tushuizen ME, Teerlink T, Heine RJ. Alanine aminotransferase as a marker of non-alcoholic fatty liver disease in relation to type 2 diabetes mellitus and cardiovascular disease. Diabetes Metab Res Rev. 2006;22:437–443. doi: 10.1002/dmrr.666. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.